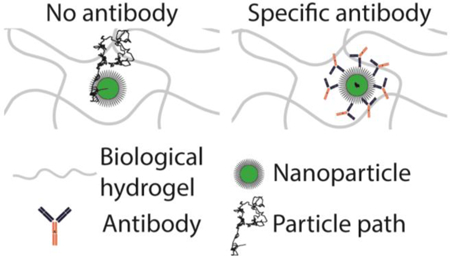
Abstract
Biological hydrogels (biogels) are selective barriers that restrict passage of harmful substances yet allow the rapid movement of nutrients and select cells. Current methods to modulate the barrier properties of biogels typically involve bulk changes in order to restrict diffusion by either steric hindrance or direct high-affinity interactions with microstructural constituents. Here, we introduce a third mechanism, based on antibody-based third party anchors that bind specific foreign species but form only weak and transient bonds with biogel constituents. The weak affinity to biogel constituents allows antibody anchors to quickly accumulate on the surface of specific foreign species and facilitates immobilization via multiple crosslinks with the biogel matrix. Using the basement membrane Matrigel® and a mixture of laminin/entactin, we demonstrate that antigen-specific, but not control, IgG and IgM efficiently immobilize a variety of individual nanoparticles. The addition of Salmonella typhimurium-binding IgG to biogel markedly reduced the invasion of these highly motile bacteria. These results underscore a generalized strategy through which the barrier properties of biogels can be readily tuned with molecular specificity against a diverse array of particulates.
Keywords: basement membrane, extracellular matrix, antibodies, laminin, particle tracking, PEG
Graphical Abstract:
1. Introduction
Biological hydrogels (biogels) are ubiquitous in living systems. In addition to providing essential structural support and mechanical function, such as lubrication in copulation and anchoring of cells to tissues [1–3], biogels serve as a selective barrier that can impede diffusion of unwanted materials yet allow the rapid movement of nutrients and select cells [2, 4–6]. The biochemical composition of biogels, such as Matrigel® [7] and mucus [3], is generally highly conserved. Matrigel®, basement membrane extract derived from the Engelbreth-Holm Swarm sarcoma, is comprised of 60% laminin, 30% collagen type IV, 8% entactin/nidogen, and 2% heparan sulfate, growth factors, and salts [8, 9]. This implies the biogel constituents are unlikely to adhesively capture the full diversity of foreign species, particularly since some pathogens have evolved to evade adhesive interactions with biogel matrices [2, 10, 11]. Thus, biogels are generally assumed to pose only a steric barrier to diffusion based on pore size, which varies based on the concentration of their matrix constituents and cannot immobilize species smaller than the mesh spacing. Indeed, steric trapping of micro- and nanoparticulates in Matrigel® and other highly glycosylated biologically derived hydrogels is well established [12–14]. Consequently, to tune the barrier properties of biogels, the predominant strategies are to either (i) alter the pore sizes of the gel matrix and impede transit through changing steric hindrance [2, 15] or (ii) conjugate specific motifs to the gel itself that in turn bind foreign species [16–20]. Both approaches involve substantial drawbacks. Bulk changes to the matrix microstructure, either by changing the crosslinker concentration in covalently linked gels or by relying on environmental stimuli (e.g. light or enzymes, respectively) to polymerize or degrade matrix constituents [20–25], only facilitate selectivity based on size and are generally irreversible. Alternatively, covalent crosslinking of antigen-specific molecules to the matrix [16–19, 25] is cumbersome in practice, is limited in the number of species that can be simultaneously immobilized, and may not efficiently immobilize diffusive species [26].
We believe the ideal method to tune the barrier properties of biogels should be antigen-specific and does not involve indiscriminate changes to the gel microstructure or direct chemical alterations of the matrix. These design requirements motivated us to explore the use of highly adaptive third-party anchors that can bind nanoparticulates with molecular specificity and crosslink them to matrix constituents. Due to the ability of Fab domains to undergo somatic hypermutation and affinity maturation to bind specific antigens with nanomolar affinities, antibodies represent a promising class of crosslinkers. We sought here to determine whether antibody Fc, which are highly conserved, would possess the appropriate affinity with biogel constituents to enable potent tuning of the barrier properties.
2. Materials and Methods
2.1. Preparation of PEG-coated nanoparticles
To produce PEGylated nanoparticles (PS-PEG), we covalently modified 200 nm fluorescent, carboxyl-modified polystyrene beads (PS-COOH; Invitrogen) with 2 kDa methoxy polyethylene glycol amine (PEG; Sigma) via a carboxyl-amine reaction, as published previously [27, 28]. Particle size and ζ-potential were determined by dynamic light scattering and laser Doppler anemometry, respectively, using a Zetasizer Nano ZS (Malvern Instruments, Southborough, MA). Size measurements were performed at 25 °C at a scattering angle of 90°. Samples were diluted in 10 mM NaCl solution, and measurements were performed according to instrument instructions. PEG conjugation was also confirmed by a near-neutral ζ-potential [27]. Dense PEG grafting (>1 PEG/nm2) was further verified using the fluorogenic compound 1 -pyrenyldiazomethane (PDAM) to quantify residual unmodified carboxyl groups on the polystyrene beads [28].
