Abstract
Fibroblasts are polymorphic cells with pleiotropic roles in organ morphogenesis, tissue homeostasis and immune responses. In fibrotic diseases, fibroblasts synthesize abundant amounts of extracellular matrix which lead to scaring and organ failure. In contrast, the hallmark feature of fibroblasts in arthritis is matrix degradation by the release of metalloproteinases and degrading enzymes, and subsequent tissue destruction. The mechanisms driving these functionally opposing pro-fibrotic and pro-inflammatory phenotypes of fibroblasts are enigmatic. We identified the transcription factor PU.1 as an essential orchestrator of the pro-fibrotic gene expression program. The interplay between transcriptional and post-transcriptional mechanisms which normally control PU.1 expression is perturbed in various fibrotic diseases, resulting in upregulation of PU.1, induction of fibrosis-associated gene sets, and a phenotypic switch in matrix-producing pro-fibrotic fibroblasts. In contrast, pharmacological and genetic inactivation of PU.1 disrupts the fibrotic network and enables re-programming of fibrotic fibroblasts into resting fibroblasts with regression of fibrosis in different organs.
Keywords: Fibrosis, matrix-producing fibrotic fibroblasts, matrix-degrading inflammatory fibroblasts, tissue repair, PU.1, Spi1
INTRODUCTION
Fibroblasts play an important role during the maintenance of tissue integrity1,2. They are also critical for the response to tissue injury, which goes far beyond deposition of extracellular matrix. In the context of inflammatory and neoplastic diseases2,3, fibroblasts can differentiate into a matrix-producing contractile phenotype that promotes progressive accumulation of extracellular matrix and the initiation of fibrotic disease4–6 In contrast, in chronic inflammatory diseases such as rheumatoid arthritis, fibroblasts acquire a matrix-degrading catabolic phenotype7. Phenotypic differences between matrix-producing pro-fibrotic fibroblasts and catabolic pro-inflammatory fibroblasts also become evident with respect to their distribution in tissues: Pro-inflammatory fibroblasts display an imprinted phenotype that forms hypertrophic lining layers in tissues such as the joints and grow similar to locally invasive tumors. In contrast, pro-fibrotic fibroblasts do not form such lining layers and establish a diffuse arrangement within the connective tissues8–11.
Studying the transcriptional network that drives polarization of fibroblasts into these two functionally opposing phenotypes revealed that the E26 transformation-specific (ETS) family transcription factor PU.1 is highly expressed in matrix-producing fibrotic fibroblasts, but silenced by epigenetic mechanisms in resting and matrix-degrading inflammatory fibroblasts. PU.1 activity acts as a genetic switch promoting the matrix-producing fibrotic fibroblastic fate.
RESULTS
PU.1 expression identifies fibrosis-associated fibroblasts
To understand the transcriptional network fostering the pro-fibrotic phenotype of fibroblasts, we extracted promoter sequences of differentially expressed genes from a published database of skin biopsy specimens from patients with systemic sclerosis compared to unaffected control subjects12. These promoter sequences were searched for occurrence of motifs of 984 human transcription factors13 using the HOMER software. For each of the selected motifs, the correlation between occurrence of the motif in the gene promoter and the respective differential expression level in fibrotic tissues was examined in comparison with healthy samples. Analysis of variance (ANOVA) highlighted 58 transcription factors with an increased presence in the regulatory sequences of pro-fibrotic genes (Fig. 1a). A considerable proportion of those transcription factors belonged to the ETS family (17.2%). Of those ETS members, PU.1 showed the highest enrichment at promoters of pro-fibrotic genes.
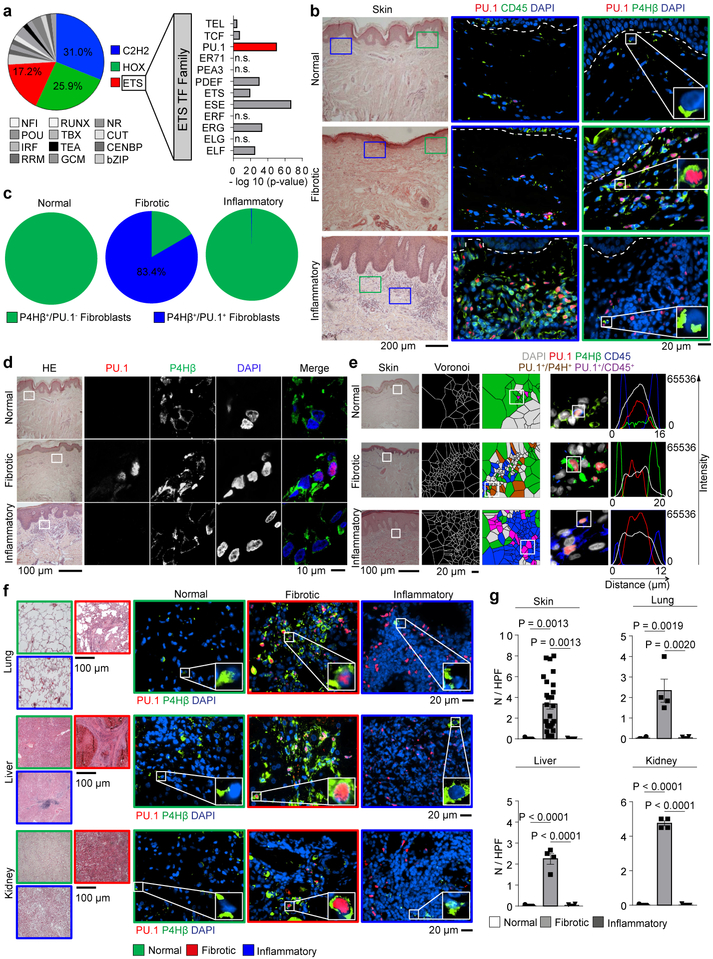
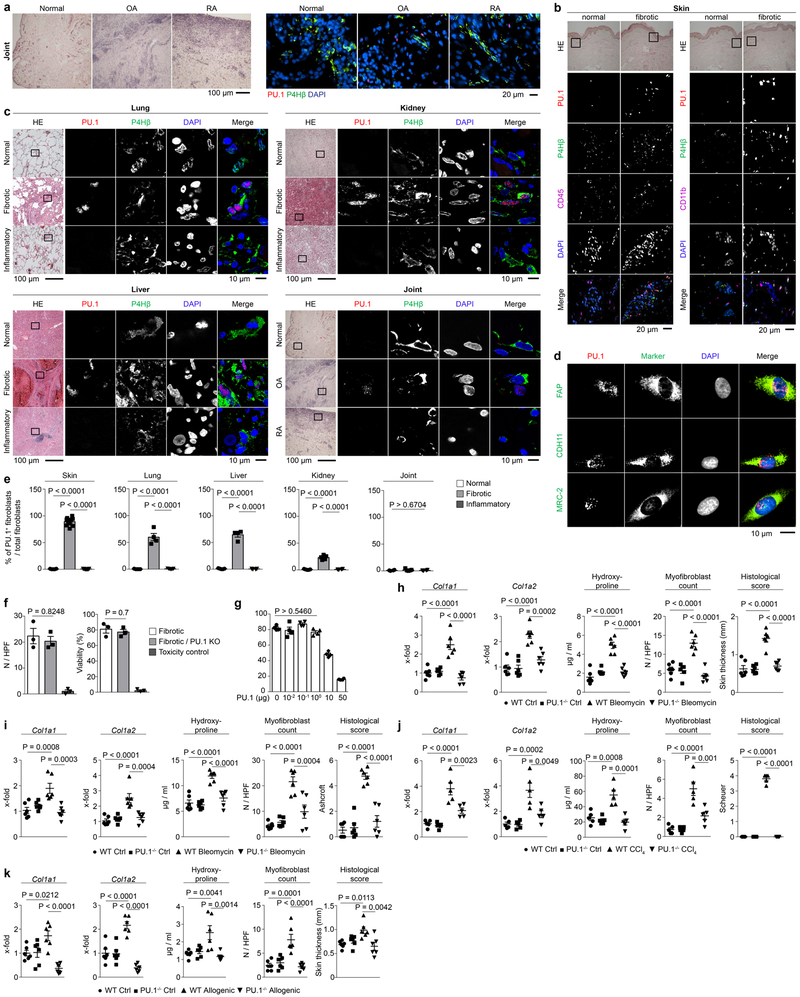
Figure 1: PU.1 expression in leukocytes and fibroblasts from normal human tissues and tissues affected by inflammatory or fibrotic diseases.
(a) Motif binding analysis of 984 transcription factors (TF) within promoter sequences of differentially expressed genes in skin biopsy specimens12 from patients with systemic sclerosis (n = 61) compared to unaffected controls (n = 36) using HOMER findMotifs. Log2(FoldChange) expression of differentially expressed genes was calculated and a linear model with the formula log2(FoldChange) ~ MotifOccurrences performed. Transcription factors with significant enhanced motif occurrence (p-value; -log 10) in pro-fibrotic genes as assessed by ANOVA. (b-f) Representative immunofluorescence (IF) of (b, e, f) widefield and (d) confocal microscopy of human skin, lung, liver and kidney biopsy specimens stained for PU.1 (red), CD45 or P4Hβ (green), and DAPI (blue); respective tissues were obtained from healthy individuals (n = 5 each), patients with systemic sclerosis (n = 25), plaque psoriasis (n = 7), idiopathic pulmonary fibrosis (IPF; n = 4), acute asthma (n = 5), alcoholic liver cirrhosis (n = 4), autoimmune hepatitis (n = 4), cirrhotic kidney (n = 4) and interstitial nephritis (n = 5). Hematoxylin & eosin (HE) stained tissue specimens are included. (c) Semi-quantification of PU.1+ fibroblasts / total P4Hβ+ fibroblasts per high-power field (HPF); (e) Voronoi mesh based tessellated pictures amenable to computational simulation, IF microscopy images and histograms of respective IF signals; (g) Semi-quantification of PU.1+ fibroblasts per high-power field (HPF); 6 randomly chosen HPF of each slide (respective n is given in b-f) were used. Data are shown as the mean ± s.e.m from biologically independent samples. P values were determined by one-way ANOVA with Tukey's multiple comparison post hoc test.
PU.1 is a key factor for the differentiation of monocytes and B cells, and deregulation of PU.1 expression has been implicated as a central mechanism in the pathogenesis of leukemia14,15. In fibrotic diseases, however, we observed prominent expression of PU.1 in prolyl 4-hydroxylase (P4H)β+ fibroblasts lacking the hematopoietic fate markers CD45 and CD11b (Fig. 1b-g, Extended Data Fig. 1a, b). PU.1 was upregulated in fibroblasts of various fibrotic diseases. We also detected PU.1-expressing lymphocytes but staining with additional fibroblast markers16–19 revealed that the majority of PU.1+ cells in fibrotic tissues were indeed fibroblasts (Fig. 1b-g, Extended Data Fig. 1b-d). In contrast to the abundant expression of PU.1 in fibrotic tissues, PU.1+ fibroblasts were not found in normal or inflamed tissues of skin, lung, liver, kidney and joints (Fig. 1b-g, Extended Data Fig. 1a, c, e).
PU.1 controls tissue fibrosis
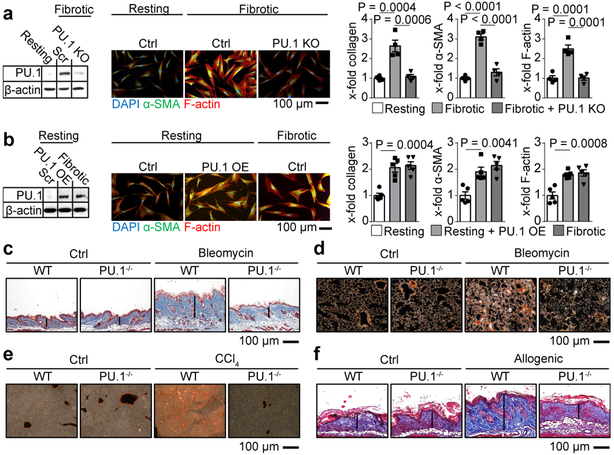
To examine the physiologic relevance of PU.1 expression in fibroblasts we used CRISPR/Cas9 technology to knockout the gene encoding PU.1 (in human: SPI1, in murine: Spi1; called ‘PU.1’ here) in human fibroblasts isolated from fibrotic tissue. PU.1 KO fibrotic fibroblasts displayed reduced collagen release, alpha smooth muscle actin (α-SMA) and F-actin expression to the levels of resting fibroblasts without affecting cell viability (Fig. 2a, Extended Data Fig. 1f). Conversely, PU.1 overexpression in human resting fibroblasts induced the transition of resting fibroblasts from healthy donors into a highly activated, pro-fibrotic phenotype with upregulation of collagen release, α-SMA and F-actin (Fig. 2b, Extended Data Fig. 1g). Next, we addressed the functional impact of PU.1-expressing fibroblasts in several murine models of fibrosis resembling different fibrotic conditions across different organs20. Similar to humans, PU.1 was expressed in fibroblasts from mouse models of fibrosis, but not in the non-fibrotic controls (Extended Data Fig. 2a-g, 3a-j). Fibroblast-specific knockout of PU.1 ameliorated fibrosis in these various models (Fig. 2c-f, Extended Data Fig. 1h-k).
Figure 2: Fibrogenic potential of PU.1-expressing fibroblasts.
