Abstract
Functional characterisation of different HIV-1 subtypes may improve understanding of viral pathogenesis and spread. Here, we evaluated the ability of 345 unique HIV-1 Nef clones representing subtypes A, B, C and D to inhibit NFAT signalling following TCR stimulation. The contribution of this Nef function to disease progression was also assessed in 211 additional Nef clones isolated from unique subtype C infected individuals in early or chronic infection. On average, subtype A and C Nef clones exhibited significantly lower ability to inhibit TCR-mediated NFAT signalling compared to subtype B and D Nef clones. While this observation corroborates accumulating evidence supporting relative attenuation of subtypes A and C that may paradoxically contribute to their increased global prevalence and spread, no significant correlations between Nef-mediated NFAT inhibition activity and clinical markers of HIV-1 infection were observed, indicating that the relationship between Nef function and pathogenesis is complex.
Keywords: HIV-1 Nef, HIV-1 subtype C, NFAT inhibition, TCR signalling, HIV-1 disease progression, HLA-associated polymorphisms, immune-driven escape mutations
Introduction
The HIV-1 accessory protein Nef plays an important role in enhancing HIV-1 pathogenesis (Deacon et al., 1995; Kestler et al., 1991) through facilitating viral immune evasion (Swigut et al., 2004) and increasing viral infectivity and replication (Iafrate et al., 2000) (reviewed in (Foster and Garcia, 2008)). Major activities of Nef include CD4 downregulation to enhance viral budding (Ross et al., 1999) and facilitate evasion of antibody-dependent cell-mediated cytotoxicity (Veillette et al., 2014), HLA-I downregulation to evade CD8+ T cells (Ali et al., 2005; Schwartz et al., 1996), and CD4-independent enhancement of virion infectivity (an important mechanism includes inhibition of SERINC5 incorporation into virions (Rosa et al., 2015; Usami et al., 2015b)). In addition, Nef tailors TCR signalling to modulate the activation state of infected cells and this has been recognised as a key function of Nef (Abraham and Fackler, 2012; Fackler et al., 2007), however there is a lack of studies defining the role of this function in HIV-1 pathogenesis.
Nef has been shown to have both activating (Fenard et al., 2005; Manninen et al., 2000) and inhibitory (Haller et al., 2007; Thoulouze et al., 2006) effects on different aspects of TCR signalling in infected cells. Following TCR stimulation, lymphocyte-specific protein tyrosine kinase (Lck) is the most proximal protein to be activated, leading to multiple downstream events that include phosphorylation of signalling mediators zeta-chain-associated protein kinase 70 (ZAP-70), linker for activation of T cells (LAT) and phospholipase C-gamma 1 (PLC-gamma1). This is followed by activation of transcription factors, such as nuclear factor of activated T cells (NFAT) and activator protein 1 (AP-1), that then result in optimal production of IL-2. In quiescent primary T cells, through distal aspects of the TCR signalling pathway Nef enhances NFAT and IL-2 production thereby increasing viral replication, while in pre-activated primary T cells Nef inhibits NFAT and IL-2 production through disrupting proximal events following TCR stimulation (Neri et al., 2011). A model has emerged to reconcile apparently contradictory findings, whereby Nef reduces the availability of Lck and LAT at the plasma membrane and retargets these to an intracellular compartment, resulting in inhibition of signalling from the plasma membrane yet stimulation of a relocalised narrow signalling response from the intracellular compartment (Abraham and Fackler, 2012). Thus, Nef disrupts proximal TCR signalling events and activates selected downstream events, and which effect predominates is determined by the activation state of the infected cell. Overall, Nef fine tunes activation to achieve an intermediate activation state where HIV replication is promoted and activation-induced cell death (which would limit production of progeny viruses) is avoided (Abraham and Fackler, 2012).
The relative significance of each Nef function for HIV-1 disease progression remains incompletely understood, although there is evidence that Nef-mediated CD4 down-regulation and enhancement of infectivity are likely the major contributors to Nef’s effect of enhancing pathogenicity (Iafrate et al., 2000; Stoddart et al., 2003; Watkins et al., 2013). Few studies have linked Nef activities with clinical outcome using patient-derived sequences and the majority of these have focused on subtype B infection, although subtype C is predominant world-wide. Nef clones derived from elite controllers with subtype B infection did not display major genetic defects however they showed impairments in HLA-I/CD4 down-regulation, CD74 upregulation, enhancement of infectivity and stimulation of replication in peripheral blood mononuclear cells (PBMCs), in comparison to normal progressors, suggesting that these Nef activities may affect clinical outcome in subtype B infection (Mwimanzi et al., 2013a). Furthermore, a study of multiple Nef activities in subtype B infection supported biological importance of Nef-mediated enhancement of virion infectivity for HIV pathogenesis (Mwimanzi et al., 2013b). Nef-mediated CD4 and HLA-I down-regulation have been associated with viral set point and rate of CD4+ T cell decline in early subtype C infection (Mann et al., 2014), however it remains unknown whether natural variation in other Nef functions, such as alteration of TCR signalling, influences HIV-1 subtype C disease progression.
Our previous observation of inter-subtype differences in the ability of Nef isolates to downregulate CD4 and HLA-I (Mann et al., 2013), which are Nef activities shown to correlate with markers of disease progression (Mann et al., 2014), raised the hypothesis that inter-subtype differences in Nef activities may contribute to differing prevalence and spread of subtypes through affecting pathogenicity. Inter-subtype functional differences have also been reported for Gag and Pol, and these have been linked to inter-subtype differences in HIV-1 disease progression and prevalence (Kiguoya et al., 2017; Ng et al., 2014). HIV-1 subtypes differ greatly in prevalence and they are expanding unevenly: the most prevalent HIV-1 genetic subtypes include subtypes A, B and C, with subtype C being the most prevalent (Hemelaar, 2012; Tebit and Arts, 2011), and subtypes A and C may be expanding more rapidly than other subtypes (Conroy et al., 2010; Gräf and Pinto, 2013; Tebit and Arts, 2011). Studies have also shown that the transmissibility and the rate of disease progression may differ between HIV-1 subtypes (Kanki et al., 1999; Kiwanuka et al., 2009; Palm et al., 2014; Renjifo et al., 2004; Silveira et al., 2012). Identifying inter-subtype differences is of interest to better understand the global distribution and prevalence of subtypes, and uncovering biological factors contributing to these differences may have influence on strategies for therapeutic or vaccine development. However, it remains unknown whether the ability of Nef to modulate TCR signalling, which may be an important function of Nef, differs between HIV-1 subtypes.
In the present study, we investigated whether there are differences in Nef-mediated alteration of TCR signalling between HIV-1 subtypes using previously published patient-derived Nef clones from subtypes A, B, C and D (Mann et al., 2013). We used a high throughput NFAT-based luciferase reporter T cell assay to measure the ability of each Nef clone to inhibit NFAT, a downstream molecule of TCR signalling, following TCR stimulation. We further analysed the impact of Nef-mediated inhibition of NFAT signalling on HIV-1 disease progression and assessed which amino acid variants may affect this Nef function by studying a large number of previously published Nef clones derived from individuals with recent and chronic HIV subtype C infection (Mann et al., 2014).
Methods
Patient-derived Nef clones
Patient-derived Nef sequences assessed in the present study were previously cloned into a modified pSELECT-GFPzeo plasmid (that expresses Nef and green fluorescent protein [GFP] from separate promoters) and assessed for their CD4 and HLA down-regulation abilities (Mann et al., 2013; Mann et al., 2014). Nef clones were derived from individuals chronically infected with subtypes A (n=94), B (n=92), C (n=74), and D (n=85) from cohorts in Uganda, Canada and South Africa (Mann et al., 2013) and from recently (n=101) and chronically (n=110) HIV-1 subtype C infected individuals from observational cohorts and clinical sites in South Africa and Botswana (Mann et al., 2014). A single clone per infected individual was analysed and all individuals were antiretroviral naive. Viral load, CD4 count and HLA class I data were available. Nef protein expression was confirmed on a subset by Western blot (Mann et al., 2013; Mann et al., 2014). Nef sequences are available under Genbank accession numbers KC906733-KC907077, KF208819, KF208821, KF208823, KF208825-KF208828, KF208831-KF208834, KF208836, KF208838-KF208839, KF208842-KF208843, KF208845, KF208847-KF208853, KF208855, KF208857-KF208861, KF208863-KF208865, KF208867, KF208870, KF208872-KF208873, KF208878-KF208879, KF208886, KF208889, KF208893-KF208895, KM262907-KM262921, KM262925-KM262926, KM262928-KM262954, KM262956-KM262985, KM263030-KM263118, and KM263120-KM263141.
NFAT-based luciferase reporter T cell assay
The ability of each Nef clone to alter TCR-mediated signalling was assessed using a transfection-based assay that measures luciferase production driven by nuclear factor of activated T cells (NFAT) following stimulation of Jurkat T cells (Jin S, accepted, in press). Briefly, 5 million Jurkat T cells, 10 μg Nef clone and 5 μg pNFAT-luciferase were combined in 400 μl of OptiMEM without phenol red (Gibco) and electroporated at 250V and 950 μF using a Gene Pulser Xcell electroporator (Biorad). Transfected Jurkat cells were mixed with 600 μl of RPMI-1640 medium without phenol red (Sigma) supplemented with 10% foetal bovine serum (Gibco), 2mM L-glutamine (Sigma), 10mM Hepes (Gibco) and 50U/ml penicillin-streptomycin (Gibco), and incubated for 18 hours. Transfection efficiency (measured as the percentage of green fluorescent protein [GFP] positive cells) and the percentage live cells were assessed after the 18 hour incubation by flow cytometry to confirm that these parameters were similar across different samples within the same experiment. Simultaneously, 100 μl of transfected Jurkat cells was transferred in triplicate to a plate coated with 0.1 μg/ml anti-CD3 antibody (eBioscience) and incubated for 6 hours to stimulate TCR signalling (Smith et al., 1997). Following the 6 hour incubation, 50 μl of stimulated cells were transferred to a half-well white plate (Greiner) and mixed with 50 μl of Steady-Glo luciferase substrate (Promega). The luminescence signal was measured following a 10 minute incubation using a GloMax-Multi Microplate Multimode Reader (Promega) with an integration time of 2000 ms and settling time of 500 ms. The 3 luminescence readings per Nef clone assessed were averaged and the ability of each Nef clone to inhibit TCR-mediated NFAT signalling was calculated relative to the positive control (SF2 Nef, representing 100% activity) and negative control (SF2 Nef mutated from G to A at codon 2, representing 0% activity) as follows: (G2A Nef luminescence – Nef clone luminescence) / (G2A Nef luminescence – SF2 Nef luminescence). For each Nef clone, normalised activity from two independent experiments was averaged.
