Abstract
SIRT3 is a major mitochondrial deacetylase, which regulates various metabolic pathways by deacetylation; however, the effect of SIRT3 on proline metabolism is not reported. Pyrroline-5-carboxylate reductase 1 (PYCR1) participates in proline synthesis process by catalyzing the reduction of P5C to proline with concomitant generation of NAD+ and NADP+. PYCR1 is highly expressed in various cancers, and it can promote the growth of tumor cells. Here, through immunoprecipitation and mass spectrometry, we found that PYCR1 is in SIRT3’s interacting network. PYCR1 directly binds to SIRT3 both in vivo and in vitro. CBP is the acetyltransferase for PYCR1, whereas SIRT3 deacetylates PYCR1. We further identified that K228 is the major acetylation site for PYCR1. Acetylation of PYCR1 at K228 reduced its enzymatic activity by impairing the formation of the decamer of PYCR1. As a result, acetylation of PYCR1 at K228 inhibits cell proliferation, while deacetylation of PYCR1 mediated by SIRT3 increases PYCR1’s activity. Our findings on the regulation of PYCR1 linked proline metabolism with SIRT3, CBP and cell growth, thus providing a potential approach for cancer therapy.
Introduction
SIRT3 is an NAD+-dependent deacetylase in mitochondria which contains 399 amino acids and two isoforms [1]. It regulates many cellular processes, including metabolism, oxidative stress, apoptosis and cell survival through deacetylation of mitochondrial proteins [2]. SIRT3 thus plays an important role in tumor-promotion or tumor-suppression [1], [2].
Proline, a unique non-essential amino acid, plays a key role in stress protection and the maintenance of the redox balance in cells [3]. The NAD(P)H-dependent reduction of 1-pyrroline-5-carboxylate (P5C) is catalyzed by pyrroline-5-carboxylate reductase (P5CR), which is the final step in proline biosynthesis [4]. This reaction exists in almost all organisms and is highly conserved [5]. In humans, P5CR is also called PYCR; the human genome contains three homologous PYCR genes, which respectively encode PYCR1, PYCR2 and PYCRL (also called PYCR3). The PYCR1 protein is composed of five homodimers and is localized in mitochondria. The five homodimers form a circular groove, which is the binding site of the cofactor and the substrate [5]. Mutations of the PYCR1 gene are associated with autosomal recessive cutis laxa (ARCL), a group of syndromal disorders characterized by wrinkled skin and a progeroid appearance [6].
PYCR1 participates in proline metabolism by catalyzing the reduction of P5C to proline with concomitant generation of NAD+ and NADP+, which may augment glycolysis and the pentose phosphate pathway, respectively [7]. The properties of proline as a compatible solute enable its anti-stress function in a variety of organisms. In addition to its natural osmolyte properties, proline can protect cells against ROS via the secondary amine of the pyrrolidine ring [8]. Proline can also minimize protein aggregation, thus playing a role in inhibiting the accumulation of misfolded proteins caused by endoplasmic reticulum stress. PYCR1, then, both combats oxidative stress and the endoplasmic reticulum stress through the biosynthesis of proline [9]. PYCR1 is overexpressed in various cancers, including prostate cancer, breast cancer, renal cell carcinoma, melanoma, non-small cell lung cancer, and tumors of the head, neck, esophagus and pancreas [4], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20]. PYCR1 can promote the growth of tumor cells, and knockout of PYCR1 shows obvious inhibition of cell proliferation [4], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20].
Post-translational modification of PYCR1 has not yet been reported and the relationship between proline metabolism and SIRT3 remains unknown. In this study, we find that SIRT3 interacts with and deacetylates PYCR1. Deacetylation of PYCR1 increases its enzymatic activity, thus enhancing cell proliferation. Our findings enrich the functions of SIRT3 and provide new insight into post-translational modification regulation of PYCR1.
Materials and methods
Cell culture and cell lines
HEK293T, H1299, MCF7 and U2OS cells were cultured in DMEM (Invitrogen) containing 10% fetal bovine serum and 1% penicillin–streptomycin at 37 °C and 5% CO2.
To generate SIRT3 stable overexpression cell lines, the SIRT3-FLAG-HA sequence was cloned into a pCIN4 vector. The plasmid was transfected into H1299 cells and the transfected cells were selected by 1 mg/ml G418 for two weeks.
CRISPR-Cas9 knockout cell lines: we cloned the sgRNA sequence:
(SIRT3:5’-CACCGCTCTACACGCAGAACATCGA-3′;
PYCR1:5’-CACCGCATGACCAACACTCCAGTCG-3′) into a LentiCRISPR V2 vector and transfected the plasmids with packaging plasmids (psPAX2 and pMD2G) into HEK293T cells. The medium was changed after 8–10 hours and we collected the viral supernatant and filtered it into the target cells (U2OS or MCF7) with a certain amount of serum 48 hours later. Then, we selected the infected cells with 1 μg/ml puromycin for two weeks.
MCF7 rescued cell lines: we cloned flag tagged PYCR1-WT/K228R/K228Q sequence into pQCXIH retrovirus vector and transfected the plasmids with packaging plasmids (vsvg and gag-pol) into HEK293T cells. The medium was changed after 6–8 hours and we collected the viral supernatant and filtered it into MCF7 PYCR1 KO cells with a certain amount of serum 24 hours later. Then we repeated the steps from transfection and selected the infected cells with 150 μg/ml hygromycin for two weeks.
Mitochondrial isolation
SIRT3-FLAG-HA stable cell lines and control cells were harvested and homogenized, the homogenate was then centrifuged for 5 min at 740 g, and then the supernatant was collected and centrifuged for 10 min at 9000 g. The pellet was collected and thus the crude mitochondria was obtained [21]. The crude mitochondria were lysed by BC100 buffer and filtered by 0.45 μm filter, thus the mitochondrial protein lysates were obtained [22].
Co-immunoprecipitation and western blotting
Whole cells were lysed by BC100 buffer (100 mM NaCl, 20 mM pH 7.3 Tris, 20% glycerol, 0.1% NP-40). The cell lysates were incubated with anti-Flag M2 (Sigma) /HA affinity gel (Roche) overnight at 4 °C. The beads were washed with BC100 6 times and eluted by Flag peptide (Sigma) at 4 °C. The elution was subjected to western blot and immunoblotted with antibodies. Flag (Sigma), HA (Pierce), α-tubulin (Santa Cruze), β-actin (Santa Cruze), GAPDH (Cell Signaling Technology), SIRT3 (Cell Signaling Technology), SIRT4 (Abiocode), PYCR1 (proteintech), pan-acetyllysine (Cell Signaling Technology /PTM Biolabs).
GST pull-down and in vitro acetylation assay
The GST fusion proteins, purified from Rosetta cells, were incubated with GST resin (Novagen) overnight at 4 °C. The beads were washed with BC100 and eluted by Glutathione (GSH). The elution was added to the 30 μl reaction system[3 μl 10 × Ac Buffer1(200 mM PH8.0 HEPES,10 mM DTT,10 mM PMSF,1 mg/ml BSA),1 μl acetyl-CoA(17 ng/μl) and 0.1 μg CBP] at 37 °C for 2 hours; the acetylation levels were detected by western blot and the amount of protein was detected by Coomassie blue staining.
In vitro deacetylation assay
The 30 μl reaction system [3 μl 10 × Ac Buffer1, 15 μl 2 × DeAc Buffer2 (8 mM MgCl2, 100 mM NaCl, 20% glycerol) and 1 μg acetylated PYCR1 (PYCR1-FLAG was co-transfected with HA-CBP in HEK293T cells; the cells were then treated with trichostatin A (TSA) and Nicotinamide (NAM) for 8 hours, and then we purified the PYCR1-FLAG)] was present or absent of 2 μg SIRT3/SIRT4 and 3 μl NAD+ (1 mM) and reacted at 37 °C for 2 hours.
Enzymatic activity assays for PYCR1
The reaction system (300 mM PH = 6.8 Tris, 0.5 mM NADH, 5 mM P5C, 0.2 mg/ml BSA, 10% glycerol, 1 mM DTT) was added with 0.01 μg different status of PYCR1 proteins on ice, and the changes of absorption in 340 nm at room temperature were detected immediately. The reduction of OD value in 7 minutes was the quantitative value of enzymatic activity of PYCR1.
