In this issue Mansfield et al. report the occurrence of inter- or intra-chromosomal rearrangements with a frequent pattern of chromothripsis or chromoplexy, in 28 treatment-naÏve cases of malignant mesothelioma (MM), irrespective of the different tumor subtypes 1. The analysis was conducted by a mate-pair sequencing (MPseq)/RNA sequencing (RNAseq) approach. The authors identified chromosomal rearrangements associated with amplification and deletions of numerous chromosomal segments affecting several genes, including CDKN2A, NF2, and BAP1 copy losses as reported in previous MM studies 2, 3. MPseq allows the identification of complex genomic rearrangements and structural variants with high resolution and accuracy, including DNA fragments larger than those analyzed by the next generation sequencing (NGS) technology. MPseq has been used to study genomic abnormalities in hematological malignancies and some other cancers 4. A limitation of MPseq is that it does not allow to identify deletions smaller than ~3 kb (i.e., minute deletions). Minute deletions can be identified using high-density array-comparative genomic hybridization (aCGH) 5.
Early cytogenetic studies showed multiple structural chromosomal abnormalities in tissues and cells from patients with MM 6. Recent NGS studies unexpectedly identified a very low mutation burden in MM compared to other adult malignancies 2, 3, estimating an average of 23 mutations/MM biopsy, with a range of 2–51 mutations per biopsy 2. This low mutational burden is similar to that identified in pediatric malignancies and it is quite unusual for an adult cancer linked to exposure to environmental carcinogens 5.
It was difficult to reconcile the low mutational burden 2, 3 detected by NGS in MM biopsies with the large number of chromosomal alterations detected by older cytogenetic studies 6. However, NGS is a technique that was specifically designed to detect nucleotide level changes, but NGS is not reliable to identify copy number changes 7.
The study by Mansfield et al. 1, and a previous study by Yoshikawa et al. 7, revealed that MMs contain numerous genetic alterations, as some of them are missed by NGS analyses because of the technical limitations of this technique that does not allow to reliably detect copy number changes 7. These studies 1, 7 used novel technical approaches that allow the identification of structural variants, including copy number changes, insertions, deletions, and rearrangements, as well as of point mutations. The MPseq based study by Mansfield et al., 1 and a previous study using a combination of high-density aCGH and NGS 7, identified a much higher number of genetic alterations (including point mutations, copy number changes, and all sorts of chromosomal rearrangements) than previously reported by NGS.
Among the mechanisms responsible for the genetic alterations in MMs, both studies identified chromothripsis 1‘ 7 as the cause of some of the detected DNA damage. Chromothripsis is a process in which accumulating DNA damage may lead to the formation of “micronuclei”. Micronuclei are extra-nuclear structures that usually contain a single chromosome surrounded by a nuclear membrane. Micronuclei can undergo disruption during interphase and consequent chromosome shattering. The resulting chromosomal fragments, or some of these fragments, can be re-incorporated in the nuclei during subsequent mitoses and, when in the nuclei, they are randomly re-ligated by the non-homologous-end-joint (NHEJ) repair pathway producing a derivative chromosome with complex, random, rearrangements 8. This process, known as chromothripsis (Figure 1), leads to the rapid formation of a very high number of genetic alterations within a few cell cycles, and to the possible emergence of tumor cells 8, in a manner that is different from the classical prevalent tumorigenesis hypothesis (current dogma) in which mutations accumulate over the course of many years until they eventually reach a tipping point and give rise to a malignancy. These two hypotheses (current dogma and chromothripsis), however, are not mutually exclusive, because the accumulation of DNA damage can favor the formation of micronuclei and chromosomal bridges that eventually may cause chromothripsis 8. Moreover, in MM, accumulation of DNA damage is enhanced by the presence of germline mutations in mesothelial cells 9 and/or by exposure to asbestos and to other carcinogenic fibers 10
Figure 1.