(a) CRISPR-Cas9 mediated knockout (KO) of PU.1 in human fibrotic fibroblasts (n = 4 each); (b) PU.1-overexpressing (OE) human resting fibroblasts (n = 5 each); (a, b) KO and OE of PU.1 was measured by Western Blot analysis. Representative immunofluorescence images of fibroblasts stained for α-SMA (green), F-actin (red) and DAPI (blue) are included. Collagen production, α-SMA and F-actin expression were quantified. (c-f) Representative images of trichrome or sirius red stained tissue sections of fibrosis models of wild-type (WT) and PU.1fl/fl X Col1a2CreER (for skin models) or PU.1fl/fl X Col6Cre mice (for lung and liver models); (c) bleomycin-induced skin (n = 6 per group) and (d) lung fibrosis model (n = 6 per group); sodium chloride (NaCl) treated mice served as controls (ctrl). (e) Carbon tetrachloride (CCl4)-induced liver fibrosis model (n = 5 per group); mice treated with oil served as controls (ctrl). (f) Sclerodermatous chronic graft-versus-host disease (scl cGvHD) model (n = 6 per group); data are shown as the mean ± s.e.m. of respective n biologically independent samples. P values were determined by one-way ANOVA with Tukey's multiple comparison post hoc test.
PU.1 expression is controlled by epigenetic and post-transcriptional mechanisms
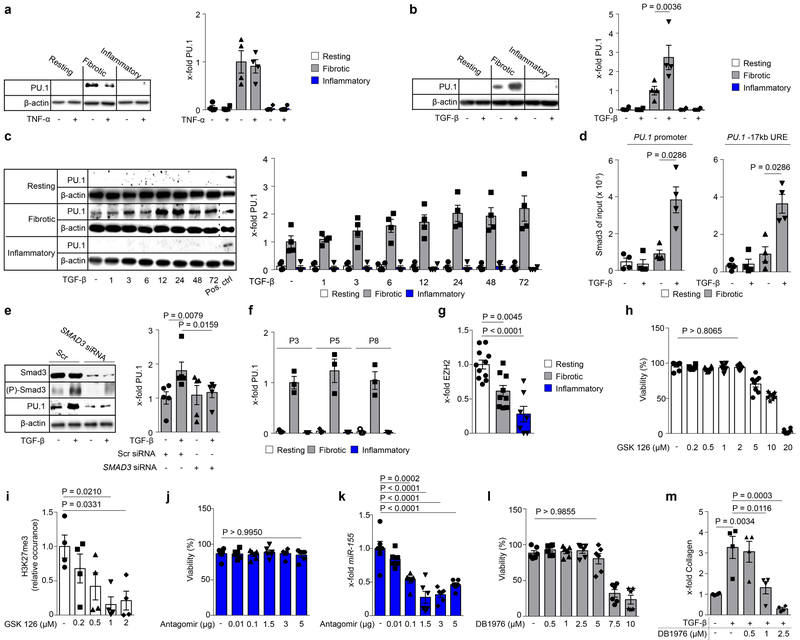
As PU.1 was required for tissue fibrosis, we next examined the potential mechanisms that account for its differential expression between fibrotic and inflammatory fibroblasts. Inflammatory stimuli such as tumor necrosis factor (TNF)-α did not influence PU.1 expression in resting, fibrotic, or inflammatory fibroblasts (Extended Data Fig. 4a). Similarly, short-term stimulation with pro-fibrotic mediators such as transforming growth factor (TGF)-β did not convert either resting nor inflammatory fibroblasts into PU.1-expressing fibroblasts (Extended Data Fig. 4b). Persistent TGF-β activity as in fibrotic diseases21–23 also failed to induce PU.1 in resting or inflammatory fibroblasts (Extended Data Fig. 4c). In fibrotic fibroblasts, however, the basal levels of PU.1 were further upregulated by TGF-β (Fig. 3b, c) in a Smad-3 dependent manner (Extended Data Fig. 4d, e). Fibrotic fibroblasts maintained constant levels of PU.1 through several passages in contrast to normal and inflammatory fibroblasts (Extended Data Fig. 4f).
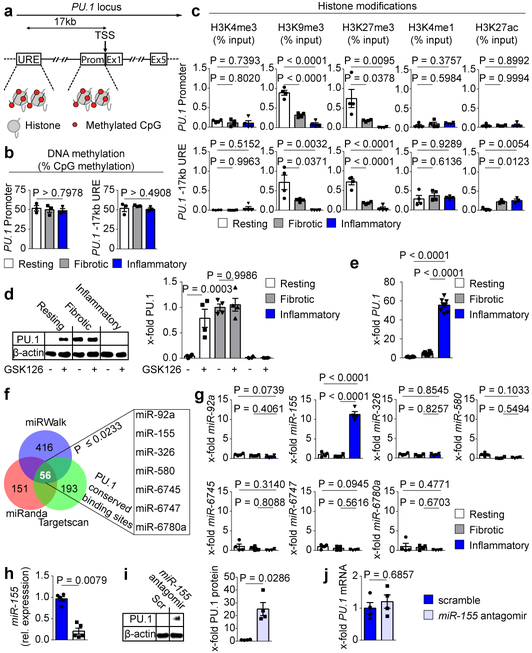
Figure 3: Epigenetic and post-transcriptional regulation of PU.1 in human fibroblasts.
(a) Schematic diagram of the PU.1 gene locus; Pr, promoter; URE, −17kb upstream regulatory element; TSS, transcriptional start site, exons 1-5 (E1 – E5); Met, methylated CpG; (b-h) Ex vivo experiments with primary human resting, fibrotic and inflammatory fibroblasts. (b) DNA methylation analysis of Pr and URE (n = 3) in respective fibroblasts; (c) Occurrence of respective histone modifications at PU.1 promoter and URE as assessed by ChIP and qPCR relative to input DNA (n = 4 each). (d) Representative Western Blot and semi-quantitative analysis of PU.1 expression in resting, fibrotic and inflammatory fibroblasts in the presence or absence of GSK126 for 96 hours (n = 4). (e) Quantitative analysis of PU.1 mRNA levels (n = 8 each). (f) Prediction of miRNA binding sites within the PU.1 mRNA by miRWalk (n = 416 hits), Targetscan (n = 193 hits) and miRanda (n = 151 hits). The overlap of possible miRNAs from all 3 tools were further restricted to p ≤ 0.0233 predicted by miRWalk73,76 (g) Respective miRNA expression levels relative to expression levels in resting fibroblasts (n = 4); (h) miR-155 reduction in inflammatory fibroblasts co-transfected with miR-155 specific or scrambled (scr) antagomirs as control (n = 5 each); (i) representative Western Blot and semi-quantitative analysis of PU.1 expression in inflammatory fibroblasts co-transfected with appropriate miR-155 specific or scr antagomirs. PU.1 expression is illustrated relative to β-actin (n = 4); (j) PU.1 mRNA expression levels of inflammatory fibroblasts in the presence and absence of miR-155 antagomirs (n = 4); data are shown as the mean ± s.e.m. of respective n biologically independent samples. P values were determined either by one-way ANOVA with Tukey's multiple comparison post hoc test or two-tailed Mann–Whitney U test if two groups were compared.
As PU.1 expression was maintained in cell culture over multiple passages we considered if epigenetic mechanisms play a major role in its regulation24,25. Differences in the epigenetic program have previously been related to the development of fibrotic diseases24,26. Therefore, we dissected epigenetic signatures of the PU.1 locus (Fig. 3a) in resting, fibrotic and inflammatory fibroblasts. Although DNA methylation has been shown to play a central role in fibroblast activation27,28, we did not observe major differences in DNA methylation at the promoter and enhancer regions of PU.1 among fibroblast phenotypes (Fig. 3b). However, the promoter and the −17kb upstream regulatory element (URE) of the PU.1 locus were dominated by the presence of repressive histone 3 lysine 9 trimethylation (H3K9me3) and H3K27me3 in resting fibroblasts. This finding is consistent with elevated expression levels of the H3K27 trimethyltransferase enhancer of zeste homolog 2 (EZH2)29 (Fig. 3c, Extended Data Fig. 4g). Resting fibroblasts showed a poised −17kb URE (H3K4me1 and H3K27me3) which became active in fibrotic and inflammatory fibroblasts by co-localized H3K27 acetylation (Fig. 3c). Exposure to GSK12630, an inhibitor of the EZH2 methyltransferase activity, induced expression of PU.1 in resting fibroblasts (Fig. 3d, Extended Data Fig. 4h, i). In contrast and consistent with the absence of H3K27me3 marks, incubation with GSK126 did not further increase PU.1 expression in fibrotic fibroblasts. In inflammatory fibroblasts, however, the repressive marks H3K9me3 and H3K27me3 were absent, but this was not sufficient for detectable amounts of PU.1 protein (Fig. 3c, d). Indeed, we observed transcriptional activity at the PU.1 locus resulting in detectable PU.1 mRNA levels (Fig. 3e), suggesting potential post-transcriptional regulation that prevented the translation of PU.1 in inflammatory fibroblasts. Micro-RNAs regulate fibroblast growth and activation10. We identified seven potential micro-RNAs with PU.1 conserved binding sites (Fig. 3f) and found miR-155 to be significantly upregulated in inflammatory fibroblasts compared to resting and fibrotic fibroblasts in line with previous reports31,32 (Fig. 3g). As in B cells33, inactivation of miR-155 by antagomirs (Fig. 3h, Extendend Data Fig. 4j, k) induced expression of PU.1 protein in inflammatory fibroblasts (Fig. 3i), suggesting that PU.1 in inflammatory fibroblasts was post-transcriptionally regulated by miR-155. However, PU.1 protein might be degraded by another factor, and protein expression restored because the expression of PU.1 was further increased by inhibition of miR-155. Therefore, we analyzed PU.1 mRNA levels during inhibition of miR-155. We detected stable expression levels of PU.1 mRNA in those cells compared to cells transfected with scrambled antagomirs, making it unlikely that PU.1 was regulated by an independent factor that is affected by miR-155 (Fig. 3j). Together, these findings suggest that two independent mechanisms regulate PU.1 expression in fibroblasts at the level of transcription and translation determining the functional state of these cells.
PU.1 induces polarization to matrix-producing fibrotic fibroblasts
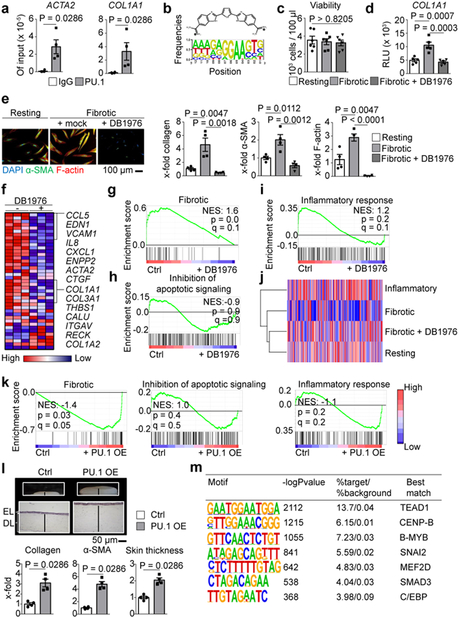
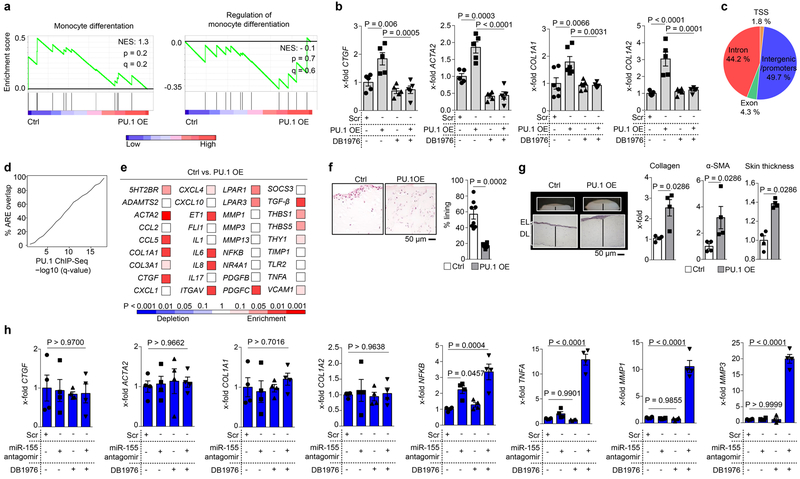
Next, we analyzed the molecular mechanisms of PU.1-induced fibroblast polarization. Chromatin immunoprecipitation (ChIP) analysis revealed binding of PU.1 at promoters of pro-fibrotic genes such as ACTA2 and COL1A1 (Fig. 4a). Like other ETS proteins, PU.1 binds to DNA sites harboring a 5´-GGAA-3´ core consensus sequence34. However, in contrast to other ETS transcription factors, PU.1 is strongly selective for binding sites in which the 5´-GGAA-3´ core is flanked by upstream AT-rich flanking sequences34. DB197635,36 is a heterocyclic diamidine that competitively blocks PU.1 binding to DNA with minimal effects on other ETS transcription factors due to its high specificity for AT-rich flanking sequences of the 5´-GGAA-3´ core (Fig. 4b). DB1976 decreased the transcription of COL1A1, reduced the expression of type I collagen and α-SMA and inhibited the expression of F-actin in fibrotic fibroblasts at least to the levels of resting fibroblasts at non-toxic concentrations (Figs. 4c-e, Extended Data Fig. 4l, m). RNASeq and subsequent gene set enrichment analysis (GSEA) demonstrated that incubation with DB1976 inhibited the pro-fibrotic gene signature of fibrotic fibroblasts37–42 without effects upon apoptosis-related and inflammatory Gene Ontology (GO)-defined gene sets (Fig. 4f-i). DB1976 induced a gene expression pattern comparable to that of resting fibroblasts (Figs. 4j). Conversely, GSEA of resting fibroblasts co-transfected with PU.1 revealed upregulation of the pro-fibrotic gene set and no effects on apoptosis-related, inflammatory and monocytic gene sets (Fig. 4k, Extended Data Fig. 5a). Additional treatment with DB1976 completely blocked the pro-fibrotic effects of PU.1 overexpression (Extended Data Fig. 5b). In 3D full-thickness skin organoids, overexpression of PU.1 in resting fibroblasts increased expression of collagen and α-SMA, and thickening of the skin organoid (Fig. 4l). RNA-seq data were validated by an integrative analysis comparing RNA-seq and PU.1 ChIP-seq data. In total 1,247 genes (8.1% of all expressed genes) were found to be significantly differentially expressed between untreated fibrotic fibroblasts and fibrotic fibroblasts treated with DB1976 (q < 0.05). A significant majority of differentially expressed genes (n = 989; 79.3%) was associated with a PU.1 ChIP-seq peak. Beside promoter regions, we also identified a substantial number of PU.1-binding sites in distal regions more than 50 kb away from known or predicted transcription start sites, reflecting the ability of PU.1 to control transcription through distal enhancers (Extended Data Fig. 5c). To address the question whether those PU.1-binding regions are implemented into regulation of the respective genes, several ENCODE datasets from DNAse-seq and histone ChIP-seq were used for unbiased identification of active regulatory elements (AREs) within the respective genes. PU.1 ChIP-seq peaks at various q-value thresholds showed a striking overlap with these predicted AREs (Extended Data Fig. 5d). These results underpin the regulatory function of PU.1 within fibrotic genes.