Real-time PCR
Total RNA was extracted by Trizol reagent (Sigma). 2 μg RNA were reverse transcribed to cDNA using the qPCR RT Kit (TOYOBO). 7500 Fast Real-time PCR System (Applied Biosystems) and qPCR SYBR green master mix (Vazyme) were used to perform real-time PCR. The amount of mRNA was calculated by △△Ct method and normalized to actin. The primer sequences were as follow:
PYCR1 forward primer: CATGACCAACACTCCAGTCG;
PYCR1 reverse primer: CCTTGGAAGTCCCATCTTCA;
PYCR2 forward primer: ATAGCCAGCTCCCCAGAAAT;
PYCR2 reverse primer: CTCCACAGAGCTGATGGTGA;
PYCRL forward primer: GCCTCATCAGAGCAGGAAAA;
PYCRL reverse primer: CACGGACACCAAGATGTGTT;
actin forward primer: TCCATCATGAAGTGTGACG;
actin reverse primer: TACTCCTGCTTGCTGATCCAC.
Cell proliferation and colony formation
5.0 × 104 cells were seeded into 6-well plates and we counted the cells every day. Cells were stained by 0.2% trypan blue and counted by Countstar IC1000.
CCK8 assay: Cell proliferation rates were measured using Cell Counting Kit 8 (CCK8, Dojindo). 2500 cells were seeded into 96-well plates and cultured for 3 days, 10 μl CCK8 was added 1 hour before we detected the changes of absorption in 450 nm at room temperature.
10000 cells were seeded into 6 cm dishes and cultured for 7 days, then fixed with 4% paraformaldehyde and stained with 0.2% crystal violet. We used 33% acetic acid to dissolve crystal violet and detected the changes of absorption at 570 nm, the quantitative value of which represented relative colony formation rates.
Statistical analysis
Statistical analysis was performed with unpaired Student's t-test, one-way ANOVA and two-way ANOVA by prism 6.0. Data were expressed as mean ± SEM and P < .05 was considered statistically significant. * denotes P < .05, ** denotes P < .01, *** denotes P < .001 and **** denotes P < .0001.
Results
Identify the SIRT3 interacting proteins
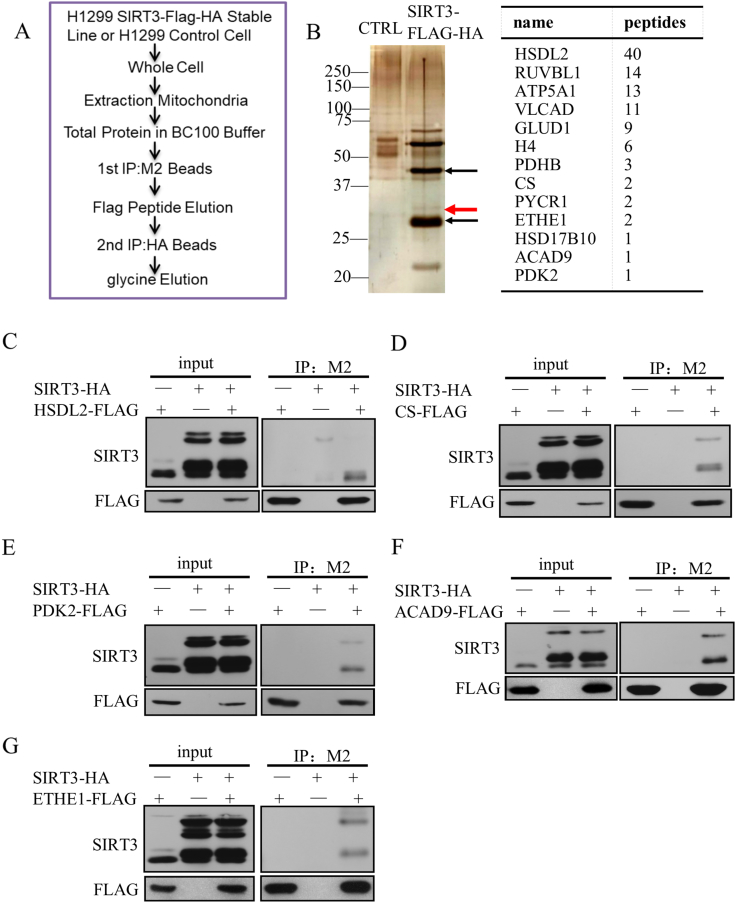
The SIRT3 stably expressed cell line was established and the mitochondria were extracted from these and the control cells [21]. The protein lysates were immunoprecipitated by M2 beads and HA beads, respectively (Figure 1A), and the elution was analyzed by SDS-PAGE to separate the proteins for observing the interacting proteins by silver staining (Figure 1B). The unique bands in SIRT3 IP were further analyzed by mass-spectrometry to identify the SIRT3 interacting proteins. There were a large number of SIRT3 reported substrates in the obtained results such as GDH and VLCAD, which proved the reliability of the results [23], [24], [25], [26]. According to the amount of peptides and function of the proteins in the mass-spectrum, we chose several proteins as candidates and identified the interaction between SIRT3 and some of those proteins to further confirm the reliability of mass-spectrometry results (Figure 1C-G). Since the relationship between SIRT3 and proline metabolism had not been reported, we chose the protein PYCR1, which involved in proline synthesis, as the object of further study. The 33KD protein PYCR1 was visible in silver staining (red arrow), which was the band above the 28KD band of SIRT3 (black arrow, Figure 1B).
Figure 1.
Identify the SIRT3 interacting proteins.
(A) Schematic diagram showed the method of identifying the SIRT3 interaction protein.
(B) Silver staining and mass-spectrometry analysis.
(C to G) Exogenous hydroxysteroid dehydrogenase-like protein 2 (HSDL2)/ citrate synthetase (CS)/ pyruvate dehydrogenase kinase isoform 2 (PDK2)/ acyl-CoA dehydrogenase family member 9 (ACAD9)/ ethylmalonic encephalopathy 1 protein (ETHE1) and SIRT3 were co-expressed in HEK293T cells, and co-immunoprecipitation assay showed the interaction between HSDL2/CS/PDK2/ACAD9/ETHE1 and SIRT3 in vivo.
SIRT3 interacts with PYCR1
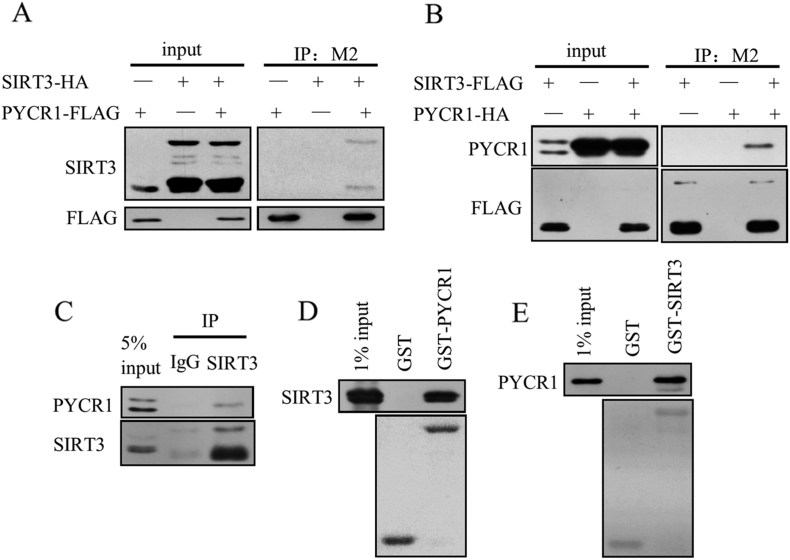
We confirmed the interaction between SIRT3 and PYCR1. SIRT3 and PYCR1 were overexpressed in HEK293T cells, SIRT3-HA was co-immunoprecipitated by PYCR1-FLAG (Figure 2A), and PYCR1-HA was also co-immunoprecipitated by SIRT3-FLAG (Figure 2B). Endogenous PYCR1 was also co-immunoprecipitated by endogenous SIRT3, indicating the interaction between PYCR1 and SIRT3 in vivo (Figure 2C). Through the GST pull down assay, we found that SIRT3-FLAG was pulled down by GST-PYCR1 (Figure 2D), and PYCR1-FLAG was also pulled down by GST-SIRT3 (Figure 2E), further demonstrating the direct interaction between PYCR1 and SIRT3 in vitro.