Generation and testing of Nef mutants
Selected mutations were introduced into patient-derived subtype C Nef sequence SK329 (GenBank accession KC906797), which displayed high similarity to the HIV-1 Nef consensus C sequence (91.7 % amino acid similarity), using overlap extension PCR. Wild-type SK329 Nef and SK329 Nef mutants were assessed in the context of the pSELECT backbone in triplicate independent experiments for the ability to inhibit TCR-mediated NFAT signalling as well as ability to down-regulate CD4, HLA-I and SERINC5. CD4 and HLA-I down-regulation activities were measured in a CEM-derived cell line expressing high levels of HLA-A*02 using flow cytometry as previously described (Mann et al., 2013). SERINC5 down-regulation was also assessed in the CEM-A*02 cell line using flow cytometry by co-transfection of 1 μg Nef and 5 μg pSELECT-SERINC5-iHA-ΔGFP, encoding an internal HA-tagged variant of SERINC5 ((Usami et al., 2015a) and Jin S, accepted, in press), followed by antibody staining the next day. Expression levels of the mutant Nef proteins were assessed by Western blot using rabbit polyclonal anti-HIV-1 Nef serum, as previously described (Mann et al., 2013), following transfection of 2.5 million Jurkat cells with 10 μg Nef.
Data analysis
The capacity of Nef to inhibit TCR-mediated NFAT signalling was compared between subtypes, as well as between mutant Nefs and the wild-type, using ANOVA with the Tukey post-hoc test since the data was normally distributed. The comparison between subtypes was done while controlling for socio-demographic and clinical factors, by performing multivariable linear regression using capacity of Nef to inhibit TCR-mediated NFAT signalling as the dependent variable and the following as independent variables: gender, log10 plasma viral load, log CD4 count, and HIV-1 subtype. For log transformed variables, model coefficients quantify the change in outcome per one log unit increase in the dependent variable of interest. The reference category for gender was female and subtype B was the reference category for HIV-1 subtypes.
The relationships between Nef-mediated inhibition of NFAT signalling in HIV-1 subtype C acute infection and subsequent viral load set point as well as the rate of CD4+ T cell decline were determined. Viral load set point was calculated as the average viral load of an infected individual between 3 to 12 months after infection, and Spearman’s correlation was used to correlate Nef-mediated inhibition of NFAT signalling function with viral load set point. The rate of CD4+ T cell decline was computed, as previously described (Wright et al., 2010), for each infected individual over a treatment-free follow-up period. A multiple linear regression was performed, taking into account follow-up time and baseline CD4+ T cell count, to determine the relationship between Nef-mediated inhibition of NFAT signalling function and the rate of CD4+ T cell decline. Since the CD4+ T cell decline data was a mixture of positive and negative values, it could not be easily transformed to normality without compromising the meaningful interpretation of coefficients. Therefore, the CD4+ T cell decline range was limited to −35 to 50 cells/mm3 per month to approximate a normal distribution and ensure that the final model met the assumption of normally distributed residuals as assessed by the Shapiro Wilk test. This resulted in 7 observations (7%) being omitted from the analysis. Distinct Nef amino acids associated with altered ability of subtype C Nef to inhibit TCR-mediated NFAT signalling were assessed using codon-by-codon Mann–Whitney U tests. Q values were used to account for multiple tests (Storey and Tibshirani, 2003). All analyses were performed using GraphPad Prism 5 and Stata 14.
Results
Measurement of Nef-mediated inhibition of NFAT signalling in a transfection-based assay
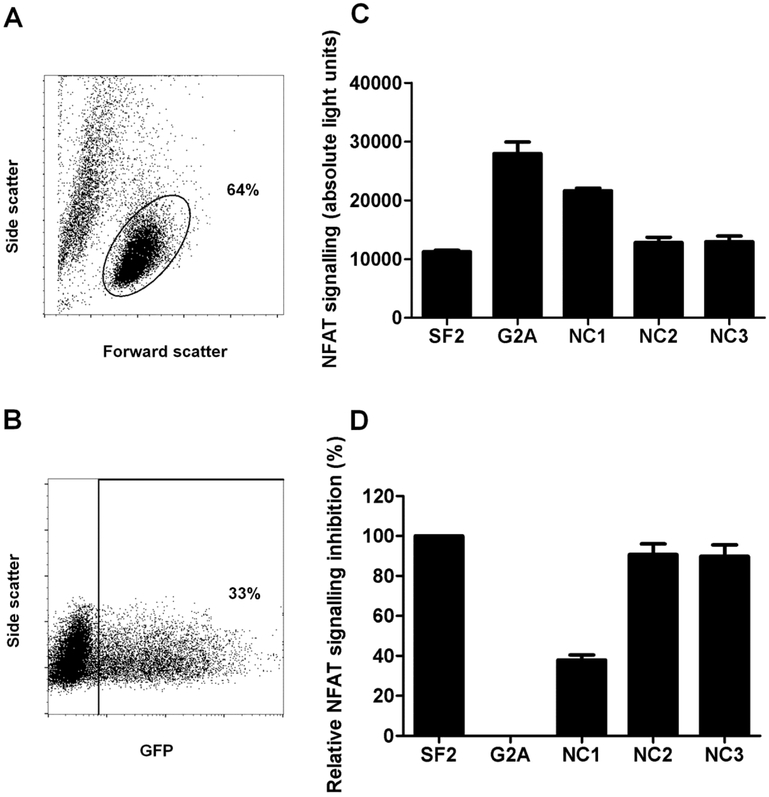
Nef-mediated inhibition of NFAT signalling activity was assessed by transfecting the Nef clones into a Jurkat T cell line, stimulating the TCR with anti-CD3 antibody, and then measuring the effect on NFAT-driven luciferase production through luminescence detection (Jin S, accepted, in press). Following transfection, flow cytometry was used to confirm similarity of live cell percentages and transfection efficiency within an experiment (representative plots shown in Figure 1A and B). Representative luminescence values are shown for the controls and patient-derived Nef clones, as well as the normalisation of these values for the patient-derived Nef clones to that of the controls (Figure 1C and D). Reproducibility of the assay was excellent with independent duplicate measurements correlating strongly (Pearson’s correlation; r = 0.86 and p < 0.0001).
Figure 1: Nef-mediated inhibition of NFAT signalling.
NFAT-driven luciferase activity was quantified in Nef-transfected Jurkat cells following TCR stimulation, to measure the ability of Nef to inhibit NFAT induction by TCR signalling. Nef was expressed in a pSELECT-GFPzeo plasmid co-expressing green fluorescent protein (GFP). (A) Flow cytometry was performed to assess the percentage of live Jurkat cells (indicated by the circular gate) following transfection. (B) Transfection efficiency of each Nef clone was assessed by flow cytometry to detect the percentage of GFP positive cells (indicated by the square gate), which represent Nef-transfected cells. We confirmed that cellular toxicity and transfection efficiency were similar across clones within an experiment and thus did not confound the assay interpretation. (C) Absolute light units measured 6 hours after TCR stimulation are shown for Jurkat cells transfected with SF2 Nef, G2A Nef and 3 patient-derived Nef clones (NC1-3). SF2 and G2A Nef clones were included as positive and negative controls in each experiment. (D) The ability of each patient-derived Nef clone to inhibit NFAT signalling was normalised to the controls such that SF2 Nef represented 100% activity while G2A Nef represented zero activity. In panels C and D, the mean and standard deviation of two independent experiments (each with triplicate measurements) is shown.
Inter-subtype comparison of Nef-mediated inhibition of NFAT signalling
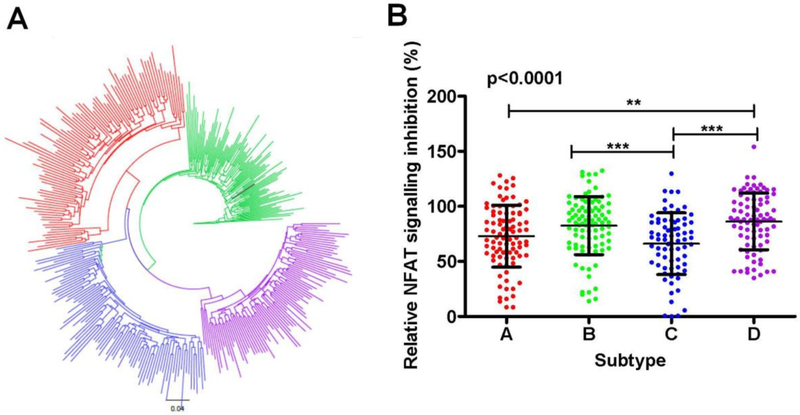
Differences in NFAT inhibition activity were investigated between Nef clones of subtypes A (n=94), B (n=92), C (n=74) and D (n=85) derived from chronically infected individuals (Mann et al., 2013). A maximum likelihood phylogenetic tree showing distinct clustering of subtypes is shown in Figure 2A. Nef’s capacity to inhibit TCR-mediated NFAT signalling was compared between subtypes using one-way ANOVA. Significant differences were observed between subtypes (mean [standard deviation], subtype A, 73% [28]; B, 82% [26]; C, 66% [28]; and D, 86% [26]) (p < 0.0001; Figure 2B). Tukey post-hoc tests indicated that subtype C was significantly less functional than subtypes B and D (p < 0.001 for both) and that subtype A was significantly less functional than subtype D (p < 0.01). Therefore, the hierarchy of Nef function was C<A<B<D, where C and A were not significantly different from one another and B and D were not significantly different from one another.