Figure 4: Regulation of pro-fibrotic genes by PU.1.
(a) ChIP of PU.1 binding at promoters of ACTA and COL1A1 analyzed by qPCR relative to input DNA (n = 4 each); (b) Chemical structure of DB1976 and the genomic consensus. (c) CCK-8 toxicity assay of human fibroblasts stimulated with DB1976 for 96h (n = 5). (d) Luciferase activity of human fibroblasts transfected with reporter vector containing the full promoter region of COL1A1 24 h after stimulation with/without DB1976 (n = 5 each). (e) Resting/fibrotic fibroblasts treated with/without DB1976 (hydroxyproline; α-SMA, F-actin per high power field (HPF) relative to control). Representative IF images (n = 4). (f-i) RNA-Seq of human fibrotic fibroblasts treated with/without DB1976 for 96h (n = 3 each); (f) heat map of pro-fibrotic gene signature. (g-i) Gene set enrichment analysis (GSEA) of RNA-Seq signals of GO-defined gene clusters; NES, normalized enrichment score; (j) hierarchical dendrogram of RNA-Seq profiles of resting/inflammatory/fibrotic fibroblasts stimulated with/without DB1976. (k) GSEA of resting fibroblasts co-transfected with PU.1 (OE) or scrambled vector (ctrl; n = 4 each); GO-defined gene signatures are assessed. (l) 3D full skin organoid model of human resting fibroblasts co-transfected with PU.1 or scrambled vector (n = 4 each); epidermal layer (EL) and dermal layer (DL) are marked. Collagen content (hydroxyproline assay); α-SMA, skin thickness quantified per HPF; data normalized to Ctrl; (m) Transcription factor binding motifs enriched in the 400 bp region surrounding the PU.1-binding sites that are specific for fibrotic fibroblasts using a random GC-corrected genomic background (n = 3 each). Data are shown as the mean ± s.e.m of respective n biologically independent samples. P values were determined either according to Subramanian et al.72 (g-i, k), Heinz et al.43 (m), by one-way ANOVA with Tukey's multiple comparison post hoc test (c-e) or two-tailed Mann–Whitney U test (a, l).
PU.1 switches inflammatory into fibrotic fibroblasts
We next investigated whether forced expression of PU.1 can re-polarize inflammatory into fibrotic fibroblasts. Indeed, ectopic PU.1 expression in inflammatory fibroblasts resulted in the upregulation of fibrosis-associated genes instead of genes for matrix-degrading proteins and inflammatory mediators (Extended Data Fig. 5e). In 3D micromass organoids resembling the synovial membrane, inflammatory fibroblasts reduced their ability to form lining layers upon PU.1 overexpression (Extended Data Fig. 5f). Instead, inflammatory fibroblasts forced to express PU.1 acquired a matrix-producing, pro-fibrotic phenotype with expression of α-SMA, increased collagen deposition and thickening of the dermal compartment in full thickness skin organoids (Extended Data Fig. 5g). Inhibition of miR-155 induced PU.1 expression in inflammatory fibroblasts and ingenuity pathway analysis (IPA) revealed that the transcriptional network of miR-155 includes several inflammatory targets such as NF-κB in addition to fibrosis-related targets (data not shown). Accordingly, inhibition of miR-155 alone was not sufficient to induce a fibrotic phenotype in inflammatory fibroblasts (Extended Data Fig. 5h). Consistent with the broader effects of miR-155, simultaneous blockade of miR-155 and PU.1 inhibited transcription of pro-fibrotic genes, and induced expression of inflammatory mediators and metalloproteinases (Extended Data Fig. 5h). These results highlight that fibrotic and inflammatory mediators are tightly balanced by a complex network of transcriptional and post-transcriptional factors. Within this network, PU.1 inhibition is sufficient to block transcription of fibrotic gene clusters.
PU.1 anchors differentiation towards fibrotic fibroblasts
Previously published reports show that PU.1 collaboratively interacts with other transcription factors at closely spaced binding sites to shape the phenotype of a cell43. To address whether PU.1 alone is sufficient to institute a fibrotic phenotype, we performed ChIP-Seq and additionally investigated transcription factor binding in the vicinity of occupied PU.1 sites. We identified binding motifs for several pro-fibrotic factors including TEAD1, CENP-B, B-MYB, SNAI2, MEF2D, SMAD3 and C/EBP in the vicinity of PU.1 peaks. Notably this represents a different set of factors from those known to collaboratively interact with PU.1 in monocytes and B cells44 (Fig. 4m). TEAD1 was further validated and showed robust expression levels in resting, fibrotic and inflammatory fibroblasts (Extended Data Fig. 6a). Key pro-fibrotic genes were screened for PU.1 ChIP-seq peaks and potential flanking TEAD1 binding sites. ChIP analysis revealed binding of TEAD1 at those predicted regions of key pro-fibrotic genes including 5HT2BR, ACTA2, COL1A1, COL1A2, CTGF, ITGAV, LPAR, PDGF, THBS1, TGFB1 (Extended Data Fig. 6b).
To confirm the necessity of the transcriptional network with PU.1 to institute a fibrotic phenotype, PU.1 transfected inflammatory fibroblasts were cultured under neutral, fibrotic and inflammatory conditions. Indeed, PU.1 induced pro-fibrotic mediators in inflammatory fibroblasts cultured under neutral conditions. However, the expression of key fibrotic factors was substantially facilitated under TGF-β-related, fibrotic culture conditions. In contrast, TNF-α-rich, inflammatory conditions interfered with the fibrotic effects of PU.1 (Extended Data Fig. 6c). These results corroborate the orchestrating role of PU.1 as susceptibility anchor within the network of factors that drive the differentiation towards a fibrotic phenotype.
Pharmacological inhibition of PU.1 controls tissue fibrosis
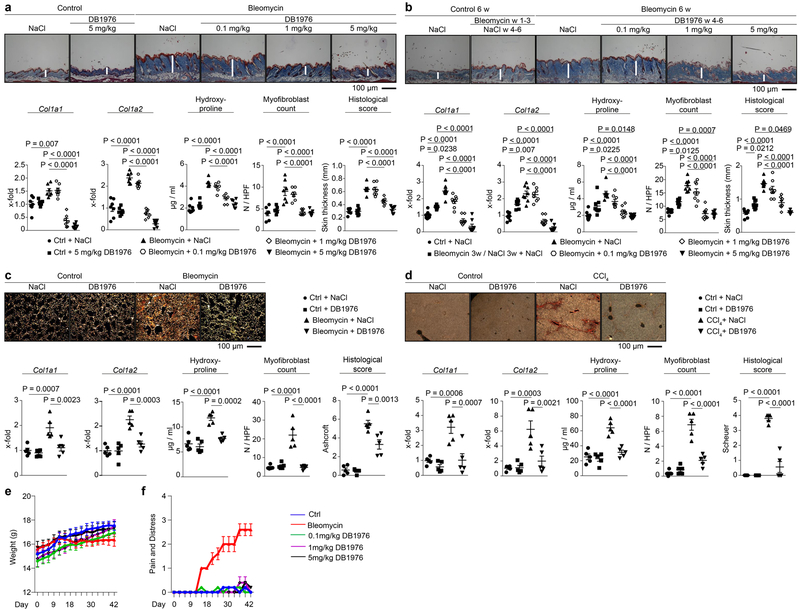
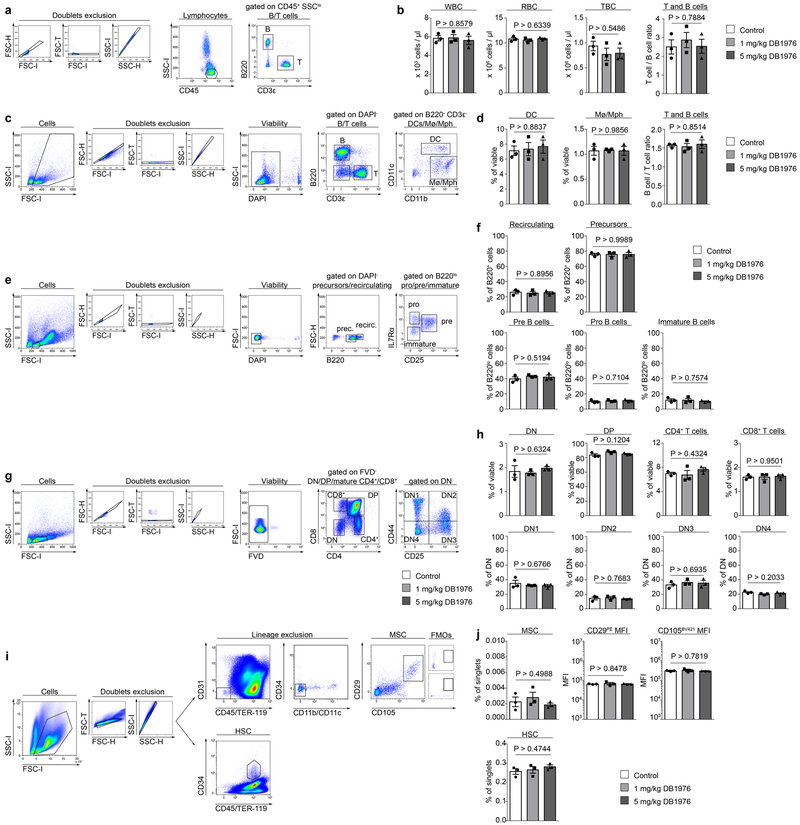
Finally, we investigated pharmacological targeting of PU.1 as potential strategy to tackle uncontrolled fibrotic tissue remodeling. Indeed, DB1976 showed anti-fibrotic effects in vivo in various fibrosis models and across several organs. Treatment with DB1976 not only prevented bleomycin-mediated skin fibrosis, but also induced regression of pre-established fibrosis (Extended Data Fig. 7a-d). Treatment with DB1976 in anti-fibrotic concentrations did not affect body weight, pain and distress levels of mice (Extended Data Fig. 7e, f). At the cellular level, we did not detect disturbance of hematopoiesis, alterations of hematopoietic and mesenchymal stem cells, defects in B cell development in the bone marrow, or in T cell maturation within the thymus upon DB1976-treatment (Extended Data Fig. 8a-j).
DISCUSSION
Our data demonstrate that expression of PU.1 is effectively silenced in fibroblasts during tissue homeostasis. When the epigenetic control of PU.1 is lost and PU.1 expression is induced, fibroblasts differentiate into a fibrotic phenotype that includes the transcription of numerous pro-fibrotic mediators. PU.1 has to date been mainly implicated in the regulation of hematopoiesis where PU.1 is indispensable for normal myeloid and lymphoid development14,15 and determines the fate of respective progenitors45–48. We found that the majority of PU.1-expressing cells in fibrotic tissues, in multiple disease settings are of a mesenchymal phenotype. PU.1 polarized resting fibroblasts and even re-polarized matrix-degrading inflammatory fibroblasts into a matrix-producing fibrotic phenotype.
PU.1 is embedded within a network of pro-fibrotic factors including members of TEAD/HIPPO, canonical TGF-β/SMAD and AP1 signaling pathways. Other transcription factors with fibrotic propensity such as SNAI2 and myocyte enhancer factor (MEF)2 bind in close vicinity to PU.1-binding sites within the genome and might contribute to the recruitment of the transcription machinery that drives the switch towards the fibrotic phenotype. Motif enrichment does not establish whether or not respective factors are required for collaborative binding. However, we show that the simultaneous induction of TGF-β-related mediators facilitated the pro-fibrotic properties of PU.1, whereas TNFα-rich, inflammatory settings interfered with them. In line with previous reports in monocytes43, these results implicate that the crosstalk between PU.1 and factors that are enriched in the vicinity of PU.1 sites drives fibroblast polarization. Also analogous to its lineage-defining function in monocytes, our results demonstrate that PU.1 has a major coordinating role within this complex network of transcription factors in fibroblasts, as the inactivation of PU.1 alone is sufficient to prevent fibrotic polarization.