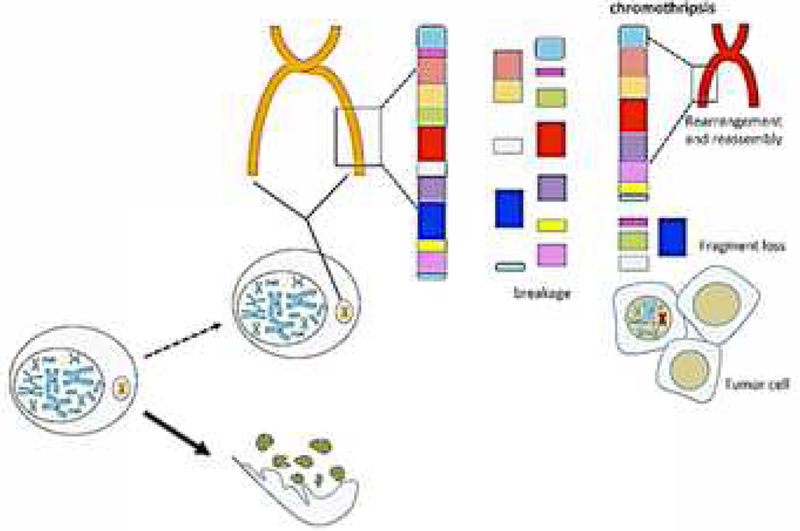
Chromothripsis and tumor development. DNA damage may lead to the formation of micronuclei, extranuclear structures containing a single chromosome, surrounded by a nuclear membrane. During the interphase, the micronuclei membrane disrupts leading to chromosome shattering because the DNA is fragmented by exposure to cytoplasmic nucleases. The resulting chromosome fragments –or some of them- can be re-localized in the nuclei during subsequent mitoses and re-ligated at random by the non-homologous-end-joint (NHEJ) repair system. This process, known as chromothripsis, generates aberrant chromosomes with complex, random, rearrangements, leading to a very high number of genetic alterations within a few cell cycles, and to the possible emergence of malignant cell clones.
In summary, these findings 1, 7 indicate that the notion that MM is a tumor with low genetic mutation burden (in which the term “mutation” is used to identify any type of genetic alteration), a notion that was based on NGS studies, has to be reconsidered. This change in perspective in MM mutational burden is relevant to understand the pathogenesis of this tumor, as well as to the design of novel diagnostics, prognostics, and patient-tailored therapies.
A high tumor mutation burden has been associated with favorable response to immune checkpoint inhibitors 11, especially in lung cancer 12. Therefore, the numerous alterations and rearrangements found in MM, i.e., the much higher mutational burden than anticipated based on NGS analyses, may account for some encouraging preliminary results obtained by a clinical trial with nivolumab (anti-programmed death-1, PD-1) in patients with MM 13.
A very recent comprehensive integrated genomic study on MM, updating a previous similar genomic analysis 3, revealed the upregulation of the member of the B7 family of negative checkpoint VISTA (V-domain Ig suppressor of T cell activation) in epithelioid MM, at a higher extent than in other solid tumors 14. The same study showed that the expression of IRF8, a mediator of interferon signals and dendritic cell differentiation, is significantly increased in BAP1-inactivated MMs 14, suggesting possible opportunities for MM immunotherapy.
Mansfield et al. investigated whether the chromosomal rearrangements detected by MPseq in MM might induce the potential expression of neoantigens. They propose that the transcription of these rearrangement-related junctions allowed them to predict the expression of many peptides with neoantigenic potential. Proteolytic processing and transport, binding to patient-specific MHC complexes, and immunogenicity were predicted for these putative peptides, using the Immune Epitope Database (IEDB) MHC-I Processing and the NetMHC-4.0 prediction tools 1.
Next, the Authors selected and synthesized three of the predicted neoantigen peptides, based on predicted HLA-peptide binding and immunogenicity. These peptides were validated for MHC binding, activation of T cells, and the ability to expand intratumoral T cell clones by an in vitro MHC binding assay, the ELISpot assay, and T cell receptor sequencing (TCRseq), respectively. The results revealed that the increase in predicted neoantigens resulting from chromosomal rearrangements in MM correlated with the clonal expansion of tumor-infiltrating T cells. The analysis of peripheral blood mononuclear cells (PBMCs) of an additional MM case, who also putatively expressed rearrangement-related neoantigens, revealed the presence of T cells responsive to the respective predicted neoantigens 1.