Figure 2: Inter-subtype comparison of HIV-1 Nef-mediated inhibition of NFAT signalling.
(A) A maximum likelihood phylogenetic tree of Nef sequences was constructed using Phyml (Guindon and Gascuel, 2003). Subtypes form distinct clusters, with subtypes A, B, C, and D shown in red, green, blue, and purple, respectively. The SF2 strain is shown in black. (B) ANOVA with the Tukey post-hoc tests was used to compare Nef’s ability to inhibit TCR-mediated NFAT signalling (measured by NFAT-driven luciferase activity through luminescence detection) between subtypes A, B, C and D. Luminescence values for the patient-derived Nef clones are normalized to SF2 Nef, which represents 100% activity. Each dot represents the average of two independent experiments (each with triplicate measurements) for one patient-derived Nef clone. The ANOVA p value is shown. The asterisks above the bar indicate significant differences between subtypes, with the bar indicating the two subtypes being compared. The level of significance is indicated by the number of asterisks: p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***). Means and standard deviations are indicated by bars and whiskers, respectively.
Sociodemographic (gender and age) and clinical (viral load and CD4 count) variables differ significantly between the cohorts described here (Mann et al., 2013) and could potentially confound the inter-subtype comparison of Nef-mediated inhibition of NFAT signalling (Addo and Altfeld, 2014; Antonio et al., 2016; Corró et al., 2012; Huang et al., 1995; Miura et al., 2008; Mwimanzi et al., 2011b; Mwimanzi et al., 2013a). To adjust for these potential confounders when comparing Nef function between subtypes, a multivariate analysis was performed. In univariable analysis, where the outcome variable of interest was Nef-mediated inhibition of NFAT signalling, we considered subtype, viral load, CD4 count, gender and age as potential predictors (Table 1). Subtype and log CD4 count were identified as significant predictors of NFAT inhibition, and subsequently considered in the multivariable analysis. Consistent with the ANOVA analysis, in the multivariable analysis subtype remained significantly associated with NFAT inhibition, after controlling for CD4 count (Table 1). Subtype A, in particular, was associated with a 10% lower NFAT inhibition ability compared to subtype B (p = 0.01). We observed 15% lower NFAT inhibition ability in subtype C compared to subtype B (p < 0.001). There was no significant difference observed between NFAT inhibition ability in subtypes B and D (p = 0.49).
Table 1:
Linear regression models assessing the effect of HIV-1 subtype, socio-demographic and clinical factors on Nef-mediated inhibition of NFAT signalling.
| Variable | Univariate (TCR) |
Multivariable (TCR)a |
|||
|---|---|---|---|---|---|
| Estimate | p-value | Estimate | p-value | ||
| Subtype | A | −0.099 | 0.02 | −0.10 | 0.01 |
| B | 0 | N/A | 0 | N/A | |
| C | −0.162 | 0.0001 | −0.15 | 0.0007 | |
| D | 0.035 | 0.4 | 0.03 | 0.49 | |
| Log viral load | 0.032 | 0.1 | - | - | |
| Log CD4 | −0.084 | 0.02 | −0.04 | 0.25 | |
| Male | 0.040 | 0.2 | - | - | |
| Age | −0.001 | 0.5 | - | - | |
CD4 counts and viral load estimates are expressed per log10 increment. Age is expressed per year increment.
Female was the reference group for gender. Subtype B was the reference group for HIV-1 subtypes.
Multivariable model for Nef’s inhibition of TCR-mediated NFAT signalling: multiple r2 = 0.079, p = 0.00002.
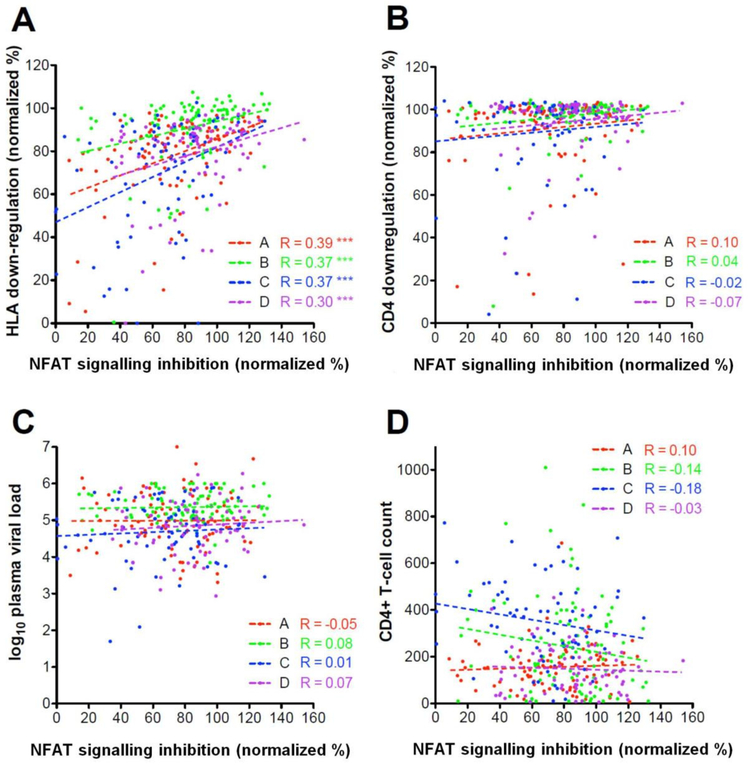
Previously, Nef-mediated CD4 down-regulation and HLA down-regulation activities were measured for the same Nef clones (Mann et al., 2013; Mann et al., 2014). Although these Nef activities are to a large extent genetically separable (Foster et al., 2011), CD4 down-regulation and HLA down-regulation activities of primary Nef clones nevertheless correlate to some extent (e.g. as previously reported, CD4 and HLA functions of the Nef clones studied here correlated weakly [Spearman’s correlation; r ≥ 0.3 and p < 0.01]) (Mann et al., 2013). Therefore, we investigated the relationship between Nef-mediated inhibition of NFAT signalling and Nef-mediated HLA down-regulation as well as CD4 down-regulation. A significant relationship between Nef-mediated inhibition of NFAT signalling and HLA down-regulation was observed within the Nef clones used for the inter-subtype comparison (Spearman’s correlation; all r ≥ 0.3 and p < 0.0001) (Figure 3A). There was no significant association between Nef-mediated inhibition of NFAT signalling and CD4 down-regulation (Spearman’s correlation; r ≥ −0.7 and ≥ 0.1 and p ≥ 0.29) (Figure 3B).
Figure 3: The relationship between Nef-mediated inhibition of NFAT signalling and other Nef functions as well as clinical markers.
Nef-mediated inhibition of NFAT signalling activity was significantly correlated with HLA down-regulation activity within each subtype with use of Spearman’s correlation (panel A). However Nef-mediated inhibition of NFAT signalling was not correlated with CD4 down-regulation ability (panel B), log10 plasma viral load (panel C), or CD4+ T cell count (panel D). Values are normalized to SF2, which represents 100% activity. Subtypes A, B, C, and D are represented in red, green, blue, and purple, respectively. Significance is indicated by asterisks: p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***).
No significant correlations were observed between Nef’s function of inhibiting TCR-mediated NFAT signalling and patient CD4 count or viral load within each subtype (Spearman’s correlation; r ≥ −0.18 and ≤ 0.1 and p ≥ 0.13) (Figure 3C and D). Although it should be noted that these cohorts largely comprised individuals with late-stage infection (e.g. average CD4 counts were < 200 cells/mm3 for subtype A and D cohorts) when associations with clinical parameters may no longer be strongly detectable.
Lack of association of Nef-mediated inhibition of NFAT signalling in HIV-1 subtype C recently infected individuals with subsequent disease progression
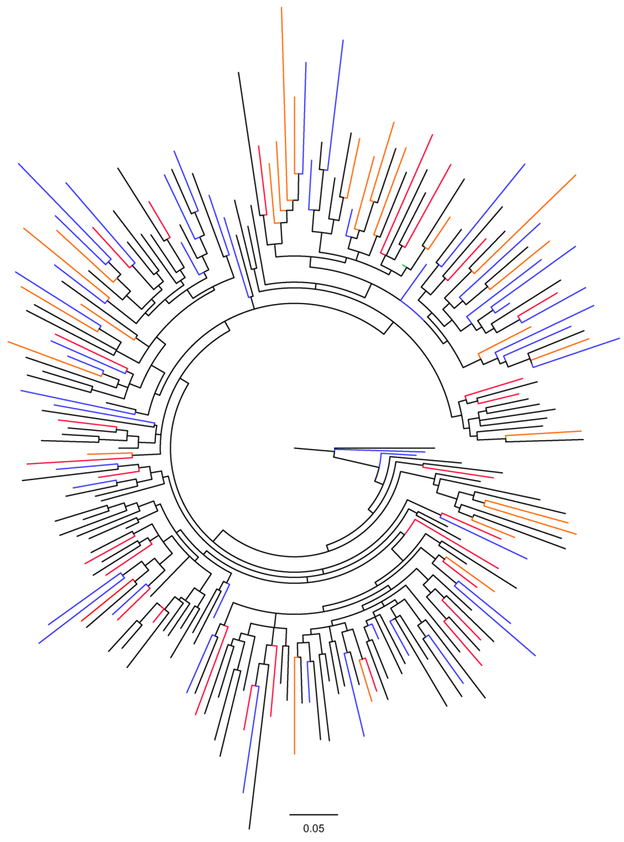
To further assess whether Nef-mediated inhibition of NFAT signalling significantly influences disease progression, we employed 101 previously constructed Nef clones of the same subtype (C) that were derived from recently infected individuals, as well as an additional 110 previously constructed subtype C Nef clones from chronic infection (Mann et al., 2014). The subtype C Nef clones from recently infected individuals were derived from different cohorts in South Africa (HPP acute infection [n=29] and TRAPS [n=49]) and Botswana (Tshedimoso [n=26]) while the chronic ones were from the Sinikithemba cohort in South Africa. Despite differences in geographic locale, all subtype C sequences co-mingled in a phylogeny (Figure 4). While there was no overall difference in the NFAT inhibition ability between Nef clones derived from acute infection versus chronic infection (Student’s T test; p = 0.89), we compared this Nef activity between acute cohorts and observed that function was significantly lower in Tshedimoso compared to HPP (estimate = −0.20, p < 0.001). The mean NFAT inhibition function was slightly lower in TRAPs versus HPP, however this effect was not statistically significant (estimate = −0.09, p = 0.10). Therefore although phylogenetic analyses indicated no major differences between sequences from difference cohorts (Mann et al., 2014), the cohort variable was taken into account in subsequent analyses correlating this Nef function with markers of disease progression.