2.2. Preparation of Matrigel®, Laminin, and Collagen for Microscopy
A mixture of the following was added to a custom-made micro-volume (~10 μL) glass chamber slide: (i) growth-factor reduced Matrigel® (MG; Corning), high-concentration laminin/entactin (LAM; Corning), or mouse collagen IV (COL; Sigma); (ii) BSA (Sigma); (iii) Eagle’s Minimum Essential Medium (EMEM, Lonza BioWhittacker); (iv) fluorescent PS-COOH (Ex: 505 nm, Em: 515 nm) and PS-PEG (Ex: 625 nm, Em: 645 nm) nanoparticles; and (v) different Ab. The mixture was incubated at 37°C for 45 minutes in a custom hydration chamber, then sealed and incubated for another 30 minutes prior to microscopy. Final concentrations of reagents in slides were as follows: 2.2 mg/mL MG, 1 mg/mL BSA, ~ 4.3*108 beads/mL for both PEG- and COOH-modified nanoparticles, antibody as listed, EMEM, q.s. In LAM experiments, the final concentration of LAM was 1.5 mg/mL (comparable to the concentration of laminin in 2.2 mg/mL MG). In COL experiments, gel did not set at the concentration of laminin corresponding to that in 2.2 mg/mL of MG, so a final concentration of 3 mg/mL of COL was required. Lyophilized COL was resuspended in 0.25% v/v acetic acid and neutralized with 1 M NaOH to facilitate gelation. Trapping of PS-COOH beads in MG was used as an internal control in all microscopy experiments as a measure of complete polymerization of MG constituents.
2.3. Antibodies
Anti-PEG IgG1 (CH2074 except excess biotin experiment, which used CH2076, Silver Lake Research) and anti-PEG IgM (AGP4, IBMS) were used as test Ab and anti-Biotin IgG1 (Z021, ThermoFisher) and anti-Vancomycin IgM (2F10, Santa Cruz) were used as control Ab. All native Ab were used as is provided by the manufacturer. To prepare deglycosylated Ab, N-glycans on anti-PEG IgG and IgM were removed with rapid non-reducing PNGase F enzyme (New England Biolabs) according to the manufacturer’s protocol. The deglycosylation reaction was confirmed with two NuPAGE 4–12% Bis-Tris gels (Novex) in MOPS buffer. The first gel was transferred to a nitrocellulose membrane (Novex) with a Semi-Dry Blotter (Novex), blocked with carbo-free buffer (Vector Labs), labeled overnight at 4°C in 2 µg/mL biotinylated concavalin A (Vector), and probed with anti-biotin peroxidase (Vector) before imaging with Clarity Western Blot ECL Substrate (BioRad) in a FluorChemE unit (Cell Biosciences). The second gel was silver stained with Pierce Silver Stain Kit (Thermo Scientific) and imaged with the same unit. The remaining deglycosylated IgG and IgM was buffer-exchanged into PBS using 50 MWCO spin-x columns (Corning) and quantified based on A280.
2.4. High-resolution multiple particle tracking
Using an EMCCD camera (Evolve 512; Photometrics, Tucson, AZ) affixed to an inverted epifluorescence microscope (AxioObserver D1; Zeiss, Thornwood, NY) outfitted with an Alpha Plan-Apo 100x/1.46 NA objective, temperature- and CO2- controlled environmental chamber, and an LED illuminator (Lumencor Light Engine DAPI/GFP/543/623/690), the motion of fluorescent nanoparticles was captured. 20s videos (512 × 512, 16-bit image depth) were recorded at a temporal resolution of 66.7 ms using MetaMorph Microscopy Automation and Imaging Software (Molecular Devices, Sunnyvale, CA). The 10 nm spatial resolution (nominal pixel resolution 0.156 µm/pixel) was established previously [29]. Particles were tracked using MATLAB software as described previously [30]. Sub-pixel tracking resolution was realized with light-intensity-weighted averaging of nearby pixels to ascertain the particle center. A mean of n ≥ 40-particles/frame (with n ≥100 total traces) were traced for each experiment with 3–4 independent experiments/condition. Time-averaged mean squared displacements (MSD), calculated as <Δr2(τ)>= [x(t + τ) – x(t)]2 + [y(t + τ) – y(t)]2 (where τ = time scale or time lag) was calculated from the coordinates of particle centers. From this, distributions of MSDs and effective diffusivities (Deff) were computed as previously shown [27]. MSD is also expressed as MSD = 4D0τα, where α, the slope of the line on a log-log plot, represents the extent of hindrance to particle diffusion (α = 1 for pure Brownian diffusion; α < 1 for sub-diffusive motion). Mobile particles were defined as those with Deff ≥ 10−1 µm2/s at τ = 0.2667 s (this τ corresponds to a trajectory length of ≥ 5 frames), based on datasets of mobile and immobile nanoparticles (e.g., PS-PEG and PS-COOH nanoparticles) in MG or LAM [14, 27]. Theoretical diffusivity in water was calculated from the Stokes-Einstein equation, as described previously [4, 31].
2.5. Dot Blotting
To determine antibody affinity to MG, 1 µL of MG (diluted 1:10 in water), rat anti-mouse laminin-1 IgG1 (AL2, Millipore), or anti-biotin antibody was slowly spotted on a nitrocellulose membrane. After the membrane dried, it was blocked in 2% BSA in PBS for 30 minutes. Membrane with MG dots was incubated for 1 hour with either anti-laminin or anti-biotin at 1 µg/ml in 0.5% BSA, then washed 3 times for 5 minutes each with 0.05% PBS-Tween 20. All membranes were incubated with goat anti-mouse IgG (HRP-conjugated, 626520, Life Technologies), at a 1:5000 dilution for 1 hour in 0.5% BSA, then washed 3 times for 5 minutes each with 0.05% PBS-Tween 20 and developed with Clarity Western ECL Substrate.
2.6. Salmonella invasion assay
To assay Salmonella invasion, MG (final concentration 2.2 mg/mL) or LAM (final concentration 1.5 mg/mL) with Luria Broth (LB, BD Falcon), BSA, and varying concentrations of anti-Salmonella typhimurium IgG1 (6331; ViroStat) were mixed and incubated for 2 hours at 37°C in the upper chamber of a HTS FluoroBlok MultiWell System with 3.0 μm pores (BD Falcon) in a custom hydration chamber. After confirming mobility in the above-mentioned epifluorescence microscope, GFP-labeled S. typhimurium in 10 μL LB was then added to the top of each well, and 200 μL LB to each well in the bottom chamber. After 2 hours of incubation at 37°C, the top chamber was removed and OD600 in the lower chamber was measured with a SpectraMax M2 (Molecular Devices). Variations included Luria Broth with or without antibody instead of Matrigel® or LAM in the upper chamber. Experiment was performed in triplicate, n=3.
2.7. Statistics
All statistical analysis was performed in GraphPad and was two-sided. MSD data were log-log-transformed and compared within groups using a repeated-measures two-way ANOVA and post hoc Šidák test. Average Deff and % mobile were compared with ANOVA and subsequent Šidák tests. Salmonella invasion data was normalized within each replicate by controlling for background (defined as OD600 of LB only) and maximal invasion of Salmonella in a given matrix. Data were compared within groups using a one-way ANOVA and subsequent Šidák test. In all analyses, global α=0.05. Error bars and ± represent SEM.
3. Results
Viruses undergo Brownian motion in biogels while evading adhesion [2, 10, 32]. We prepared densely PEGylated, virus-sized polymeric nanoparticles (PS-PEG) that evade adhesion to biogel constituents to serve as a synthetic mimic of viruses. Using high resolution multiple particle tracking to quantify the diffusion of hundreds of individual nanoparticles in each specimen, we confirmed that nearly all PS-PEG exhibited diffusive Brownian motion in Matrigel®, slowed only ~1.6- fold compared to their theoretical diffusivity in water (Movie S1; Fig. S1). In contrast, similarly sized carboxyl-modified polystyrene nanoparticles (PS-COOH) were extensively immobilized in the same specimen, with geometrically averaged ensemble effective diffusivities (<Deff>) that are ~4000-fold reduced on average compared to PS-PEG (Movie S2; Fig. S1; p < 0.0018). Only 0.2% ± 0.09% of PS-COOH beads were classified as mobile (possessing <Deff> in excess of 10−1 μm2/s) vs. 97% ± 1.6% for PS-PEG beads. The effective immobilization of PS-COOH but not PS-PEG nanoparticles confirms Matrigel® affords a sufficiently rigid matrix that can immobilize virus-sized nanoparticles by adhesive interactions, and that PS-PEG nanoparticles can effectively evade adhesive interactions with the biogel constituents.
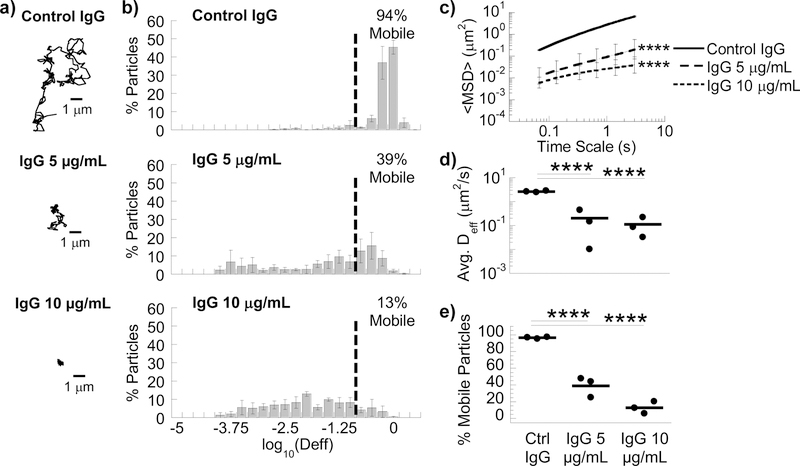
We next assessed whether we can selectively tune the barrier properties of Matrigel® simply by introducing antigen-specific antibodies, using IgG that specifically bind PEG as model antibody. In contrast to the typical high affinity of antibody-antigen bonds (e.g. anti-laminin and laminin), the affinity between typical antibodies with Matrigel® appears very low, virtually undetectable in dot blot assays (Fig. S2). Despite this seemingly negligible affinity, in Matrigel® that contains a final concentration of 10 µg/mL anti-PEG IgG, the <Deff> for PS-PEG (Movie S3) was reduced ~167-fold compared to control anti-biotin IgG (Movie S4; Fig. 1d), and the mobile nanoparticle fraction was reduced from 96 ± 0.7% in control IgG to 13 ± 4.2% (p=0.018; Fig. 1e). Anti-PEG IgG enabled substantial trapping of PS-PEG at 5 µg/mL (Fig. 1), but not at 1 µg/mL (data not shown). The appearance of the PS-PEG nanoparticles, as seen in microscopy videos, remained identical between control IgG and anti-PEG IgG conditions, indicating that the impeded motion of the nanoparticles was not attributed to anti-PEG IgG crosslinking multiple nanoparticles together.
Figure 1.
Anti-PEG IgG antibodies trap 200 nm PEG-coated nanoparticles in MG. (A) Representative traces in MG with 10 µg/mL control IgG or anti-PEG IgG (5 or 10 µg/mL). (B) Distribution of effective diffusivities of nanoparticles under different conditions. (C) Ensemble averaged mean squared displacements (<MSD>) over time. **** p < 0.0001 compared to Control IgG by two-way RM ANOVA with post hoc Šidák test. (D) Average ensemble effective diffusivities (<Deff>) at a time scale of 1 s. (E) Fraction of mobile nanoparticles. For (D) & (E) **** p < 0.0001 compared to Control IgG by one-way ANOVA with post hoc Šidák test. Error bars represent SEM.
To further confirm the antigen specificity and ability to reinforce barrier properties against different antigens, we mixed in nanoparticles conjugated with biotin-PEG, and found they were effectively immobilized in Matrigel® treated with anti-biotin IgG (Fig. S3). To demonstrate the ability of antigen-specific IgG to mediate trapping even in the presence of large quantities of other IgG crosslinkers, we further evaluated the mobility of PS-PEG in Matrigel® simultaneously treated with both 10 µg/mL anti-PEG IgG and 100-fold excess anti-biotin IgG (i.e. 1 mg/mL) that trapped biotin-PEG beads. We found no appreciable reduction in the trapping potency of anti-PEG IgG despite the excess levels of other antigen-specific IgG (Fig. S4). These results underscore that antigen-specific IgG can robustly immobilize virus-sized nanoparticles in Matrigel® and that this strategy can likely accommodate trapping of a large number of diverse nanoparticle species without compromising the ability to trap any individual foreign species.
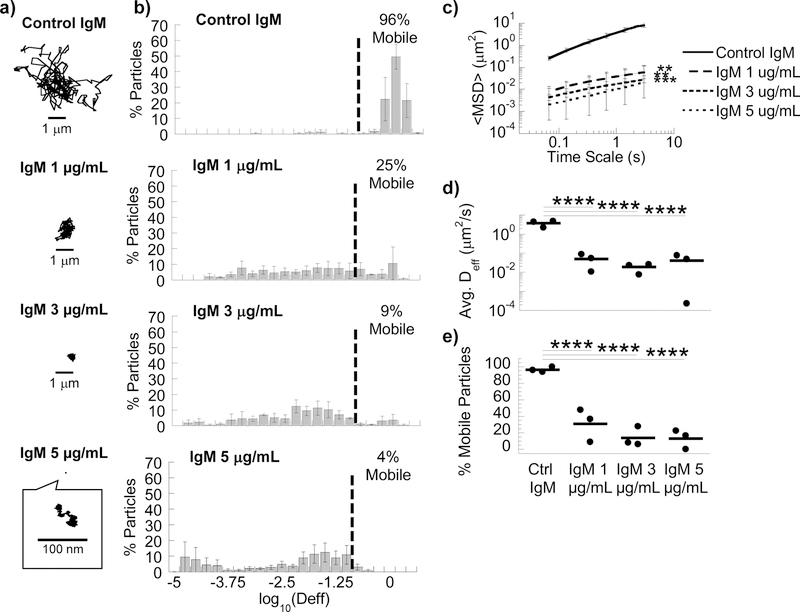
We hypothesized that crosslinkers with slightly greater antigen avidity as well as slightly greater affinity to the matrix constituents should facilitate more potent immobilization. IgM, due to its pentameric structure, possesses markedly greater overall binding avidity than IgG [33]. IgM was previously shown to possess slightly greater affinity to mucins than IgG [10]. In good agreement with our expectation, anti-PEG IgM exhibited greater trapping potency, immobilizing a similar fraction of PS-PEG as anti-PEG IgG but at substantially lower concentrations (Fig. 2a and 2b; Movie S5). The <Deff> of PS-PEG was reduced by ~90-fold and 137-fold at 5 μg/mL and 3 μg/mL anti-PEG IgM, respectively, compared to control (Fig. 2c; Movie S6). Surprisingly, despite the common assumption that IgM can mediate efficient agglutination, we did not observe appreciable nanoparticle agglutination in Matrigel® with anti-PEG IgM (Movie S5).
Figure 2.
Anti-PEG IgM antibodies trap 200 nm PEG-coated nanoparticles in MG. (A) Representative traces in MG with 5 µg/mL control IgM or anti-PEG IgM (1, 3 or 5 µg/mL). (B) Distribution of effective diffusivities of nanoparticles under different conditions. (C) Ensemble averaged mean squared displacements (<MSD>) over time. **p<0.01; ***p<0.001 compared to Control IgM by two-way RM ANOVA with post hoc Šidák test. (D) Average ensemble effective diffusivities (<Deff>) at a time scale of 1 s. (E) Fraction of mobile nanoparticles. For (D) & (E), ****p<0.0001 compared to Control IgM by one-way ANOVA with post hoc Šidák test. Error bars represent SEM.
Since many antibody-effector functions are influenced by N-glycans on IgG-Fc [33, 34], we speculate that N-glycans on Fc may be responsible for IgG’s affinity with Matrigel® constituents. We deglycosylated anti-PEG IgG and IgM with PNGase F and found that the removal of N-glycans substantially reduced the trapping potency of IgG (Movie S7) and IgM (Movie S8). The fraction of mobile nanoparticles increased from 13 ± 4.2% to 57 ± 5.3% for IgG (p=0.0006) and from 13 ± 6.7% to 67 ± 9.4% for IgM (0<0.0001; Fig. S5).
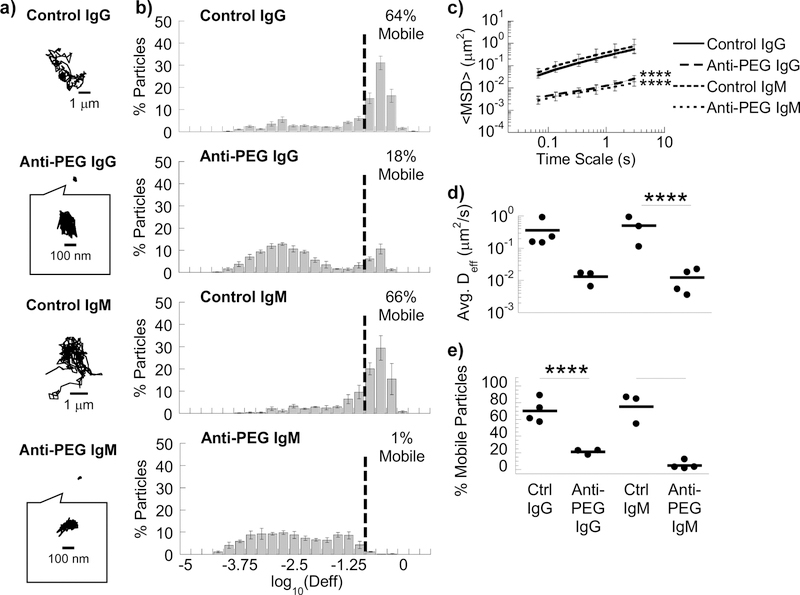
We next sought to evaluate the extent to which specific components of Matrigel® participate in antibody-mediated trapping. By mass, Matrigel® is composed 60% of laminin, 30% of collagen IV, and 8% of entactin (or nidogen). Laminin is a high molecular weight glycoprotein that is crosslinked primarily by entactin (or nidogen) in a 1:1 molar ratio [7]. In biogels composed of laminin/entactin (LAM), we observe comparable trapping of nanoparticles by anti-PEG IgG (Movies S9 and S10) and IgM antibodies (Movies S11 and S12): the <Deff> of PS-PEG was reduced ~20-fold and ~40-fold in anti-PEG IgG- and IgM-treated LAM compared to corresponding controls, respectively (Fig. 3). The collagen IV (COL) found in Matrigel® is a triple helix composed of α1 and α2 chains, components of the undifferentiated basement membrane found from early in development [35], and does not contain the more highly cross-linked α3α4α5 heterotrimer of the glomerular basement membrane, leading to reduced rigidity and larger pore sizes [36]. In COL biogels, anti-PEG IgG failed to trap nanoparticles (Supplemental Fig. 5): there was no appreciable difference in <Deff> in Collagen IV with and without antibody, suggesting Collagen IV does not directly participate in IgG-matrix interactions.
Figure 3.
Anti-PEG IgG and IgM antibodies trap 200 nm PEG-coated nanoparticles in LAM. (A) Representative traces in LAM with control IgG/IgM or anti-PEG IgG/IgM. (B) Distribution of effective diffusivities of nanoparticles under different conditions. (C) Ensemble averaged mean squared displacements (<MSD>) over time. **** p < 0.0001 compared to corresponding control by two-way RM ANOVA with post hoc Šidák test at every time point. (D) Average ensemble effective diffusivities (<Deff>) at a time scale of 1 s. (E) Fraction of mobile nanoparticles. For (D) & (E), ***p<0.001; ****p<0.0001 compared to corresponding control by one-way ANOVA with post hoc Šidák test. Error bars represent SEM.
Finally, we evaluated whether specific IgG can reinforce the barrier properties of Matrigel® against even highly motile Salmonella typhimurium. We first added Matrigel® or LAM along with different IgG to the upper chamber of a transwell system and allowed the gel to form, followed by the addition of motile fluorescent Salmonella, and monitored the amount of bacteria that entered the bottom chamber. Anti-LPS IgG markedly reduced the invasion of Salmonella through MG in a dose-dependent manner, with >70% reduction in the fraction of Salmonella that entered the bottom compartment at an IgG doses of 10 µg/mL and >93% reduction at an IgG dose of 25 µg/mL (Fig. 4). We observed a similar extent of reduction of bacterial invasion by Salmonella-binding IgG in LAM gels.
Figure 4.
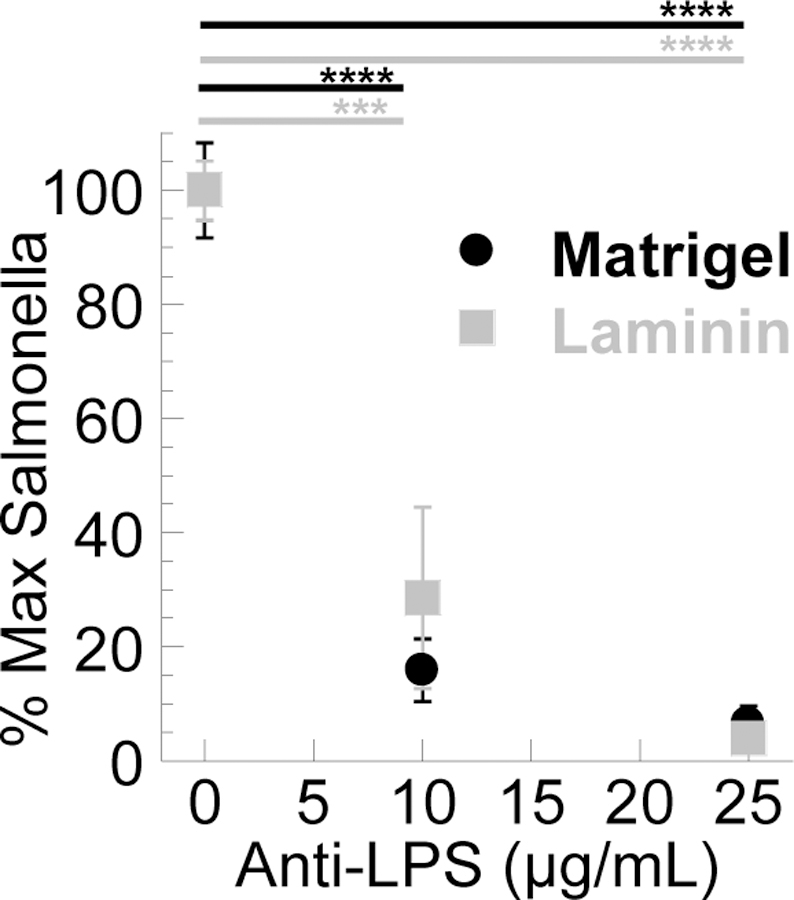
IgG anti-Salmonella typhimurium in MG and LAM impairs the invasion of Salmonella in a transwell experiment. OD600 of Salmonella in bottom well was normalized to OD600 of media alone and of Salmonella through biogel without antibody. MG: n=4 independent experiments done in triplicate; LAM: n= 3 independent experiments done in triplicate. *** p<0.001; **** p < 0.0001 compared to control by one-way ANOVA with post hoc Šidák test. Error bars represent SEM.
4. Discussion
By using third-party crosslinkers that can be adapted to specific antigens of interest, we demonstrate for the first time that the barrier properties of extracellular matrices can be tuned without perturbing the underlying matrix itself. When coupled to our earlier elucidation of IgG interactions with mucin [32, 37], our findings suggest antibodies may broadly interact with the matrices of various biogels to create a universal mechanism enabling an effective and highly tunable diffusional barrier against a diverse array of pathogens and foreign particulates. Indeed, this study marks the first example of characterizing IgG and IgM-mediated trapping in native, unmodified extracellular matrix. Previously, we used biotinylated Matrigel to demonstrate that high affinity bonds between antibodies and matrix surprisingly reduce overall trapping potency [26] The current work is markedly distinct on several fronts, including: 1) work with native, non-mucosal extracellular matrices; 2) comparison of trapping potency of IgM vs. IgG; 3) discovery that laminin, not collagen, is the predominant matrix element facilitating Ab-mediated trapping in Matrigel; and 4) the finding that this trapping mechanism is potent against even highly motile bacteria. The modular nature of antibody-matrix interactions allows the barrier properties of hydrogels to be dynamically tuned with molecular specificity despite the relatively static biochemistries of most biogels, simply through tailoring specific Ab crosslinkers. The principles laid down here substantially expand our ability to alter the barrier properties of hydrogels.
The observed trapping by IgG and IgM is not caused by non-specific accumulation of antibodies on nanoparticles or non-specific alterations of the biogel microstructure, since control IgG and IgM did not slow the mobility of nanoparticles. While antibodies accumulated on the surface of nanoparticles will invariably increase the hydrodynamic diameter of the antibody-nanoparticle complex, the small increase cannot account for the 10–100+-fold reduction in nanoparticle mobility. Indeed, smaller antibody-nanoparticle complexes were slowed to a much greater extent than larger nanoparticles in biogel, as shown by the potent trapping of ~100 nm PS-PEG by anti-PEG IgG (Fig. 1) compared to the relatively unimpeded motion of ~200 nm PS-PEG in Matrigel® treated with control IgG (Fig. S4). Antibodies can also agglutinate nanoparticles into aggregates far larger than the mesh spacing of the hydrogel. However, no agglutination was observed. Thus, the observed immobilization of nanoparticles must be attributed to adhesive interactions between the array of nanoparticle-bound antibodies and the biogel matrix. Such interactions also limit the ability for particles to collide, a necessary condition to form aggregates.
The notion that antibodies can be harnessed to selectively tune the barrier properties of biogels was likely overlooked in part because interactions between antibodies and biogel constituents are exceedingly weak and transient. For example, the diffusion of IgG and IgM in mucus is slowed only ~10–20 and ~30– 50% relative to their speeds in water [10, 38], respectively, which suggests that the bonds between individual antibodies and mucins are sufficiently weak that they are readily broken by thermal excitation. Such weak bonds imply a single antibody is likely incapable of crosslinking a pathogen or particle to mucus. Nevertheless, this weak affinity allows the antibodies to undergo rapid diffusion in hydrogel [26, 39–41], and thus quickly accumulate on the surface of nanoparticles and pathogens. In turn, once a critical threshold of antibodies is reached, the array of bound antibodies ensures there is at least one bond between the biogel matrix and nanoparticle-antibody complex at any moment in time, resulting in nanoparticle trapping with permanent avidity [26]. With such a mechanism of immobilization, high affinity bonds between antibodies and the matrix constituents can actually limit the rate of antibody accumulation on the particle surface, and consequently reduce the overall immobilization potency.
The minimal number of nanoparticle-bound antibodies remains unclear, as does the corresponding nanoparticle size needed to facilitate effective immobilization in different biogels. We recently found that HIV virus-like particles, which have few surface Env glycoproteins [42], could be effectively immobilized by monoclonal anti- gp120 IgG antibodies (unpublished observations), suggesting that as few as one- to two- dozen bound antibodies may be sufficient for immobilization. It is possible that even smaller nanoparticles (diameter < ~100 nm) can be effectively immobilized as long as there is sufficient number of bound antibodies. We hypothesize that optimization of crosslinkers used to mediate trapping, a concept we have discussed in more detail elsewhere [26, 42], could reduce the minimum threshold for bound antibodies. Finally, we are actively investigating the specific chemical moieties on the biogel matrix that participate in antibody-mediated trapping to further motivate and support the use of third-party antibody-based crosslinkers for tuning the barrier properties of hydrogels with molecular precision.
Supplementary Material
Statement of Significance:
Biological hydrogels (biogels) are essential in living systems to control the movement of cells and unwanted substances. However, current methods to control transport within biogels rely on altering the microstructure of the biogel matrix at a gross level, either by reducing the pore size to restrict passage through steric hindrance or by chemically modifying the matrix itself. Both methods are either nonspecific or not scalable. Here, we offer a new approach, based on weakly adhesive third-party molecular anchors, that allow for a variety of foreign entities to be trapped within a biogel simultaneously with exceptional potency and molecular specificity, without perturbing the bulk properties of the biogel. This strategy greatly increases our ability to control the properties of biogels at the nanoscale, including those used for wound healing or tissue engineering applications.