In summary, the most recent literature suggests that in contrast to hypotheses based on NGS studies, MM may be an immunogenic tumor, an interpretation supported by the study of Mansfield et al. predicting a possible abundance of neoantigens in MM.
It should be noted, however, that so far, the advantage of immunotherapy over standard chemotherapy in MM remains to be demonstrated 15. Moreover, the evidence of neoantigen formation remains to be proven, as, so far, has been only hypothesized using sophisticated prediction software. Therefore, the demonstration that genetic alterations actually generate neoantigens and that these peptides are really expressed by MM cells is still only indirect and it is based on the recognition of synthetic peptides by T cells in the ELISpot assay, as acknowledged by the authors 1. This same approach was used in recent papers published in highly regarded journals 11, 12, however, the actual presence of these neo-antigens in this and in previous studies remains to be proven. It is hoped that in the near future the direct demonstration that these neoantigens are produced will be provided: until then, it remains an intriguing theoretical hypothesis that such neoantigens actually exist and that they can generate anti-tumor-immune responses.
References
- 1.Mansfield AS, Peikert T, Smadbeck JB, et al. Neoantigenic potential of complex chromosomal rearrangements in mesothelioma. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo G, Chmielecki J, Goparaju C, et al. Whole-exome sequencing reveals frequent genetic alterations in BAP1, NF2, CDKN2A, and CUL1 in malignant pleural mesothelioma. Cancer research 2015;75:264–269. [DOI] [PubMed] [Google Scholar]
- 3.Bueno R, Stawiski EW, Goldstein LD, et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nature genetics 2016;48:407–416. [DOI] [PubMed] [Google Scholar]
- 4.Aypar U, Smoley SA, Pitel BA, et al. Mate pair sequencing improves detection of genomic abnormalities in acute myeloid leukemia. European journal of haematology 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carbone M, Kanodia S, Chao A, et al. Consensus Report of the 2015 Weinman International Conference on Mesothelioma. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 2016;11:1246–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taguchi T, Jhanwar SC, Siegfried JM, et al. Recurrent deletions of specific chromosomal sites in 1p, 3p, 6q, and 9p in human malignant mesothelioma. Cancer research 1993;53:4349–4355. [PubMed] [Google Scholar]
- 7.Yoshikawa Y, Emi M, Hashimoto-Tamaoki T, et al. High-density array-CGH with targeted NGS unmask multiple noncontiguous minute deletions on chromosome 3p21 in mesothelioma. Proc Natl Acad Sci U S A 2016;113:13432–13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ly P, Cleveland DW. Rebuilding Chromosomes After Catastrophe: Emerging Mechanisms of Chromothripsis. Trends in cell biology 2017;27:917–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bononi A, Giorgi C, Patergnani S, et al. BAP1 regulates IP3R3-mediated Ca2+ flux to mitochondria suppressing cell transformation. Nature 2017;546:549–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carbone M, Bedrossian CW. The pathogenesis of mesothelioma. Semin Diagn Pathol 2006;23:56–60. [DOI] [PubMed] [Google Scholar]
- 11.Turajlic S, Litchfield K, Xu H, et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. The Lancet Oncology 2017;18:1009–1021. [DOI] [PubMed] [Google Scholar]
- 12.Hellmann MD, Callahan MK, Awad MM, et al. Tumor Mutational Burden and Efficacy of Nivolumab Monotherapy and in Combination with Ipilimumab in Small-Cell Lung Cancer. Cancer cell 2018;33:853–861.e854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quispel-Janssen J, van der Noort V, de Vries JF, et al. Programmed Death 1 Blockade With Nivolumab in Patients With Recurrent Malignant Pleural Mesothelioma. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 2018;13:1569–1576. [DOI] [PubMed] [Google Scholar]
- 14.Hmeljak J, Sanchez-Vega F, Hoadley KA, et al. Integrative Molecular Characterization of Malignant Pleural Mesothelioma. Cancer discovery 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mutti L, Peikert T, Robinson BWS, et al. Scientific Advances and New Frontiers in Mesothelioma Therapeutics. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 2018;13:1269–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]