Figure 4: Phylogenetic tree of subtype C Nef sequences from different cohorts.
A maximum likelihood phylogenetic tree of 211 subtype C Nef sequences was constructed using Phyml (Guindon and Gascuel, 2003). Nef sequences from the chronic infection cohort (n=110) are shown in black, while 101 sequences from the recent infection cohorts HPP acute, Tshedimoso, and TRAPs are shown in red, orange, and blue, respectively. Sequences from different cohorts are intermingled. The consensus C sequence (Los Alamos HIV sequence database, compiled in 2004) is shown in green.
To assess the relationship between Nef-mediated inhibition of NFAT signalling in recently infected individuals and subsequent disease progression, this activity of the Nef clones from the earliest time-point post-infection in subtype C infected individuals was correlated with subsequent viral load set point and rate of CD4+ T cell decline. We observed no significant correlation between viral load set point and Nef-mediated ability inhibit NFAT signalling (Spearman’s correlation; r=0.03 and p=0.79). Due to the functional differences observed between the acute cohorts, a linear regression analysis was performed to check for an effect of the interaction of cohorts and this Nef function on viral load set point. No significant effect was found (Table 2), indicating that the impact of this Nef function on viral load set point was similar between cohorts.
Table 2:
Linear regression models assessing the effect of Nef-mediated inhibition of NFAT activity and cohort differences on viral load set point.
| Variable | Univariate | Multivariablea | |||
|---|---|---|---|---|---|
| Estimate | p-value | Estimate | p-value | ||
| Main effects | |||||
| HPP | 0 | N/A | 0 | N/A | |
| Cohorts | Tshedimoso | 0.08 | 0.72 | 0.26 | 0.77 |
| TRAPS | −0.32 | 0.13 | −0.65 | 0.38 | |
| NFAT inhibition | 0.19 | 0.58 | 0.09 | 0.89 | |
| Interaction effect | |||||
| NFAT inhibition X cohort | HPP | 0 | N/A | 0 | N/A |
| Tshedimoso | - | - | −0.19 | 0.84 | |
| TRAPS | - | - | 0.36 | 0.67 | |
The HPP cohort is the reference group for cohorts.
Multivariable model for viral load set point: multiple r2 = 0.05, p = 0.41.
Next, a multiple linear regression was performed to determine the relationship between Nef-mediated inhibition of NFAT signalling and the rate of CD4+ T cell decline while controlling for baseline log viral load and confounding factors, namely baseline CD4+ T cell count (transformed by square root) and follow-up time, which were both known to affect the rate of CD4+ T cell decline (Mann et al., 2014). Cohort was not included as a variable in the final model since there was no significant difference in the effect of Nef-mediated inhibition of NFAT signalling on CD4 decline in the TRAPS and Tshedimoso cohorts compared to the HPP cohort (p = 0.64 and p = 0.67, respectively). In both univariate and multivariate analyses there was no significant relationship between Nef-mediated inhibition of NFAT signalling and rate of CD4 decline (Table 3).
Table 3:
Linear regression models assessing the effect of Nef-mediated inhibition of NFAT signalling on rate of CD4 decline
| Variable | Univariate | Multivariablea | ||
|---|---|---|---|---|
| Estimate | p-value | Estimate | p-value | |
| NFAT inhibition | −3.68 | 0.26 | −2.91 | 0.31 |
| Square-root baseline CD4+ count | −0.47 | 0.006 | −0.88 | 0.0001 |
| Follow-up time | 0.003 | 0.009 | 0.01 | <0.0001 |
| Log baseline viral load | 0.4 | 0.57 | −0.37 | 0.61 |
CD4+ counts are expressed per one square root unit increase and viral load estimates are expressed per log10 increment. Follow-up time is expressed per day increment.
Multivariable model for rate of CD4+ T cell decline: multiple r2 = 0.27, p < 0.00001.
Furthermore, in agreement with the recent infection analysis, in the chronic infection cohort there was no significant correlation between Nef-mediated inhibition of NFAT signalling and log viral load (Spearman’s correlation; r = 0.1 and p = 0.29) or CD4+ T cell count (Spearman’s correlation; r = −0.05 and p = 0.62).
Sequence-function analysis: Amino acids associated with increased or decreased ability of subtype C Nef to inhibit TCR-mediated NFAT signalling
A secondary aim of this study was to identify amino acid variants that significantly affect Nef-mediated inhibition of NFAT signalling through performing sequence-function analyses. We used the larger dataset of the additional 211 subtype C clones to identify these variants. Taking into account each amino acid variant at each codon present at least 3 times or more in the combined dataset, we identified 11 amino acids variants at 6 different codons significantly associated with increased or decreased Nef-mediated inhibition of NFAT signalling (Table 4). Several of these amino acid associations (3N, 3G, and 71K) corresponded with codons previously described in the literature to be involved in modulation of TCR signalling (Arold et al., 1997; Foster et al., 2011; Geyer et al., 2001; Lee et al., 1996), while others were adjacent to such previously described residues (9S, 9R, 108D and 108E) (Table 4). In addition, 57W, 71K, 71R, 108D and 108E were previously reported as HLA-associated polymorphisms (Carlson et al., 2014) and 71K was confirmed to confer escape from CD8+ T cell responses to the 68FPVRPQVPLR77 epitope (Du et al., 2016), indicating that immune-driven polymorphisms can impact the ability of Nef to modulate TCR signalling.
Table 4:
Amino acids in HIV-1 subtype C Nef significantly associated with altered Nef-mediated NFAT inhibition activity
| Codona | AAb | Cons.c | Functional difference (%)d |
No. of samplese | p value | q value | Motif-Function in TCR signallingf |
HLA-associated polymorphismsg |
CTL escape mutationsh |
|
|---|---|---|---|---|---|---|---|---|---|---|
| with AA | without AA | |||||||||
| 3 | N | G | +21 | 24 | 187 | <0.0001 | 0.004 | 1MGxxxS6 Myristoylation | - | - |
| 3 | G | G | −14 | 151 | 60 | 0.0001 | 0.01 | 1MGxxxS6 Myristoylation | - | - |
| 9 | S | S | −24 | 175 | 14 | 0.004 | 0.1 | - | - | - |
| 9 | R | S | 25 | 8 | 181 | 0.007 | 0.19 | - | - | - |
| 57 | W | w | −34 | 206 | 5 | 0.006 | 0.19 | A*68 | - | |
| 71 | K | R | −32 | 18 | 193 | 0.0004 | 0.03 |
72PxxPxR77 Nef-SH3 interaction/ PAK2 activation |
C*07:02 C*17 |
Yes (Du et al., 2016) |
| 71 | R | R | 28 | 191 | 20 | 0.001 | 0.09 |
72PxxPxR77 Nef-SH3 interaction/ PAK2 activation |
B*44 | - |
| 108 | E | E | 13 | 117 | 94 | 0.006 | 0.19 | 105RR106 PAK2 activation | B*57:02 | - |
| 108 | D | E | −13 | 94 | 117 | 0.006 | 0.19 | 105RR106 PAK2 activation | B*44/ B*18 | - |
| 161 | D | N | 12 | 39 | 171 | 0.003 | 0.1 | - | - | - |
| 161 | N | N | −12 | 162 | 48 | 0.004 | 0.1 | - | - | - |
Numbered according to HXB2
Amino acid associated with increased or decreased inhibition of NFAT activity.
Consensus amino acid at a particular codon from the reference 2004 consensus C Nef sequence from the Los Alamos HIV sequence database
Median functional difference in percentage inhibition of NFAT activity (expressed relative to SF2 control) between Nef clones with and without the amino acid.
Number of sequences with (+) and without (−) the amino acid. Amino acid totals vary, as gaps in the alignment are considered missing data.
Motif/residue previously reported to be involved in Nef-mediated alteration of TCR signalling.
HLA-associated polymorphisms are polymorphisms positively associated with a specific HLA allele, as previously described (Carlson et al., 2014), and are likely to be immune escape mutations.