These findings also suggest that PU.1 inhibition may represent a novel and effective therapeutic approach to treat a wide range of fibrotic diseases. Inactivation of PU.1 effectively reverted the fibrotic phenotype of fibroblasts to a resting state and induced the regression of tissue fibrosis. Furthermore, the level of PU.1 inhibition necessary to revert the functional phenotype of fibroblasts and alleviate fibrosis appears to be substantially lower than the one necessary for inhibition of hematopoietic cell differentiation. Targeting of PU.1 might thus provide a novel therapeutic option to efficiently but also safely interfere with excessive matrix deposition and allow restoring tissue homeostasis in fibrotic diseases.
Online Methods
Ethical compliance and experimental approaches
This project complied with all relevant ethical regulations regarding animal research and human studies. Experiments were done in a blinded fashion except when specifically indicated. There were no exclusion criteria for the human and animal experiments. Mice were stratified according to sex and then randomized into the different groups. Cells from human donors were also randomized.
Patient characteristics
Skin biopsies were obtained from 25 patients with systemic sclerosis according to the 2013 American College of Rheumatology (ACR) / European League Against Rheumatism (EULAR) criteria49, seven patients with plaque psoriasis and 21 age- and sex-matched healthy volunteers. Lung tissue was obtained from four patients with idiopathic pulmonary fibrosis (IPF), five patients with asthma and five matched non-inflammatory/non-fibrotic controls. Liver samples were obtained from four patients with alcoholic liver cirrhosis50, four samples from patients with autoimmune hepatitis (AIH)51 and five matched non-inflammatory/non-fibrotic controls. To investigate fibrotic kidney tissue we used cirrhotic kidneys from four patients with end-stage renal disease after renal transplantation or hydronephrosis. Kidney tissues from five patients with interstitial nephritis served as controls. Normal kidney tissues were obtained from macroscopically normal portions of kidneys surgically excised due to the presence of a localized neoplasm (n = 5). Synovial tissue specimens were obtained from five patients with rheumatoid arthritis (RA) who fulfilled the 2010 ACR classification criteria for RA52 as well as five patients with osteoarthritis (OA). Normal synovium was used as control tissue, which was obtained from surgery specimen of patients with no articular disease (n = 4). Written informed consent was obtained from all subjects. The study was approved by the ethical committee of the University of Erlangen-Nürnberg.
Mice
Wildtype C57/BL6NRj mice were purchased from Janvier. PU.1fl/fl mice46 were bred in house. To selectively inactivate PU.1 in fibroblasts, PU.1fl/fl mice were crossbred with either Col1a2CreER mice53 or Col6Cre mice54 to generate PU.1fl/fl X Col6Cre or PU.1fl/fl X Col1a2CreER mice. Cre-mediated recombination was induced by repeated intraperitoneal (i.p.) injections of tamoxifen over 5 days (d). Control groups were injected with corn oil. PU.1GFP reporter mice55 were kindly provided by Dr. S. Nutt. All mice were bred under specific pathogen-free conditions, and all studies were approved by the animal ethical committee of the government of Unterfranken, Wurzburg, Germany. The study has complied with all relevant ethical regulations.
Cell culture
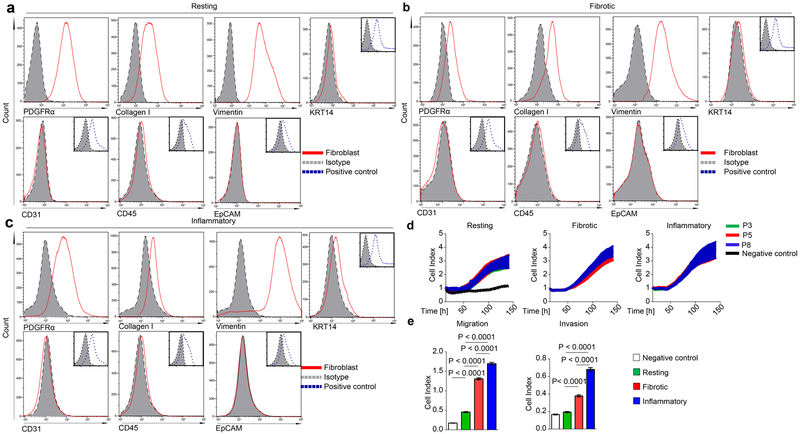
Human dermal fibroblasts were isolated from ten systemic sclerosis patients (fibrotic fibroblasts) and ten age- and sex-matched healthy volunteers (resting fibroblasts). After enzymatic digestion of the skin biopsies with collagenase type II (Merck, Darmstadt, Germany) for 3h at 37°C, the digested tissues were filtered using 100 mm nylon filter and centrifuged at 1400 rpm for 5 min. The pellet was resuspended in DMEM/F-12 medium containing 10% heat inactivated fetal bovine serum (FBS), 25 mM HEPES, 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine, 2.5 μg/ml amphotericin B (Thermo Fisher Scientific, Dreieich, Germany) and 0.2 mM ascorbic acid (Sigma-Aldrich, Steinheim, Germany). Synovial fibroblasts (inflammatory fibroblasts) were isolated from inflamed joints of six RA patients. Fibroblasts were cultured for 3 passages and then quality checked for a pure fibroblast population before using them in experiments. Fibroblasts were negative for CD31, CD45, CD326 (EpCAM) and KRT14 and positive for collagen-1, PDGFRα and vimentin. Representative FACS plots are presented in Extended Data Fig. 9a-c. Resting, fibrotic and inflammatory fibroblasts from passages 3 – 8 showed homogeneous characteristics respectively with regard to proliferation, migration and invasion capacity (Extended Data Fig. 9d, e) as assessed by xCELLigence Real-Time Cell Analyzer (RTCA) instrument (Acea Biosciences, San Diego, USA). As indicated, fibroblasts were transfected with 0.1 µg of either pUNO1 empty vector (control) or pUNO1-hSPI1a (both from InvivoGen, Toulouse, France) plasmids using the 4D-Nucleofector (Lonza, Cologne, Germany). Gene silencing was achieved using either 3 μg control CRISPR/Cas9 plasmid or 3 µg of PU.1 CRISPR/Cas9 KO Plasmid (h2) (Santa Cruz Biotechnology, Heidelberg, Germany). miR-155 silencing was performed using the anti-hsa-miR-155–5p miScript miRNA inhibitor or the miScript negative control inhibitor (1500 ng) (both from QIAGEN, Hilden, Germany). In selective experiments, cells were incubated with either recombinant TGF-β (5 ng/ml) (PeproTech, Hamburg, Germany), recombinant human TNF-α (10 ng/ml) (ImmunoTools, Friesoythe, Germany) and/or a combination of one or several of the following: GSK126 (1 µM) (Selleck Chemicals, Houston, USA) or DB1976 (2.5 µM; provided by Drs. Boykin/Poon, Georgia State University, Atlanta, USA).
Cell viability and cytotoxicity assays
Cell viability of cultured cells was quantified using the Cell Counting Kit (CCK)-8 (Dojindo Molecular Technologies, Maryland, USA) and an MRX ELISA reader (Dynex Technologies, Chantilly, USA).
Preparation of micro-mass cultures
Micro-mass organ culture experiments were performed as described elsewhere9. Synovial or dermal fibroblasts were released from the culture dish using TrypLE (Thermo Fisher Scientific) following transfection with plasmids as described above. Cells were re-suspended in ice-cold Matrigel Matrix (BD Biosciences, Heidelberg, Germany) at a density of 5 × 106 cells/ml. 40 µl droplets of the cell suspension were placed onto non-adherent 12-well culture dishes (Thermo Fisher Scientific). Gelation was allowed for 45 minutes at 37°C. Afterwards, the gel was overlaid with basal culture medium (DMEM, supplemented with penicillin, streptomycin, L-glutamine, nonessential amino acid solution, insulin–transferrin–selenium (Thermo Fisher Scientific), 0.1 mmol/l of ascorbic acid, 10 ng/ml of TNF-α and 10% heat-inactivated FBS. The floating 3-D culture was maintained for 3 weeks; medium was changed twice a week.
3D full thickness skin organoids
3D full thickness skin organoids were generated by submerging transfected fibroblasts (1 × 105 cells/ml) in neutralization solution (232.5 ml DMEM/F-12, 7.5 ml FBS, 7.5 ml 3 M HEPES, 2.5 ml chondroitin sulfate, 10 mg/ml rat tail collagen type 1). This mixture (500 µl) was filled in cell culture inserts with porous membranes (8 µm) and a 15 mm diameter providing a growth area of 1.13 cm2 (Greiner Bio One, Kremsmünster, Austria). The dermal components were cultured for one day in DMEM supplemented with 10% FBS, 5 ng/ml TGF-β and 1% penicillin-streptomycin at 37°C, 5% CO2 and atmospheric O2. To build up full thickness skin models (FTSM), the epidermal component was generated by seeding 5 x 105 normal human epidermal keratinocytes re-suspended in Epilife medium with 1% human keratinocyte growth supplement (E1 medium; Gibco), and with extra 1.44 mM CaCl2 (denoted as E2 medium) on the apical surface of the dermal components on the following day. After a submersed incubation of the models in E2 medium for 16 hours at 37°C, 5% CO2 and atmospheric O2 the medium was aspirated and the airlift-interface culture was initiated. FTSM were cultured in E2 medium supplemented with 0.125 mM L-ascorbic acid 2-phosphate and 10 ng/mL keratinocytes growth factor (Sigma Aldrich, St. Louis, Missouri, USA) for additional 5–10 days and the level of the culture medium was adjusted to the meniscus of the skin models56.
Real-time monitoring of cell proliferation, migration and invasion
Real-time proliferation assay was performed using the xCELLigence Real-Time Cell Analyzer (RTCA) system (ACEA Biosciences, San Diego, USA) according to the manufacturer’s instruction. Cells were seeded at a density of 25 % in an E-plate in cell culture medium (10% FBS) and measured every hour for 7 days. As negative control serum-starved (0.1% FBS) medium was used. For assessment of cell migration and invasion, CIM-plates 16 were used according to the manufacturer’s instruction. Briefly, 2 × 104 cells were plated in serum-starved (0.1% FBS) medium in the upper chamber. The lower chambers were filled with cell culture medium containing 10% FBS or with serum-starved medium as control. For invasion assays, the experimental setup of the migration assay was slightly modified as the upper chambers were loaded with 20 µL of a 1:10 dilution of Matrigel to create a 3D biomatrix film in each well prior to cell loading. Cell status is measured by electrical impedance and the relative change between impedance measured at any time (t) and baseline; respective values are displayed as the dimensional parameter “Cell Index” (CI). The obtained data were analyzed using the xCELLigence RTCA software. Results are presented as curve over time.
Reporter assays
Cells were transfected with a COL1A1 luciferase reporter plasmid (Active Motif, La Hulpe, Belgium) using the 4D-Nucleofector. Luciferase assays were performed using the Renilla Luciferase Assay System according to the manufacturer’s instructions (Progema, Madison, USA). Relative light units (RLUs) were obtained with a Luminoskan Ascent instrument with automated well-wise injection (Thermo Fisher Scientific). RLUs were normalized to the protein concentration, as determined by Bradford Protein Assay according to the manufacturer’s protocol (Bio-Rad).
Quantification of collagen protein
The amount of soluble collagen in cell culture supernatants was quantified using the SirCol collagen assay (Biocolor, Belfast, Northern Ireland). The total collagen content of tissue samples was determined by hydroxyproline assays57.
Histological analysis
Formalin-fixed, paraffin-embedded skin sections (2–5µm) were deparaffinized and stained with either hematoxylin and eosin (HE), sirius red or trichrome. Dermal thickness was analyzed at four different sites in each mouse in a blinded manner. For direct visualization of collagen fibers, sirius red staining was performed (Sigma-Aldrich, Steinheim, Germany). For evaluation of lung tissue, Ashcroft score was used as described elsewhere58. Liver cirrhosis evaluation (Scheuer Score) was performed as described elsewhere59.
Fluorescence imaging
Epitopes were retrieved from deparaffinized sections using a heat-induced method. Briefly, sections were alternately bathed in boiling sodium citrate buffer (10 mM sodium citrate, pH 6.0) or Tris-EDTA buffer (10 mM Tris base, 1 mM EDTA, 0.05 % Tween 20, pH 9.0). Each bathing step was repeated five times for 2 min following a washing step in distilled water for 5 min.
For cryo-sections, tissues were placed in OCT compound (Tissue-Tek) (Newcomer Supply, Middleton, USA) and then snap-frozen in liquid nitrogen and cut to 7 µm slices. Sections were washed in distilled water after thawing and fixed in 4% PBS-buffered formaldehyde for 10 min following another washing step in PBS.