Figure 2.
SIRT3 interacts with PYCR1.
(A and B) Exogenous PYCR1 and SIRT3 were co-expressed in HEK293T cells, and co-immunoprecipitation assay showed the interaction between PYCR1 and SIRT3 in vivo.
(C) The HEK293T whole cell lysates were incubated with anti-SIRT3 antibody or isotype control IgG antibody, and then immunoprecipitated by protein A/G. The elution was detected by western blot.
(D and E) GST-pull down assays were performed with GST fusion protein and Flag-tagged protein purified from HEK293T cells, showed the interaction between PYCR1 and SIRT3 in vitro.
CBP is the acetyltransferase of PYCR1
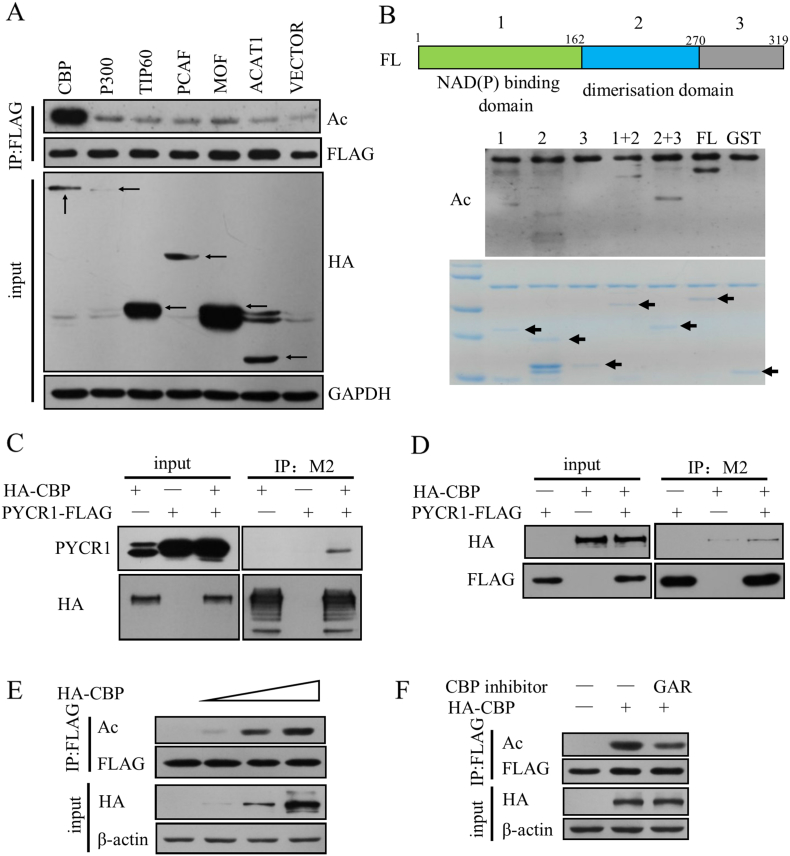
The interaction of PYCR1 and SIRT3 prompted us to examine if PYCR1 is a novel target for SIRT3. We first tested if PYCR1 could be modified by acetylation. The first step was to find the acetyltransferase for PYCR1. We examined several common acetyltransferases, such as CBP, P300, TIP60, PCAF, MOF and ACAT1, and only CBP dramatically increased the acetylation levels of PYCR1 (Figure 3A). We divided PYCR1 into three fragments according to its structure: the first fragment is the NAD (P) binding domain at the N-terminal, the second fragment the dimerization domain in the middle and the third fragment the C-terminal of PYCR1. Using these fragments, we examined their acetylation levels through in vitro acetylation assay. The acetylation levels of full-length PYCR1 was the strongest; the first and second fragments were weaker. However, it was hard to detect the acetylation levels of the third fragment, indicating that CBP directly acetylated PYCR1 in vitro, and the acetylation sites mainly existed in the first and the second fragments (Figure 3B). We found that PYCR1-FLAG was co-immunoprecipitated by HA-CBP (Figure 3C), and HA-CBP was also co-immunoprecipitated by PYCR1-FLAG (Figure 3D),further verifying the interaction of PYCR1 and CBP in vivo. CBP acetylated PYCR1 in a dose-dependent manner (Figure 3E). GAR, an inhibitor of CBP/P300, reduced the acetylation levels of PYCR1 by inhibiting the acetyltransferase activity of CBP (Figure 3F). All these results validate that CBP is the acetyltransferase of PYCR1, and it can acetylate PYCR1 both in vivo and in vitro.
Figure 3.
CBP is the acetyltransferase of PYCR1.
(A) PYCR1 was co-transfected with several acetyltransferases (black arrows highlighted the molecular weight of the proteins) in HEK293T cells and the acetylation levels were detected by western blot.
(B) Upper panel: Schematic diagram of PYCR1 structure. Lower panel: In vitro acetylation assay was performed with each fragment of PYCR1 and the arrows highlighted the molecular weight of the proteins.
(C and D) PYCR1-FLAG and HA-CBP were co-expressed in HEK293T cells, and co-immunoprecipitation assay showed the interaction between PYCR1-FLAG and HA-CBP in vivo.
(E) CBP acetylated PYCR1 in a dose-dependent manner. PYCR1-FLAG was co-transfected with increasing amount of HA-CBP in HEK293T cells and the acetylation levels were detected by western blot.
(F) PYCR1-FLAG and HA-CBP were co-expressed in HEK293T cells and the cells were treated with 20 μM CBP/P300 inhibitor GAR for 8 hours. The acetylation levels were detected.
SIRT3 is the deacetylase of PYCR1
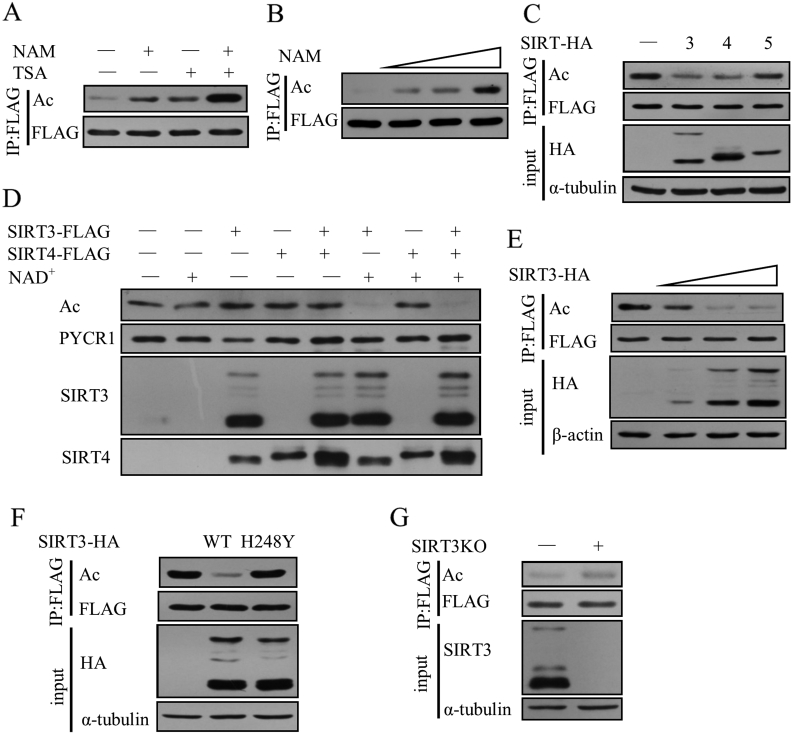
To investigate whether SIRT3 is able to deacetylate PYCR1 in cells, we first treated cells with HDACI, II family inhibitor TSA and the Sirtuin family inhibitor nicotinamide and found that both TSA and nicotinamide increased the acetylation levels of PYCR1, suggesting that PYCR1 was deacetylated by both HDACI, II family and the Sirtuin family. Nicotinamide increased the acetylation levels of PYCR1 more significantly than TSA (Figure 4A) and in a dose-dependent manner (Figure 4B), indicating that PYCR1 acetylation levels were mainly regulated by the Sirtuin family. As PYCR1 is located in mitochondria, we examined PYCR1 acetylation levels with SIRT3, SIRT4 or SIRT5 co-expressed, which are the Sirtuins located in mitochondria, and found that both SIRT3 and SIRT4 deacetylated PYCR1. We compared the amount of SIRT3 with SIRT4 in input and found that much more SIRT4 was required to produce the same deacetylation effect on PYCR1 (Figure 4C), indicating that SIRT3 was more efficient in deacetylating PYCR1. To further confirm the major deacetylase of PYCR1, we used in vitro deacetylation assay to compare the deacetylation levels of SIRT3 and SIRT4 for PYCR1. The acetylation levels of PYCR1 decreased only in the group where both NAD + and SIRT3 were added but not in the group where both NAD+ and SIRT4 were added (Figure 4D). This result suggests that SIRT3 is the major deacetylase for PYCR1 and SIRT4 deacetylates PYCR1 in an indirect manner, perhaps through interaction with proteins such as SIRT3. Furthermore, SIRT3 deacetylated PYCR1 in a dose-dependent manner in cells (Figure 4E). SIRT3-H248Y, a mutant that abolishes the enzymatic activity of SIRT3 but does not affect the expression and its localization in mitochondria, failed to deacetylate PYCR1 (Figure 4F). The acetylation levels of PYCR1 were increased in SIRT3 knockout U2OS cells (Figure 4G). All these results indicate that SIRT3 is the major deacetylase for PYCR1, and it can directly deacetylate PYCR1 both in cells and in vitro.