CTL escape mutations listed in HIV Los Alamos immunology database
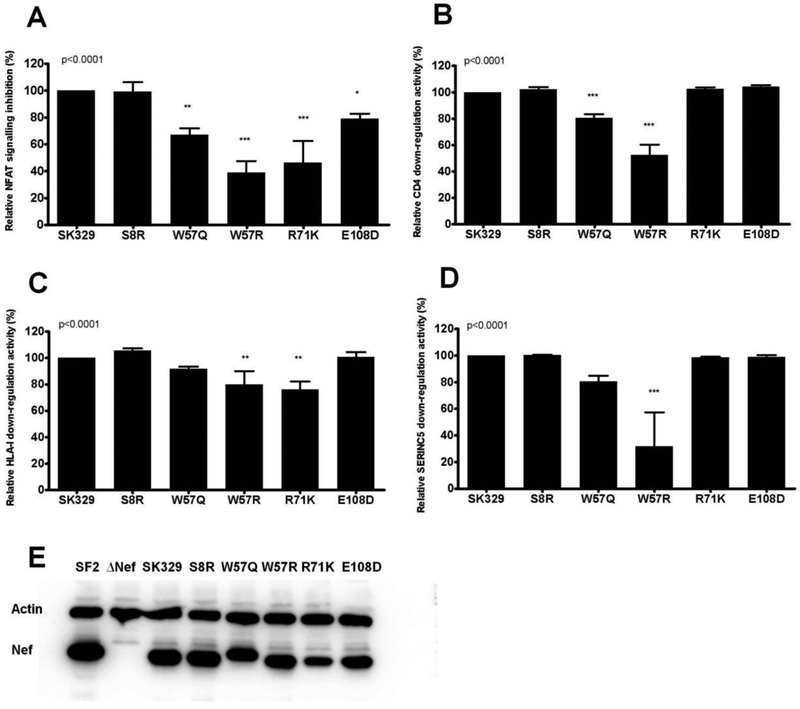
To further explore the contribution of amino acid variants to Nef’s ability to modulate TCR signalling, the same codon-by-codon analysis was performed on the independent panel of chronic subtype C Nef clones (n=74) that were used in the cross-clade comparison. Although, no amino acid variants were identified that met the q value cutoff for significance in this smaller sample subset (q<0.2), three variants that were identified in the previous analysis were associated with altered NFAT inhibition function at p ≤ 0.07, namely 57W (p = 0.06), 108D (p = 0.07) and 108E (p = 0.07). Furthermore, 8S (p = 0.002), which was associated with decreased Nef activity, was the most significant association in the smaller dataset and was present at p = 0.03 and q = 0.28 in the larger analysis. We therefore introduced non-consensus mutations at codons 8, 57 and 108 into a patient-derived sequence, SK329, of high similarity to the consensus C sequence to further assess their functional consequences. In addition, since 71K was associated with the most pronounced decrease in ability to inhibit TCR-mediated NFAT signalling in the larger dataset, this mutation was also included in the mutant panel for functional assessment. As expected, mutants 71K and 108D displayed significantly lower ability to inhibit NFAT signalling (46 % and 79 % relative to the wildtype, respectively) (ANOVA; p < 0.001 and p < 0.05, respectively) (Figure 5A). The negative consequence of 108D was specific to NFAT inhibition activity as this mutation did not influence Nef’s ability to down-regulate CD4, HLA or SERINC5. In contrast, 71K significantly impaired Nef-mediated HLA downregulation ability (p < 0.01) but it had no effect on CD4 or SERINC5 down-regulation (Figure 5B-D). It was predicted that 8R, 57Q and 57R would increase Nef’s ability to inhibit TCR-mediated NFAT signalling. However, while 8R displayed similar function as the wildtype (99 % of the wildtype), 57Q and 57R significantly decreased this Nef function (67 % and 39 % of the wildtype, respectively) (p < 0.01 and p < 0.001, respectively; Figure 5A) as well as Nef’s ability to down-regulate CD4 (both p < 0.001; Figure 5B). Furthermore, W57R was impaired for HLA-I down-regulation and SERINC5 down-regulation, respectively (p < 0.01 and p < 0.001, respectively; Figure 5C and D). The expression of all Nef mutants was readily detected by Western blot, although the expression of 71K was modestly reduced when compared to the wildtype SK329 (Figure 5E). This could partly account for the lower ability of 71K to down-regulate HLA-I and inhibit NFAT as these activities both require high intracellular Nef concentrations relative to that required for CD4 down-regulation (Liu et al., 2001). Consistent with this, in primary Nef isolates both HLA-I down-regulation ability and NFAT inhibition ability correlate positively with Nef protein expression levels by Western blot (Jin S, accepted, in press). In summary, these results confirm the impact of 71K and 108D on Nef-mediated inhibition of NFAT signalling, but suggest that unidentified secondary polymorphisms may alter the impact of 8R, 57Q, and 57R on this Nef function. Additional studies will be needed to address this question.
Figure 5: Functional consequences of selected mutations in Nef.
Mutations (S8R, W57Q, W57R, R71K and E108D) were introduced into the patient-derived Nef sequence SK329 (47.5% NFAT inhibition activity relative to SF2 Nef) and the Nef-mediated abilities to inhibit NFAT (A), down-regulate CD4 (B), down-regulate HLA-I (C) and down-regulate SERINC5 (D) were measured in triplicate. Activity relative to the wild-type SK329 (representing 100% activity) is shown. The ANOVA p value is shown and asterisks indicate significant differences between the mutants and the wild-type SK329: p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***). (E) Western blot analyses were used to assess the steady-state protein expression of Nef mutants. SF2 Nef and empty vector (ΔNef) were included as controls. Beta-actin protein was included as a cellular loading control.
Discussion
HIV-1 subtypes are uneven in their prevalence and spread globally (Buonaguro et al., 2007; Hemelaar, 2012) and the biological reasons for this are not fully understood. Nef-mediated alteration of TCR signalling may be important for virus survival and persistence, since it optimizes the activation state of infected cells for optimal virus replication. We aimed to compare the Nef-mediated alteration of TCR signalling between subtypes A-D, through measuring the ability of Nef to inhibit NFAT, and to relate this to differences in subtype prevalence and expansion.
Inter-subtype differences in Nef-mediated inhibition of NFAT signalling were observed with a hierarchy of Nef functions C<A<B<D, where A and C, as well as B and D, were not significantly different from one another. Furthermore, this hierarchy was supported by a multivariate analysis controlling for gender, age, viral load and CD4 count, wherein it was estimated that, on average, subtype C Nef clones displayed 15% lower function and subtype A Nef clones displayed 10% lower function relative to subtype B Nef clones.
Consistent with the inter-subtype differences observed here, we previously found that subtypes C and A were significantly less functional in Nef-mediated CD4 and HLA-I down-regulation when compared to subtype B (Mann et al., 2013). Similarly, it was demonstrated that subtypes A and C displayed significantly lower Gag-protease-driven replication capacity than subtype D and inter-subtype recombinants, and that subtype C Gag-protease was less functional than that of subtype B (Kiguoya et al., 2017). Furthermore, subtype D Pol isolates were reported to have increased replication capacity compared to subtype A Pol isolates (Ng et al., 2014) and subtype C whole isolates had lower ex vivo replicative fitness compared to isolates from other group M HIV-1 subtypes (Abraha et al., 2009). Taken together, these studies and the present study suggest that subtypes A and C are attenuated compared to other M group subtypes.
The Nef functional hierarchy observed in the present study is also consistent with reported inter-subtype differences in disease progression, prevalence and expansion. In Eastern Africa, subtype A is reported to result in slower disease progression when compared to subtype D (Kanki et al., 1999; Ssemwanga et al., 2013). There are conflicting reports of the rate of disease progression of subtype C relative to other M group subtypes (Ariën et al., 2007; Easterbrook et al., 2010; Neilson et al., 1999; Silveira et al., 2012; Vasan et al., 2006). However, a recent study controlling for clinical factors showed that subtype C infection was associated with slower rate of CD4 decline relative to subtype A or D infection (Venner et al., 2016). Subtypes A and C are the most prevalent subtypes (Buonaguro et al., 2007; Hemelaar, 2012; Tebit and Arts, 2011) which may also be expanding more rapidly than other subtypes, as evidenced by expansion of subtype C in Brazil, India and sub-Saharan Africa (Gräf and Pinto, 2013; Rodriguez et al., 2009; Venner et al., 2016) and increased prevalence of subtype A in Uganda (Conroy et al., 2010). Overall, the most attenuated subtypes – A and C – which were the least functional in the present study of Nef function, display slower disease progression yet are the most prevalent and are expanding rapidly. The increased dominance of subtypes A and C may be due to longer asymptomatic periods (Venner et al., 2016) and/or the low replication capacity of these subtypes may favour transmission due to increased half-life of infected cells in genital fluids, although this hypothesis still requires experimental verification (Kiguoya et al., 2017).
Given the inter-subtype differences observed in Nef-mediated inhibition of NFAT signalling, we then evaluated this Nef activity using a large dataset of clinically derived HIV-1 subtype C Nef clones isolated from recently and chronically infected individuals to analyse the impact of Nef-mediated NFAT inhibition on HIV-1 disease progression. We also used this high-powered dataset to assess which naturally occurring amino acid variants affect this Nef activity.
We observed no relationship between Nef-mediated NFAT inhibition activity in early infection and subsequent viral load set point as well as rate of CD4+ T cell decline. HLA-I down-regulation and CD4 down-regulation functions measured for the same Nef clones did significantly correlate with the rate of CD4+ T cell decline and viral load set point, respectively (Mann et al., 2014), suggesting that these Nef activities have greater impact on disease progression than Nef-mediated inhibition of NFAT signalling. However, Nef clones derived from elite controllers were shown to have a lower ability to inhibit NFAT signalling when compared with those derived from chronic progressors (Jin S, accepted, in press), suggesting that this Nef activity may contribute to clinical outcome in some HIV-1 controllers. Watkins et al. (2013) observed that CD4 down-regulation and one or more Nef activities undefined in their study contribute to viral replication and pathogenesis in a substantial way while activities that rely on the SH3 binding motif have less effect (Watkins et al., 2013). Mwimanzi et al. (2013) reported that Nef sequences from chronically infected individuals have a poly-functional and co-dependent nature, indicating that there is a selective pressure for multiple Nef functions during chronic infection (Mwimanzi et al., 2013b). Taken together, certain Nef activities have a stronger contribution to pathogenesis, yet there is selection for multiple functions. This study suggests that Nef-mediated inhibition of NFAT signalling is not one of the stronger contributors to HIV-1 pathogenesis.
We also aimed to identify amino acid variants associated with altered ability of subtype C Nef to inhibit NFAT signalling to better understand the determinants of this Nef activity. Several amino acid associations identified (3N, 3G, 71K and 71R) corresponded with codons previously described in the literature to be involved in modulation of TCR signalling (Arold et al., 1997; Foster et al., 2011; Geyer et al., 2001; Lee et al., 1996), while others were adjacent to such previously described residues (8S, 8R, 9S, 9R, 108D and 108E). Residue 3 plays a role in myristoylation, which is required for all Nef’s membrane localization and is thus important for all Nef functions (Geyer et al., 2001), and residues 8 and 9 are adjacent to the myristoylation motif. Residue 71 contributes Nef-SH3 domain interaction that allows Nef to translocate Lck to intracellular compartments and also allows interaction with other signalling kinases (Geyer et al., 2001; Lee et al., 1996). Residue 108 is adjacent to residues involved in PAK2 activation, namely 105RR106 (Foster et al., 2011). Consistent with previous reports of the important role of the Nef N-terminus in Nef-mediated alteration of TCR signalling (Baur et al., 1997) (Jin S, accepted, in press), we identified residues 3, 8, 9, and 57 in Nef’s N-terminal domain as associated with differential ability of Nef to inhibit NFAT signalling.