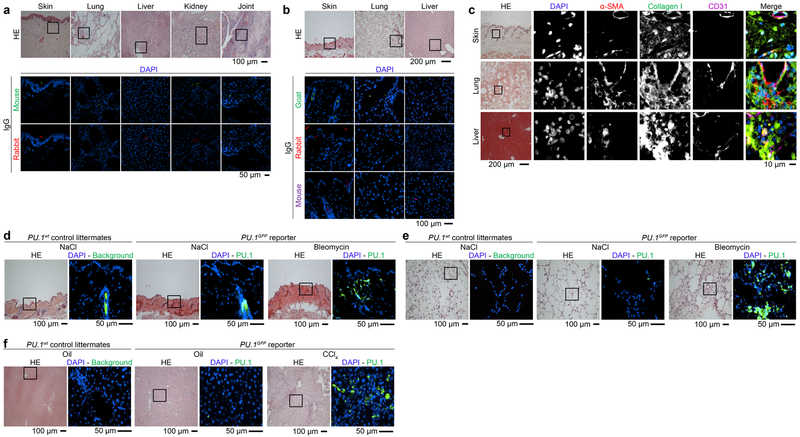
Next, sections were blocked for 1 h in PBS supplemented with 5 % BSA and 2 % horse serum. Primary antibodies were incubated over night at 4°C, secondary antibodies and DAPI after an intense washing step for 2h at ambient temperature. Consecutive staining was performed to minimize cross reactivity. Cross reactivity was blocked by pre-incubation with species specific Igs. The following antibodies were used: α-Smooth muscle clone 1A4 (1/500, Sigma-Aldrich), Cadherin 11 polyclonal (LS-B2308, 1/100, LS-Bio), CD45R/B220 clone RA3–6B2 (1/500, eBioscience), Collagen I clone COL1 (1/200), Collagen I polyclonal (ab21286, 1/500), CD11c clone N418 (1/100), CD45 polyclonal (ab10558, 1/500), F4/80 polyclonal (ab100790, 1/200), F4/80 clone CI:A3–1 (1/100), fibroblast activation protein polyclonal (ab28244, 1/5000), MRC2 polyclonal (ab70132, 1/1000), Vimentin clone VI-10 (1/500) (all abcam), CD3ε clone 145–2C11 (1/100), CD45-BV421 clone 30-F11 (1/500), CD49f-APC clone GoH3 (1/1000), CD117-APC clone 2B8 (1/1000), EpCAM-APC/Cy7 clone G8.8 (1/1000) , KRT14 polyclonal (905301, 1/1000) (all Biolegend), CD11b clone M1/70 (1/100), CD31 polyclonal (AF3628, 1/20), Ly6G/GR-1 clone RB6–8C5 (1/200) (all R&D Systems), PDGFRα-PE/Cy7 clone APA5 (1/100, Thermo Fisher Scientific), Prolyl 4-hydroxylase subunit beta clone 3–2B12 (1/50, Acris), PU.1 polyclonal (2266, 1/200, Cell Signaling) and Vimentin-Alexa 647 clone V9 (1/50, Santa Cruz Biotechnology). As secondary antibodies in IHC Rabbit-Alexa 594 polyclonal (A-11037, 1/200), Rabbit-Alexa 488 polyclonal (A-11034, 1/200), Rabbit-Alexa 647 polyclonal (A-21443, 1/500), Rat-Alexa 647 polyclonal (A-21472, 1/500), Mouse-Alexa 488 polyclonal (A11001, 1/200), Mouse-Alexa 647 polyclonal (A-21236, 1/500) and Goat-Alexa 647 polyclonal (A-21447, 1/500) were used (all Thermo Fisher Scientific). For IgG controls in IHC Goat IgG (sc-2028), Rabbit IgG (sc-2027), Rat IgG (sc-2026) and Mouse IgG (sc-2025) were used (all Santa Cruz Biotechnology). The specificity of the respective antibodies was controlled by corresponding IgG staining (Extended Data Fig. 10a, b). F-actin cytoskeleton was visualized with rhodamine-conjugated phalloidin (no. R415, Thermo Fisher Scientific, dilution 1:250). In addition, cell nuclei were stained using DAPI (no. sc-3598, Santa Cruz Biotechnology, dilution 1:800). Six randomly chosen high-power fields (HPF, 0.125 mm2) at 200-fold magnification per patient or healthy volunteer were evaluated by two experienced researchers in a blinded manner. Stained cells were visualized either using a Nikon Eclipse Ni-U microscope (Nikon, Badhoeve dorp, Netherlands) or using a CLSM-1P Leica SP5 II (Inverted) (Leica, Wetzlar, Germany). Representative images were reconstructed using the ImageJ distribution Fiji60,61. Voronoi tessellated pictures were generated as described elsewhere62. For quantification of F-actin, same microscope settings were used for each HPF. The mean intensity of rhodamine-conjugated phalloidin was measured as raw integrated density (RawIntDen) divided by the area of the cell using ImageJ. Myofibroblasts were identified as single cells double-positive for α-SMA and collagen and not directly adjacent to CD31 positive endothelial cells (Extended Data Fig. 10c); α-SMA and collagen double-positive cells were counted in three randomly chosen HPF of n specimens per mouse in a blinded manner at 200-fold magnification.
Chromatin immunoprecipitation (ChIP) assays
ChIP assays were performed using the ChIP-IT® Express Kit (Active Motif). 10 μg of sonificated chromatin extract was incubated with antibodies against H3K27me3, H3K27ac, H3K9me3, H3K4me1, H3K4me3 (no. 39155, 39297, 39161, 39135, 39915 all from Active Motif), PU.1, Smad3 (no. 2266, 9523 both from Cell Signaling Technology, Danvers, USA) and TEAD1 (no. 610923 from BD Biosciences) or normal rabbit or mouse IgG antibody (no. sc-2027 X, sc-2025, both from Santa Cruz Biotechnology). Purification was performed using the Chromatin IP DNA Purification Kit (Active Motif) and bound sequences were determined by quantitative real-time PCR using primers listed in supplementary table.
ChIP sequencing
Single end reads were generated from PU.1 precipitated (no. 2266 from Cell Signaling), input and IgG control DNA on an Illumina HiSeq 2500 system. Alignment to the GRCh37 reference genome was performed with bwa mem v0.7.14-r113663. MACS version 2.1.1.2016030964 was used to call peaks for each sample, using both input and IgG alignments as controls. In preparation for a motif enrichment search, two different region files were generated from the peak files as output by MACS: First, a file with the union of all three peak files, then a file with only the flanking regions (200 bp in either direction) of each peak region in the union file (vicinity analysis). Motif enrichment analysis was then performed using HOMER software version 4.9.143. For region of interest (ROI)-based ChIP-seq peak and RNA-seq overlap analysis, ROI were defined as 225kb upstream and downstream of differentially expressed genes65. Within each ROI, the most significantly enriched peak from ChIP-seq analysis was determined. Differentially expressed genes from RNA-seq were mapped to the annotation of PU.1 ChIP-seq peaks (q < 0.05). For identification of active regulatory elements (AREs), 11 ENCODE datasets from DNAse-Seq and Histone ChIP-Seq of human dermal and lung fibroblast (ENCFF128ARX, ENCFF148DHA, ENCFF195SIN, ENCFF328XNN, ENCFF350PQN, ENCFF392WNX, ENCFF524YEK, ENCFF811YTI, ENCFF965XKX, ENCFF073ILZ, wgEncodeBroadHistoneNhlfH3k27acStdSig) were used. For each file, the 99th percentile of all enrichment values was set as threshold. The regions beyond this threshold from all files were merged to determine the percentage of ChIP-seq peaks at various q-value thresholds overlapping these regions.
RNA sequencing
RNA sequencing was performed on an Illumina HiSeq 2500 system with a read length of 100 bp (forward only). After adapter trimming and filtering using cutadapt v1.9.166, reads were mapped to the Ensembl GRCh37 human reference using STAR v2.5.2a67. Features were counted with subread featureCounts v1.5.168 (count > 5 as threshold) on the Ensembl GRCh37 release 85 genome annotation. All further analysis was performed in R version 2.15.3 using the DESeq2 package69,70. Gene set enrichment analysis (GSEA) was performed using GSEA v3.0 software (Broad Institute)71,72. The statistical significance was assessed using 10000 random permutations of the gene set with a signal2noise metric for ranking genes. An FDR q-value < 0.25 was considered significant. Gene sets were obtained from the Molecular Signatures Database (MSigDB) v6.1 or created based on published signature genes (fibrotic cluster).
Bisulfite pyrosequencing
Genomic DNA was prepared from fibroblasts using the QIAamp DNA Blood Mini kit (QIAGEN). The DNA (1 µg) was bisulfite modified using the EpiTect bisulfite kit (QIAGEN). PCR amplifications of bisulfite modified DNA (2 µl) were performed using AmpliTaq Gold polymerase (Thermo Fisher Scientific). The PCR program was 95°C x 4 min; 95°C x 30 s, 52°C x 90 s, 72°C x 2 min x 5; 95°C x 30 s, 52°C x 90 s, 72°C x 90 s x 25; 72°C x 4 min. The following primers were used: PU.1 promoter: FW-5’-TAGTAAGTTAGGAGGGTAGTGGGTG, REV-biot 5’-CCCCATCCTAAAAAACTCTACATTA, PyroSeq FW 5’-GTTGGGTTGGTGGA GGAGT, PU.1 enhancer: FW 5’-GGTTGTAGTTGTTTTTGTTTTTATAT, REV-biot 5’-CTAAACATCCCCCTAAAACCTAAC, Pyro-Seq FW 5’-AGTTATTATAGGAAGTAT GTG. The PCR products were visualized with agarose gel electrophoresis. Afterwards, they were directly sequenced using the PyroMark Q48 Autoprep according to the manufacture instructions (QIAGEN).
Cell isolation and flow cytometry
Mice were sacrificed by cervical dislocation under anesthesia and dissected to generate single cell suspensions from the lung, spleen, thymus and/or bone marrow. Fat was thoroughly removed from the dissected organs and their capsules were opened to ensure good drainage of the digestive solution consisting of RPMI 1640 medium supplemented with 1 mg/ml Collagenase D from Clostridium histolyticum and 0.2 mg/ml DNase I, grade II from bovine pancreas (both from Roche Diagnostics, Mannheim, Germany). For the digestion of liver and lung samples, the digestive solution was enriched of 0.1 mg/ml Dispase II (Roche Diagnostics). Lung, spleen and thymus were digested in 1 ml digestion medium at 37 ° C for 1 h on a thermo shaker at 500 rpm (Eppendorf, Hamburg, Germany). Pipetting after every 20 min ensured good dissociation of the tissue. Tibia was cut off at both ends and bones were flushed with PBS to collect bone marrow. The resulting single cell suspensions were filtered through 70 µm cell strainers and washed in a larger volume of RPMI 1640 supplemented with 10 mM EDTA and 10 %FBS. Red blood cells were lysed after digest applying self-made ACK buffer for 1 min. Lysis was stopped adding a qs of 10X PBS to generate a 1X solution. Cells were then washed in PBS supplemented with 5 mM EDTA and 2 % FBS and filtered through 40 µm cell strainers. Skin and liver samples were centrifuged through a gradient to remove debris (Debris Removal Kit, Miltenyi Biotec). For flow cytometric analysis, 1 × 106 cells of the resulting single cell suspensions were incubated in 100 µl of diluted antibody solution in V-shaped plates for 20 min on ice. The following antibodies were used: CD3ε-Pacific Blue or -PE/Cy7 clone 145–2C11 (1/500 or 1/100), CD4-FITC clone RM4–5 (1/1,500), CD8a-APC clone 53–6.7 (1/300), CD11b-PE/Cy7 or -APC clone M1/70 (each 1/1000), CD11c-PE/Cy7 or -APC/Cy7 clone N418 (each 1/200), CD25-PE or PE/Cy7 clone PC61 (each 1/500), CD29-PE clone HMβ1–1 (1/1000), CD31-APC clone WM59 (1/1000), CD31-PerCP/Cy5.5 clone 390 (1/1000), CD34-PerCP/Cy5.5 clone HM34 (1/500), CD44-PE clone IM7 (1/2000), CD45-BV421 clone 30-F11 (1/2000), CD45-PerCP/Cy5.5 clone HI30 (1/1000), CD45R/B220-FITC or -APC/Cy7 clone RA3–6B2 (each 1/500), CD49f-APC clone GoH3 (1/1000), CD115-PE clone AFS98 (1/100), CD117-APC or -BV480 clone 2B8 (each 1/100), CD117-PE clone 104D2 (1/500), CD127/IL7R-PE/Cy7 clone A7R34 (1/100), EpCAM-APC/Cy7 clone G8.8 (1/500), EpCAM-FITC clone 9C4 (1/200), PDGFRα-PE clone 16A1 (1/100), PU.1-PE clone 7C2C34 (1/1000), TER119-APC/Cy7 clone TER119 (1/100) (all Biolegend), CD45-V500 clone 30F11 (1/1000), CD105-BV421 clone MJ7/18 (1/100) (all BD Biosciences), COL1A1-FITC clone 5D8-G9 (1/200, Merck), KRT14-PE clone LL002 (1/1000, Novus Biologicals), PDGFRα-PE/Cy7 clone APA5 (1/1000, eBiosciences) and Vimentin-Alexa 647 clone V9 (1/2000, Santa Cruz Biotechnology). For viability staining in flow cytometry Zombie Violet (423113, 1/1000, Biolegend), DAPI (D9542, 0.1µg/ml, Sigma-Aldrich) and eFluor780 (65–0865-14, 1/4000, eBiosciences) were used. Blocking of Fc-Receptors was performed prior to staining with fluorophore labelled antibodies. Murine blood (50 µl collected in EDTA) was incubated with the respective antibodies for 20 minutes at 4 ° C. Afterwards, 450 µl of RBC Lysis/Fixation Buffer (1X) (Biolegend, London, UK) was added for another 15 minutes at room temperature. After washing two times with PBS, cells were re-suspended in PBS supplemented with 5 mM EDTA and 2 % FBS and filtered through 40 µm cell strainers. All flow cytometric analysis was performed on a Gallios or Cytoflex-S flow cytometer (both Beckmann Coulter, Krefeld, Germany) equipped with 3 Laser (405nm, 488nm, 633nm) and 10 fluorescence detection channels and analyzed using Beckmann’s proprietary software Kaluza version1.5 or CytExpert. Gating strategy was performed as shown in Extended Data Fig. 8.