Figure 4.
SIRT3 is the deacetylase of PYCR1.
(A) PYCR1-FLAG and HA-CBP were co-expressed in HEK293T cells and the cells were treated with 1 μM TSA or 5 mM NAM for 8 hours.
(B) PYCR1-FLAG was co-transfected with HA-CBP in HEK293T cells, and the cells were treated with increasing amount of NAM.
(C) HEK293T cells were co-transfected with PYCR1-FLAG, HA-CBP and SIRT3-HA, SIRT4-HA or SIRT5-HA and the acetylation levels of PYCR1 were detected.
(D) In vitro deacetylation assay was performed and the acetylation levels of PYCR1 were detected.
(E) PYCR1-FLAG and HA-CBP were co-transfected with increasing amount of SIRT3-HA in HEK293T cells and the acetylation levels of PYCR1 were detected.
(F) HEK293T cells were co-transfected with PYCR1-FLAG, HA-CBP and SIRT3-HA or SIRT3-H248Y-HA and the acetylation levels of PYCR1 were detected.
(G) PYCR1-FLAG and HA-CBP were overexpressed in SIRT3 KO U2OS cells or U2OS control cells. The acetylation levels were detected.
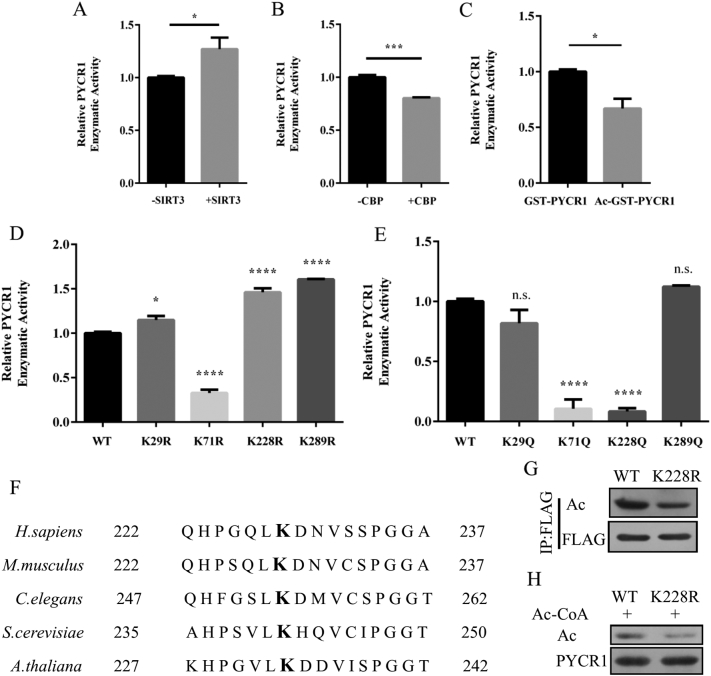
K228 is the major acetylation site of PYCR1
In order to study the effect of acetylation on the physiological function of PYCR1, we first examined its influence on enzymatic activity. We found that overexpression of SIRT3 enhanced the enzymatic activity of PYCR1, indicating that PYCR1 with deacetylation status possessed higher enzymatic activity, and that SIRT3 thus increased the activity of PYCR1 through deacetylation (Figure 5A). Conversely, CBP reduced the enzymatic activity of PYCR1, which manifested that CBP mediated acetylation reduced the enzymatic activity of PYCR1 (Figure 5B). The enzymatic activity of GST-PYCR1 decreased after in vitro acetylation, which further verified that the enzymatic activity of PYCR1 was affected by its acetylation levels (Figure 5C). We further mapped the acetylation sites of PYCR1 by purifying highly acetylated PYCR1 from HEK293T cells and identified the acetylation sites of PYCR1 by mass-spectrometry. Four lysine residues (K29, K71, K228 and K289) were identified. We mutated all the sites to R (mimicking the deacetylation status) or Q (mimicking the acetylation status) and examined their respective enzymatic activities (Figure 5D and E). We found that the mutations to R showed increased enzymatic activities everywhere but the K71 site, further verifying that PYCR1 with deacetylation status possessed a higher enzymatic activity. However, the mutations to Q showed basically unchanged or decreased activities, in which the activities of K71Q and K228Q showed dramatic reduction (Figure 5E). We noticed that both K71R and K71Q showed reduced enzymatic activities. The structural analysis showed that K71 is the key point at the NAD (P) H binding domain: the mutation on K71 may have led to the decrease of NAD (P) H binding, thus reducing PYCR1 activity. The enzymatic activity of K228R increased and the activity of K228Q decreased significantly, suggesting that acetylation of K228 had a great impact on the activity of PYCR1. The K228 site is highly conserved in evolution, indicating that K228 is very important for biological functions (Figure 5F). K228 mutation decreased the acetylation levels of PYCR1 in vivo (Figure 5G). Moreover, the mutation of K228 reduced the binding of acetyl-CoA in vitro (Figure 5H). These results confirm that K228 is the major acetylation site of PYCR1 to regulate its enzymatic activity.
Figure 5.
K228 is the major acetylation site of PYCR1.
(A and B) PYCR1-WT was co-transfected with SIRT3, CBP or vector in HEK293T cells, then we purified different status of PYCR1 and detected the enzymatic activities. Error bars represent ±SEM (n = 3).
(C) GST-PYCR1 was incubated with or without 1.5 mM acetyl-CoA, the enzymatic activities of the reaction products were detected. Error bars represent ±SEM (n = 3).
(D and E) The mutants of PYCR1 and PYCR1-WT were purified and the enzymatic activities were detected. Error bars represent ±SEM (n = 3).
(F) The sequences of PYCR1 in different organisms.
(G) PYCR1-WT and K228R were overexpressed in HEK293T cells and the acetylation levels of PYCR1 were detected.
(H) PYCR1-WT and K228R purified from HEK293T cells were incubated with 1.5 mM acetyl-CoA through in vitro acetylation assays. The acetylation levels of PYCR1 were detected.
