ABSTRACT
It has long been established that neuronal growth cone navigation depends on changes in microtubule (MT) and F-actin architecture downstream of guidance cues. However, the mechanisms by which MTs and F-actin are dually coordinated remain a fundamentally unresolved question. Here, we report that the well-characterized MT polymerase, XMAP215 (also known as CKAP5), plays an important role in mediating MT–F-actin interaction within the growth cone. We demonstrate that XMAP215 regulates MT–F-actin alignment through its N-terminal TOG 1–5 domains. Additionally, we show that XMAP215 directly binds to F-actin in vitro and co-localizes with F-actin in the growth cone periphery. We also find that XMAP215 is required for regulation of growth cone morphology and response to the guidance cue, Ephrin A5. Our findings provide the first strong evidence that XMAP215 coordinates MT and F-actin interaction in vivo. We suggest a model in which XMAP215 regulates MT extension along F-actin bundles into the growth cone periphery and that these interactions may be important to control cytoskeletal dynamics downstream of guidance cues.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: Growth cones, +TIP, Plus-end tracking proteins, CKAP5, Cytoskeleton, Super resolution, F-actin alignment
Highlighted Article: XMAP215 binds F-actin in vitro and promotes microtubule–F-actin alignment in growth cones, which could be controlling cytoskeletal dynamics downstream of guidance cues, and thus, steering the axon during neuronal development.
INTRODUCTION
The growth cone is a structure at the tip of the growing axon that is responsible for directing neuronal extension and guidance. Growth cone motility depends upon actin filament (F-actin) dynamics in the periphery, while growth cone steering occurs as microtubules (MTs) from the axon shaft advance in the direction of new outgrowth (Coles and Bradke, 2015; Lowery et al., 2013; Vitriol and Zheng, 2012). There are also dynamic MTs that explore the F-actin-rich periphery, and these MTs appear to probe along filopodial F-actin bundles as if they were tracks to follow (Letourneau, 1983; Schaefer et al., 2002). There is a complicated interplay between these two cytoskeletal systems during axonal extension and guidance, and coordination between MTs and F-actin is essential for the axon to mechanically respond to extracellular guidance cues. Thus, crosstalk between these two systems is a critical component of axonal pathfinding.
Recent work has increasingly shown the importance of canonically described MT-associated proteins in facilitating regulation of interactions between MTs and F-actin within the growth cone. These studies have ascribed these roles in cytoskeletal coordination to well-characterized MT regulators TAU (also known as MAPT) and CLASP family proteins, among others, and sought to identify motifs and binding partners that facilitate MT–F-actin coordination (Bearce et al., 2015; Biswas and Kalil, 2018; Cammarata et al., 2016; Geraldo et al., 2008; Hur et al., 2011; Marx et al., 2013; Neukirchen and Bradke, 2011; Prokop, 2013). Nevertheless, many of the molecular mechanisms underlying this cytoskeletal crosstalk during axonal growth are still largely unresolved. Elucidating these mechanisms is essential for understanding how neuronal development occurs in both normal and pathological conditions.
We previously demonstrated that Xenopus microtubule associated protein 215 (XMAP215, also known as CKAP5) is important for promoting axon outgrowth as well as regulating MT trajectories within the growth cone (Lowery et al., 2013). However, it was unclear how changes in XMAP215 activity impacted MT behaviors within the growth cone, although it was apparent that XMAP215 must have additional functions beyond catalyzing MT polymerization. Given that MT advancement into the growth cone periphery depends upon MT extension along F-actin bundles, we wondered whether XMAP215 might be specifically involved in the regulation of MT–F-actin interactions within the growth cone. XMAP215 family members have received significant attention as critical regulators of MT polymerization and nucleation (Ayaz et al., 2012; Brouhard et al., 2008; Flor-Parra et al., 2018; Milunović-Jevtić et al., 2018; Thawani et al., 2018; Widlund et al., 2011; Zanic et al., 2013), but there are no previous studies that examine whether XMAP215 can bind directly to F-actin or mediate MT–F-actin interactions in any system.
In this study, we used a combination of approaches, ranging from super-resolution microscopy of cultured neurons to in vitro assays with purified proteins. Here, we have uncovered a critical role for XMAP215, and specifically the N-terminal TOG 1–5 domains, in regulating MT–F-actin interactions within the growth cone in Xenopus laevis embryos. In addition, we show that XMAP215 is required for maintaining normal growth cone morphology and its accurate response to guidance cues. Finally, we demonstrate that XMAP215 can directly bind to F-actin. Thus, our work highlights a newly discovered role for XMAP215 in cytoskeletal coordination and steering of the axonal growth cone during embryonic development.
RESULTS
XMAP215 knockdown leads to global morphological changes in the growth cone and defects in axon guidance
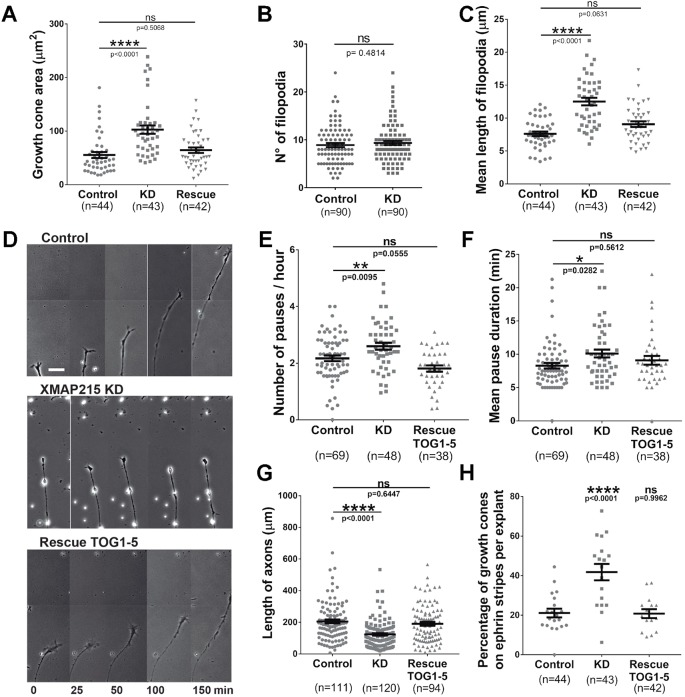
Our previous studies demonstrated that XMAP215 is important for promoting axon outgrowth but did not provide thorough characterization of the growth cone phenotype after XMAP215 knockdown (Lowery et al., 2013). Here, when we knocked down XMAP215 levels by 70% in X. laevis embryos (XMAP215 KD hereafter) (Fig. S1A), we observed distinct and unexpected phenotypic changes in the growth cone (Fig. S1B). The most noticeable effect of XMAP215 KD was increased area of the growth cone by 52% (Fig. 1A; Fig. S1B,C). Additionally, while the number of growth cone filopodia was not significantly altered (Fig. 1B), XMAP215 KD resulted in increased filopodia mean length by 43% (Fig. 1C). These phenotypes were rescued by concomitant expression of XMAP215–GFP mRNA in the KD condition (Fig. 1A,C), suggesting that the XMAP215 is necessary and sufficient to maintain normal growth cone morphology.
Fig. 1.
XMAP215 KD leads to global growth cone phenotypic changes and guidance defects. (A–C). XMAP215 KD induces growth cone pausing-like morphology. XMAP215 KD increases growth cone area (A), has no effect on number of filopodia (B), and increases the mean length of filopodia (C). See Fig. S1C for more information regarding how the quantifications were performed. (D–F). XMAP215 KD increases number and duration of growth cone pauses. (D) Time-lapse montage of representative axons from control, XMAP215 KD and XMAP215 KD rescued by expression of TOG1–5. Scale bar: 25 µm. XMAP215 KD increases the number of pauses per hour (E) and the mean pause duration (F). (G,H) TOG1–5 rescues the axon outgrowth parameters affected by XMAP215 KD. (G) XMAP215 KD decreases the length of axons. See Fig. S2A for more information regarding how the quantifications were performed. (H) XMAP215 KD increases the percentage of growth cones on Ephrin A5 stripes per explant. See Fig. S2C for representative images. *P<0.05; **P<0.01; ****P<0.0001; ns, not significant, from a one-way ANOVA analysis comparing multiple conditions, and from a Student's t-test when comparing XMAP215 KD to control. Data presented as mean±s.e.m.
Axon pathfinding is a dynamic process consisting of growth cone exploratory movements, in which the growth cone pauses before advance, retraction or turning. Growth cone pauses are characterized by an increase in growth cone area and filopodia length, compared to rapidly advancing growth cones (Kalil et al., 2000; Tanaka et al., 1995). As XMAP215 KD also results in larger growth cones and longer filopodia, we next investigated whether XMAP215 KD leads to changes in parameters of growth cone pausing (Fig. 1D–F). We determined that XMAP215 KD led to a 20% increase in the number of growth cone pauses per hour (Fig. 1E) and a 22% increase in the duration of the pauses (Fig. 1F) compared to a control condition. In addition, we were able to rescue these phenotypes by concomitant expression of the XMAP215 N-terminal TOG1–5 domains (TOG1–5 hereafter) to the KD condition (Fig. 1E,F). Furthermore, we determined that expression of the XMAP215 N-terminus could rescue the decrease in overall axon length and axon outgrowth that is observed with XMAP215 KD (Fig. 1G; Fig. S2A,B).
We next wanted to determine whether XMAP215 KD might affect axon guidance in addition to outgrowth. Stripe assays have been widely used to study the responsiveness of growing axons to external cues (Knöll et al., 2006; Knoll et al., 2001; Monnier et al., 2003; Wizenmann and Bähr, 1997). We investigated the effect of XMAP215 KD on axon guidance by determining whether cultured axons avoided stripes containing the repellent guidance cue Ephrin A5 (Fig. S2C). We observed a twofold increase in the number of growth cones per explant that were trespassing on Ephrin A5 stripes when comparing XMAP215 KD cells to controls (Fig. 1H). Importantly, this guidance defect was rescued by concomitant expression of exogenous TOG1–5 along with XMAP215 KD (Fig. 1H; Fig. S2C). These results suggest that normal levels of XMAP215 expression are necessary for axons to be responsive to Ephrin A5 and, thus, for correct axon guidance.