Gene-expression analysis
Total RNA was extracted from single cell suspensions using either the Nucleo Spin RNA isolation kit (Macherey-Nagel, Düren, Germany) or the miRNeasy Mini Kit (QIAGEN). 1 µg of RNA was used to transcribe mRNA to cDNA following standard protocols. Real-time PCR was performed in triplicates using either the SYBR® Select Master Mix (Thermo Fisher Scientific) or the miScript SYBR Green PCR Kit and miScript Primer Assay (both from QIAGEN) and a QuantStudio™ 6 Flex System (Thermo Fisher Scientific). Expression of target genes was calculated by the ΔCT comparative method for relative quantification after normalization. Samples without enzyme in the reverse transcription reaction (non-RT controls) were used as negative controls. Unspecific signals caused by primer dimers were excluded by non-template controls and by dissociation curve analysis. ACTB, let-7b or miR-15a were used to normalize for the amounts of cDNA within each sample. The following miScript primer assays were used: let-7b (MS00003122), miR-15a (MS00003178), miR-92a-2 (MS00032137), miR-155 (MS00031486), miR-326 (MS00003948), miR-580 (MS00010227), miR-6745 (MSC0075916), miR-6747 (MS00046515), miR-6780a (MS00046872) were used (all from QIAGEN). Primer sequences are listed in supplementary table.
In silico analysis of potential microRNA binding sites to PU.1
For predictions of potential microRNA binding to PU.1, miRWalk73, miRanda74 and Targetscan75 were used. The overlap of possible miRNAs from all 3 tools were further restricted to p < 0.0233 as well as PU.1 conserved binding sites predicted by miRWalk73,76.
Western Blot analysis
Protein concentrations were determined using the Bradford Protein Assay according to the manufacturer’s protocol (Bio-Rad). Same amounts of protein were loaded on a Tris-Glycine buffered gel. Proteins were separated by SDS-PAGE and transferred to PVDF membrane. The membrane was incubated with the appropriate primary antibody and HRP-conjugated secondary antibodies (Dako, Glostrup, Denmark). The following antibodies were used: β-Actin clone A5441 (1/10000, Sigma-Aldrich), Collagen I clone COL-1 (1/1000, abcam), TEAD1 clone 610923 (1/500, BD Biosciences), EZH2 polyclonal (4905, 1/2000), total histone H3 polyclonal (9715, 1/1000), Tri-Methyl-Histone H3(Lys27) polyclonal (9733, 1/1000), total polyclonal (9513, 1/1000) or phospho-Smad3 polyclonal (9520, 1/1000) and PU.1 polyclonal (2266, 1/500) (all Cell Signaling). As secondary antibodies in western blot anti-mouse polyclonal (P0447, 1/1500) or anti-rabbit polyclonal (P0448, 1/2000) HRP-conjugated secondary antibodies (all Dako) were used. Blots were visualized using enhanced chemiluminescence (ECL). β-Actin was used as a loading control. Western Blots were quantified using ImageJ Software (version 1.46r).
Animal studies of fibrosis
The role of PU.1 in fibrosis was investigated in five different mouse models. Fibrosis was induced in six to eight weeks old littermates of the stated background. (i) Bleomycin-induced skin fibrosis was induced by local injections of bleomycin at a concentration of 0.5 mg/ml in defined areas of 1cm2 at the upper back every other day for 4 weeks (6 weeks of age, mixed genders)77. Subcutaneous injections of 0.9% NaCl served as controls. (ii) In the Tsk-1 model (10 weeks of age, mixed genders), a dominant mutation of gene that encodes fibrillin-1 results in activated TGF-β signaling in Tsk-1 fibroblasts and progressive, generalized hypodermal thickening within 10 weeks after birth67. (iii) Bleomycin-induced pulmonary fibrosis was induced by a single intratracheal application of bleomycin (0.025 U, 8 weeks of age, males) using a high pressure syringe (Penn-Century, Wyndmoor, PA, USA). Instillation of equal volumes of 0.9% NaCl served as a control78. (iv) CCl4-induced hepatic fibrosis was induced by i.p. injections of CCl4 diluted in sunflower oil (week 1: 1:31 dilution; week 2: 1:15 dilution; week 3: 1:7 dilution; week 4–6: 1:3 dilution) in mice (8 weeks of age, mixed genders)79 three times per week. Sunflower oil was used in the control group. (v) The LP/J-->C57BL/6 minor histocompatibility antigen-mismatched model, which reflects clinical and pathologic symptoms of human sclerodermatous cGVHD, was used80. Recipient C57BL/6 mice underwent total body irradiation with 1 dose of 9.5 Gy. Each recipient mouse received 5 × 107 splenocytes dissolved in 100 μL PBS within 6 hours post irradiation from either C57BL/6 in a syngeneic or LP/J in an allogeneic, multiple minor mismatched transplantation via eye vein injection. As indicated, mice were treated with DB197681. DB1976 was solved in water and applied i.p. Controls received NaCl. In the (i) preventive model of bleomycin-induced skin fibrosis, DB1976 was injected i.p. simultaneously to bleomycin or NaCl applications for 4 weeks. (ii) In the therapeutic model of bleomycin-induced skin fibrosis, mice were pre-challenged with bleomycin for 3 weeks to induce robust skin fibrosis. After 3 weeks, treatment with DB1976 or NaCl as control was initiated, while injections with bleomycin were continued. After a total of 6 weeks of bleomycin and 3 weeks of treatment with DB1976/NaCl, mice were sacrificed and the extent of fibrosis was compared to control mice. (iii) In the bleomycin-induced lung fibrosis model, DB1976 was injected for 4 weeks. (iv) In the model of CCl4-induced liver fibrosis, mice were treated for 6 weeks. ADVIA 120 analyzer (version 3.1.8.0-MS; Siemens Healthcare Diagnostics) was used for analysis of red blood count (RBC), white blood count (WBC), thrombocytes and reticulocyte count. In all murine experiments, body weight as well as pain and distress levels were monitored every second day. Pain and distress was evaluated as followed: 0 = no signs of stress, mouse is active in good condition calm and has normal appetite; 1 = no/mild signs of stress, mouse is active but shows some signs of restlessness; 2 = mild pain and distress, mouse is not well groomed and slightly hunched, less appetite; 3 = moderate stress, mouse moves slowly and shows signs of depression; 4 = severe pain, mouse loses significant weight and shows non-response reaction when touched, if symptoms become worse, mice were excluded from analysis and euthanized82.
Statistical analysis
Results were visualized and analyzed with Prism version 7 (GraphPad Software, La Jolla, USA) and are depicted as the mean ± SEM if not stated otherwise. For a two-group comparison, a Mann-Whitney U-test for nonparametric data was used. When two groups of samples were compared for iterating parameters or more than two groups of samples were compared, one-way ANOVA was used. Tukey’s range test was used as post-hoc analysis of ANOVA. Significance levels are indicated as suggested by Prism Software: n.s. p > 0.5, * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001.
Data availability
The RNA-seq data used in this study have been deposited in the Gene Expression Omnibus (GEO) under the accession number GSE122334. The sequencing data from the ChIP-seq expriments have been submitted to the National Center for Biotechnology Information (NCBI) database under BioProject PRJNA480591; the project includes following biosamples: SUB4300598, SUB4300595, SUB4300592, SUB4300591, SUB4300589, SUB4300587, SUB4300586, SUB4300583 and SUB4300579; the FASTQ data was uploaded to the NCBI Sequence Read Archive (SRA).
Extended Data
Extended Data Figure 1: PU.1-expressing fibroblasts control tissue fibrosis.
Representative images of (a, b) immunofluorescence (IF) and (c) confocal microscopy of human skin, lung, liver, kidney and joint biopsy specimens stained for PU.1 (red), prolyl 4-hydroxylase (P4H)β (green), CD45 or CD11b (cyan), and DAPI (blue); respective tissues were obtained from healthy individuals (n = 5 each), idiopathic pulmonary fibrosis (IPF; n = 4), acute asthma (n = 5), alcoholic liver cirrhosis (n = 4), autoimmune hepatitis (n = 4), cirrhotic kidney (n = 4), interstitial nephritis (n = 5), osteoarthritis (OA; n = 5) and rheumatoid arthritis (RA; n = 5). Hematoxylin & eosin (HE) stained tissue specimens are included. (d) Representative IF images (n = 4) of explanted fibrotic fibroblasts stained for PU.1 (red) and one of the following markers (green): fibroblast activation protein (FAP), cadherin 11 (CDH11) or MRC-2; nuclei were stained with DAPI (blue). (e) Semi-quantification of PU.1+ fibroblasts / total P4Hβ+ fibroblasts per high-power field (HPF); respective tissues were obtained from healthy individuals (n = 5 each), patients with systemic sclerosis (n = 10), plaque psoriasis (n = 7), idiopathic pulmonary fibrosis (IPF; n = 4), acute asthma (n = 5), alcoholic liver cirrhosis (n = 4), autoimmune hepatitis (n = 4), cirrhotic kidney (n = 4) and interstitial nephritis (n = 5), osteoarthritis (OA; n = 5) and rheumatoid arthritis (RA; n = 5). (f) Cell counts and viability of CRISPR-Cas9 mediated PU.1 knockout in human fibrotic fibroblasts compared to unaffected control fibroblasts and fibroblasts treated with 50% DMSO as toxic control (n = 3 each). Cells were counted per HPF. (g) Resting fibroblasts co-transfected with different amounts of PU.1 plasmid as indicated (n = 4 each); cell viability of fibroblasts was determined by CCK-8 toxicity assay. (h-k) Relative Col1a1, Col1a2 mRNA levels, hydroxyproline concentration, myofibroblast counts / HPF and respective histological scores (skin thickness, Ashcroft, Scheuer) of (h) bleomycin-induced skin (n = 6 per group), (i) bleomycin-induced lung fibrosis (n = 6 per group), (j) carbon tetrachloride (CCl4)-induced liver fibrosis model (n = 5 per group) and (k) sclerodermatous chronic graft-versus-host disease (scl cGvHD) model (n = 6 per group); data are shown as the mean ± s.e.m. of respective n independent experiments. Respective P values were determined by one-way ANOVA with Tukey's multiple comparison post hoc test.
Extended Data Figure 2: PU.1-expressing fibroblasts in different mouse models of fibrosis.
Representative hematoxylin & eosin (HE) and immunofluorescence (IF) stainings of (a) bleomycin-induced skin fibrosis model (n = 8 per group); injections of the solvent, sodium chloride (NaCl), served as controls. (b) Mouse model of sclerodermatous chronic graft versus host disease (scl cGvHD; n = 8 per group); syngeneic transplanted mice were used as controls. (c) Fibrosis model of tight skin 1 (Tsk-1) mice (n = 11 per group); (d) model of bleomycin-induced pulmonary fibrosis (n = 6 per group); controls received intratracheal application of NaCl. Representative HE and IF images of respective tissues stained for PU.1 (red), vimentin (green), DAPI (blue) are included. Total Pu.1 mRNA in the respective tissues was measured by qPCR. Absolute counts of PU.1-expressing fibroblasts were analyzed per high power field (HPF). (e, f) Mouse model of bleomycin-induced skin fibrosis (n = 5 per group); controls received NaCl. Representative HE and IF images of frozen serial tissue sections; boxed areas in the HE stained sections indicate the representative histological regions (yellow, orange, purple) of the correspondingly framed IF panels; (e) control littermates or PU.1GFP reporter mice stained for DAPI (blue) and the respective antibody as indicated in the figure (red); (f) IgG control of NaCl treated control littermates of PU.1GFP reporter mice (n = 3 per group) (g) Semi-quantitative analysis of PU.1 (GFP)-expressing fibroblasts. Absolute counts of PU.1-expressing fibroblasts were analyzed per HPF (respective n is given in e). Control images of GFP+ tissue sections are presented in Extended Data Fig. 10d. Data are shown as the mean ± s.e.m. of respective n independent experiments. P values were determined by either one-way ANOVA with Tukey's multiple comparison post hoc test or two-tailed Mann–Whitney U test if two groups were compared.
Extended Data Figure 3: PU.1-expressing fibroblasts in bleomycin-induced lung and CCl4-induced liver fibrosis.
(a-e) Mouse model of bleomycin-induced lung fibrosis (n = 4 per group); controls received sodium chloride (NaCl). (a, b) Representative hematoxylin & eosin (HE) and immunofluorescence (IF) images of (a) frozen serial tissue sections of control littermates or PU.1GFP reporter mice stained for DAPI (blue) and the respective antibody as indicated in the figure (red); (a, b, f, g) boxed areas in the HE stained sections indicate the representative histological regions of the corresponding IF panels; experiments were repeated three-times independently with similar results; (b) IgG control of NaCl treated control littermates of PU.1GFP reporter mice (n = 3 per group). (c) Semi-quantitative analysis of PU.1 (GFP)-expressing fibroblasts (n = 4 each). Absolute counts of PU.1-expressing fibroblasts were analyzed per high-power field (HPF). Control images of GFP+ tissue sections are presented in Extended Data Fig. 10e. (d, e) Flow cytometric analysis of digested lungs; (d) respective gating strategy to characterize GFP+ cells; (e) quantitative analysis of PU.1 (GFP)-expressing fibroblasts (n = 3 each). Percentage of CD45−vimentin+ PU.1-expressing fibroblasts per lung sample. (f-j) Mouse model of CCl4-induced liver fibrosis (n = 4); controls received oil. (f, g) Representative HE and IF images of frozen serial tissue sections of (f) control littermates or PU.1GFP reporter mice stained for DAPI (blue) and the respective antibody as indicated in the figure (red); (g) IgG control of sunflower oil treated control littermates of PU.1GFP reporter mice (n = 4 per group). (h) Semi-quantitative analysis of PU.1 (GFP)-expressing fibroblasts (n = 3 each). Absolute counts of PU.1-expressing fibroblasts were analyzed per HPF. Control images of GFP+ tissue sections are presented in Extended Data Fig. 10f. (i, j) Flow cytometric analysis of digested livers; (i) respective gating strategy to characterize GFP+ cells; (j) quantitative analysis of PU.1 (GFP)-expressing fibroblasts (n = 4 each). Percentage of CD31−CD45−vimentin+ PU.1-expressing fibroblasts per liver sample; data are shown as the mean ± s.e.m. of respective n biologically independent samples. P values were determined by one-way ANOVA with Tukey's multiple comparison post hoc test.