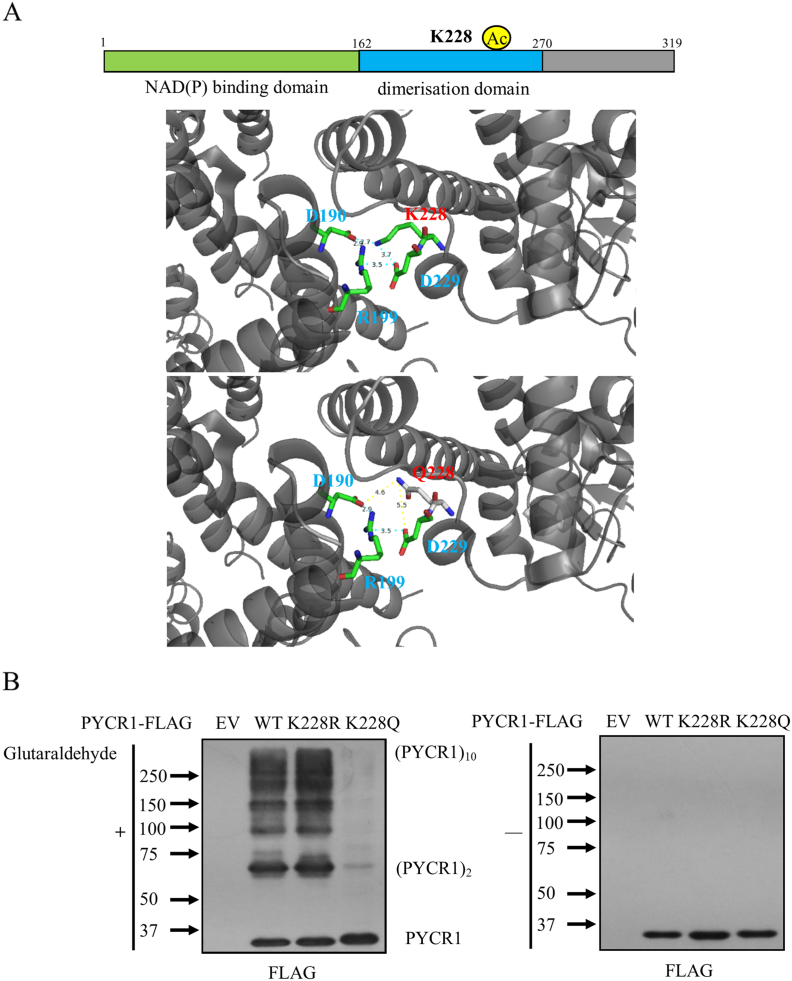
Acetylation of K228 impairs the formation of decamers thus inhibiting enzymatic activity of PYCR1
Since K228 is located in the dimerization domain and only the decamer form of PYCR1 possesses catalytic activity and the ability to bind to cofactors, we further analyzed the impact of K228 acetylation on the structure of PYCR1. As shown in Figure 6A, the negatively charged D190 and the positively charged R199 on one monomer formed a stable pocket with D229 and K228 on another monomer, which is important for the two monomers to form a dimer [5]. When K228 binds to an acetyl group or mutates to K228Q, the positive charge there is neutralized, thus enlarging the distances between the residues and destroying the pocket. As a result, the mutation of K228Q makes the dimer unstable and disturbs the formation of decamers. We further compared the polymerization of WT, K228R and K228Q in vivo, and found that K228Q reduced the formation of homodimers and decamers, but K228R slightly increased the formation (Figure 6B). It verifies that acetylation of K228 impairs decamer formation of PYCR1, the active form of PYCR1, thus inhibiting its enzymatic activity and revealing the mechanism by which acetylation reduces the activity of PYCR1. These results confirm that acetylation of K228 impairs the formation of decamers, thus inhibiting enzymatic activity of PYCR1.
Figure 6.
Acetylation of K228 impairs the formation of decamers thus inhibiting enzymatic activity of PYCR1.
(A) Crystal structure of PYCR1.
(B) PYCR1-WT, K228R or K228Q were overexpressed in HEK293T cells and the cell lysates were treated with or without 0.025% glutaraldehyde for 3 hours. The polymerizations of PYCR1 were detected by western blot.
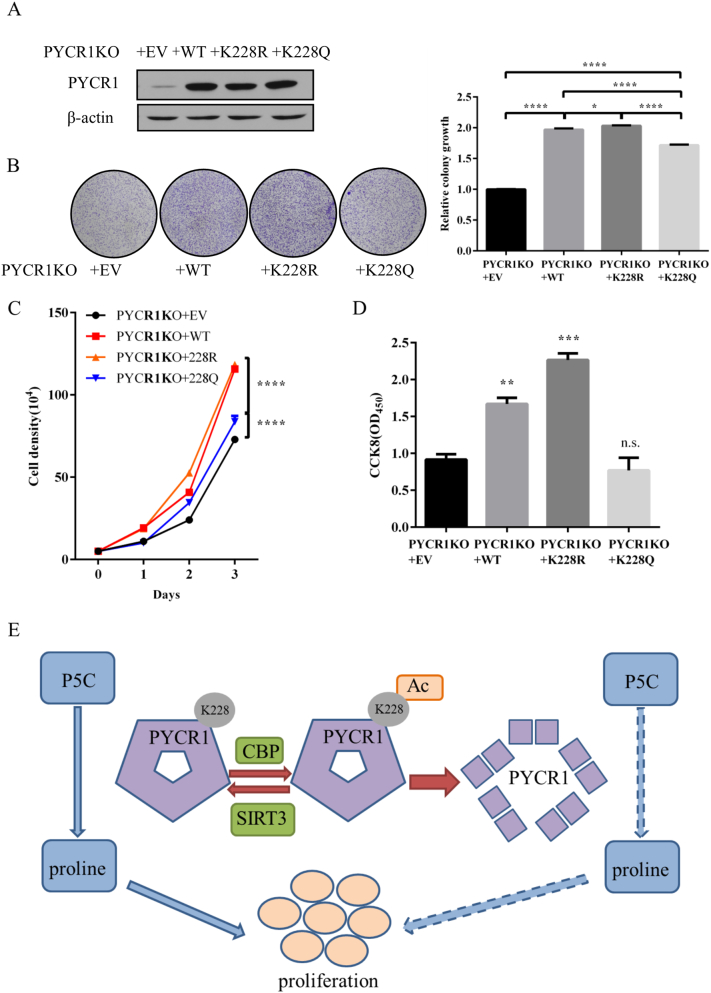
Acetylation of PYCR1 at K228 inhibits cell proliferation
As knockout of PYCR1 affected the growth of various tumor cells, we further examined the effect of its acetylation on cell proliferation. First, we generated PYCR1 KO MCF7 cell lines through CRISPR-Cas9 technique. The mRNA and protein expression levels of PYCR1 in knockout cells were obviously reduced, but not those of PYCR2 and PYCRL (Supplementary Figure S1A and S1B), suggesting that the PYCR1 KO stable cell lines were reliable. The number of clones of PYCR1 KO cells was significantly fewer than those of the PYCR1 WT cells (Supplementary Figure S1C), and PYCR1 KO cells showed decreased proliferation rates (Supplementary Figure S1D). These results confirm that knockout of PYCR1 affects cell growth and PYCR1 plays an important role in cell proliferation of tumor cells.
In order to identify the effect of acetylation of PYCR1 at K228 on cell growth, we re-expressed empty vector (EV), flag-tagged WT, K228R and K228Q mutant of PYCR1 to the aforementioned PYCR1 KO cells. As shown in Figure 7A, the rescued cell lines were generated successfully and the expression levels of PYCR1 WT, K228R and K228Q were nearly the same. The number of clones of PYCR1 K228R cells was significantly more than those of PYCR1 K228Q cells (Figure 7B). Furthermore, MCF7 rescued PYCR1 K228Q cells showed clearly decreased cell proliferation rates compared to MCF7 rescued PYCR1 K228R and PYCR1 WT cells (Figure 7C). By conducting CCK8 assays that reflect cell proliferation, we found that PYCR1 K228R cells grew most rapidly; however, PYCR1 K228Q cells grew slowly and PYCR1 WT cells grew at a rate just between the other two cell lines (Figure 7D). All the results above indicate that acetylation of PYCR1 at K228 inhibits cell growth and that deacetylation of PYCR1 at K228 by SIRT3 produces the opposite effect.
Figure 7.
Acetylation of PYCR1 at K228 inhibits cell proliferation.
(A) The protein expression levels of MCF7 rescued EV/PYCR1 WT/PYCR1 K228R/PYCR1 K228Q cells were detected by western blot.
(B) Clonogenic growth of MCF7 rescued EV/PYCR1 WT/PYCR1 K228R/PYCR1 K228Q cells was observed and the statistical chart was as shown. Error bars represent ±SEM (n = 3).
(C) MCF7 rescued EV/PYCR1 WT/PYCR1 K228R/PYCR1 K228Q cells were counted every day. Error bars represent ±SEM (n = 3).