Mutagenesis experiments testing the functional consequences of non-consensus residues 8R, 57Q, 57R, 71K and 108D, supported that 71K, and to a lesser extent, 108D decrease the ability of Nef to inhibit NFAT signalling. 8R, 57Q and 57R were expected to increase activity, however while 8R displayed a similar function to the wild-type, 57Q and 57R were impaired to different extents for NFAT inhibition activity and all other Nef activities measured. Residue 57 is in the Nef protease cleavage domain and it is essential for CD4 down-regulation (Geyer et al., 2001). It is a highly conserved residue and therefore it not surprising that mutation of residue 57 negatively affected several Nef functions, although it was previously reported to not affect HLA-I down-regulation activity (Geyer et al., 2001). Additional mutations potentially arose prior to 57Q/R, facilitating its development, in the few patient-derived sequences encoding this mutation, and additional experiments would be required to test this possibility.
We then assessed whether any of the variants identified as associated with impaired Nef function in this study were likely CTL escape mutations, as this may have relevance for an HIV-1 attenuation based vaccine, an approach that has been proposed for the highly mutable virus (Allen and Altfeld, 2008; Mann and Ndung'u, 2015). While Nef is a critical virulence factor in HIV infection (Kestler et al., 1991) and highly immunogenic (Radebe et al., 2011), it is also highly variable and largely excluded from the leading conserved T-cell based vaccine immunogens in testing (Borthwick et al., 2014; Mothe et al., 2015). Nevertheless, growing evidence suggests that a limited number of mutations in Nef, particularly specific mutation combinations, can impair the virus (Kuang et al., 2014; Shahid et al., 2015; Ueno et al., 2008), and that CTL targeting of selected Nef epitopes could lead to better clinical outcome (Adland et al., 2013). In the present analysis, two HLA-associated polymorphisms, 71K and 108D, were associated with decreased ability of Nef to inhibit TCR-mediated NFAT signalling. 71K was associated with HLA-C*07:02 and HLA-C*17 (Carlson et al., 2014) and confirmed to confer escape from CD8+ T cell responses to the 68FPVRPQVPLR77 epitope (Du et al., 2016), and 108D was associated with HLA-B*44 and HLA-B*18 (Carlson et al., 2014). Therefore, we found limited evidence for immune-driven impairment of Nef-mediated inhibition of NFAT signalling. In addition, residue 71 is immediately adjacent to the 72PxxPxR77 motif, which is important for Nef-mediated HLA-I down-regulation function (Foster and Garcia, 2008), and we confirmed by mutagenesis that 71K also decreases HLA-I down-regulation ability, a Nef function correlated with the rate of CD4+ T cell decline in subtype C infection (Mann et al., 2014). Therefore, residue 71 could represent a vulnerable immune target where escape impairs at least two known functions of Nef, including its immune evasion ability. In support of this, an overlapping peptide (OLP) spanning Nef residue 71 is the only Nef OLP previously identified as a viral target associated with relative control (Mothe et al., 2011) that is included in a human immune data-informed vaccine (Mothe et al., 2015).
Although different Nef functions are reported to be largely genetically separate (Foster et al., 2011), we investigated whether there was any overlap between Nef-mediated inhibition of NFAT signalling and HLA down-regulation as well as CD4 down-regulation. It was observed that HLA-I down-regulation activity significantly correlated with NFAT inhibition activity. This was not surprising as critical residues for both Nef functions include P72, P75 and R77 of the 68PxxP78 motif (Foster et al., 2011; Geyer et al., 2001). These residues are important in Nef-mediated alteration of TCR signalling because they are the primary binding site of the SH3 domain of signalling kinases (Geyer et al., 2001). In HLA-I down-regulation, these residues play a role through securing the binding of the HLA-I cytoplasmic tail to AP-1 (Collins and Collins, 2014).
It should be noted that when Nef interacts with other viral proteins its function might change and therefore the analysis of Nef in isolation is not a full representation of the microenvironment (Shrivastava et al., 2016). In addition, functions of Nef may differ depending on the infected cell type (Mwimanzi et al., 2011a; Mwimanzi et al., 2013a). Another limitation is the assessment of Nef’s ability to alter TCR signalling using Jurkat cells, which have some physiological differences (e.g. defective expression of PTEN) from primary cells that may affect TCR signalling (Abraham and Weiss, 2004). Nevertheless, Jurkat cells have yielded many insights into TCR signalling (Abraham and Weiss, 2004), and have the advantage of uniformity and consistency. Furthermore, the Jurkat-based assay used in the current study and companion report (Jin S, accepted, in press) was consistent with the effect of Nef to inhibit NFAT through impairing early TCR signalling events in pre-activated primary T cells (Neri et al., 2011). Similarly, in Jurkat cells, Nef inhibited NFAT production and this corresponded with the ability of Nef to co-localise with Lck, a proximal TCR signalling protein (Jin S, accepted, in press).
In summary, it was observed that Nef-mediated inhibition of NFAT activity differed among HIV-1 M group subtypes, with a hierarchy of C<A<B<D. Subtypes A and C were the least functional and this may correspond with the slower disease progression, increased expansion and high prevalence of these subtypes worldwide. Although we did not find evidence to support that Nef-mediated inhibition of NFAT signalling has a strong impact on disease progression, other studies indicate that there is selective pressure to maintain multiple Nef functions and that decreased ability to inhibit NFAT may contribute to elite control in some cases, supporting that it nevertheless plays a role in disease. Overall, we found limited evidence that CTL escape mutations attenuate this Nef function; however, CTL escape at residue 71 attenuates both Nef-mediated HLA-I down-regulation and inhibition of NFAT, and is therefore a potentially useful antiviral target.
Acknowledgements:
This work was supported by a Sullivan Early Career Scientist Award from the Sullivan Family Foundation to JM. JM received additional funding from the National Research Foundation. TN received additional funding from the South African Department of Science and Technology through the National Research Foundation (South African Research Chairs Initiative), the Victor Daitz Foundation and the Howard Hughes Medical Institute. This work was also partially supported by the Bill and Melinda Gates Foundation, the International AIDS Vaccine Initiative (IAVI) (UKZNRSA1001), and the NIAID (R37AI067073).
Additional funding was provided by the Sub-Saharan African Network for TB/HIV Research Excellence (SANTHE), a DELTAS Africa Initiative [grant # DEL-15-006]. The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS)’s Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust [grant # 107752/Z/15/Z] and the UK government. The views expressed in this publication are those of the author(s) and not necessarily those of AAS, NEPAD Agency, Wellcome Trust or the UK government. ZLB is supported by a Scholar Award from the Michael Smith Foundation for Health Research. MAB holds a Canada Research Chair in Viral Pathogenesis and Immunity. ZLB and MAB received additional funding from the Canadian Institutes of Health Research (CIHR; PJT-148621). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We acknowledge Dr. Chanson Brumme for statistical support. We thank Prof. David Bangsberg, Prof. Peter Hunt, Prof. Jeff Martin, Prof. Richard Harrigan, Dr. Vladimir Novitsky, Prof. Bruce Walker, and Prof. Philip Goulder for providing access to the samples. We are grateful to all study participants and support staff.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abraha A, Nankya IL, Gibson R, Demers K, Tebit DM, Johnston E, Katzenstein D, Siddiqui A, Herrera C, Fischetti L, Shattock RJ, Arts EJ, 2009. CCR5- and CXCR4-tropic subtype C HIV-1 isolates have lower pathogenic fitness as compared to the other dominant group M subtypes: Implications for the epidemic. J Virol 83(11), 5592–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham L, Fackler OT, 2012. HIV Nef: a multifaceted modulator of T cell receptor signaling. Cell Commun Signal. 10(1), 39. doi: 10.1186/1478-1811X-1110-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham RT, Weiss A, 2004. Jurkat T cells and development of the T-cell receptor signalling paradigm. Nat Rev Immunol 4, 301–308. [DOI] [PubMed] [Google Scholar]
- Addo MM, Altfeld M, 2014. Sex-based differences in HIV type 1 pathogenesis. J Infect Dis. 209 Suppl 3, S86–92. doi: 10.1093/infdis/jiu1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adland E, Carlson JM, Paioni P, Kløverpris H, Shapiro R, Ogwu A, Riddell L, Luzzi G, Chen F, Balachandran T, Heckerman D, Stryhn A, Edwards A, Ndung'u T, Walker BD, Buus S, Goulder P, Matthews PC, 2013. Nef-specific CD8+ T cell responses contribute to HIV-1 immune control. PLoS One 8(9), e73117. doi: 73110.71371/journal.pone.0073117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali A, Ng HL, Dagarag MD, Yang OO, 2005. Evasion of cytotoxic T lymphocytes is a functional constraint maintaining HIV-1 Nef expression. Eur J Immunol. 35(11), 3221–3228. [DOI] [PubMed] [Google Scholar]
- Allen TM, Altfeld M, 2008. Crippling HIV one mutation at a time. J Exp Med 205(5), 1003–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonio MJ, Chaparro-Sánchez A, Hernández-López JC, Huerta-García G, Domínguez-Hermosillo JC, Cruz-Domínguez P, 2016. Clinical and epidemiological differences between women and men with HIV infection in Mexico. J AIDS Clin Res 7(551), doi: 10.4172/2155-6113.1000551. [DOI] [Google Scholar]
- Ariën KK, Vanham G, Arts EJ, 2007. Is HIV-1 evolving to a less virulent form in humans? Nat Rev Microbiol. 5(2), 141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arold S, Franken P, Strub M-P, Hoh F, Benichou S, Benarous R, Dumas C, 1997. The crystal structure of HIV-1 Nef protein bound to the Fyn kinase SH3 domain suggests a role for this complex in altered T cell receptor signaling. Structure 5, 1361–1372. [DOI] [PubMed] [Google Scholar]
- Baur AS, Sass G, Laffert B, Willbold D, Cheng-Mayer C, Peterlin BM, 1997. The N-Terminus of Nef from HIV-1/SIV Associates with a Protein Complex Containing Lck and a Serine Kinase. Immunity 6, 283–291. [DOI] [PubMed] [Google Scholar]
- Borthwick N, Ahmed T, Ondondo B, Hayes P, Rose A, Ebrahimsa U, Hayton EJ, Black A, Bridgeman A, Rosario M, Hill AV, Berrie E, Moyle S, Frahm N, Cox J, Colloca S, Nicosia A, Gilmour J, McMichael AJ, Dorrell L, Hanke T, 2014. Vaccine-elicited human T cells recognizing conserved protein regions inhibit HIV-1. Mol Ther. 22(2), 464–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonaguro L, Tornesello ML, Buonaguro FM, 2007. Human Immunodeficiency Virus Type 1 Subtype Distribution in the Worldwide Epidemic: Pathogenetic and Therapeutic Implications. Journal of Virology 81, 10209–10219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson JM, Schaefer M, Monaco DC, Batorsky R, Claiborne DT, Prince J, Deymier MJ, Ende ZS, Klatt NR, DeZiel CE, Lin TH, Peng J, Seese AM, Shapiro R, Frater J, Ndung'u T, Tang J, Goepfert P, Gilmour J, Price MA, Kilembe W, Heckerman D, Goulder PJ, Allen TM, Allen S, Hunter E, 2014. HIV transmission. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science 345(6193), 1254031. doi: 1254010.1251126/science.1254031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins DR, Collins KL, 2014. HIV-1 accessory proteins adapt cellular adaptors to facilitate immune evasion. PLoS Pathogens 10, e1003851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy SA, Laeyendecker O, Redd AD, Collinson-Streng A, Kong X, Makumbi F, Lutalo T, Sewankambo N, Kiwanuka N, Gray RH, Wawer MJ, Serwadda D, Quinn TC, Rakai Health Sciences Program, 2010. Changes in the distribution of HIV type 1 subtypes D and A in Rakai District, Uganda between 1994 and 2002. AIDS Res Hum Retroviruses 26(10), 1087–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corró G, Rocco CA, De Candia C, Catano G, Turk G, Mangano A, Aulicino PC, Bologna R, Sen L, 2012. Genetic and Functional Analysis of HIV Type 1 nef Gene Derived from Long-Term Nonprogressor Children: Association of Attenuated Variants with Slow Progression to Pediatric AIDS. AIDS Res Hum Retroviruses 28(12), 1617–1626. [DOI] [PubMed] [Google Scholar]
- Deacon NJ, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker DJ, McPhee DA, Greenway AL, Ellett A, Chatfield C, Lawson VA, Crowe S, Maerz A, Sonza S, Learmont J, Sullivan JS, Cunningham A, Dwyer D, Dowton D, Mills J, 1995. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science 270(5238), 988–991. [DOI] [PubMed] [Google Scholar]
- Du VY, Bansal A, Carlson J, Salazar-Gonzalez JF, Salazar MG, Ladell K, Gras S, Josephs TM, Heath SL, Price DA, 2016. HIV-1–Specific CD8 T Cells Exhibit Limited Cross-Reactivity during Acute Infection. The Journal of Immunology 196, 3276–3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easterbrook PJ, Smith M, Mullen J, O’Shea S, Chrystie I, de Ruiter A, Tatt ID, Geretti AM, Zuckerman M, 2010. Impact of HIV-1 viral subtype on disease progression and response to antiretroviral therapy. J Int AIDS Soc 13, 4. doi: 10.1186/1758-2652-1113-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fackler OT, Alcover A, Schwartz O, 2007. Modulation of the immunological synapse: a key to HIV-1 pathogenesis? Nat Rev Immunol. 7(4), 310–317. [DOI] [PubMed] [Google Scholar]
- Foster JL, Denial SJ, Temple BRS, Garcia JV, 2011. Mechanisms of HIV-1 Nef Function and Intracellular Signaling. J Neuroimmune Pharmacol 6, 230–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster JL, Garcia JV, 2008. HIV-1 Nef: at the crossroads. Retrovirology 5, 84. doi: 10.1186/1742-4690-1185-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer M, Fackler OT, Peterlin BM, 2001. Structure-function relationships in HIV-1 Nef. EMBO reports 21(7), 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräf T, Pinto AR, 2013. The increasing prevalence of HIV-1 subtype C in Southern Brazil and its dispersion through the continent. Virology 435(1), 170–178. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O, 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52(5), 696–704. [DOI] [PubMed] [Google Scholar]
- Hemelaar J, 2012. The origin and diversity of the HIV-1 pandemic. Trends Mol Med. 18(3), 182–192. [DOI] [PubMed] [Google Scholar]
- Huang Y, Zhang L, Ho DD, 1995. Characterization of nef sequences in long-term survivors of human immunodeficiency virus type 1 infection. Journal of Virology 69, 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate AJ, Carl S, Bronson S, Stahl-Hennig C, Swigut T, Skowronski J, Kirchhoff F, 2000. Disrupting surfaces of nef required for downregulation of CD4 and for enhancement of virion infectivity attenuates simian immunodeficiency virus replication in vivo. J Virol 74(21), 9836–9844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki PJ, Hamel DJ, Sankalé JL, Hsieh C, Thior I, Barin F, Woodcock SA, Guèye-Ndiaye A, Zhang E, Montano M, Siby T, Marlink R, NDoye I, Essex ME, MBoup S, 1999. Human Immunodeficiency Virus Type 1 Subtypes Differ in Disease Progression. J Infect Dis 179(1), 68–73. [DOI] [PubMed] [Google Scholar]
- Kestler HW, Ringler DJ, Mori K, Panicali DL, Sehgal PK, Daniel MD, Desrosiers RC, 1991. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell 65(4), 651–662. [DOI] [PubMed] [Google Scholar]
- Kiguoya MW, Mann JK, Chopera D, Gounder K, Lee GQ, Hunt PW, Martin JN, Ball TB, Kimani J, Brumme ZL, Brockman MA, Ndung'u T, 2017. Subtype-Specific Differences in Gag-Protease-Driven Replication Capacity are Consistent with Inter-Subtype Differences in HIV-1 Disease Progression. J Virol, pii: JVI.00253-00217. doi: 00210.01128/JVI.00253-00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiwanuka N, Laeyendecker O, Quinn TC, Wawer MJ, Shepherd J, Robb M, Kigozi G, Kagaayi J, Serwadda D, Makumbi FE, Reynolds SJ, Gray RH, 2009. HIV-1 subtypes and differences in heterosexual HIV transmission among HIV-discordant couples in Rakai, Uganda. AIDS 23(18), 2479–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang XT, Li X, Anmole G, Mwimanzi P, Shahid A, Le AQ, Chong L, Qian H, Miura T, Markle T, Baraki B, Connick E, Daar ES, Jessen H, Kelleher AD, Little S, Markowitz M, Pereyra F, Rosenberg ES, Walker BD, Ueno T, Brumme ZL, Brockman MA, 2014. Impaired Nef function is associated with early control of HIV-1 viremia. J Virol 88(17), 10200–10213. doi: 10210.11128/JVI.01334-10214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C-H, Saksela K, Mirza UA, Chait BT, Kuriyan J, 1996. Crystal structure of the conserved core of HIV-1 Nef complexed with a Src family SH3 domain. Cell 85, 931–942. [DOI] [PubMed] [Google Scholar]
- Liu X, Schrager JA, Lange GD, Marsh JW, 2001. HIV Nef-mediated cellular phenotypes are differentially expressed as a function of intracellular Nef concentrations. J Biol Chem. 276(35), 32763–32770. [DOI] [PubMed] [Google Scholar]
- Mann JK, Byakwaga H, Kuang XT, Le AQ, Brumme CJ, Mwimanzi P, Omarjee S, Martin E, Lee GQ, Baraki B, Danroth R, McCloskey R, Muzoora C, Bangsberg DR, Hunt PW, Goulder PJ, Walker BD, Harrigan PR, Martin JN, Ndung'u T, Brockman MA, Brumme ZL, 2013. Ability of HIV-1 Nef to downregulate CD4 and HLA class I differs among viral subtypes. Retrovirology 10(1), 100. doi: 110.1186/1742-4690-1110-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann JK, Chopera D, Omarjee S, Kuang XT, Le AQ, Anmole G, Danroth R, Mwimanzi P, Reddy T, Carlson J, Radebe M, Goulder PJ, Walker BD, Abdool Karim S, Novitsky V, Williamson C, Brockman MA, Brumme ZL, Ndung'u T, 2014. Nef-mediated down-regulation of CD4 and HLA class I in HIV-1 subtype C infection: association with disease progression and influence of immune pressure. Virology 468-470, 214–225. doi: 210.1016/j.virol.2014.1008.