Extended Data Figure 4: Regulation of PU.1 expression in fibroblasts.
(a-c) PU.1 expression levels of primary human fibroblasts. Representative Western Blot and semi-quantitative analysis of PU.1 protein expression in resting (isolated from normal skin), fibrotic (isolated from fibrotic skin of SSc patients) and inflammatory (isolated from inflamed joints of RA patients) fibroblasts stimulated (a) with/without TNF-α for 24 hours, (b) with/without TGF-β for 24 hours or (c) for up to 72 hours (n = 4 each). Protein extracts of fibrotic fibroblasts were used as positive control in each lane. (d) ChIP analysis (n = 4 each) assessing binding of Smad3 to PU.1 promoter and its −17 kb upstream regulatory element (URE) is shown. (e) siRNA mediated knockdown of SMAD3 in fibrotic fibroblasts stimulated with/without TGF-β for 24 hours (n = 5). Scrambled (scr) siRNA was used as control. (f) Expression levels of PU.1 protein in primary human resting, fibrotic and inflammatory fibroblasts (n = 3 each) cultured ex vivo for several passages. (g) Expression levels of the enhancer of zeste homolog 2 (EZH2) in resting (n = 11), fibrotic (n = 9) and inflammatory fibroblasts (n = 7) relative to β-actin as assessed by Western blot analysis; results are presented relative to resting fibroblasts. (h, i) Resting fibroblasts treated with different concentrations of GSK126 as indicated (n = 3 each); (h) cell viability of fibroblasts was determined by CCK-8 toxicity assay. (i) Expression levels of H3K27me3 relative to total H3 as assessed by Western blot analysis; results are presented relative to untreated control. (j, k) Inflammatory fibroblasts treated with different concentrations of miR-155 antagomirs as indicated (n = 3 each) to investigate (j) cell viability by CCK-8 toxicity assay; (k) miR-155 expression levels relative to let-7b as assessed by qPCR; results are presented relative to cells co-transfected with scrambled antagomirs. (l, m) Fibrotic fibroblasts treated with different concentrations of DB1976 to analyze (l) cell viability by CCK-8 toxicity assay (n = 6); (m) DB1976 dose dependent effects upon TGF-β-induced collagen production (n = 4 each); results are presented relative to untreated control. Data are shown as the mean ± s.e.m. of respective n independent experiments. P values were determined by one-way ANOVA with Tukey's multiple comparison post hoc test.
Extended Data Figure 5: Profibrotic potential of PU.1.
(a) Gene set enrichment analysis (GSEA) of quantitative RNA-Seq signals of Gene Ontology (GO)-defined monocytic related gene clusters in human resting fibroblasts co-transfected with PU.1 (n = 4). Resting fibroblasts co-transfected with control plasmid served as controls (n = 4). NES, normalized enrichment score; (b) mRNA expression levels of indicated transcripts in human resting fibroblasts treated with or without DB1976 and simultaneously co-transfected with or without PU.1 (pUNO.1-hSPI1, called ‘PU.1 OE’ here, n = 5 each) as assessed by qPCR; co-transfection with scrambled (scr) plasmid served as control. Results are presented relative to cells co-transfected with scr. (c) Genomic annotation of PU.1-binding sites defined by ChIP-seq analysis in primary human fibrotic fibroblasts. TSS, transcription start site. (d) Annotation of PU.1 ChIP-seq peaks (n = 3 each) at various q-value treshholds to active regulatory elements (AREs). For unbiased identification of AREs 11 ENCODE datasets from DNAse-seq and histone ChIP-seq were used as described in materials & methods; q-values are those provided by MACS2 call-peak64. (e) Differentially expressed genes from gene sets of inflammatory fibroblasts co-transfected with PU.1 (PU.1 OE) or scr vector as control (ctrl); gene sets include fibrosis-associated, inflammatory and matrix-degrading pathways determined by qPCR (n = 4 each). Colors represent the significance levels of the observed changes of the respective expression levels in PU.1 OE compared to ctrl. (f) Micro-mass organoids of inflammatory fibroblasts co-transfected with PU.1 (PU.1 OE) or scr vector as control in the presence of TNF-α for 21 days (n = 8 each). Sections of micro-mass organoids were stained with hematoxylin & eosin. Lining fibroblasts were quantified relative to total number of cells per high power field (HPF). (g) 3D full skin organoid model of inflammatory fibroblasts co-transfected with PU.1 (PU.1 OE) or scr vector as control; EL: epidermal layer, DL: dermal layer; collagen content was measured by hydroxyproline assay; α-SMA and skin thickness were quantified per HPF (n = 4 each). (h) mRNA expression levels of indicated transcripts in primary human inflammatory fibroblasts treated with or without DB1976 and simultaneously co-transfected with or without miR-155 antagomirs (n = 4 each); results are presented relative to cells co-transfected with scrambled antagomirs (scr). Data are shown as the mean ± s.e.m. of respective n independent experiments. P values were determined either according to Subramanian et al.72 (a), by one-way ANOVA with Tukey's multiple comparison post hoc test (b, h) or two-tailed Mann–Whitney U test (e-g).
Extended Data Figure 6: PU.1 anchors differentiation towards fibrotic fibroblasts in a network of flanking factors including TEAD1.
(a) TEAD1 expression levels of primary human fibroblasts. Representative Western Blot and semi-quantitative analysis of TEAD1 protein expression in resting, fibrotic and inflammatory fibroblasts (n = 4 each). (b) ChIP analysis of TEAD1 binding at fibrotic signature gene regions in the vicinity of PU.1 binding sites; DNA fragments of human fibrotic fibroblasts were immunoprecipitated with anti-TEAD1 and analyzed by qPCR relative to input DNA (n = 4 each); results are compared to IgG control. Signature pro-fibrotic genes were screened for PU.1 ChIP-seq peaks and potential flanking TEAD1 binding site. (c) mRNA expression levels of indicated transcripts in primary human inflammatory fibroblasts co-transfected with PU.1 (PU.1 OE) or scrambled (scr) plasmid (n = 4 each); cells were cultured under neutral conditions (serum-starved medium only) or in the presence of TGF-β (fibrotic) or TNF-α (‘inflam’, inflammatory). Results are presented relative to scr under neutral culture conditions. Data are shown as mean ± s.e.m. of respective n biologically independent samples. P values were determined either by one-way ANOVA with Tukey's multiple comparison post hoc test or two-tailed Mann–Whitney U test if two groups were compared.
Extended Data Figure 7: PU.1-silencing in experimental fibrosis.
(a-f) Experimental fibrosis models; representative images of (a, b) trichrome or (c, d) sirius red stained tissue sections; mRNA levels of Col1a1 Col1a2, hydroxyproline content, myofibroblast counts, and respective histological scores (skin thickness, Ashcroft, Scheuer) of mice treated with/without DB1976. Mice treated with sodium chloride (NaCl) or oil served as controls (ctrl). (a, b) Bleomycin-induced skin fibrosis model with (a) preventive (n = 7) and (b) therapeutic (n = 8) treatment; in the latter regression of pre-established fibrosis was evaluated since mice were challenged with bleomycin for 3 weeks to induce robust skin fibrosis before treatment with DB1976 was initiated, while injections with bleomycin were continued. As additional control, mice were injected with bleomycin for 3 weeks followed by injections of NaCl for another 3 weeks. (c) Bleomycin-induced lung fibrosis model (n = 5); (d) CCl4-induced liver fibrosis model (n = 5); (e) body weight, (f) pain and distress levels of DB1976-treated mice as monitored every second day (n = 5 each); mice challenged with subcutaneous injections of bleomycin served as positive controls. Data are shown as the mean ± s.e.m. of respective n biologically independent samples. P values were determined by one-way ANOVA with Tukey's multiple comparison post hoc test.
Extended Data Figure 8: Effects of DB1976 in anti-fibrotic concentrations on hematopoietic cells and bone marrow derived stem cells.
(a, c, e, g, i) Flow cytometric gating strategy to identify (a) different peripheral blood cells and (c) different splenic cell populations, (e) B cell precursors and mature B cells in the bone marrow, (g) T cell precursors and mature T cells in the thymus or (i) bone marrow derived mesenchymal stem cells (MSC) and hematopoietic stem cells (HSC) in mice treated with different concentrations of DB1976 or NaCl (n = 3 each) as control for 6 weeks; FMO = fluorescence minus one controls. (b) White blood count (WBC), red blood count (RBC), numbers of thrombocytes (TBC) and T-cell/B-cell ratio in the peripheral blood; (d) quantification of splenic monocytes (Mo), macrophages (Mph), dendritic cells (DC), and the T-cell/B-cell ratio; (f) frequencies of respective B cell populations as indicated; (h) frequency of respective thymocyte subsets; DN, double-negative thymocytes; DP, double-positive thymocytes; DN thymocytes according to the expression of CD25 and CD44; (j) percentage of Lin−CD29+CD105+ MSCs in the bone marrow; mean fluorescence intensity (MFI) of CD29+ MSCs; MFI of CD105+ MSCs; percentage of CD45+CD34+ HSCs in the bone marrow; data are shown as the mean ± s.e.m. of respective n biologically independent samples. P values were determined by one-way ANOVA with Tukey's multiple comparison post hoc test.
Extended Data Figure 9: Characterization of cultured fibroblast phenotypes.
(a-c) Gating strategy of cultured human (a) resting, (b) fibrotic and (c) inflammatory fibroblasts stained for PDGFRα, Collagen I, vimentin (fibroblast markers) and for KRT14, CD31, CD45, CD326 (control markers). Respective isotype and corresponding positive controls for KRT14 (human keratinocytes), CD31 (human umbilical vein endothelial cells), CD45 (human peripheral blood mononuclear cells) and EpCAM (human kidney tubular epithelial cells) are included. (d, e) Proliferation, migration and invasion of different passages (P3, P5, P8) of human resting, fibrotic and inflammatory fibroblasts (n = 3 each) as assessed by xCELLigence Real Time Cell Analysis Instrument. Resting fibroblasts cultured in the absence of a gradient of chemoattractants served as controls. Data are shown as the mean ± s.e.m. of respective n biologically independent samples. P values were determined by one-way ANOVA with Tukey's multiple comparison post hoc test.
Extended Data Figure 10. Control stainings of human and murine tissues.
Representative hematoxylin & eosin (HE) and immunofluorescence (IF) images of (a-c) paraffin-embedded human (a) skin, lung, liver, kidney and joint tissues or murine (b) skin, lung and liver tissues stained for DAPI and Ig controls as indicated (n = 5 each). (c) Representative images of HE stained and IF images of murine biopsy specimens (n = 5 each) of fibrotic skin, lung and liver stained for DAPI, α-SMA, Collagen I and CD31. (d-f) Representative HE and IF images of frozen tissue sections of control littermates or PU.1GFP reporter mice stained for DAPI (blue) in the mouse model of (d) bleomycin-induced skin fibrosis (n = 5 per group); controls received NaCl; (e) mouse model of bleomycin-induced lung fibrosis (n = 4 per group); controls received NaCl; (f) mouse model of CCl4-induced liver fibrosis (n = 4 per group); controls received oil.
Supplementary Material
Acknowledgements
The authors thank to Mónica Pascual, Katja Dreißigacker, Kai-Ting Yang, Regina Kleinlein and Matthias Spiller-Becker for excellent technical assistance. We thank Ralf Palmisano and Philipp Tripal of the Optical Imaging Center Erlangen (OICE) for helping with confocal images.
Grant support
Grants RA 2506/3–1, RA 2506/4–1, DI 1537/9–1, DI 1537/11–1, DE 2414/2–1, AK 144/2–1, DFG FOR2438, SCHE 1583/7–1, SPP1468, CRC1181 (C01) and SFB TR221 (B04) of the German Research Foundation, the Bundesministerium für Bildung und Forschung (BMBF; project Metharthros), the Marie Curie project OSTEOIMMUNE, the TEAM project of the European Union (305549) and the IMI funded project RTCure (115142), Else Kröner-Fresenius-Stiftung 2014_A184, grants A64, D28 and J40 of the IZKF in Erlangen, grant 16–10-05–1-Ramming of the ELAN-Foundation Erlangen, grant 2017.129.1 of the Wilhelm Sander-Stiftung, NIH (R01 A1057459 and AI095282 to MHK; H1.129063 to GMKP).
Footnotes
Conflict of Interest
None
REFERENCES
- 1.Driskell RR et al. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature 504, 277–281, doi: 10.1038/nature12783 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalluri R & Zeisberg M Fibroblasts in cancer. Nat Rev Cancer 6, 392–401 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Flavell SJ et al. Fibroblasts as novel therapeutic targets in chronic inflammation. British Journal of Pharmacology 153, S241–S246, doi: 10.1038/sj.bjp.0707487 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palumbo-Zerr K et al. Orphan nuclear receptor NR4A1 regulates transforming growth factor-beta signaling and fibrosis. Nat Med 21, 150–158, doi: 10.1038/nm.3777 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Ramming A, Dees C & Distler JH From pathogenesis to therapy - Perspective on treatment strategies in fibrotic diseases. Pharmacol Res 100, 93–100, doi: 10.1016/j.phrs.2015.06.012 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Chakraborty D et al. Activation of STAT3 integrates common profibrotic pathways to promote fibroblast activation and tissue fibrosis. Nat Commun 8, 017–01236 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McInnes IB & Schett G Cytokines in the pathogenesis of rheumatoid arthritis. Nat Rev Immunol 7, 429–442, doi: 10.1038/nri2094 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Parsonage G et al. Global gene expression profiles in fibroblasts from synovial, skin and lymphoid tissue reveals distinct cytokine and chemokine expression patterns. Thrombosis and haemostasis 90, 688–697, doi: 10.1160/th03-04-0208 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Kiener HP et al. Synovial fibroblasts self-direct multicellular lining architecture and synthetic function in three-dimensional organ culture. Arthritis & Rheumatism 62, 742–752, doi: 10.1002/art.27285 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Wynn TA & Ramalingam TR Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat Med 18, 1028–1040, doi: 10.1038/nm.2807 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank-Bertoncelj M et al. Epigenetically-driven anatomical diversity of synovial fibroblasts guides joint-specific fibroblast functions. Nat Commun 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Assassi S et al. Dissecting the heterogeneity of skin gene expression patterns in systemic sclerosis Arthritis & rheumatology (Hoboken, N.J.) 67, 3016–3026 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jolma A et al. DNA-binding specificities of human transcription factors. Cell 152, 327–339, doi: 10.1016/j.cell.2012.12.009 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Scott EW, Simon MC, Anastasi J & Singh H Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages Science (New York, N.Y.) 265, 1573–1577 (1994). [DOI] [PubMed] [Google Scholar]
- 15.McKercher SR et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. Embo J 15, 5647–5658 (1996). [PMC free article] [PubMed] [Google Scholar]
- 16.Scanlan MJ et al. Molecular cloning of fibroblast activation protein alpha, a member of the serine protease family selectively expressed in stromal fibroblasts of epithelial cancers. Proc Natl Acad Sci U S A 91, 5657–5661 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu M et al. Identification of cadherin 11 as a mediator of dermal fibrosis and possible role in systemic sclerosis Arthritis & rheumatology (Hoboken, N.J.) 66, 1010–1021 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uccelli A, Moretta L & Pistoia V Mesenchymal stem cells in health and disease. Nat Rev Immunol 8, 726–736 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Sheikh H, Yarwood H, Ashworth A & Isacke CM Endo180, an endocytic recycling glycoprotein related to the macrophage mannose receptor is expressed on fibroblasts, endothelial cells and macrophages and functions as a lectin receptor. J Cell Sci 113, 1021–1032 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Soare A, Ramming A, Avouac J & Distler JHW Updates on animal models of systemic sclerosis. Journal of Scleroderma and Related Disorders, 1–11, doi: 10.5301/jsrd.5000220 (2016). [DOI] [Google Scholar]
- 21.Gabrielli A, Avvedimento EV & Krieg T Scleroderma. N Engl J Med 360, 1989–2003, doi: 10.1056/NEJMra0806188 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Sonnylal S et al. Postnatal induction of transforming growth factor beta signaling in fibroblasts of mice recapitulates clinical, histologic, and biochemical features of scleroderma. Arthritis Rheum 56, 334–344 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Gyorfi AH, Matei AE & Distler JHW Targeting TGF-beta signaling for the treatment of fibrosis. Matrix Biol 69, 8–27 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Zeybel M et al. Multigenerational epigenetic adaptation of the hepatic wound-healing response. Nat Med 18, 1369–1377 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramming A, Druzd D, Leipe J, Schulze-Koops H & Skapenko A Maturation-related histone modifications in the PU.1 promoter regulate Th9-cell development. Blood 119, 4665–4674, doi: 10.1182/blood-2011-11-392589 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Van De Wiel MA et al. Bayesian analysis of RNA sequencing data by estimating multiple shrinkage priors. Biostatistics 14, doi: 10.1093/biostatistics/kxs031 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Bechtel W et al. Methylation determines fibroblast activation and fibrogenesis in the kidney. Nature medicine 16, 544–550, doi: 10.1038/nm.2135 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noda S et al. Simultaneous downregulation of KLF5 and Fli1 is a key feature underlying systemic sclerosis. 5, 5797, doi: 10.1038/ncomms6797 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao R et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing Science (New York, N.Y.) 298, 1039–1043, doi: 10.1126/science.1076997 (2002). [DOI] [PubMed] [Google Scholar]
- 30.McCabe MT et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492, 108, doi: 10.1038/nature11606 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Christmann RB et al. miR-155 in the progression of lung fibrosis in systemic sclerosis. Arthritis research & therapy 18, 016–1054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Migita K et al. TNF-α-induced miR-155 regulates IL-6 signaling in rheumatoid synovial fibroblasts. BMC Research Notes 10, 403, doi: 10.1186/s13104-017-2715-5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vigorito E et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity 27, 847–859, doi: 10.1016/j.immuni.2007.10.009 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pham TH et al. Mechanisms of in vivo binding site selection of the hematopoietic master transcription factor PU.1. Nucleic Acids Res 41, 6391–6402 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stephens DC et al. Pharmacologic efficacy of PU.1 inhibition by heterocyclic dications: a mechanistic analysis. Nucleic Acids Research 44, 4005–4013, doi: 10.1093/nar/gkw229 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munde M et al. The unusual monomer recognition of guanine-containing mixed sequence DNA by a dithiophene heterocyclic diamidine. Biochemistry 53, 1218–1227, doi: 10.1021/bi401582t (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farina G, Lafyatis D, Lemaire R & Lafyatis R A FOUR-GENE BIOMARKER PREDICTS SKIN DISEASE IN PATIENTS WITH DIFFUSE CUTANEOUS SYSTEMIC SCLEROSIS. Arthritis and rheumatism 62, 580–588, doi: 10.1002/art.27220 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanchez-Lopez E et al. Targeting colorectal cancer via its microenvironment by inhibiting IGF-1 Receptor-insulin receptor substrate and STAT3 signaling. Oncogene 35, 2634–2644, doi: 10.1038/onc.2015.326 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scholten D et al. Migration of Fibrocytes in Fibrogenic Liver Injury. The American Journal of Pathology 179, 189–198, doi: 10.1016/j.ajpath.2011.03.049 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vollmann EH et al. Identification of Novel Fibrosis Modifiers by In Vivo siRNA Silencing. Molecular Therapy. Nucleic Acids 7, 314–323, doi: 10.1016/j.omtn.2017.04.014 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henderson NC et al. Targeting of [alpha]v integrin identifies a core molecular pathway that regulates fibrosis in several organs. Nat Med 19, 1617–1624 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakai N et al. LPA(1)-induced cytoskeleton reorganization drives fibrosis through CTGF-dependent fibroblast proliferation. The FASEB Journal 27, 1830–1846, doi: 10.1096/fj.12-219378 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell 38, 576–589, doi: 10.1016/j.molcel.2010.05.004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gosselin D et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 159, 1327–1340 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bakri Y et al. Balance of MafB and PU.1 specifies alternative macrophage or dendritic cell fate. Blood 105, 2707–2716 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Chang HC et al. The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nat Immunol 11, 527–534, doi: 10.1038/ni.1867 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anderson KL et al. Transcription factor PU.1 is necessary for development of thymic and myeloid progenitor-derived dendritic cells. J Immunol 164, 1855–1861 (2000). [DOI] [PubMed] [Google Scholar]
- 48.Rosenbauer F & Tenen DG Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol 7, 105–117 (2007). [DOI] [PubMed] [Google Scholar]
- 49.van den Hoogen F et al. 2013 classification criteria for systemic sclerosis: an American college of rheumatology/European league against rheumatism collaborative initiative. Annals of the rheumatic diseases 72, 1747–1755, doi: 10.1136/annrheumdis-2013-204424 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Singal AK, Bataller R, Ahn J, Kamath PS & Shah VH ACG Clinical Guideline: Alcoholic Liver Disease. The American journal of gastroenterology 113, 175–194, doi: 10.1038/ajg.2017.469 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.EASL Clinical Practice Guidelines: Autoimmune hepatitis. Journal of hepatology 63, 971–1004 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Aletaha D et al. 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Annals of the rheumatic diseases 69, 1580–1588, doi: 10.1136/ard.2010.138461 (2010). [DOI] [PubMed] [Google Scholar]
- 53.Zheng B, Zhang Z, Black CM, de Crombrugghe B & Denton CP Ligand-dependent genetic recombination in fibroblasts : a potentially powerful technique for investigating gene function in fibrosis. Am J Pathol 160, 1609–1617, doi: 10.1016/s0002-9440(10)61108-x (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Armaka M et al. Mesenchymal cell targeting by TNF as a common pathogenic principle in chronic inflammatory joint and intestinal diseases. The Journal of Experimental Medicine 205, 331–337, doi: 10.1084/jem.20070906 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nutt SL, Metcalf D, D’Amico A, Polli M & Wu L Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. The Journal of Experimental Medicine 201, 221–231, doi: 10.1084/jem.20041535 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rossi A, Appelt-Menzel A, Kurdyn S, Walles H & Groeber F Generation of a three-dimensional full thickness skin equivalent and automated wounding. Journal of visualized experiments : JoVE, doi: 10.3791/52576 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Akhmetshina A et al. Dual inhibition of c-abl and PDGF receptor signaling by dasatinib and nilotinib for the treatment of dermal fibrosis. FASEB J 22, 2214–2222, doi: 10.1096/fj.07-105627 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Ashcroft T, Simpson JM & Timbrell V Simple method of estimating severity of pulmonary fibrosis on a numerical scale. J Clin Pathol 41, 467–470 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scheuer PJ Classification of chronic viral hepatitis: a need for reassessment. Journal of hepatology 13, 372–374 (1991). [DOI] [PubMed] [Google Scholar]
- 60.Schneider CA, Rasband WS & Eliceiri KW NIH Image to ImageJ: 25 years of image analysis. Nature methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nature methods 9, 676–682, doi: 10.1038/nmeth.2019 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jarjour M et al. Fate mapping reveals origin and dynamics of lymph node follicular dendritic cells. The Journal of Experimental Medicine 211, 1109–1122, doi: 10.1084/jem.20132409 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. doi: arXiv:1303.3997 (2013). [Google Scholar]
- 64.Zhang Y et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137, doi: 10.1186/gb-2008-9-9-r137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen Y et al. A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–120, doi: 10.1038/nature11243 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martin M Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011 17, doi: 10.14806/ej.17.1.200pp. 10–12 (2011). [DOI] [Google Scholar]
- 67.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, doi: 10.1093/bioinformatics/bts635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liao Y, Smyth GK & Shi W featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930, doi: 10.1093/bioinformatics/btt656 (2014). [DOI] [PubMed] [Google Scholar]
- 69.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 550, doi: 10.1186/s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.R: A language and environment for statistical computing (R Foundation for Statistical Computing, 2013). [Google Scholar]
- 71.Mootha VK et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics 34, 267–273, doi: 10.1038/ng1180 (2003). [DOI] [PubMed] [Google Scholar]
- 72.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550, doi: 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dweep H, Sticht C, Pandey P & Gretz N miRWalk--database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. Journal of biomedical informatics 44, 839–847, doi: 10.1016/j.jbi.2011.05.002 (2011). [DOI] [PubMed] [Google Scholar]
- 74.Enright AJ et al. MicroRNA targets in Drosophila. Genome Biology 5, R1–R1 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP & Burge CB Prediction of mammalian microRNA targets. Cell 115, 787–798 (2003). [DOI] [PubMed] [Google Scholar]
- 76.Dweep H & Gretz N miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nature methods 12, 697, doi: 10.1038/nmeth.3485 (2015). [DOI] [PubMed] [Google Scholar]
- 77.Dees C et al. Platelet-derived serotonin links vascular disease and tissue fibrosis. J Exp Med 208, 961–972, doi: 10.1084/jem.20101629 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hecker L et al. NADPH oxidase-4 mediates myofibroblast activation and fibrogenic responses to lung injury. Nat Med 15, 1077–1081 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Popov Y et al. Tissue transglutaminase does not affect fibrotic matrix stability or regression of liver fibrosis in mice. Gastroenterology 140, 1642–1652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.DeClerck Y, Draper V & Parkman R Clonal analysis of murine graft-vs-host disease. II. Leukokines that stimulate fibroblast proliferation and collagen synthesis in graft-vs. host disease. J Immunol 136, 3549–3552 (1986). [PubMed] [Google Scholar]
- 81.Munde M et al. Structure-dependent inhibition of the ETS-family transcription factor PU.1 by novel heterocyclic diamidines. Nucleic Acids Res 42, 1379–1390 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Burkholder T, Foltz C, Karlsson E, Linton CG & Smith JM Health Evaluation of Experimental Laboratory Mice. Curr Protoc Mouse Biol 2, 145–165 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data used in this study have been deposited in the Gene Expression Omnibus (GEO) under the accession number GSE122334. The sequencing data from the ChIP-seq expriments have been submitted to the National Center for Biotechnology Information (NCBI) database under BioProject PRJNA480591; the project includes following biosamples: SUB4300598, SUB4300595, SUB4300592, SUB4300591, SUB4300589, SUB4300587, SUB4300586, SUB4300583 and SUB4300579; the FASTQ data was uploaded to the NCBI Sequence Read Archive (SRA).