XMAP215 KD disrupts MT organization in the growth cone
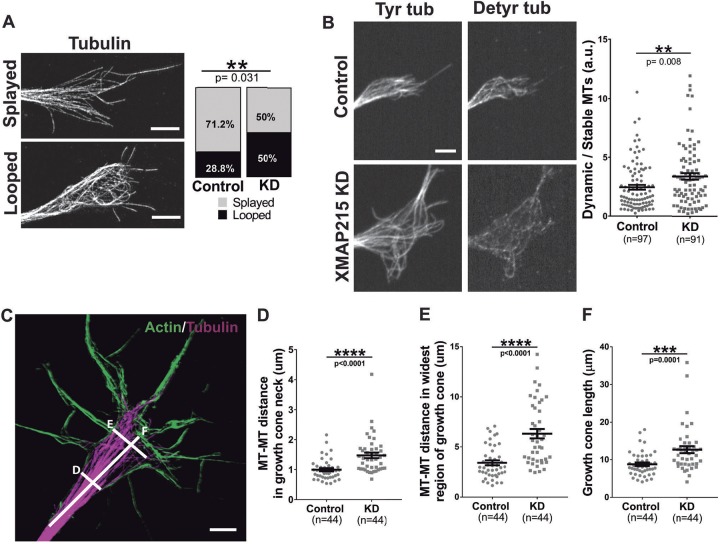
We previously demonstrated that partial XMAP215 KD led to abnormal MT trajectories within the neuronal growth cone (Lowery et al., 2013). The mechanism by which this occurred was unclear, but it seemed likely that XMAP215 might display another function in growth cones other than simply acting as a catalytic MT polymerase (Lowery et al., 2013). Thus, we sought to further investigate and quantify several MT parameters in axonal growth cones after XMAP215 KD. Because X. laevis neurons express very high levels of XMAP215 compared to other cell types, knocking down XMAP215 by 70% still allows for sufficient XMAP215 to facilitate MT polymerization, while also resulting in abnormal MT behaviors specifically in growth cones (Lowery et al., 2013). MT dynamics vary depending on axonal behaviors (Voelzmann et al., 2016); during pausing periods, growth cones present looped or bundled MTs, which then reorganize into splayed MTs during growth cone advance (Dent et al., 1999; Hendricks and Jesuthasan, 2009). In order to analyze MT morphology, we quantified the percentage of growth cones presenting ‘splayed’ versus ‘looped’ MTs after tubulin staining, as previously described (Tanaka and Kirschner, 1991). XMAP215 KD axons displayed a significant increase in the number of growth cones presenting a looped morphology (almost twice as many as in controls) (Fig. 2A). Next, we examined MT post-translational modifications using antibodies against tyrosinated and detyrosinated tubulin as a proxy for MT dynamicity versus stability. XMAP215 KD led to a 37% increase in the dynamic:stable ratio compared to controls (Fig. 2B). Finally, we examined the distribution of MTs within the growth cone using structured illumination microscopy (SIM) imaging. We observed a 48% increase in MT spreading within the growth cone neck after XMAP215 KD and an 85% increase in the widest part of the central domain (Fig. 2C–E). Moreover, XMAP215 KD resulted in a 44% increase in the overall length of the growth cone, from axon shaft to end of the central domain (Fig. 2F). These results demonstrate that XMAP215 regulates MT organization in terms of morphology, dynamicity and distribution throughout the growth cone.
Fig. 2.
XMAP215 KD disrupts MT organization in the growth cone. (A) Representative SIM images of growth cone MTs considered as ‘splayed’ and ‘looped’. XMAP215 KD increases the MT looped morphology compared to controls. Scale bar: 3 µm. (B) Representative growth cone images of control and XMAP215 KD neural explants immunostained for tyrosinated tubulin (Tyr tub) and detyrosinated tubulin (Detyr tub) to label dynamic versus stable MTs, respectively. XMAP215 KD increases the dynamic:stable MT ratio. Scale bar: 5 µm. a.u., arbitrary units. (C) Representative SIM image of growth cones stained for actin and tubulin. The labeled lines letter indicate how the MT measurements were performed for the corresponding plots in D–F. Scale bar: 8 µm. (D–F) Quantification of the different MT parameters. XMAP215 KD increases the MT–MT distance in growth cone neck (D), the MT–MT distance in the growth cone's widest region (E), as well as the growth cone length (F). **P<0.01; ***P<0.001; ****P<0.0001; ns, not significant, from a Student's t-test comparing XMAP215 KD to control. Data presented as mean±s.e.m.
XMAP215 promotes penetration of MTs into growth cone filopodia
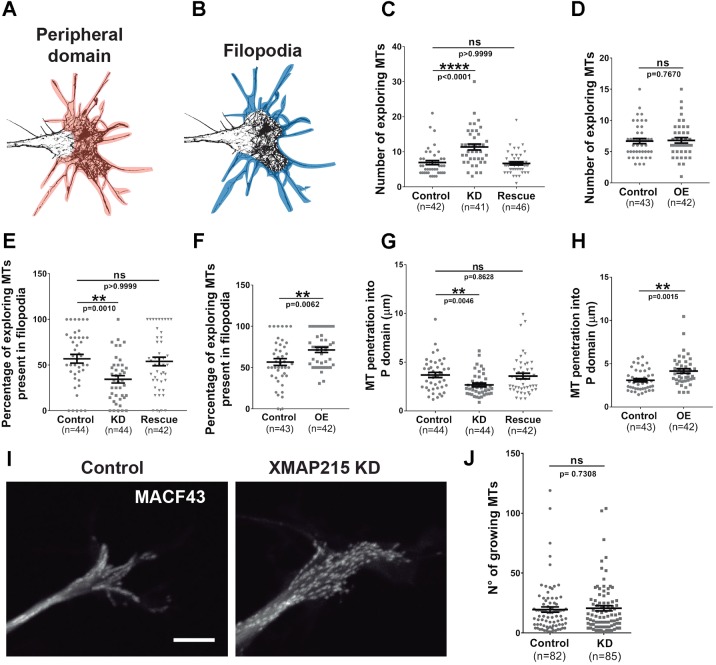
We next investigated whether XMAP215 regulates MT penetration into the peripheral domain of the growth cone. We overexpressed (OE) and knocked down XMAP215 in X. laevis embryos and analyzed axonal growth cone MTs and F-actin. We collected SIM images after immunostaining for F-actin and tubulin and counted the number of ‘exploring’ MTs, which are defined as MTs that extend beyond the central domain of the growth cone, into the actin-rich peripheral domain (Fig. 3A; Fig. S3A,B). In the case of XMAP215 KD, we observed a 64% increase in the number of exploring MTs, which was rescued by co-expression of a KD-resistant XMAP215 mRNA (Fig. 3C). No changes were observed in the total number of exploring MTs in the growth cone when XMAP215 was overexpressed (Fig. 3D). To determine whether these exploring MTs were able to enter filopodia, we quantified the number of MTs present in filopodia (Fig. 3B) as well as measured the extent of penetration of these MTs into the peripheral domain (Fig. S3A,B). With XMAP215 KD, although there were more exploring MTs within the entire growth cone periphery compared to controls, there was also a 23% decrease in the number of MTs that were present specifically within filopodia (Fig. 3E). By contrast, we observed a 26% increase in the number of MTs within filopodia upon XMAP215 OE (Fig. 3F). We also determined that MT penetration into the peripheral domain was 28% less with XMAP215 KD (Fig. 3G) and 35% greater with XMAP215 OE (Fig. 3H).
Fig. 3.
XMAP215 promotes penetration of MTs into growth cone filopodia. (A,B) Growth cone mask images outlining the growth cone peripheral domain colored in orange (A), and the filopodia colored in blue (B). (C–H). Quantification of MT penetration into the actin-rich peripheral domain of the growth cone after XMAP215 knockdown (KD) (C,E,G) or overexpression (OE) (D,F,H). XMAP215 KD increases the number of exploring MTs (C), while decreasing the percentage of exploring MTs present in filopodia (E), as well as MT penetration into the peripheral (P) domain (G), compared to controls. XMAP215 KD effects were rescued by co-expression of KD-resistant XMAP215–GFP mRNA (C,E,G). See Fig. S3 for more information regarding how the quantifications were performed. **P<0.01; ****P<0.0001; ns, not significant, from a one-way ANOVA analysis comparing XMAP215 KD, XMAP215 rescue and control, and from a Student's t-test comparing XMAP215 OE to control. (I) Micrograph overlays of GFP–MACF43 tracks from a 1 m time-lapse image series in control and XMAP215 KD neural tube explant growth cones. Scale bar: 8 µm. (J) No difference was observed between XMAP215 KD and control when comparing the number of growing MTs. ns, not significant from a Student's t-test analysis. Data presented as mean±s.e.m.
As XMAP215 is a well-characterized MT polymerase, we then examined whether observed differences in the number of exploring MTs might be due to a change in the number of total growing MTs within the growth cone or whether the difference was restricted to MTs within the peripheral domain. For this purpose, along with XMAP21 KD or OE, we concomitantly expressed GFP–MACF43, which binds growing MT plus-ends (Honnappa et al., 2009), and used automated imaging analysis to quantify the number of GFP–MACF43 comets in high-resolution live images. Despite observing more broadly distributed GFP–MACF43 trajectories within the XMAP215 KD growth cones compared to controls (Fig. 3I), there were no significant differences in the number of growing MTs in XMAP215 KD growth cones (Fig. 3J). Thus, with the levels of XMAP215 protein that are present in our partial KD condition, the number of MTs undergoing polymerization is not negatively affected, although MTs are unable to properly penetrate into filopodia. It is also important to note that this lack of MT penetration is not due to reduction in MT polymerization rate, as we previously found that MT polymerization rate was not significantly reduced with partial XMAP215 KD in these growth cones (Lowery et al., 2013).
XMAP215 is required for normal MT–F-actin alignment in growth cones
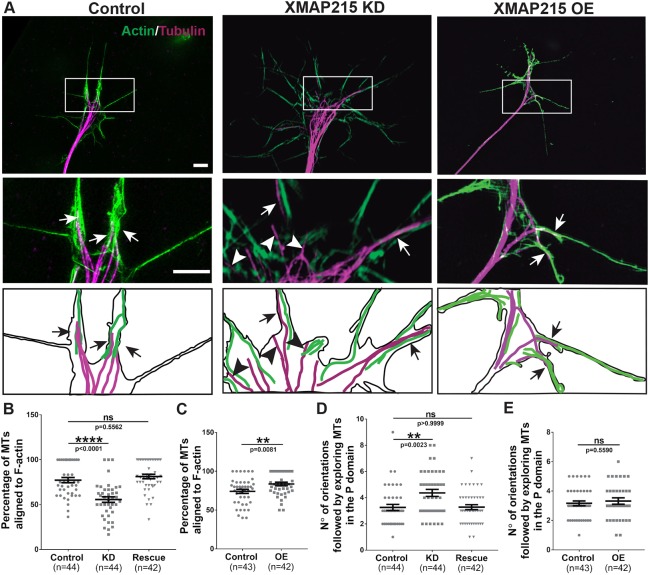
MT trajectories and penetration into the peripheral domain depend upon MT extension along F-actin bundles. Thus, we wondered whether XMAP215 might play a role in MT alignment to F-actin bundles in the growth cone periphery. Upon XMAP215 KD, there was a 28% decrease in the percentage of F-actin-aligned MTs, compared to controls (Fig. 4A,B), while there was a 16% increase in F-actin-aligned MTs in the case of XMAP215 OE (Fig. 4C). We also noted that there were some MT plus-ends that were nowhere close to bundled F-actin in the XMAP215 KD condition (Fig. 4A, arrowheads). These results suggest that XMAP215 promotes MT–F-actin alignment in the growth cone. As trajectories of MTs within the growth cone are normally restricted to following F-actin bundles (Letourneau, 1983; Schaefer et al., 2002), we quantified the number of different MT orientations within the growth cone after XMAP215 KD. We defined the orientations as being different if the direction followed by the tip of a MT forms an angle greater than 30° with other neighboring MTs, and angles were measured with respect to a central line that follows the growth direction of the axon. We observed that XMAP215 KD led to a 34% increase in the number of unique orientations followed by exploring MTs in the peripheral domain in comparison to controls (Fig. 4D), while XMAP215 OE did not generate any changes (Fig. 4E). Taken together, these results suggest that XMAP215 is important for MT spatiotemporal guidance along F-actin tracks within growth cones. Additionally, the MT–F-actin alignment defects were rescued by co-expression of KD-resistant XMAP215–GFP mRNA (Fig. 4B,D), attributing these effects specifically to XMAP215. Furthermore, we performed all the aforementioned experiments using another antisense oligonucleotide to knock down XMAP215 and obtained similar results (data not shown), suggesting that the observed effects of XMAP215 KD on MT–F-actin alignment was not because of off-target effects of the antisense oligonucleotide.
Fig. 4.
XMAP215 is required for normal MT–F-actin alignment in growth cones. (A) Top: representative SIM images of growth cones from control, XMAP215 KD and XMAP215 OE neural explants stained for actin and tubulin. Scale bar: 8 µm. Middle insets: magnification of the boxed region of interest in the upper panels. Scale bar: 3 µm. Arrows point to MTs that are aligned with F-actin, while arrowheads point to exploring MTs that are not aligned with F-actin. Bottom insets: ‘camera lucida’-type depictions with subtracted background and highlighted MTs and F-actin. (B,C) XMAP215 KD decreases (B) while XMAP215 OE increases (C) the percentage of exploring MTs aligned to F-actin, compared to controls. (D,E). XMAP215 KD increases (D) while XMAP215 OE does not affect (E) the number of orientations followed by exploring MTs in the growth cone peripheral (P) domain. **P<0.01; ****P<0.0001; ns, not significant, from a one-way ANOVA analysis comparing multiple conditions, and from Student's t-test comparing XMAP215 OE to control. Data presented as mean±s.e.m.
The N-terminal of XMAP215 is necessary and sufficient to promote MT–F-actin alignment
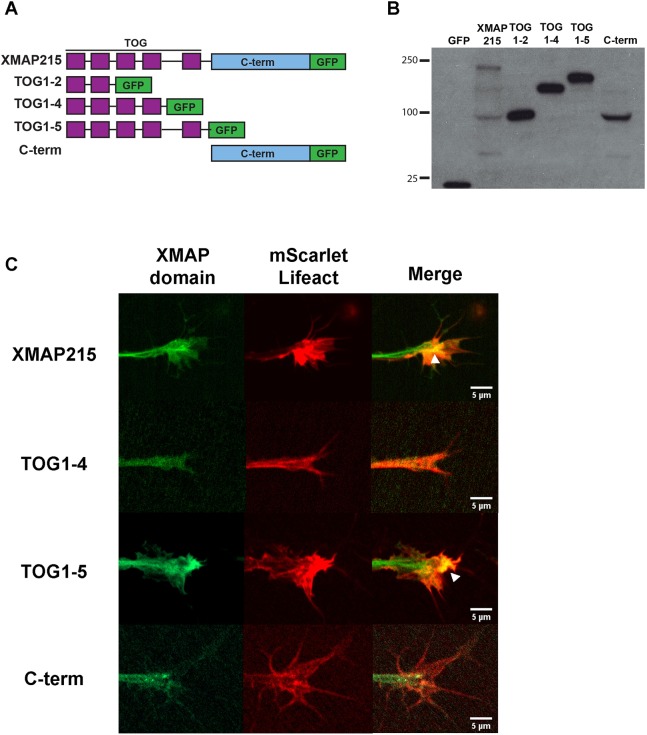
Several previous studies have determined that the XMAP215 N-terminal domain [TOG1–5, amino acids (aa) 1–1460] is composed of five TOG domains that bind tubulin dimers (Al-Bassam and Chang, 2011; Brouhard et al., 2008; Widlund et al., 2011) and a MT lattice-binding domain that comprises the region between TOG4 and TOG5 as well as part of TOG5 (Widlund et al., 2011). Additionally, the TOG1–2 region is known for being responsible for the MT polymerase activity (Widlund et al., 2011). The C-terminal domain (aa 1461–2070) is conserved and known to bind the protein TACC3 (Mortuza et al., 2014; Thakur et al., 2013), and is important for XMAP215 localization at centrosomes (Lee et al., 2001; O'Brien et al., 2005; Popov et al., 2002). It was also recently shown to be required for MT nucleation and direct binding to gamma-tubulin (Thawani et al., 2018). To investigate which domain(s) of XMAP215 are responsible for promoting MT–F-actin alignment, we generated a series of XMAP215 deletion constructs (similar to those previously described in Widlund et al., 2011) (Fig. 5A) and expressed them in X. laevis embryos (Fig. 5B). We first compared their localizations in the growth cone in conjunction with F-actin (mScarlet–Lifeact) using time-lapse imaging, and we observed that each XMAP215 deletion mutant displayed unique localizations (Fig. 5C; Movies 1–4). Both XMAP215 full-length (FL) and the TOG1–5 construct displayed characteristic MT lattice binding, which has previously been observed in these growth cones (Lowery et al., 2013). However, there was little to no MT plus-end tracking seen with the TOG1–5 construct (Movie 3). While the XMAP215 C-terminal domain did display distinct MT plus-end tracking localization (likely due to C-terminal domain interaction with endogenous TACC3), the localization did not overlap with F-actin in the periphery (Movie 4). Most strikingly, there was a clear co-localization pattern of XMAP215-FL and TOG1–5 with F-actin in the growth cone periphery (Fig. 5C, white arrowheads), while the TOG1–2 and TOG1–4 constructs did not show this co-localization with F-actin (they displayed only diffuse localization throughout the growth cone). These results were further validated through three-channel immunofluorescence and confocal microscopy, labeling microtubules, actin and XMAP215 deletion mutants. Similar to the live imaging data, XMAP215-FL and TOG1–5 showed distinct lattice binding and F-actin co-localization, while the XMAP215 C-terminal domain showed MT plus-end tracking localized primarily to the central domain (Fig. S4). This localization data suggests that a region within TOG1–5 is required for XMAP215 co-localization with F-actin in growth cones.
Fig. 5.
The N-terminal TOG1–5 domains of XMAP215 are necessary and sufficient to localize to F-actin in the growth cone periphery. (A) Schematic representation of the GFP-tagged XMAP215 domain constructs used, showing full-length XMAP215, XMAP215 domains TOG1–2, TOG1–4, TOG1–5 and C-terminal region (C-term). (B) Western blot showing the expression of the different XMAP215 domain constructs in Xenopus embryo lysates. (C) Representative confocal images showing localization of GFP-tagged XMAP215 mutants (green) and F-actin (red) in the growth cone. White arrowhead in full-length XMAP215 and TOG1–5 merged panels denote XMAP215 and actin fluorescence co-localization. Scale bar: 5 µm. (See Movies 1–4).
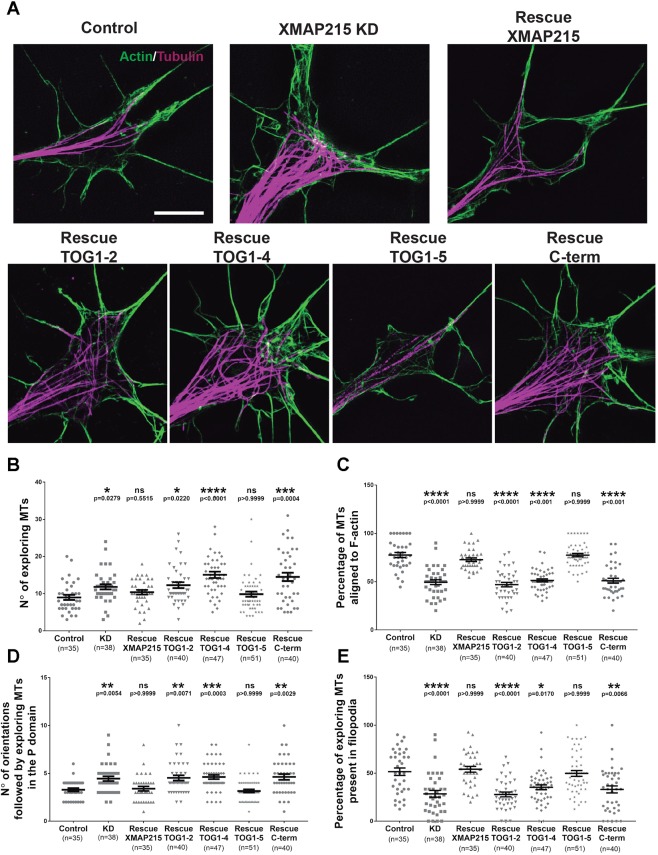
We then determined whether expression of each mutant could rescue the MT–F-actin alignment phenotype of the XMAP215 KD condition (Fig. 6A). Of all the deletions, only the TOG1–5 rescue attempt had a similar growth cone MT phenotype to that of the control and the rescue with XMAP215 full length (Fig. 6A). In addition, expression of TOG1–5 was able to rescue the number of exploring MTs (Fig. 6B), the percentage of MTs aligned to F-actin (Fig. 6C), the number of MT orientations in the peripheral domain (Fig. 6D), and the percentage of MTs present in filopodia (Fig. 6E), to a similar extent to that of XMAP215 full length (Fig. 6B–E). These results are consistent with the localization data and suggest that the region comprising all five TOG domains is both necessary and sufficient for XMAP215 to promote MT–F-actin alignment in the growth cone.