Acknowledgements
This work was supported by Eshelman Institute for Innovation, National Institutes of Health (http://www.nih.gov/;Grants R21EB017938 (S.K.L.); the National Science Foundation CAREER Award DMR-1810168 (S.K.L.); the David and Lucile Packard Foundation (http://www.packard.org/;Grant 2013–39274; S.K.L.); Eshelman Institute of Innovation; and PhRMA Foundation Predoctoral Fellowship (J.L.S.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code Availability
Particle trajectories were analyzed using a MATLAB version of open source particle tracking code, originally developed in IDL by Crocker and Hoffman30. We have adapted this code to analyze particle trajectories on a ‘frame-by-frame’ basis, as described previously31.
Disclosures
Mucommune LLC seeks to harness antibody-mucin interactions to improve protection against or treatment of infections at mucosal surfaces, and has licensed intellectual property from the University of North Carolina - Chapel Hill (UNC-CH). SKL is a founder of Mucommune and owns company stock. SKL’s relationship with Mucommune is subject to certain restrictions under University policy. The terms of this arrangement are being managed by UNC-CH in accordance with its conflict of interest policies.
References
- [1].Lai SK, Wang Y-Y, Wirtz D, Hanes J, Micro- and macrorheology of mucus, Adv. Drug Del. Rev 61(2) (2009) 86–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lieleg O, Ribbeck K, Biological hydrogels as selective diffusion barriers, Trends Cell Biol 21(9) (2011) 543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bansil R, Turner BS, The biology of mucus: Composition, synthesis and organization, Adv. Drug Del. Rev 124 (2018) 3–15. [DOI] [PubMed] [Google Scholar]
- [4].Cu Y, Saltzman WM, Mathematical modeling of molecular diffusion through mucus, Adv. Drug Del. Rev 61(2) (2009) 101–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hansing J, Ciemer C, Kim WK, Zhang X, DeRouchey JE, Netz RR, Nanoparticle filtering in charged hydrogels: Effects of particle size, charge asymmetry and salt concentration, Eur. Phys. J. E Soft Matter 39(5) (2016) 53. [DOI] [PubMed] [Google Scholar]
- [6].Stylianopoulos T, Poh M-Z, Insin N, Bawendi MG, Fukumura D, Munn LL, Jain RK, Diffusion of particles in the extracellular matrix: The effect of repulsive electrostatic interactions, Biophys. J 99(5) (2010) 1342–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kleinman HK, Martin GR, Matrigel: Basement membrane matrix with biological activity, Seminars in Cancer Biology 15(5) (2005) 378–386. [DOI] [PubMed] [Google Scholar]
- [8].Kleinman HK, McGarvey ML, Hassell JR, Star VL, Cannon FB, Laurie GW, Martin GR, Basement membrane complexes with biological activity, Biochemistry 25(2) (1986) 312–318. [DOI] [PubMed] [Google Scholar]
- [9]. Corning® Matrigel® Matrix: Frequently Asked Questions.
- [10].Olmsted SS, Padgett JL, Yudin AI, Whaley KJ, Moench TR, Cone RA, Diffusion of macromolecules and virus-like particles in human cervical mucus, Biophys. J 81 (2001) 1930–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lai SK, Hida K, Shukair S, Wang Y-Y, Figueiredo A, Cone R, Hope TJ, Hanes J, Human immunodeficiency virus type 1 is trapped by acidic but not by neutralized human cervicovaginal mucus, J. Virol 83(21) (2009) 11196–11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tomasetti L, Breunig M, Preventing obstructions of nanosized drug delivery systems by the extracellular matrix, Advanced Healthcare Materials 7(3) (2018) 1700739. [DOI] [PubMed] [Google Scholar]
- [13].Dancy JG, Wadajkar AS, Schneider CS, Mauban JRH, Woodworth GF, Winkles JA, Kim AJ, Non-specific binding and steric hindrance thresholds for penetration of particulate drug carriers within tumor tissue, J. Control. Release 238 (2016) 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lai SK, Wang Y-Y, Hida K, Cone R, Hanes J, Nanoparticles reveal that human cervicovaginal mucus is riddled with pores larger than viruses, Proc. Natl. Acad. Sci. U. S. A 107(2) (2010) 598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chaudhuri O, Koshy ST, Branco da Cunha C, Shin J-W, Verbeke CS, Allison KH, Mooney DJ, Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium, Nature Materials 13(10) (2014) 970–978. [DOI] [PubMed] [Google Scholar]
- [16].Bodenberger N, Kubiczek D, Trösch L, Gawanbacht A, Wilhelm S, Tielker D, Rosenau F, Lectin-mediated reversible immobilization of human cells into a glycosylated macroporous protein hydrogel as a cell culture matrix, Sci. Rep 7(1) (2017) 6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Updyke TV, Nicolson GL, Immunoaffinity isolation of membrane antigens with biotinylated monoclonal antibodies and immobilized streptavidin matrices, J. Immunol. Methods 73 (1984) 83–95. [DOI] [PubMed] [Google Scholar]
- [18].Miyata T, Asami N, Uragami T, A reversibly antigen-responsive hydrogel, Nature 399 (1999) 766–769. [DOI] [PubMed] [Google Scholar]
- [19].Soto CM, Patterson CH, Charles PT, Martin BD, Spector MS, Immobilization and hybridization of DNA in a sugar polyacrylate hydrogel, Biotechnol. Bioeng 92(7) (2005) 934–942. [DOI] [PubMed] [Google Scholar]