(D) CCK8 assay was performed with MCF7 rescued EV/PYCR1 WT/PYCR1 K228R/PYCR1 K228Q cells. Error bars represent ±SEM (n = 3).
(E) Work model illustrates the relationship between PYCR1 acetylation and tumor growth.
Discussion
There are only a few studies about SIRT3 regulation of amino acid metabolism. SIRT3 participates in ornithine and glutamate metabolism by activating OTC and GDH, and the regulation of proline metabolism has not been reported [2], [24], [25], [26], [27]. In current study, we found that SIRT3 participated in the regulation of proline metabolism through the deacetylation of PYCR1 and consequently enriched the function of SIRT3 on amino acid metabolism. CBP acetylates PYCR1, and the acetylation at K228 inhibits the formation of decamers, thus reducing the enzymatic activity of PYCR1. As PYCR1 plays an important role in proline biosynthesis and tumor cell proliferation, the acetylation of PYCR1 at K228 results in the inhibition of tumor cell growth. On the contrary, SIRT3 directly deacetylates PYCR1, and the deacetylation at K228 promotes the formation of decamers and enhances the enzymatic activity of PYCR1, thereby producing more proline and promoting tumor cell growth (Figure 7E).
SIRT3 regulates tumor cell growth by eliminating ROS, regulating metabolism, proliferative or apoptotic pathways. SIRT3 plays a dual role in the development of cancers, acting either as an oncogene or as a tumor suppressor in different types of tumor cells. Even in the same type of tumor cells, with different metabolic microenvironments, SIRT3 can act as a tumor-promotor or a tumor-suppressor by regulating different metabolic substrates [2], [28], [29]. SIRT3 is highly expressed in thyroidal, esophageal, melanoma, renal and oral squamous cancer, and knocking down SIRT3 inhibits the proliferation of cancer cells [1], [2], [30]. In ovarian, pancreatic, hepatocellular and basal cell cancers, overexpression of SIRT3 reduces cell proliferation and alleviates cancer progression [1], [2], [30]. SIRT3 exhibits conflicting effects in gastric cancer, lung cancer, colon cancer, and head and neck squamous cancer [1], [2], [30]. SIRT3 represses glycolysis in and proliferation of breast cancer cells by decreasing ROS and destabilizing HIF1α [28], [29]. However, SIRT3 overexpression is related with poorer prognosis of patients with grade 3 breast cancer and SIRT3 is found to be up-regulated in breast cancer [31], [32]. There may be different conditions for SIRT3 to act as different role. According to our hypothesis, SIRT3 promotes cell growth through deacetylation of PYCR1; in order to verify this, we overexpressed SIRT3 in MCF7 cells and found that overexpression of SIRT3 clearly increased cell proliferation (Supplementary Figure S2A-D).
CBP acetylates many proteins in regulation of DNA repair, cell growth, apoptosis and other cellular processes [33]. Mutations or deletions of CBP has been found in breast, lung, colon and ovarian cancers, suggesting its role as a tumor suppressor [33]. In hematological malignancies, CBP participates in chromosomal translocation, which results in aberrant growth of tumor cells, thus acting as an oncogene protein [34]. In our findings, CBP reduced the enzymatic activity of PYCR1 through acetylation, thereby playing a role as a tumor suppressor.
PYCR1 was identified from the SIRT3 interaction network, and SIRT3 directly deacetylated PYCR1 both in cells and in vitro. Notably, acetylation of PYCR1 was also regulated by SIRT4 in an indirect way (Figure 4C). SIRT3 can interact with SIRT4 and SIRT5 to regulate protein acetylation levels in mitochondria [22], [24], [25], [26]. GDH was the first found substrate of both SIRT3 and SIRT4 [24], [26]. CS and HSD17B10 are targets for both SIRT3 and SIRT5 [22]. Our data support this notion.
Although proline is a non-essential amino acid, it plays a key role in stress protection and the maintenance of the redox balance in cells [3]. PYCR1 rescues endoplasmic reticulum stress and oxidative stress through the biosynthesis of proline [8], [9]. Outside of these functions, proline, degraded from collagen in extracellular matrix, also supplies ATP for cellular energy needs [35]. Under hypoxic or hypoglycemic conditions, proline utilization contributes to ATP generation or autophagy caused by ROS, which is necessary for tumor survival [35]. These functions of proline may relate to the high expression of PYCR1 in tumor cells. The concomitant NAD+ and NADP+ in proline biosynthesis catalyzed by PYCR1 augments glycolysis and the pentose phosphate pathway, respectively [7], which may be the reason that PYCR1 promotes tumor proliferation. PYCR1 is highly expressed in a variety of tumor cells including prostate cancer, breast cancer, renal cell carcinoma, melanoma, non-small cell lung cancer, and tumors of the head, neck, esophagus and pancreas [4], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20]. The PYCR1 knockout MCF7 cells showed significant inhibition of cell proliferation and the formation of clones, further verifying the functions of PYCR1 in promoting tumor growth. Our findings on SIRT3 regulating PYCR1 enzymatic activity provides a new insight on proline production in tumor cell growth.
The structural analysis of PYCR1 manifests that the K71 is the key point at the NAD (P) H binding domain and that K228 is located in the dimerization domain. We speculate that mutation of K71 may lead to the decrease of NAD (P) H binding, thus reducing the activity of PYCR1. Since K71R and K71Q showed the same change of enzymatic activity (Figure 5D, E), K71 is likely not the major acetylation site to regulate PYCR1 enzymatic activity. In humans, the monomers of PYCR1 form a decamer to activate its enzymatic activity [5]. Deacetylation of K228 increased the formation of decamers and the enzymatic activity of PYCR1, whereas acetylation significantly impaired the polymerization of PYCR1 and reduced the enzymatic activity of PYCR1 (Figure 6B). K228 maintains deacetylation status in tumor cells, allowing the polymerization of PYCR1 and eventually forming the active form of decamers to biosynthesize the required proline, satisfying the needs of tumor cells to grow rapidly. When an acetyl group binds to the K228 site, it neutralizes the positive charge of the site and changes the charge characteristics of the dimerization domain (Figure 6A). As a result, the dimer becomes unstable and the formation of decamers is disturbed, which is the reason why acetylation reduces the enzymatic activity of PYCR1. As PYCR1 plays a pivotal role in proline biosynthesis and tumor cell proliferation, the acetylation of PYCR1 at K228 may result in the decrease in the amount of proline and the inhibition of tumor cell growth. These results indicate that the K228 site of PYCR1 may be a potential therapeutic target for cancer.
In general, our findings prove that SIRT3 and CBP participate in the regulation of proline metabolism through acetylation/deacetylation of PYCR1 and affect tumor cell growth by targeting the K228 site of PYCR1, thereby providing a new option for cancer treatment.
Financial Support
This work was supported by grants from National Natural Science Foundation of China (81671389, 81874147, 81621063, 81471405) and Innovative Fund for Doctoral Students of Peking University Health Science Center (2018).
Acknowledgements
We thank Thomas Luo for editorial assistance. We thank the core facility at Peking University Health Science Center for mass-spectrometry analysis. We thank Dr. Jiadong Wang (Peking University) for generously providing LentiCRISPRv2 plasmid.
Footnotes
Conflicts of interest: The authors declare no potential conflicts of interest.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2019.04.008.