Fig. 6.
The N-terminal of XMAP215 is necessary and sufficient to promote MT–F-actin alignment. (A) Representative SIM images of growth cones from control, XMAP215 KD and XMAP215 KD rescue with the XMAP215 domain constructs as indicated. Scale bar: 5 µm. (B–E) Determination of the XMAP215 domain responsible for MT–F-actin alignment. Expression of TOG1–5, but not TOG1–2, TOG1–4 or C-term, rescues the increase in the number of exploring MTs (B), the decrease in the percentage of MTs aligned to F-actin (C), the increase in the number of orientations followed by exploring MTs in the peripheral (P) domain (D), and the decrease in the percentage of exploring MTs present in filopodia (E), induced by XMAP215 KD. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001; ns, not significant, from a one-way ANOVA analysis. Data presented as mean±s.e.m.
XMAP215 can interact with F-actin directly
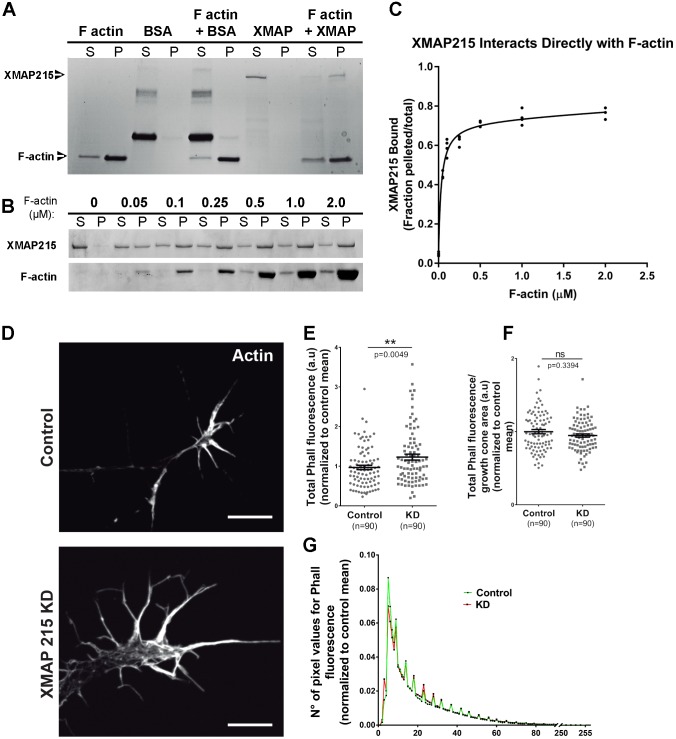
Given our findings that XMAP215 promotes the ability of MTs to enter filopodia (Fig. 3) and promote MT alignment to F-actin bundles (Fig. 4), we wondered whether XMAP215 might directly bind to F-actin. Thus, we first tested for XMAP215– F-actin binding using an F-actin co-sedimentation assay, and, intriguingly, we observed that XMAP215 is able to co-sediment with F-actin in vitro (Fig. 7A). While using BSA as a negative control showed little to no sedimentation with F-actin, XMAP215 presence switched from supernatant to pelleted fractions upon addition of F-actin. Next, by adding increasing concentrations of F-actin (Fig. 7B), we were able to obtain a binding curve for XMAP215 against F-actin. XMAP215 was shown to bind F-actin with a high affinity and quick saturation. Fitting the data to the binding curve gave an XMAP215 Kd value of 0.032±0.0051 μM (mean±s.e.m.), and XMAP215 maximum binding corresponded to a Bmax of 0.682±0.0283 when the data was non-linearly fit to a curve (Fig. 7C). These results indicate the novel finding that XMAP215 is able to directly bind F-actin in addition to MTs. We next wanted to determine if XMAP215 KD led to specific changes in the amount and distribution of F-actin within the growth cone (Fig. 7D). We measured phalloidin fluorescence intensity within the growth cone as a read-out for F-actin and observed a 27% increase in overall fluorescence intensity within the growth cone in XMAP215 KD compared to controls (Fig. 7E). However, when we examined F-actin levels after normalizing the total fluorescence intensity by the growth cone area (since XMAP215 KD growth cones are larger than controls), we observed no difference in the XMAP215 KD compared to controls (Fig. 7F). To investigate this further, we examined the number of pixel values for phalloidin fluorescence as a means to determine if Phalloidin was accumulating more or less in the different regions of the growth cone. We observed no differences when comparing XMAP215 KD to controls (Fig. 7G), suggesting that the F-actin distribution within the growth cone was not specifically affected. These results demonstrate that XMAP215 is capable of a direct interaction with F-actin, although the increase in overall F-actin levels is not due to denser F-actin bundles but correlates with the increased size of the growth cone. It remains to be determined how XMAP215 KD leads to this increase.
Fig. 7.
XMAP215 can directly bind to F-actin but does not affect local F-actin density. (A,B). XMAP215–F-actin co-sedimentation assays. Coomassie Blue-stained gels from binding assays containing a constant F-actin concentration (A) or increasing F-actin concentrations (B), show direct binding of XMAP215 to F-actin. BSA was used as a negative control. S, supernatant; P, pellet. (C) XMAP215–F-actin binding curve obtained by plotting the percentage of XMAP215 present in the pelleted fraction over the total, against the F-actin concentration. Data points from multiple experiments plotted for each concentration. Non-linearly fit binding curve displays a Kd value at a sub-micromolar binding affinity, 0.032±0.0051 µM (mean±s.e.m.). Bmax=0.682±0.0283. (D) Representative phalloidin-stained images of growth cones from control and XMAP215 KD neural explants. Scale bars: 10 µm. (E–G) Quantification of F-actin amount and distribution. XMAP215 KD increases total phalloidin fluorescence (E), while it does not affect phalloidin fluorescence normalized by growth cone area (F) or the number of pixel values for phalloidin fluorescence (G). a.u., arbitrary units; Phall, phalloidin. **P<0.01; ns, not significant, from Student's t-test comparing XMAP215 KD to control. Data presented as mean±s.e.m.
DISCUSSION
In this study, we reveal new functions for the MT polymerase XMAP215 in promoting MT–F-actin interaction in both spinal cord explants from X. laevis embryos and in vitro biochemical assays. We utilized knockdown and overexpression studies to determine that XMAP215 is required for the axon to respond to the repellent guidance cue Ephrin A5 and affects axonal extension by impacting growth cone pausing events. In addition, XMAP215 promotes MT alignment with F-actin bundles in the growth cone, and XMAP215 is important for determination of growth cone size, filopodia length, and MT morphology and distribution throughout the growth cone. Additionally, we find that XMAP215 is able to bind F-actin in vitro and co-localize with F-actin in the growth cone peripheral domain.
XMAP215 and its family members have been some of the most intensively studied and well-characterized MT polymerases over the last decade (Al-Bassam et al., 2007; Ayaz et al., 2012; Brouhard et al., 2008; Fox et al., 2014; Widlund et al., 2011; Zanic et al., 2013). Moreover, they have received recent attention for having additional MT-related functions, including MT nucleation (Flor-Parra et al., 2018; Thawani et al., 2018), mitotic spindle scaling (Milunović-Jevtić et al., 2018; Reber et al., 2013) and MT destabilization (Humphrey et al., 2018). Our study, using F-actin co-sedimentation assays and growth cone imaging, adds a new element to the arsenal of XMAP215 functions, as we demonstrate that XMAP215 also exhibits the potential to directly interact with F-actin. This provides the first strong evidence that XMAP215 may function as a MT–F-actin crosslinker in growth cones, although it is still unclear whether XMAP215 is sufficient to bind both cytoskeletal structures simultaneously, or whether additional protein complexes are required for mediating MT–F-actin interaction in growth cones.
Interestingly, we observed that XMAP215 KD leads to changes in growth cone area and filopodial length (Fig. 1). While it is unclear exactly how XMAP215 KD could lead to such increases, it is likely through an indirect mechanism. As there are complicated feedback mechanisms between MTs and F-actin within growth cones, it is possible that when alignment of MTs with F-actin is reduced, the overall cytoskeletal architecture and mechanical response of the growth cone is disrupted, thus leading to growth cone pausing and a corresponding increase in growth cone area. Early studies showed that the local loss of F-actin bundles on one side of the growth cone drives subsequent remodeling of MT organization, where MTs are no longer present in the F-actin bundle depleted zone, and instead reorient towards the remaining F-actin bundles (Zhou et al., 2002). Additionally, F-actin in filopodia are critical for appropriate guidance cue sensing (McConnell et al., 2016), while separately, the absence of F-actin bundles can act as a repulsive signal, inducing growth cone turning (Zhou et al., 2002). Our findings from XMAP215-depleted growth cones replicate a similar scenario in that a decrease in MT–F-actin alignment randomizes the directionality of exploring MTs, indicative of a loss in MT guidance by the F-actin bundles in the growth cone peripheral domain. This loss in MT guidance and resultant MT–F-actin cytoskeletal discoordination could facilitate a loss in growth cone directionality, and prompt the growth cone to pause and await cues that would recalibrate it to its environment. Mechanically, growth cone extension may also demonstrate decreased efficiency if MT–F-actin interactions are reduced around the area of transient focal point contacts, as dynamic MTs are required at sites of adhesion remodeling (Ezratty et al., 2005; Juanes et al., 2017).
Importantly, our results demonstrated that XMAP215 affects several MT phenotypes such as MT organization, extension into growth cone periphery and filopodia, and MT–F-actin alignments (Figs 2–4). When XMAP215 levels were reduced, MT behaviors were severely impacted throughout the growth cone. MT trajectories into the peripheral domain showed increased randomization, while central domain MTs demonstrated an increase in looping. Additionally, we also note an increased widening of the MT array throughout the growth cone neck and central domain that occurs with XMAP215 KD. This phenotype is strikingly reminiscent of the growth cone MT bundling defect previously observed upon myosin II inhibition (Burnette et al., 2008). These studies suggested that MT arrays must be spatially consolidated to form the core of the axon as the growth cone progresses forward, and that actin- and myosin II-dependent compressive forces are required for MT bundling (Burnette et al., 2008; Schaefer et al., 2008). Interestingly, we previously observed that the reduction of axon outgrowth that occurs with XMAP215 KD can be partially rescued by myosin II inhibition (Lowery et al., 2013). Thus, it is tempting to speculate that XMAP215 might somehow interact with myosin II, in addition to F-actin, during the regulation of MT dynamics within growth cones. However, while MT bundling defects caused by partial XMAP215 KD may account for some of the MT phenotypes that we observe, such as growth cone neck widening and changes in growth cone size, it does not explain the changes in peripheral MTs aligning to F-actin in filopodia. In addition, the N-terminal portion of XMAP215, capable of rescuing the MT–F-actin alignment defect, is also the region that is necessary to co-localize with F-actin in the growth cone.
It has been well-established that MTs follow along F-actin bundles in growth cones (Letourneau, 1983; Zhou and Cohan, 2004), and that when MTs lose the ability to interact with F-actin (for example, by filopodial F-actin bundle removal), this induces lateral excursions by MTs that penetrate the peripheral domain at various angles (Burnette et al., 2007) and promote formation of random MT loops (Zhou et al., 2002). A similar phenotype is observed when the MT–F-actin linker Spectraplakin is knocked out (Alves-Silva et al., 2012). Consistently, increasing MT–F-actin interaction can lead to areas that are devoid of MTs in S2 Drosophila cells (Adikes et al., 2018). Thus, the MT phenotypes that we observe with XMAP215 KD are consistent with previous studies in which MT–F-actin interaction is disrupted.
Our deletion construct analysis and rescue experiments demonstrate that the XMAP215 N-terminus, comprising the TOG1–5 domains, is both necessary and sufficient for promoting MT–F-actin alignment (Figs 5–6). Many previous studies have dissected the specific functions of the various domains of XMAP215 (for example, Ayaz et al., 2012; Thawani et al., 2018; Widlund et al., 2011), and the N-terminal TOG1–5 domains are known to bind to α/β-tubulin heterodimers and promote MT polymerization (Widlund et al., 2011). In our experiments, the deletion comprising only the TOG1–4 domains, which lacks the MT lattice-binding domain, was not able to rescue any of the MT–F-actin phenotypes within XMAP215 KD growth cones. In addition, the cytoplasmic localization of GFP-tagged TOG1–4 did not show distinct MT lattice-binding phenotypes or co-localization with actin in the periphery of the growth cone, unlike TOG1–5 or full-length XMAP215 (Fig. 5C; Fig. S4A, Movies 1–4). The C-terminal portion, while not showing distinct co-localization to actin in the growth cone, did show MT plus-end tracking (most likely through coiled-coil homodimerization or C-terminal interactors TACC3 and SLAIN1/2) (Mortuza et al., 2014; Van der Vaart et al., 2011). Thus, it seems likely that the N-terminal portion of XMAP215, specifically the area between its TOG4 and TOG5 domains, is important in determining a close MT–F-actin interaction in the growth cone periphery.
In order to determine if XMAP215 can bind directly to F-actin, we perform an F-actin co-sedimentation assay with purified XMAP215 protein. We determined that XMAP215 and F-actin can indeed bind directly, the binding saturates quickly, and the binding affinity is in the sub-micromolar range in vitro (Fig. 7). Given that it was previously shown that MT plus-end-tracking protein CLASP can likely bind F-actin through its N-terminal Dis1 (TOG) domain (Tsvetkov et al., 2007), it is possible that one or more of the TOG domains of XMAP215 can also bind to F-actin. Thus, with TOG domains of XMAP215 binding to F-actin while the MT lattice-binding domain interacts with MTs, MTs would be able to extend along F-actin into the growth cone periphery. While augmenting XMAP215 levels affected growth cone morphology, MT distributions and MT–F-actin alignment in the growth cone, these effects were not the result of altered F-actin densities or levels in the growth cone (Fig. 7D–G). Additionally, we previously reported that F-actin retrograde flow rates were not affected after partial XMAP215 KD (Lowery et al., 2013). Given that the N-terminus of XMAP215 was also sufficient to rescue axonal responsiveness to Ephrin A5, we speculate that there may be phosphorylation sites within the N-terminal that are phosphorylated downstream of guidance signals and modulate the ability of XMAP215 to crosslink MTs and F-actin in a spatially-restricted manner.
While this study is the first to demonstrate that XMAP215 can directly bind F-actin, promote MT–F-actin alignment in growth cones, and mediate axon guidance, it is notable that two well-known XMAP215 interactors have also been implicated in processes that are consistent with this function. The most frequent binding partner of XMAP215, TACC3, was previously described to be necessary for stabilizing XMAP215 on the MT plus-end (Mortuza et al., 2014; Nwagbara et al., 2014; O'Brien et al., 2005; Thakur et al., 2013) and for the growth cone response to the guidance cue Slit2 (Erdogan et al., 2017). Furthermore, the Drosophila ortholog of XMAP215, Mini spindles (MSPS) genetically and biochemically interacts with the plus-end-tracking protein CLASP (also known as Orbit, MAST and CHB) (Lowery et al., 2010), which was one of the first MT-associated proteins identified to participate downstream of axon guidance cue signals (Lee et al., 2004). In that seminal study, CLASP was shown to specifically bind to MT plus-ends that entered the growth cone periphery and could regulate MT entry into this domain (Lee et al., 2004). Additional studies on CLASP function have since demonstrated that changing levels of this MT-associated protein can result in striking changes in actin architecture throughout the growth cone (Engel et al., 2014; Marx et al., 2013).
Taken together, our results show how XMAP215 can regulate MT polymerization and extension along F-actin bundles into the growth cone periphery, and that these interactions can be modulated downstream of a repulsive guidance cue. Partial KD of XMAP215 leads to decreased MT–F-actin alignment, resulting in an inability to exert proper coordination between MTs and F-actin during growth cone extension, increased pausing and desensitization to guidance cues. This study explores and defines a novel function of XMAP215 in MT–F-actin behavioral coordination and axonal guidance, in addition to its classical and recently emerged roles in MT polymerization and nucleation. Future work should seek to address whether there are other participants in this functional coupling and how MT–F-actin interaction may be spatially and temporally regulated. A better understanding of how MT-associated proteins can dynamically tune their behaviors to regulate MT dynamicity and stability, and separately or additionally facilitate MT–F-actin cytoskeletal coordination downstream of environmental cues, is necessary to fully describe the mechanistic underpinnings of proper neuronal pathfinding.
MATERIALS AND METHODS
Xenopus embryonic neural tube explants
Eggs collected from female X. laevis frogs were fertilized in vitro, dejellied and cultured following standard methods (Sive et al., 2010). Embryos were grown to stage 22–24 (Nieuwkoop and Faber, 1994), and neural tubes were dissected as described (Lowery et al., 2012). All experiments were approved by the Boston College Institutional Animal Care and Use Committee and performed according to national regulatory standards.
Constructs, RNA and antisense oligonucleotides
Capped mRNAs were transcribed in vitro using SP6 or T7 mMessage mMachine Kit (Ambion). RNA was purified with LiCl precipitation. Constructs used were: XMAP215–GFP (a gift from the Hyman lab; Widlund et al., 2011) sub-cloned into pT7TS; GFP–MACF43 (a gift from the Hoogenraad Lab) and GFP in pCS2+. XMAP215 domain constructs TOG1–2–GFP, TOG1–4–GFP, TOG1–5–GFP, and the C-terminus were obtained by PCR from the XMAP215–GFP plasmid, and sub-cloned into pT7TS for generation of mRNA or into pFB HTA for protein purification. Previously validated morpholino antisense oligonucleotides targeted to the translation start site of X. laevis XMAP215 (5′-TCATCCACTCGCTGTCATCCCCCAT-3′), and another antisense oligo targeted to a splice site (5′-GGCTTTCCAAACCTACCATGAAACA-3′) validated herein (Fig. S1A), in addition to standard control antisense oligo (5′-cctcttacctcagttacaatttata-3′) (Gene Tools, LLC) were used. The embryos were injected four times in dorsal blastomeres at two-to-four cell stage in 0.1× Marc's Modified Ringer's (MMR) solution containing 5% Ficoll. Total mRNA amounts injected per embryo were: 400 pg of GFP; 700 pg of XMAP215–GFP for rescue experiments and 4000 pg of XMAP215–GFP for overexpression experiments; 300 pg of GFP–MACF43; 2000 pg of TOG1–2, 1000 pg of TOG1–4, 2000 pg of TOG1–5 and 2000 pg of C-terminus (these concentrations are molar equivalents to 4000 pg of XMAP215 and 400 pg of GFP). Antisense oligonucleotides were injected in a concentration of 8 ng/embryo (5′-tcatccactcgctgtcatcccccat-3′) and 20 ng/embryo (5′-ggctttccaaacctaccatgaaaca-3′), in both cases equivalent to a 70% knockdown (Lowery et al., 2013).
Immunocytochemistry
Embryonic explant cultures were fixed with 0.2% glutaraldehyde and labeled as described (Challacombe et al., 1997). Immunostaining was done using rat anti-tyrosinated tubulin [YL1/2] (1:1000, ab6160, Abcam), rabbit anti-de-tyrosinated tubulin (1:1000, AB3201, Millipore), mouse anti-DM1a (1:100, T6199, Sigma-Adrich), mouse anti-GFP (1:200, 3E6, Invitrogen) or rabbit anti-GFP (1:500, AB290, Abcam) antibodies, Alexa Fluor 488–phalloidin, Alexa Fluor 568–phalloidin and Alexa Fluor 633–phalloidin (1:500, Molecular Probes). The secondary antibodies used were goat anti-rat Alexa Fluor 568 (1:500, ab175476, Abcam), goat anti-rabbit Alexa Fluor 488 (1:500, A-11008, Life Technologies), goat anti-mouse Alexa Fluor 488 (1:500, A32723, Life Technologies), and goat anti-rabbit Alexa Fluor 647 F(ab′)2 fragment specific (1:500, 111-605-047, Jackson ImmunoResearch).
Image acquisition
High-resolution images of cultured spinal cord explants were obtained with a CSU-X1M 5000 spinning-disk confocal (Yokogawa) on a Zeiss Axio Observer inverted motorized microscope with a Zeiss Plan-Apochromat 63×/1.40 numerical aperture lens, or with a phase contrast 20× objective (Zeiss). Images were acquired with an ORCA R2 charge-coupled device camera (Hamamatsu) controlled with Zen software. For analyzing the number of growing MTs, time-lapse images were collected every 2 s for 1 min using the 63×/1.40 oil objective. The number of growth cone pauses and duration of the pauses were obtained by analyzing time-lapse images collected every 5 min for 4 h using a phase contrast 20× objective. This objective was also utilized to collect images for quantifying axon length, number of axons per explant and guidance cue assays. Structured illumination super-resolution images were collected on a Zeiss Axio Observer.Z1 for super-resolution microscope with Elyra S.1 system, utilizing an Objective Plan-Apochromat 63×/1.40 oil (DIC). Images were acquired with a PCO-Tech Inc. pco.edge 4.2 sCMOS camera. The images were obtained in a chamber at approximately 28°C and utilizing the immersion oil Immersol 518F 30°. Stacked images were obtained with a z-step of 100 nm per growth cone. Channel alignment and structured illumination processing were applied to the super-resolution images using the Zeiss Black program. The fluorochromes Alexa Fluor 488–phalloidin, Alexa Fluor 633–phalloidin, Alexa Fluor 568 and Alexa Fluor 488 were used. For supplemental immunofluorescent fixed images (Fig. S4), images were acquired using a Leica TCS SP5 scanning confocal microscope (Wetzlar) using a Leica Plan-Apochromat 63×/1.40 numerical aperture lens. Images were captured using Leica software.
Image analysis
For the following quantifications, the growth cone was defined as the region contained between the growth cone neck and the tip of filopodia (Fig. S1C). The number of growth cone growing MTs was analyzed from GFP–MACF43 movies using plusTipTracker, as previously described (Applegate et al., 2011; Stout et al., 2014). All the rest of the quantifications were done using Fiji software. To determine the growth cone area, the region starting on the growth cone neck and extending to the tip of the filopodia was manually selected using the drawing/selection tool of Fiji (Fig. S1A). To determine the number of filopodia, all the filopodia extending from the growth cone central domain into the periphery, contained within the growth cone area, were manually quantified (Fig. S1B), and the length of each filopodia was measured using the Simple Neurite Tracer ImageJ plugin (Longair et al., 2011). The quantification of number of exploring MTs and MTs present in the filopodia were performed manually. To determine the number of exploring MTs, all of the MTs entering into the peripheral domain (trespassing the F-actin mesh, orange line in Fig. S3A,B) were quantified; for the number of MTs present in filopodia, only the MTs present in the peripheral domain that were extending into the filopodia (dotted yellow line in Fig. S3A,B) were considered. An example quantification of Fig. S3A is shown in Fig. S3C,D. For the quantification of MT penetration into the peripheral domain, the region of the MT present in the F-actin mesh and extending throughout the peripheral domain was manually selected using the segmented line tool, and the length was measured (Fig. S3A,B). For the quantification of MT spreading, the distance between MTs was measured in the growth cone neck at the widest region of the growth cone (spreading through the central domain); and for the central domain width, the distance occupied by MTs, starting at the growth cone neck, was measured (Fig. 2C). The quantifications of fluorescence intensity for tubulin were performed on the manually selected regions of interest using the drawing/selection tool in ImageJ. Intensity outside of the growth cone area was measured with the same area drawn for growth cone and was considered as background. The percentage of MTs aligned to F-actin was quantified by measuring the distance and angle of separation between the MT and F-actin bundle. MT and F-actin were considered aligned if closer than 300 nm and separated by an angle not greater than 15°. For MT orientation quantification, a line was drawn following the axon growth (central line) and the central line was intersected by arrows showing the direction followed by each MT. The angle between the central line and the arrows were measured and every 30° was considered as a different orientation. The length of the axons was measured using the Simple Neurite Tracer ImageJ plugin (Longair et al., 2011) (Fig. S2A). For the quantification of the number of growth cone pauses and duration of the pauses, a pause was defined as when the center of the growth cone movement at each time point was less than the control mean movement minus the standard deviation, or when the growth cone changed the orientation of the movement by more than 180°, as growth cones pause before turning. For the duration of the pauses, the total number of frames where the growth cone was paused was divided by the number of pauses, and then multiplied by the number of minutes between two consecutive frames. For the guidance cue assay, the number of growth cones on and off Ephrin A5 stripes was manually quantified. Only growth cones with the soma localized off the stripes were quantified. Additionally, a growth cone was considered ‘off stripe’ when it had not been on an Ephrin A5 stripe at any time. All the quantifications that were manually performed were quantified by two different individuals in double blind experiments.
Protein purification
XMAP215 was purified using a bac-to-bac purification system as previously described (Brouhard et al., 2008; Kinoshita et al., 2001). The pFB HTA XMAP215 vector was used to create bacmid and amplified baculovirus, which was subsequently used to transfect cultures of Hi-5 insect cells (Trichoplusia ni) (Thermo Fisher Scientific, B85502) at a density of 2.0×106 cells ml−1. Following 48 h incubation, cells were centrifuged, and lysates were filtered and applied to Nickel NTA affinity columns (Qiagen). Protein elution was done using a 250 mM imidazole elution buffer (50 mM HEPES pH 7.25, 200 mM NaCl, 250 mM imidazole, 0.1% Triton-X, 1 mM DTT, 0.5 mM EDTA, 5% glycerol, 1 mM PMSF, 1× complete protease inhibitor cocktail). Buffer was exchanged using Amicon ultracentrifugation 100,000 MWCO column (storage buffer: 10 mM K-HEPES pH 7.25, 100 mM KCl, 5.0 mM EGTA, 2.0 mM MgCl2, 0.1 mM CaCl2, 10% glycerol), then snap frozen and stored at −80°C. Purified monomeric actin (Cytoskeleton Inc., APHL99) was diluted to 0.4 mg/ml in a general actin buffer (5 mM Tris pH 8.0, 0.2 mM CaCl2, 0.2 mM ATP, 0.5 mM DTT) and polymerized by the addition of a 10× actin polymerization buffer (500 mM KCl, 20 mM MgCl2, 10 mM ATP) at room temperature.
F-actin co-sedimentation assays
F-actin co-sedimentation assays were performed as described previously (Srivastava and Barber, 2008). Polymerized F-actin or an equivalent amount of F-actin control buffer were incubated with either BSA protein (as a negative control) or purified XMAP215 in injection buffer (10 mM Tris pH 7.0, 150 mM NaCl, 1 mM ATP, 0.1 mM CaCl2, 2.0 mM MgCl2, 0.2 mM DTT and 1.0 mM EGTA) for 30 min at room temperature. Protein was precleared to remove any aggregates. Actin was added to XMAP215 purified protein in excess. Following incubation, samples were centrifuged at 100,000× g for 20 min at 22°C using a Beckman TLA100.4 ultracentrifuge. For F-actin binding curves, a constant concentration of 0.1 µM of XMAP215 was incubated with increasing concentrations of 0, 0.05, 0.1, 0.25, 0.5, 1.0 and 2.0 µM of phalloidin-stabilized F-actin. Samples were run on SDS-PAGE gels and stained with Coomassie Brilliant Blue G-250 before being imaged. ImageJ was used to calculate densitometry of protein bands on SDS-PAGE gels, and plotted using GraphPad Prism. Experiments were repeated 3–4 times before curves were plotted (curves fit by GraphPad Prism non-linear regression assuming a one-site saturation model).
Stripe assay
Stripe assays were performed following the modified version of the stripe assay described previously (Knöll et al., 2007). The Ephrin A5-Fc solution was prepared by mixing 10 µg ml−1 of the Ephrin A5-Fc, human (R&D Systems, 374-EA) with 2.5 µg ml−1 of anti-human IgG, Fc-specific Cy3-conjugated (Sigma-Adrich, C2571), and the Fc control solution was generated by mixing 10 µg ml−1 Fc (Calbiochem, 401104) with 2.5 µg ml−1 of anti-human IgG, Fc-specific (Sigma-Adrich, I2136). Both solutions were kept for 30 min on a rotator, covered from light. Zig-zag striped silicon matrices attached to glass coverslips were used. The Ephrin A5-Fc solution was applied to the matrices and incubated for 30 min at 37°C. After three washes in 1× PBS, the Fc control solution was applied on the glass coverslips and incubated for 30 min at 37°C. Following that, three washes in 1× PBS were done and the glass coverslips were coated with 20 µg ml−1 laminin. X. laevis spinal cord explants were plated and imaged 24 h later.
Experimental design and statistical analysis
All graphs and statistical analyses were performed using the statistical software Prism 5 (GraphPad Software Inc.). Three to four independent experiments were performed for each condition to ensure reproducibility. Images of 8–10 explants acquired from 2–3 different embryos were obtained for each condition in every independent experiment. All data sets were analyzed for normal distribution using the D’Agostino normality test. When comparing two conditions, the unpaired Student's t-test was performed, and when comparing multiple conditions, one-way ANOVA was used, by comparing the mean of each column with the mean of the control column under the multiple comparison option. In the cases that the data did not pass the normality test, Mann–Whitney test was used for comparing two conditions, and Kruskal–Wallis test for multiple comparisons. To analyze MT morphology, as it is a categorical variable, a χ2 test was performed. The α-value was set at 0.05 for all statistical tests, and the P-values are represented as follows: *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001. Values are expressed as mean±s.e.m.
Supplementary Material
Acknowledgements
We thank members of the Lowery Lab for helpful discussions, suggestions and editing, especially Beth Bearce, Sangmook Lee and Burcu Erdogan. We also thank Nancy McGilloway and Todd Gaines for excellent Xenopus husbandry. We thank Anthony Koleske for expert advice and assistance in the XMAP215 protein purifications. We also thank the Harvard Center for Biological Imaging, as well as Bret Judson and the Boston College Imaging Core for infrastructure and support. This material is based upon work supported by the National Science Foundation under Grant No. 1626072. We also thank the National Xenopus Resource RRID:SCR_013731 and XenBase RRID:SCR 003280 for their support.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: P.G.S., L.A.L.; Methodology: P.G.S., L.A.L.; Validation: A.G.S., A.M.; Formal analysis: P.G.S., G.M.C., A.G.S., A.M.; Investigation: P.G.S., G.M.C., A.G.S., A.M.; Resources: G.M.C., Y.H.; Writing - original draft: P.G.S.; Writing - review & editing: P.G.S., L.A.L., G.M.C.; Visualization: P.G.S.; Supervision: L.A.L.; Project administration: L.A.L.; Funding acquisition: L.A.L.
Funding
This work was supported by the National Institutes of Health [R00 MH095768 and R01 MH109651 to L.A.L.], and a postdoctoral fellowship from Comisión Nacional de Investigación Científica y Tecnológica (CONICYT) to P.G.S. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.224311.supplemental
References
- Adikes R. C., Hallett R. A., Saway B. F., Kuhlman B. and Slep K. C. (2018). Control of microtubule dynamics using an optogenetic microtubule plus end–F-actin cross-linker. J. Cell Biol. 217, 779-793. 10.1083/jcb.201705190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J. and Chang F. (2011). Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol. 21, 604-614. 10.1016/j.tcb.2011.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J., Larsen N. A., Hyman A. A. and Harrison S. C. (2007). Crystal structure of a TOG domain: conserved features of XMAP215/Dis1-family TOG domains and implications for tubulin binding. Structure 15, 355-362. 10.1016/j.str.2007.01.012 [DOI] [PubMed] [Google Scholar]
- Alves-Silva J., Sánchez-Soriano N., Beaven R., Klein M., Parkin J., Millard T. H., Bellen H. J., Venken K. J. T., Ballestrem C. and Kammerer R. A. (2012). Spectraplakins promote microtubule-mediated axonal growth by functioning as structural microtubule-associated proteins and EB1-dependent +TIPs (tip interacting proteins). J. Neurosci. 32, 9143-9158. 10.1523/JNEUROSCI.0416-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applegate K. T., Besson S., Matov A., Bagonis M. H., Jaqaman K. and Danuser G. (2011). plusTipTracker: quantitative image analysis software for the measurement of microtubule dynamics. J. Struct. Biol. 176, 168-184. 10.1016/j.jsb.2011.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayaz P., Ye X., Huddleston P., Brautigam C. A. and Rice L. M. (2012). A TOG:alphabeta-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase. Science 337, 857-860. 10.1126/science.1221698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearce E. A., Erdogan B. and Lowery L. A. (2015). TIPsy tour guides: how microtubule plus-end tracking proteins (+TIPs) facilitate axon guidance. Front. Cell Neurosci. 9, 241 10.3389/fncel.2015.00241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S. and Kalil K. (2018). The microtubule-associated protein tau mediates the organization of microtubules and their dynamic exploration of actin-rich Lamellipodia and Filopodia of cortical growth cones. J. Neurosci. 38, 291-307. 10.1523/JNEUROSCI.2281-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard G. J., Stear J. H., Noetzel T. L., Al-Bassam J., Kinoshita K., Harrison S. C., Howard J. and Hyman A. A. (2008). XMAP215 is a processive microtubule polymerase. Cell 132, 79-88. 10.1016/j.cell.2007.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette D. T., Schaefer A. W., Ji L., Danuser G. and Forscher P. (2007). Filopodial actin bundles are not necessary for microtubule advance into the peripheral domain of Aplysia neuronal growth cones. Nat. Cell Biol. 9, 1360-1369. 10.1038/ncb1655 [DOI] [PubMed] [Google Scholar]
- Burnette D. T., Ji L., Schaefer A. W., Medeiros N. A., Danuser G. and Forscher P. (2008). Myosin II activity facilitates microtubule bundling in the neuronal growth cone neck. Dev. Cell 15, 163-169. 10.1016/j.devcel.2008.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammarata G. M., Bearce E. A. and Lowery L. A. (2016). Cytoskeletal social networking in the growth cone: how +TIPs mediate microtubule-actin cross-linking to drive axon outgrowth and guidance. Cytoskeleton (Hoboken) 73, 461-476. 10.1002/cm.21272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challacombe J. F., Snow D. M. and Letourneau P. C. (1997). Dynamic microtubule ends are required for growth cone turning to avoid an inhibitory guidance cue. J. Neurosci. 17, 3085-3095. 10.1523/JNEUROSCI.17-09-03085.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles C. H. and Bradke F. (2015). Coordinating neuronal actin-microtubule dynamics. Curr. Biol. 25, R677-R691. 10.1016/j.cub.2015.06.020 [DOI] [PubMed] [Google Scholar]
- Dent E. W., Callaway J. L., Szebenyi G., Baas P. W. and Kalil K. (1999). Reorganization and movement of microtubules in axonal growth cones and developing interstitial branches. J. Neurosci. 19, 8894-8908. 10.1523/JNEUROSCI.19-20-08894.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel U., Zhan Y., Long J. B., Boyle S. N., Ballif B. A., Dorey K., Gygi S. P., Koleske A. J. and VanVactor D. (2014). Abelson phosphorylation of CLASP2 modulates its association with microtubules and actin. Cytoskeleton 71, 195-209. 10.1002/cm.21164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdogan B., Cammarata G. M., Lee E. J., Pratt B. C., Francl A. F., Rutherford E. L. and Lowery L. A. (2017). The microtubule plus-end-tracking protein TACC3 promotes persistent axon outgrowth and mediates responses to axon guidance signals during development. Neural Dev. 12, 3 10.1186/s13064-017-0080-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezratty E. J., Partridge M. A. and Gundersen G. G. (2005). Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat. Cell Biol. 7, 581 10.1038/ncb1262 [DOI] [PubMed] [Google Scholar]
- Nieuwkoop P. and Faber J. (1994). Normal table of Xenopus laevis (Daudin). A Systematical & Chronological Survey of the Development from the Fertilized Egg till the End of Metamorphosis, New York, USA: Garland Science. [Google Scholar]
- Flor-Parra I., Iglesias-Romero A. B. and Chang F. (2018). The XMAP215 ortholog Alp14 promotes microtubule nucleation in fission yeast. Curr. Biol. 28, 1681-1691.e4. 10.1016/j.cub.2018.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J. C., Howard A. E., Currie J. D., Rogers S. L. and Slep K. C. (2014). The XMAP215 family drives microtubule polymerization using a structurally diverse TOG array. Mol. Biol. Cell 25, 2375-2392. 10.1091/mbc.e13-08-0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldo S., Khanzada U. K., Parsons M., Chilton J. K. and Gordon-Weeks P. R. (2008). Targeting of the F-actin-binding protein drebrin by the microtubule plus-tip protein EB3 is required for neuritogenesis. Nat. Cell Biol. 10, 1181-1189. 10.1038/ncb1778 [DOI] [PubMed] [Google Scholar]
- Hendricks M. and Jesuthasan S. (2009). PHR regulates growth cone pausing at intermediate targets through microtubule disassembly. J. Neurosci. 29, 6593-6598. 10.1523/JNEUROSCI.1115-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnappa S., Gouveia S. M., Weisbrich A., Damberger F. F., Bhavesh N. S., Jawhari H., Grigoriev I., van Rijssel F. J. A., Buey R. M., Lawera A. et al. (2009). An EB1-binding motif acts as a microtubule tip localization signal. Cell 138, 366-376. 10.1016/j.cell.2009.04.065 [DOI] [PubMed] [Google Scholar]
- Humphrey L., Felzer-Kim I. and Joglekar A. P. (2018). Stu2 acts as a microtubule destabilizer in metaphase budding yeast spindles. Mol. Biol. Cell 29, 247-255. 10.1091/mbc.E17-08-0494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur E. M., Saijilafu E.-M., Lee B. D., Kim S.-J., Xu W.-L. and Zhou F.-Q. (2011). GSK3 controls axon growth via CLASP-mediated regulation of growth cone microtubules. Genes Dev. 25, 1968-1981. 10.1101/gad.17015911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juanes M. A., Bouguenina H., Eskin J. A., Jaiswal R., Badache A. and Goode B. L. (2017). Adenomatous polyposis coli nucleates actin assembly to drive cell migration and microtubule-induced focal adhesion turnover. J. Cell Biol. 216, 2859-2875. 10.1083/jcb.201702007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalil K., Szebenyi G. and Dent E. W. (2000). Common mechanisms underlying growth cone guidance and axon branching. J. Neurobiol. 44, 145-158. [DOI] [PubMed] [Google Scholar]
- Kinoshita K., Arnal I., Desai A., Drechsel D. N. and Hyman A. A. (2001). Reconstitution of physiological microtubule dynamics using purified components. Science 294, 1340-1343. 10.1126/science.1064629 [DOI] [PubMed] [Google Scholar]
- Knoll B., Zarbalis K., Wurst W. and Drescher U. (2001). A role for the EphA family in the topographic targeting of vomeronasal axons. Development 128, 895-906. [DOI] [PubMed] [Google Scholar]
- Knöll B., Kretz O., Fiedler C., Alberti S., Schütz G., Frotscher M. and Nordheim A. (2006). Serum response factor controls neuronal circuit assembly in the hippocampus. Nat. Neurosci. 9, 195-204. 10.1038/nn1627 [DOI] [PubMed] [Google Scholar]
- Knöll B., Weinl C., Nordheim A. and Bonhoeffer F. (2007). Stripe assay to examine axonal guidance and cell migration. Nat. Protoc. 2, 1216 10.1038/nprot.2007.157 [DOI] [PubMed] [Google Scholar]
- Lee M. J., Gergely F., Jeffers K., Peak-Chew S. Y. and Raff J. W. (2001). Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour. Nat. Cell Biol. 3, 643-649. 10.1038/35083033 [DOI] [PubMed] [Google Scholar]
- Lee H., Engel U., Rusch J., Scherrer S., Sheard K. and Van Vactor D. (2004). The microtubule plus end tracking protein Orbit/MAST/CLASP acts downstream of the tyrosine kinase Abl in mediating axon guidance. Neuron 42, 913-926. 10.1016/j.neuron.2004.05.020 [DOI] [PubMed] [Google Scholar]
- Letourneau P. C. (1983). Differences in the organization of actin in the growth cones compared with the neurites of cultured neurons from chick embryos. J. Cell Biol. 97, 963-973. 10.1083/jcb.97.4.963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longair M. H., Baker D. A. and Armstrong J. D. (2011). Simple Neurite Tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics 27, 2453-2454. 10.1093/bioinformatics/btr390 [DOI] [PubMed] [Google Scholar]
- Lowery L. A., Lee H., Lu C., Murphy R., Obar R. A., Zhai B., Schedl M., Van Vactor D. and Zhan Y. (2010). Parallel genetic and proteomic screens identify Msps as a CLASP-Abl pathway interactor in Drosophila. Genetics 185, 1311-1325. 10.1534/genetics.110.115626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery L. A., Faris A. E. R., Stout A. and Van Vactor D. (2012). Neural explant cultures from Xenopus laevis. J. Vis. Exp. 68, e4232 10.3791/4232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery L. A., Stout A., Faris A. E., Ding L., Baird M. A., Davidson M. W., Danuser G. and Van Vactor D. (2013). Growth cone-specific functions of XMAP215 in restricting microtubule dynamics and promoting axonal outgrowth. Neural Dev. 8, 22 10.1186/1749-8104-8-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx A., Godinez W. J., Tsimashchuk V., Bankhead P., Rohr K. and Engel U. (2013). Xenopus cytoplasmic linker–associated protein 1 (XCLASP1) promotes axon elongation and advance of pioneer microtubules. Mol. Biol. Cell 24, 1544-1558. 10.1091/mbc.e12-08-0573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell R. E., van Veen J. E., Vidaki M., Kwiatkowski A. V., Meyer A. S. and Gertler F. B. (2016). A requirement for filopodia extension toward Slit during Robo-mediated axon repulsion. J. Cell Biol. 213, 261-274. 10.1083/jcb.201509062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milunović-Jevtić A., Jevtić P., Levy D. L. and Gatlin J. C. (2018). In vivo mitotic spindle scaling can be modulated by changing the levels of a single protein: the microtubule polymerase XMAP215. Mol. Biol. Cell 29, 1311-1317. 10.1091/mbc.E18-01-0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnier P. P., Sierra A., Schwab J. M., Henke-Fahle S. and Mueller B. K. (2003). The Rho/ROCK pathway mediates neurite growth-inhibitory activity associated with the chondroitin sulfate proteoglycans of the CNS glial scar. Mol. Cell. Neurosci. 22, 319-330. 10.1016/S1044-7431(02)00035-0 [DOI] [PubMed] [Google Scholar]
- Mortuza G. B., Cavazza T., Garcia-Mayoral M. F., Hermida D., Peset I., Pedrero J. G., Merino N., Blanco F. J., Lyngsø J., Bruix M. et al. (2014). XTACC3–XMAP215 association reveals an asymmetric interaction promoting microtubule elongation. Nat. Commun. 5, 5072 10.1038/ncomms6072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neukirchen D. and Bradke F. (2011). Cytoplasmic linker proteins regulate neuronal polarization through microtubule and growth cone dynamics. J. Neurosci. 31, 1528-1538. 10.1523/JNEUROSCI.3983-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwagbara B. U., Faris A. E., Bearce E. A., Erdogan B., Ebbert P. T., Evans M. F., Rutherford E. L., Enzenbacher T. B. and Lowery L. A. (2014). TACC3 is a microtubule plus end-tracking protein that promotes axon elongation and also regulates microtubule plus end dynamics in multiple embryonic cell types. Mol. Biol. Cell 25, 3350-3362. 10.1091/mbc.e14-06-1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien L. L., Albee A. J., Liu L., Tao W., Dobrzyn P., Lizarraga S. B. and Wiese C. (2005). The Xenopus TACC homologue, maskin, functions in mitotic spindle assembly. Mol. Biol. Cell 16, 2836-2847. 10.1091/mbc.e04-10-0926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov A. V., Severin F. and Karsenti E. (2002). XMAP215 is required for the microtubule-nucleating activity of centrosomes. Curr. Biol. 12, 1326-1330. 10.1016/S0960-9822(02)01033-3 [DOI] [PubMed] [Google Scholar]
- Prokop A. (2013). The intricate relationship between microtubules and their associated motor proteins during axon growth and maintenance. Neural Dev. 8, 17 10.1186/1749-8104-8-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reber S. B., Baumgart J., Widlund P. O., Pozniakovsky A., Howard J., Hyman A. A. and Jülicher F. (2013). XMAP215 activity sets spindle length by controlling the total mass of spindle microtubules. Nat. Cell Biol. 15, 1116 10.1038/ncb2834 [DOI] [PubMed] [Google Scholar]
- Schaefer A. W., Kabir N. and Forscher P. (2002). Filopodia and actin arcs guide the assembly and transport of two populations of microtubules with unique dynamic parameters in neuronal growth cones. J. Cell Biol. 158, 139-152. 10.1083/jcb.200203038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A. W., Schoonderwoert V. T. G., Ji L., Mederios N., Danuser G. and Forscher P. (2008). Coordination of actin filament and microtubule dynamics during neurite outgrowth. Dev. Cell 15, 146-162. 10.1016/j.devcel.2008.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive H. L., Grainger R. M. and Harland R. M. (2010). Microinjection of Xenopus oocytes. Cold Spring Harb. Protoc. 2010, prot5536 10.1101/pdb.prot5536 [DOI] [PubMed] [Google Scholar]
- Srivastava J. and Barber D. (2008). Actin co-sedimentation assay; for the analysis of protein binding to F-actin. J. Vis. Exp. 13, e690 10.3791/690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout A., D'Amico S., Enzenbacher T., Ebbert P. and Lowery L. A. (2014). Using plusTipTracker software to measure microtubule dynamics in Xenopus laevis growth cones. J. Vis. Exp. 91, e52138 10.3791/52138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka E. M. and Kirschner M. W. (1991). Microtubule behavior in the growth cones of living neurons during axon elongation. J. Cell Biol. 115, 345-363. 10.1083/jcb.115.2.345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka E., Ho T. and Kirschner M. W. (1995). The role of microtubule dynamics in growth cone motility and axonal growth. J. Cell Biol. 128, 139-155. 10.1083/jcb.128.1.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur H. C., Singh M., Nagel-Steger L., Kremer J., Prumbaum D., Fansa E. K., Ezzahoini H., Nouri K., Gremer L. and Abts A. (2013). The centrosomal adaptor TACC3 and the microtubule polymerase chTOG interact via defined C-terminal subdomains in an Aurora-A kinase independent manner. J. Biol. Chem. 289, 74-88. 10.1074/jbc.M113.532333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thawani A., Kadzik R. S. and Petry S. (2018). XMAP215 is a microtubule nucleation factor that functions synergistically with the gamma-tubulin ring complex. Nat. Cell Biol. 20, 575-585. 10.1038/s41556-018-0091-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov A. S., Samsonov A., Akhmanova A., Galjart N. and Popov S. V. (2007). Microtubule-binding proteins CLASP1 and CLASP2 interact with actin filaments. Cell Motil. Cytoskeleton 64, 519-530. 10.1002/cm.20201 [DOI] [PubMed] [Google Scholar]
- Van der Vaart B., Manatschal C., Grigoriev I., Olieric V., Gouveia S. M., Bjelic S., Demmers J., Vorobjev I., Hoogenraad C. C., Steinmetz M. O. et al. (2011). SLAIN2 links microtubule plus end-tracking proteins and controls microtubule growth in interphase. J. Cell Biol. 193, 1083-1099. 10.1083/jcb.201012179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitriol E. A. and Zheng J. Q. (2012). Growth cone travel in space and time: the cellular ensemble of cytoskeleton, adhesion, and membrane. Neuron 73, 1068-1081. 10.1016/j.neuron.2012.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelzmann A., Hahn I., Pearce S. P., Sánchez-Soriano N. and Prokop A. (2016). A conceptual view at microtubule plus end dynamics in neuronal axons. Brain Res. Bull. 126, 226-237. 10.1016/j.brainresbull.2016.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widlund P. O., Stear J. H., Pozniakovsky A., Zanic M., Reber S., Brouhard G. J., Hyman A. A. and Howard J. (2011). XMAP215 polymerase activity is built by combining multiple tubulin-binding TOG domains and a basic lattice-binding region. Proc. Natl. Acad. Sci. USA 108, 2741-2746. 10.1073/pnas.1016498108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wizenmann A. and Bähr M. (1997). Growth characteristics of ganglion cell axons in the developing and regenerating retino-tectal projection of the rat. Cell Tissue Res. 290, 395-403. 10.1007/s004410050946 [DOI] [PubMed] [Google Scholar]
- Zanic M., Widlund P. O., Hyman A. A. and Howard J. (2013). Synergy between XMAP215 and EB1 increases microtubule growth rates to physiological levels. Nat. Cell Biol. 15, 688-693. 10.1038/ncb2744 [DOI] [PubMed] [Google Scholar]
- Zhou F.-Q. and Cohan C. S. (2004). How actin filaments and microtubules steer growth cones to their targets. J. Neurobiol. 58, 84-91. 10.1002/neu.10278 [DOI] [PubMed] [Google Scholar]
- Zhou F.-Q., Waterman-Storer C. M. and Cohan C. S. (2002). Focal loss of actin bundles causes microtubule redistribution and growth cone turning. J. Cell Biol. 157, 839-849. 10.1083/jcb.200112014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.