- [20].Kim H-J, Zhang K, Moore L, Ho D, Diamond nanogel-embedded contact lenses mediate lysozyme-dependent therapeutic release, ACS Nano 8(3) (2014) 2998–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Best JP, Cui J, Müllner M, Caruso F, Tuning the mechanical properties of nanoporous hydrogel particles via polymer cross-linking, Langmuir 29(31) (2013) 9824–9831. [DOI] [PubMed] [Google Scholar]
- [22].Kim Y, Abuelfilat AY, Hoo SP, Al-Abboodi A, Liu B, Ng T, Chan P, Fu J, Tuning the surface properties of hydrogel at the nanoscale with focused ion irradiation, Soft Matter 10(42) (2014) 8448–8456. [DOI] [PubMed] [Google Scholar]
- [23].Santhanam S, Liang J, Struckhoff J, Hamilton PD, Ravi N, Biomimetic hydrogel with tunable mechanical properties for vitreous substitutes, Acta Biomater 43 (2016) 327–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Toh WS, Lim TC, Kurisawa M, Spector M, Modulation of mesenchymal stem cell chondrogenesis in a tunable hyaluronic acid hydrogel microenvironment, Biomaterials 33(15) (2012) 3835–3845. [DOI] [PubMed] [Google Scholar]
- [25].Mabry KM, Schroeder ME, Payne SZ, Anseth KS, Three-dimensional high-throughput cell encapsulation platform to study changes in cell-matrix interactions, ACS Applied Materials & Interfaces 8(34) (2016) 21914–21922. [DOI] [PubMed] [Google Scholar]
- [26].Newby J, Schiller JL, Wessler T, Edelstein J, Forest MG, Lai SK, A blueprint for robust crosslinking of mobile species in biogels with weakly adhesive molecular anchors, Nature Communications 8(1) (2017) 833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lai SK, O’Hanlon DE, Harrold S, Man ST, Wang Y-Y, Cone R, Hanes J, Rapid transport of large polymeric nanoparticles in fresh undiluted human mucus, Proc. Natl. Acad. Sci. U. S. A 104(5) (2007) 1482–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yang Q, Jones SW, Parker CL, Zamboni WC, Bear JE, Lai SK, Evading immune cell uptake and clearance requires PEG grafting at densities substantially exceeding the minimum for brush conformation, Mol. Pharm 11(4) (2014) 1250–8. [DOI] [PubMed] [Google Scholar]
- [29].Apgar J, Tseng Y, Federov E, Herwig MB, Almo SC, Wirtz D, Multiple-particle tracking measurements of heterogeneities in solutions of actin filaments and actin bundles, Biophys. J 79(2) (2000) 1095–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang Y-Y, Nunn KL, Harit D, McKinley SA, Lai SK, Minimizing biases associated with tracking analysis of submicron particles in heterogeneous biological fluids, J. Control. Release 220(Pt A) (2015) 37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Dawson M, Wirtz D, Hanes J, Enhanced viscoelasticity of human cystic fibrotic sputum correlates with increasing microheterogeneity in particle transport, J. Biol. Chem 278(50) (2003) 50393–50401. [DOI] [PubMed] [Google Scholar]
- [32].Wang Y-Y, Kannan A, Nunn KL, Murphy MA, Subramani DB, Moench TR, Cone RA, Lai SK, IgG in cervicovaginal mucus traps HSV and prevents vaginal herpes infections, Mucosal Immunol 7(5) (2014) 1036–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lu LL, Suscovich TJ, Fortune SM, Alter G, Beyond binding: antibody effector functions in infectious diseases, Nat. Rev. Immunol 18(1) (2018) 46–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Russell A, Adua E, Ugrina I, Laws S, Wang W, Unravelling immunoglobulin G Fc N-glycosylation: A dynamic marker potentiating predictive, preventive and personalised medicine, International Journal of Molecular Sciences 19(390) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Robey PG, Martin GR, Type IV collagen contains two distinct chains in separate molecules, Coll. Relat. Res 1(1) (1980) 27–38. [DOI] [PubMed] [Google Scholar]
- [36].Brown KL, Cummings CF, Vanacore RM, Hudson BG, Building collagen IV smart scaffolds on the outside of cells, Protein Sci 26(11) (2017) 2151–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Henry CE, Wang Y-Y, Yang Q, Hoang T, Chattopadhyay S, Hoen T, Ensign LM, Nunn KL, Schroeder H, McCallen J, Moench T, Cone R, Roffler SR, Lai SK, Anti-PEG antibodies alter the mobility and biodistribution of densely PEGylated nanoparticles in mucus, Acta Biomater 43 (2016) 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Saltzman WM, Radomsky ML, Whaley KJ, Cone RA, Antibody diffusion in human cervical mucus, Biophys. J 66 (1994) 506–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Chen A, McKinley SA, Wang S, Shi F, Mucha PJ, Forest MG, Lai SK, Transient antibody-mucin interactions produce a dynamic molecular shield against viral invasion, Biophys. J 106(9) (2014) 2028–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].McKinley SA, Chen A, Shi F, Wang S, Mucha PJ, Forest MG, Lai SK, Modeling neutralization kinetics of HIV by broadly neutralizing monoclonal antibodies in genital secretions coating the cervicovaginal mucosa, PLoS One 9(6) (2014) e100598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Schroeder HA, Nunn KL, Schaefer A, Henry CE, Lam F, Pauly MH, Whaley KJ, Zeitlin L, Humphrys MS, Ravel J, Lai SK, Herpes simplex virus-binding IgG traps HSV in human cervicovaginal mucus across the menstrual cycle and diverse vaginal microbial composition, Mucosal Immunol (2018). [DOI] [PMC free article] [PubMed]
- [42].Wessler T, Chen A, McKinley SA, Cone R, Forest MG, Lai SK, Using computational modeling to optimize the design of antibodies that trap viruses in mucus, ACS Infectious Diseases 2(1) (2016) 82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.