Appendix A. Supplementary data
Supplementary figures
References
- 1.Alhazzazi TY, Kamarajan P, Verdin E, Kapila YL. SIRT3 and cancer: Tumor promoter or suppressor? BBA - Reviews on Cancer. 2011;1816:80–88. doi: 10.1016/j.bbcan.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; Alhazzazi TY, Kamarajan P, Verdin E, Kapila YL (2011). SIRT3 and cancer: Tumor promoter or suppressor? BBA - Reviews on Cancer 1816, 80-88. [DOI] [PMC free article] [PubMed]
- 2.Xiong Y, Wang M, Zhao J, Han Y, Jia L. Sirtuin 3: A Janus face in cancer (Review) Int J Oncol. 2016;49:2227. doi: 10.3892/ijo.2016.3767. [DOI] [PubMed] [Google Scholar]; Xiong Y, Wang M, Zhao J, Han Y, Jia L (2016). Sirtuin 3: A Janus face in cancer (Review) International Journal of Oncology 49, 2227. [DOI] [PubMed]
- 3.Tanner JJ. Structural biology of proline catabolism. Amino Acids. 2008;35:719–730. doi: 10.1007/s00726-008-0062-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tanner JJ (2008). Structural biology of proline catabolism Amino Acids 35, 719-730. [DOI] [PMC free article] [PubMed]
- 4.Christensen EM, Patel SM, Korasick DA, Campbell AC, Krause KL, Becker DF, Tanner JJ. Resolving the cofactor-binding site in the proline biosynthetic enzyme human pyrroline-5-carboxylate reductase 1. J Biol Chem. 2017;292:7233–7243. doi: 10.1074/jbc.M117.780288. [DOI] [PMC free article] [PubMed] [Google Scholar]; Christensen EM, Patel SM, Korasick DA, Campbell AC, Krause KL, Becker DF, Tanner JJ (2017). Resolving the cofactor-binding site in the proline biosynthetic enzyme human pyrroline-5-carboxylate reductase 1 Journal of Biological Chemistry 292, 7233-7243. [DOI] [PMC free article] [PubMed]
- 5.Zhaohui M, Zhiyong L, Zhe L, Ming L, Xiaodong Z, Mark B, Zihe R. Crystal structure of human pyrroline-5-carboxylate reductase. J Mol Biol. 2006;359:1364–1377. doi: 10.1016/j.jmb.2006.04.053. [DOI] [PubMed] [Google Scholar]; Zhaohui M, Zhiyong L, Zhe L, Ming L, Xiaodong Z, Mark B, Zihe R (2006). Crystal structure of human pyrroline-5-carboxylate reductase Journal of Molecular Biology 359, 1364-1377. [DOI] [PubMed]
- 6.Reversade B, Escande BNA. Mutations in PYCR1 cause cutis laxa with progeroid features. Nat Genet. 2009;41:1016. doi: 10.1038/ng.413. [DOI] [PubMed] [Google Scholar]; Reversade B, Escande BNA (2009). Mutations in PYCR1 cause cutis laxa with progeroid features Nature Genetics 41, 1016. [DOI] [PubMed]
- 7.Liu W, Hancock CN, Fischer JW, Harman M, Phang JM. Proline biosynthesis augments tumor cell growth and aerobic glycolysis: involvement of pyridine nucleotides. Sci Rep. 2015;5 doi: 10.1038/srep17206. [DOI] [PMC free article] [PubMed] [Google Scholar]; Liu W, Hancock CN, Fischer JW, Harman M, Phang JM (2015). Proline biosynthesis augments tumor cell growth and aerobic glycolysis: involvement of pyridine nucleotides Sci Rep 5, 17206. [DOI] [PMC free article] [PubMed]
- 8.Vaxevanou AZ. Proline modulates the intracellular redox environment and protects mammalian cells against oxidative stress. Free Radic Biol Med. 2008;44:671–681. doi: 10.1016/j.freeradbiomed.2007.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]; Vaxevanou AZ (2008). Proline modulates the intracellular redox environment and protects mammalian cells against oxidative stress Free Radical Biology & Medicine 44, 671-681. [DOI] [PMC free article] [PubMed]
- 9.Xinwen L, Dickman MB, Becker DF. Proline biosynthesis is required for endoplasmic reticulum stress tolerance in Saccharomyces cerevisiae. J Biol Chem. 2014;289:27794–27806. doi: 10.1074/jbc.M114.562827. [DOI] [PMC free article] [PubMed] [Google Scholar]; Xinwen L, Dickman MB, Becker DF (2014). Proline biosynthesis is required for endoplasmic reticulum stress tolerance in Saccharomyces cerevisiae Journal of Biological Chemistry 289, 27794-27806. [DOI] [PMC free article] [PubMed]
- 10.Cai F, Miao Y, Liu C, Wu T, Shen S, Su X, Shi Y. Pyrroline-5-carboxylate reductase 1 promotes proliferation and inhibits apoptosis in non-small cell lung cancer. Oncol Lett. 2018;15:731–740. doi: 10.3892/ol.2017.7400. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cai F, Miao Y, Liu C, Wu T, Shen S, Su X, Shi Y (2018). Pyrroline-5-carboxylate reductase 1 promotes proliferation and inhibits apoptosis in non-small cell lung cancer Oncology Letters 15, 731-740. [DOI] [PMC free article] [PubMed]
- 11.Li L, Ye Y, Sang P, Yin Y, Hu W, Wang J, Zhang C, Li D, Wan W, Li R (2017). Effect of R119G Mutation on Human P5CR1 Dynamic Property and Enzymatic Activity Biomed Res Int 2017, 4184106. [DOI] [PMC free article] [PubMed]
- 12.Phang JM, Liu W, Hancock C, Christian KJ. The proline regulatory axis and cancer. Front Oncol. 2012;2:60. doi: 10.3389/fonc.2012.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]; Phang JM, Liu W, Hancock C, Christian KJ (2012). The proline regulatory axis and cancer Front Oncol 2, 60. [DOI] [PMC free article] [PubMed]
- 13.Yosuke T, Tokuzo A, Hiroaki K, Kazuko M, Masato T, Hidetoshi H, Velasco MA, De Yoshihiko F, Hideharu K, Takushi Y. Frequent amplification of ORAOV1 gene in esophageal squamous cell cancer promotes an aggressive phenotype via proline metabolism and ROS production. Oncotarget. 2014;5:2962–2973. doi: 10.18632/oncotarget.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yosuke T, Tokuzo A, Hiroaki K, Kazuko M, Masato T, Hidetoshi H, Velasco MA, De, Yoshihiko F, Hideharu K, Takushi Y (2014). Frequent amplification of ORAOV1 gene in esophageal squamous cell cancer promotes an aggressive phenotype via proline metabolism and ROS production Oncotarget 5, 2962-2973. [DOI] [PMC free article] [PubMed]
- 14.Nilsson R, Jain M, Madhusudhan N, Sheppard NG, Strittmatter L, Kampf C, Huang J, Asplund A, Mootha VK. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat Commun. 2014;5:3128. doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nilsson R, Jain M, Madhusudhan N, Sheppard NG, Strittmatter L, Kampf C, Huang J, Asplund A, Mootha VK (2014). Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer Nature Communications 5, 3128. [DOI] [PMC free article] [PubMed]
- 15.Zeng T, Zhu L, Liao M, Zhuo W, Yang S, Wu W, Wang D. Knockdown of PYCR1 inhibits cell proliferation and colony formation via cell cycle arrest and apoptosis in prostate cancer. Med Oncol. 2017;34:27. doi: 10.1007/s12032-016-0870-5. [DOI] [PubMed] [Google Scholar]; Zeng T, Zhu L, Liao M, Zhuo W, Yang S, Wu W, Wang D (2017). Knockdown of PYCR1 inhibits cell proliferation and colony formation via cell cycle arrest and apoptosis in prostate cancer Medical Oncology 34, 27. [DOI] [PubMed]
- 16.Loayza-Puch F, Rooijers K, Buil LC, Zijlstra J, Oude Vrielink JF, Lopes R, Ugalde AP, Van BP, Hofland I, Wesseling J. Tumour-specific proline vulnerability uncovered by differential ribosome codon reading. Nature. 2016;530:490–494. doi: 10.1038/nature16982. [DOI] [PubMed] [Google Scholar]; Loayza-Puch F, Rooijers K, Buil LC, Zijlstra J, Oude Vrielink JF, Lopes R, Ugalde AP, Van BP, Hofland I, Wesseling J (2016). Tumour-specific proline vulnerability uncovered by differential ribosome codon reading Nature 530, 490-494. [DOI] [PubMed]
- 17.Richard P, Marks KM, Shaul YD, Pacold ME, Dohoon K, K?Van? B, Shalini S, Hin-Koon W, Jang HG, Jha AK. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]; Richard P, Marks KM, Shaul YD, Pacold ME, Dohoon K, K?Van? B, Shalini S, Hin-Koon W, Jang HG, Jha AK (2011). Functional genomics reveal that the serine synthesis pathway is essential in breast cancer Nature 476, 346-350. [DOI] [PMC free article] [PubMed]