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann JK, Ndung'u T, 2015. HIV-1 vaccine immunogen design strategies. Virology journal 12(1), 3. doi: 10.1186/s12985-12014-10221-12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura T, Brockman MA, Brumme CJ, Brumme ZL, Carlson JM, Pereyra F, Trocha A, Addo MM, Block BL, Rothchild AC, Baker BM, Flynn T, Schneidewind A, Li B, Wang YE, Heckerman D, Allen TM, Walker BD, 2008. Genetic Characterization of Human Immunodeficiency Virus Type 1 in Elite Controllers: Lack of Gross Genetic Defects or Common Amino Acid Changes. J Virol 82(17), 8422–8430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothe B, Hu X, Llano A, Rosati M, Olvera A, Kulkarni V, Valentin A, Alicea C, Pilkington GR, Sardesai NY, Rocafort M, Crespo M, Carrillo J, Marco A, Mullins JI, Dorrell L, Hanke T, Clotet B, Pavlakis GN, Felber BK, Brander C, 2015. A human immune data-informed vaccine concept elicits strong and broad T-cell specificities associated with HIV-1 control in mice and macaques. J Transl Med 13(1), 60. doi: 10.1186/s12967-12015-10392-12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothe B, Llano A, Ibarrondo J, Daniels M, Miranda C, Zamarreño J, Bach V, Zuniga R, Pérez-Álvarez S, Berger CT, Puertas MC, Martinez-Picado J, Rolland M, Farfan M, Szinger JJ, Hildebrand WH, Yang OO, Sanchez-Merino V, Brumme CJ, Brumme ZL, Heckerman D, Allen TM, Mullins JI, Gómez G, Goulder PJ, Walker BD, Gatell JM, Clotet B, Korber BT, Sanchez J, Brander C, 2011. Definition of the viral targets of protective HIV-1-specific T cell responses. J Transl Med. 9, 208. doi: 210.1186/1479-5876-1189-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwimanzi P, Hasan Z, Hassan R, Suzu S, Takiguchi M, Ueno T, 2011a. Effects of naturally-arising HIV Nef mutations on cytotoxic T lymphocyte recognition and Nef's functionality in primary macrophages. Retrovirology 8, 50. doi: 10.1186/1742-4690-1188-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwimanzi P, Markle T, Otsuka H, Ogata Y, Tokunaga M, Miura T, Martin E, Pereyra F, Walker B, Brumme Z, Brockman M, Ueno T, 2011b. Impairment of viral replication capacity by nef alleles from HIV elite controllers. Retrovirology 8(Suppl2), P53. doi: 10.1186/1742-4690-1188-S1182-P1153.21729311 [DOI] [Google Scholar]
- Mwimanzi P, Markle TJ, Martin E, Ogata Y, Kuang XT, Tokunaga M, Mahiti M, Pereyra F, Miura T, Walker BD, Brumme ZL, Brockman MA, Ueno T, 2013a. Attenuation of multiple Nef functions in HIV-1 elite controllers. Retrovirology 10, 1. doi: 10.1186/1742-4690-1110-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwimanzi P, Markle TJ, Ogata Y, Martin E, Tokunaga M, Mahiti M, Kuang XT, Walker BD, Brockman MA, Brumme ZL, Ueno T, 2013b. Dynamic range of Nef functions in chronic HIV-1 infection. Virology 439(2), 74–80. [DOI] [PubMed] [Google Scholar]
- Neilson JR, John GC, Carr JK, Lewis P, Kreiss JK, Jackson S, Nduati RW, Mbori-Ngacha D, Panteleeff DD, Bodrug S, Giachetti C, Bott MA, Richardson BA, Bwayo J, Ndinya-Achola J, Overbaugh J, 1999. Subtypes of human immunodeficiency virus type 1 and disease stage among women in Nairobi, Kenya. J Virol. 73(5), 4393–4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri F, Giolo G, Potestà M, Petrini S, Doria M, 2011. The HIV-1 Nef protein has a dual role in T cell receptor signaling in infected CD4+ T lymphocytes. Virology 410(2), 312–326. doi: 310.1016/j.virol.2010.1011.1018. [DOI] [PubMed] [Google Scholar]
- Ng OT, Laeyendecker O, Redd AD, Munshaw S, Grabowski MK, Paquet AC, Evans MC, Haddad M, Huang W, Robb ML, Reynolds SJ, Gray RH, Wawer MJ, Serwadda D, Eshleman SH, Quinn TC, 2014. HIV type 1 polymerase gene polymorphisms are associated with phenotypic differences in replication capacity and disease progression. J Infect Dis. 209(1), 66–73. doi: 10.1093/infdis/jit1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm AA, Esbjörnsson J, Månsson F, Kvist A, Isberg PE, Biague A, da Silva ZJ, Jansson M, Norrgren H, Medstrand P, 2014. Faster progression to AIDS and AIDS-related death among seroincident individuals infected with recombinant HIV-1 A3/CRF02_AG compared with sub-subtype A3. J Infect Dis. 209(5), 721–728. [DOI] [PubMed] [Google Scholar]
- Radebe M, Nair K, Chonco F, Bishop K, Wright JK, van der Stok M, Bassett IV, Mncube Z, Altfeld M, Walker BD, Ndung'u T, 2011. Limited Immunogenicity of HIV CD8+ T-Cell Epitopes in Acute Clade C Virus Infection. J Infect Dis 204(5), 768–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renjifo B, Gilbert P, Chaplin B, Msamanga G, Mwakagile D, Fawzi W, Essex M, Tanzanian Vitamin and HIV Study Group, 2004. Preferential in-utero transmission of HIV-1 subtype C as compared to HIV-1 subtype A or D. AIDS 18(12), 1629–1636. [DOI] [PubMed] [Google Scholar]
- Rodriguez MA, Ding M, Ratner D, Chen Y, Tripathy SP, Kulkarni SS, Chatterjee R, Tarwater PM, Gupta P, 2009. High replication fitness and transmission efficiency of HIV-1 subtype C from India: Implications for subtype C predominance. Virology 385(2), 416–424. [DOI] [PubMed] [Google Scholar]
- Rosa A, Chande A, Ziglio S, De Sanctis V, Bertorelli R, Goh SL, McCauley SM, Nowosielska A, Antonarakis SE, Luban J, Santoni FA, Pizzato M, 2015. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 526(7572), 212–217. doi: 210.1038/nature15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross TM, Oran AE, Cullen BR, 1999. Inhibition of HIV-1 progeny virion release by cell-surface CD4 is relieved by expression of the viral Nef protein. Curr Biol 9(12), 613–621. [DOI] [PubMed] [Google Scholar]
- Schwartz O, Marechal V, Le Gall S, Lemonnier F, Heard JM, 1996. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. . Nat Med 2, 338–342. [DOI] [PubMed] [Google Scholar]
- Shahid A, Olvera A, Anmole G, Kuang XT, Cotton LA, Plana M, Brander C, Brockman MA, Brumme ZL, 2015. Consequences of HLA-B*13-Associated Escape Mutations on HIV-1 Replication and Nef Function. J Virol 89(22), 11557–11571. doi: 11510.11128/JVI.01955-11515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava S, Trivedi J, Mitra D, 2016. Gene expression profiling reveals Nef induced deregulation of lipid metabolism in HIV-1 infected T cells. Biochemical and Biophysical Research Communications 472, 169–174. [DOI] [PubMed] [Google Scholar]
- Silveira J, Santos AF, Martínez AM, Góes LR, Mendoza-Sassi R, Muniz CP, Tupinambás U, Soares MA, Greco DB, 2012. Heterosexual transmission of human immunodeficiency virus type 1 subtype C in southern Brazil. J Clin Virol. 54(1), 36–41. [DOI] [PubMed] [Google Scholar]
- Smith JA, Tso JY, Clark MR, Cole MS, Bluestone JA, 1997. Nonmitogenic anti-CD3 monoclonal antibodies deliver a partial T cell receptor signal and induce clonal anergy. J Exp Med. 185(8), 1413–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ssemwanga D, Nsubuga RN, Mayanja BN, Lyagoba F, Magambo B, Yirrell D, Van der Paal L, Grosskurth H, Kaleebu P, 2013. Effect of HIV-1 subtypes on disease progression in rural Uganda: a prospective clinical cohort study. PloS one 8, e71768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddart CA, Geleziunas R, Ferrell S, Linquist-Stepps V, Moreno ME, Bare C, Xu W, Yonemoto W, Bresnahan PA, McCune JM, Greene WC, 2003. Human Immunodeficiency Virus Type 1 Nef-Mediated Downregulation of CD4 Correlates with Nef Enhancement of Viral Pathogenesis. J Virol. 77(3), 2124–2133. doi: 2110.1128/JVI.2177.2123.2124-2133.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R, 2003. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences 100, 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigut T, Alexander L, Morgan J, Lifson J, Mansfield KG, Lang S, Johnson RP, Skowronski J, Desrosiers R, 2004. Impact of Nef-mediated downregulation of major histocompatibility complex class I on immune response to simian immunodeficiency virus. J Virol 78(23), 13335–13344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebit DM, Arts EJ, 2011. Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect Dis 11, 45–56. [DOI] [PubMed] [Google Scholar]
- Ueno T, Motozono C, Dohki S, Mwimanzi P, Rauch S, Fackler OT, Oka S, Takiguchi M, 2008. CTL-Mediated Selective Pressure Influences Dynamic Evolution and Pathogenic Functions of HIV-1 Nef. J Immunol 180, 1107–1116. [DOI] [PubMed] [Google Scholar]
- Usami Y, Wu Y, Göttlinger HG, 2015a. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 526, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami Y, Wu Y, Gottlinger HG, 2015b. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 526(7572), 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasan A, Renjifo B, Hertzmark E, Chaplin B, Msamanga G, Essex M, Fawzi W, Hunter D, 2006. Different rates of disease progression of HIV type 1 infection in Tanzania based on infecting subtype. Clin Infect Dis 42(6), 843–852. [DOI] [PubMed] [Google Scholar]
- Veillette M, Désormeaux A, Medjahed H, Gharsallah NE, Coutu M, Baalwa J, Guan Y, Lewis G, Ferrari G, Hahn BH, Haynes BF, Robinson JE, Kaufmann DE, Bonsignori M, Sodroski J, Finzi A, 2014. Interaction with cellular CD4 exposes HIV-1 envelope epitopes targeted by antibody-dependent cell-mediated cytotoxicity. J Virol 88(5), 2633–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venner CM, Nankya I, Kyeyune F, Demers K, Kwok C, Chen P-L, Rwambuya S, Munjoma M, Chipato T, Byamugisha J, 2016. Infecting HIV-1 subtype predicts disease progression in women of sub-Saharan Africa. EBioMedicine 13, 305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins RL, Zou W, Denton PW, Krisko JF, Foster JL, Garcia JV, 2013. In vivo analysis of highly conserved Nef activities in HIV-1 replication and pathogenesis. Retrovirology 10(125), doi: 10.1186/1742-4690-1110-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JK, Brumme ZL, Carlson JM, Heckerman D, Kadie CM, Brumme CJ, Wang B, Losina E, Miura T, Chonco F, van der Stok M, Mncube Z, Bishop K, Goulder PJR, Walker BD, Brockman MA, Ndung’u T, 2010. Gag-Protease-Mediated Replication Capacity in HIV-1 Subtype C Chronic Infection: Associations with HLA Type and Clinical Parameters. J Virol 84(20), 10820–10831. [DOI] [PMC free article] [PubMed] [Google Scholar]