- 18.Jessica DI, Boris R, Richardson AD, Scott DA, Pedro AB, De SK, Marat K, Maurizio P, Ze'Ev R, Osterman AL. Functional specialization in proline biosynthesis of melanoma. Plos One. 2012;7 doi: 10.1371/journal.pone.0045190. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jessica DI, Boris R, Richardson AD, Scott DA, Pedro AB, De SK, Marat K, Maurizio P, Ze'Ev R, Osterman AL (2012). Functional specialization in proline biosynthesis of melanoma Plos One 7, e45190. [DOI] [PMC free article] [PubMed]
- 19.Ratnikov BI, Scott DA, Osterman AL, Smith JW, Ronai ZA. Metabolic rewiring in melanoma. Oncogene. 2016;36:147–157. doi: 10.1038/onc.2016.198. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ratnikov BI, Scott DA, Osterman AL, Smith JW, Ronai ZA (2016). Metabolic rewiring in melanoma Oncogene 36, 147-157. [DOI] [PMC free article] [PubMed]
- 20.Ding J, Kuo ML, Su L, Xue L, Luh F, Zhang H, Wang J, Lin TG, Zhang K, Chu P. Human mitochondrial pyrroline-5-carboxylate reductase 1 promotes invasiveness and impacts survival in breast cancers. Carcinogenesis. 2017;38:519–531. doi: 10.1093/carcin/bgx022. [DOI] [PubMed] [Google Scholar]; Ding J, Kuo ML, Su L, Xue L, Luh F, Zhang H, Wang J, Lin TG, Zhang K, Chu P (2017). Human mitochondrial pyrroline-5-carboxylate reductase 1 promotes invasiveness and impacts survival in breast cancers Carcinogenesis 38, 519-531. [DOI] [PubMed]
- 21.MR W, C G ML, J D PP. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat Protoc. 2009;4:1582. doi: 10.1038/nprot.2009.151. [DOI] [PubMed] [Google Scholar]; MR W, C G, M L, J D, P P (2009). Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells Nature Protocols 4, 1582. [DOI] [PubMed]
- 22.Yang X, Wang Z, Li X, Liu B, Liu M, Liu L, Chen S, Ren M, Wang Y, Yu M (2017). SHMT2 desuccinylation by SIRT5 drives cancer cell proliferation Cancer Research78, canres.1912.2017. [DOI] [PubMed]
- 23.Eri Maria S, Wagner SA, Weinert BT, Amit K, Hyun-Seok K, Chu-Xia D, Chunaram C. Proteomic investigations of lysine acetylation identify diverse substrates of mitochondrial deacetylase sirt3. Plos One. 2012;7 doi: 10.1371/journal.pone.0050545. [DOI] [PMC free article] [PubMed] [Google Scholar]; Eri Maria S, Wagner SA, Weinert BT, Amit K, Hyun-Seok K, Chu-Xia D, Chunaram C (2012). Proteomic investigations of lysine acetylation identify diverse substrates of mitochondrial deacetylase sirt3 Plos One 7, e50545. [DOI] [PMC free article] [PubMed]
- 24.Lei Z, Mostoslavsky R. Fine Tuning Our Cellular Factories: Sirtuins in Mitochondrial Biology. Cell Metab. 2011;13:621–626. doi: 10.1016/j.cmet.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lei Z, Mostoslavsky R (2011). Fine Tuning Our Cellular Factories: Sirtuins in Mitochondrial Biology Cell Metabolism 13, 621-626. [DOI] [PMC free article] [PubMed]
- 25.Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates Carbamoyl Phosphate Synthetase 1 and Regulates the Urea Cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nakagawa T, Lomb DJ, Haigis MC, Guarente L (2009). SIRT5 Deacetylates Carbamoyl Phosphate Synthetase 1 and Regulates the Urea Cycle Cell 137, 560-570. [DOI] [PMC free article] [PubMed]
- 26.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Molecular & Cellular Biology. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A (2007). Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation Molecular & Cellular Biology 27, 8807-8814. [DOI] [PMC free article] [PubMed]
- 27.Hallows WC, Wei Y, Smith BC, Devries MK, Devires MK, Ellinger JJ, Shinichi S, Shortreed MR, Tomas P, Markley JL. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell. 2011;41:139–149. doi: 10.1016/j.molcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hallows WC, Wei Y, Smith BC, Devries MK, Devires MK, Ellinger JJ, Shinichi S, Shortreed MR, Tomas P, Markley JL (2011). Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction Molecular Cell 41, 139-149. [DOI] [PMC free article] [PubMed]
- 28.Herrera KNG, Finley LW, Haigis MC. 2012. The role of SIRT3 in regulating cancer cell metabolism Bmc Proceedings6; p. P18. [Google Scholar]; Herrera KNG, Finley LW, Haigis MC (2012). The role of SIRT3 in regulating cancer cell metabolism Bmc Proceedings 6, P18.
- 29.Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell. 2011;19:416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB (2011). SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization Cancer cell 19, 416-428. [DOI] [PMC free article] [PubMed]
- 30.Alhazzazi TY, Kamarajan P, Verdin E, Kapila YL. Sirtuin-3 (SIRT3) and the Hallmarks of Cancer. Genes Cancer. 2013;4:164–171. doi: 10.1177/1947601913486351. [DOI] [PMC free article] [PubMed] [Google Scholar]; Alhazzazi TY, Kamarajan P, Verdin E, Kapila YL (2013). Sirtuin-3 (SIRT3) and the Hallmarks of Cancer Genes Cancer 4, 164-171. [DOI] [PMC free article] [PubMed]
- 31.Torrens-Mas M, Pons DG, Sastre-Serra J, Oliver J, Roca P. SIRT3 Silencing Sensitizes Breast Cancer Cells to Cytotoxic Treatments Through an Increment in ROS Production. J Cell Biochem. 2017;118:397–406. doi: 10.1002/jcb.25653. [DOI] [PubMed] [Google Scholar]; Torrens-Mas M, Pons DG, Sastre-Serra J, Oliver J, Roca P (2017). SIRT3 Silencing Sensitizes Breast Cancer Cells to Cytotoxic Treatments Through an Increment in ROS Production Journal of Cellular Biochemistry 118, 397-406. [DOI] [PubMed]
- 32.Igci M, Kalender ME, Borazan E, Bozgeyik I, Bayraktar R, Bozgeyik E, Camci C, Arslan A. High-throughput screening of Sirtuin family of genes in breast cancer. Gene. 2016;586:123–128. doi: 10.1016/j.gene.2016.04.023. [DOI] [PubMed] [Google Scholar]; Igci M, Kalender ME, Borazan E, Bozgeyik I, Bayraktar R, Bozgeyik E, Camci C, Arslan A (2016). High-throughput screening of Sirtuin family of genes in breast cancer Gene 586, 123-128. [DOI] [PubMed]
- 33.Di Martile M, Bufalo Donatella Del, Trisciuoglio Daniela. The multifaceted role of lysine acetylation in cancer prognostic biomarker and therapeutic target. Oncotarget. 2016;7:55789–55810. doi: 10.18632/oncotarget.10048. [DOI] [PMC free article] [PubMed] [Google Scholar]; Di Martile M, Donatella Del Bufalo, Daniela Trisciuoglio (2016). The multifaceted role of lysine acetylation in cancer prognostic biomarker and therapeutic target Oncotarget 7, 55789-55810. [DOI] [PMC free article] [PubMed]
- 34.Dutta R, Tiu B, Sakamoto KM. CBP/p300 acetyltransferase activity in hematologic malignancies. Molecular Genetics & Metabolism. 2016;119:37–43. doi: 10.1016/j.ymgme.2016.06.013. [DOI] [PubMed] [Google Scholar]; Dutta R, Tiu B, Sakamoto KM (2016). CBP/p300 acetyltransferase activity in hematologic malignancies Molecular Genetics & Metabolism 119, 37-43. [DOI] [PubMed]
- 35.Wei L, Phang JM. Proline dehydrogenase (oxidase), a mitochondrial tumor suppressor, and autophagy under the hypoxia microenvironment. Autophagy. 2012;8:1407–1409. doi: 10.4161/auto.21152. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wei L, Phang JM (2012). Proline dehydrogenase (oxidase), a mitochondrial tumor suppressor, and autophagy under the hypoxia microenvironment Autophagy 8, 1407-1409. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures