Abstract
Experience-dependent changes in the strength of connections between neurons in the hippocampus (HPC) are critical for normal learning and memory consolidation, and disruption of this process drives a variety of neurological and psychiatric diseases. Proper HPC function relies upon discrete changes in gene expression driven by transcription factors (TFs) induced by neuronal activity. Here, we describe the induction and function of many of the most well-studied HPC TFs, including cyclic-AMP response element binding protein, serum-response factor, AP-1, and others, and describe their role in the learning process. We also discuss the known target genes of many of these TFs and the purported mechanisms by which they regulate long-term changes in HPC synaptic strength. Moreover, we propose that future research in this field will depend upon unbiased identification of additional gene targets for these activity-dependent TFs and subsequent meta-analyses that identify common genes or pathways regulated by multiple TFs in the HPC during learning or disease.
Keywords: hippocampus, memory, plasticity, transcription
Introduction
The hippocampus (HPC) is critical for a variety of human behaviors, including episodic learning and responses to stress, and dysfunction or death of HPC neurons underlies diseases like Alzheimer’s dementia, major depressive disorder, and posttraumatic stress disorder. Proper function of the HPC relies upon discrete changes in gene expression driven by transcription factors (TFs) engaged by neuronal activity. However, as these TFs are also critical for the function of many other brain regions and nonnervous tissues, using them as pharmacological targets for the treatment of diseases involving the HPC is not feasible. We therefore propose that the future of research in this field will depend upon unbiased identification of gene targets for these activity-dependent TFs and subsequent meta-analyses that identify common genes or pathways regulated by multiple TFs in the HPC during learning or disease. Such research may uncover factors or pathways whose regulation is unique to the HPC and which may therefore serve as viable pharmacological targets for therapeutic intervention in neurological and psychiatric diseases involving HPC dysfunction. Here, we review the nature and mechanisms of activity-dependent TFs that are critical for HPC function, including their role in memory consolidation and long-term plasticity, as well as their currently known gene targets.
The HPC in memory consolidation
Memory consolidation underlies experience-dependent changes in our behavior (McGaugh, 2000), driving adaptation to novel circumstances and setting the stage for expected behavior in familiar environments. Experiences induce a cascade of events across brain regions, starting with sensory information processing in cortical regions. The HPC is a unique brain region that consolidates this experiential input into memory traces that can be shunted into other regions, such as the cortex, for long-term storage. HPC is particularly important for consolidation of explicit, declarative memories, such as contextual and spatial information.
A period of stabilization is required before a short-term memory is preserved for long-term storage. Synaptic plasticity within the HPC underlies this stabilization, or consolidation, of a labile memory. The consolidation of long-term memories requires connectivity within HPC subregions (dentate gyrus [DG], CA3, CA1) forming a trisynaptic DG–CA3–CA1 loop (Figure 1). Specifically, excitatory glutamatergic inputs convey experiential, sensory information into entorhinal cortex (EC), from both cortical and limbic regions, such as amygdala. EC’s primary projections are to DG, known as the perforant pathway, although EC also projects to CA3, CA1, and subiculum. Glutamatergic DG granule cells project to pyramidal neurons in CA3 (the mossy fiber pathway), which send glutamatergic Schaffer collateral projections to pyramidal neurons in CA1. Connectivity within the trisynaptic loop is modulated by enduring forms of activity-dependent synaptic plasticity called long-term potentiation (LTP) and long-term depression (LTD). Synaptic plasticity is hypothesized to encode a memory trace by forming networks of strongly connected neurons, or memory engrams (Govindarajan et al., 2006; Neves et al., 2008; Silva et al., 2009). There-after, the CA1 sends projections through subiculum and back to EC, where memories can be distributed throughout cortex for long-term storage. The trisynaptic loop of DG–CA3–CA1, as well as the plasticity that occurs in these HPC synapses, underlies nearly all forms of explicit memory consolidation, and these changes are driven by transactivation of gene expression.
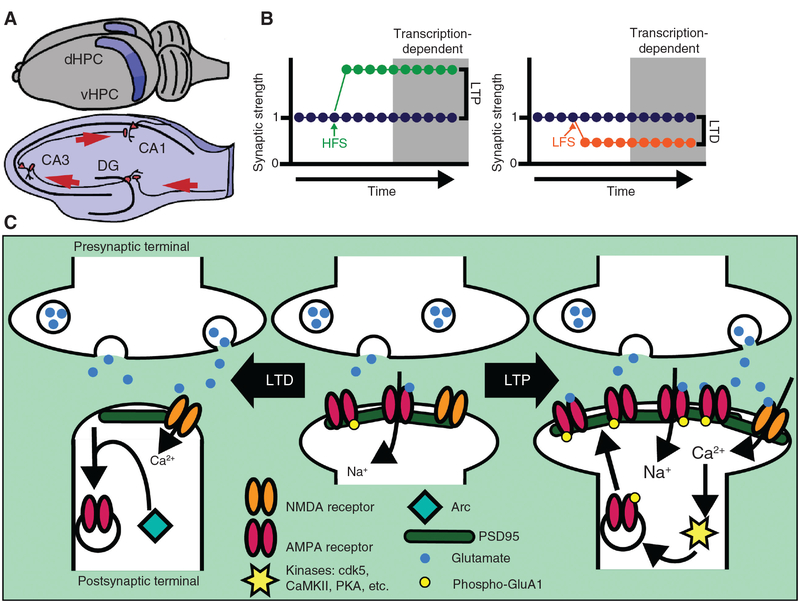
Figure 1: Hippocampal synaptic plasticity relies on activity-dependent transcription.
(A) Depiction of bilateral location of dorsal (dHPC) and ventral (vHPC) mouse HPC (top) and of a coronal slice of dHPC (bottom) showing the connections between the subregions and the general direction of glutamatergic projections (red arrows). (B) Graphical representation of LTP (left) and LTD (right) induced by HFS or LFS, respectively. Gray region indicates long-lasting change in synaptic strength requiring activity-dependent transcription. (C) Select molecular mechanisms of LTP and LTD at CA3–CA1 glutamatergic synapses involving products of genes regulated by activity-dependent TFs (see Table 1).
LTP at CA3–CA1 synapses occurs via postsynaptic signaling mechanisms, resulting in a long-lasting increase in the amplitude of the excitatory response in the postsynaptic cell (Figure 1). This process is dependent on activation of ionotropic N-methyl-d-aspartate receptors (NMDARs), and the subsequent influx of Ca2+ results in an increased synaptic response mediated by changes in the surface expression and function of a-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPARs) and/or structural alterations in the postsynaptic element, the dendritic spine. LTP can be further divided into an early phase (that does not require gene transcription) and a transcription-dependent late phase. The process of LTD also contributes to memory consolidation and occurs in both an N-methyl-d-aspartate-dependent manner and/or through a process requiring postsynaptic metabotropic glutamate receptors (mGluRs). Whereas LTP is induced in CA3–CA1 synapses by high-frequency stimulation (HFS), LTD is induced by low-frequency stimulation (LFS) or stimulation of mGluRs. The postsynaptic signaling induced by LFS leads to AMPAR endocytosis, and like LTP, late phases of LTD are also dependent on gene expression (Huber et al., 2000; Manahan-Vaughan et al., 2000). Thus, the long-term changes in synaptic strength underlying memory result from changes in HPC transcription.
Transcription in the HPC
Transcription is a complex process whereby RNA polymerase II (RNApolII) transcribes a sequence of DNA into corresponding messenger RNA (mRNA). An assortment of molecular machinery regulates transcription: promoter regions that direct transcription; TFs that are bound to DNA and facilitate RNApolII binding to promoter regions; coregulators that act to promote or suppress transcription via their interaction with TFs; cofactors that mediate protein–protein interactions between TFs and coregulators; and chromatin regulators that act to remodel chromatin to enhance or suppress TF binding to DNA. The products of transcription include mRNAs that encode proteins and noncoding RNAs that can contribute to cellular complexes (like ribosomes), regulate the translation of mRNAs (i.e. microRNAs), or perform other less well-characterized functions. While each aspect of this complex transcriptional regulatory machinery may contribute to HPC function and learning, here, we will describe the specific role of TFs induced by cellular activity in the HPC and their role in memory consolidation.
Activity-dependent TFs are induced by repeated action potentials that drive the expression of immediate early genes (IEG), many of which encode TFs. Because many of these TFs have additional mechanisms of induction that do not rely on cell activity (Wisden et al., 1990), we will focus on two states: (1) a basal condition in which these TFs are regulating mRNA synthesis to prepare neurons for rapid RNA translation in response to activity and (2) activity-dependent induction of TFs leading to down-stream synthesis of mRNA from genes that are critical for long-term changes in neuronal function and connectivity underlying memory formation, such as potentiation or depression of synaptic activity. This system is complex, tightly regulated, and stable.
Activity-dependent gene transcription is necessary for the consolidation of memories (Alberini, 2009). Transcriptional regulation underlies cellular changes induced by de novo protein and mRNA synthesis critical for memory consolidation (Alberini and Kandel, 2014), and indeed, we have known for decades that infusion of transcriptional inhibitors into HPC prevents late-phase LTP (Nguyen et al., 1994). Specifically, this decrease occurs only when inhibitors are administered during the induction of LTP, and not after induction, e.g. during late phase. This highlights that early-phase LTP is de novo mRNA synthesis independent, whereas late-phase LTP requires transcription to induce RNA synthesis for later translation. It has been recently hypothesized that transcriptional regulation controls the expression of many synaptic proteins that support plasticity (Alberini and Kandel, 2014), and it appears likely that activity-dependent transcription regulated by a subset of TFs is necessary for plasticity underlying the consolidation of an active memory trace. A series of activity-dependent TFs have been described that are critical for this transcription-dependent HPC plasticity and learning. We have divided these into (1) extracellular signal-activated TFs, including cyclic-AMP (cAMP) response element binding protein (CREB), serum-response factor (SRF), and E26 (Ets)-like TF 1 (Elk-1), and (2) IEG TFs associated with plasticity and learning.
Extracellular signaling-activated TFs associated with hippocampal plasticity and learning
CREB
CREB is one of the most well-characterized TFs in learning and memory. It was originally described due to its association with the cAMP response element (CRE) DNA sequence (Montminy and Bilezikjian, 1987), and it is widely expressed throughout the brain, playing a critical role in plasticity in many brain regions, including HPC. There are multiple CREB proteins, most notably CREB1 and CREB2 (activating TF 4; ATF4), which may have opposing actions on plasticity, with CREB1 associated with enhancement or facilitation of long-term plasticity (see below) and ATF4/CREB2 associated with constraints on plasticity (Chen et al., 2003). However, the remainder of this section will focus solely on CREB1, hereafter referred to solely as CREB. CREB activity is mediated by its phosphorylation state (Bito et al., 1996), which lends to its stability, as unphosphorylated CREB is targeted for degradation (Mouravlev et al., 2007). CREB is induced in HPC during spatial learning (Mizuno et al., 2002) and HPC NMDAR-dependent avoidance learning (Cammarota et al., 2000) and is critical for LTP (Yin et al., 1994). Furthermore, HPC CREB inhibition disrupts long-term memory consolidation (Bourtchuladze et al., 1994; Kida et al., 2002; Pittenger et al., 2002), and constitutively active CREB over-expression in HPC increases contextual memory (Restivo et al., 2009). CREB transcriptional activity is also mediated by its binding partners, including CREB binding protein (CBP) (Chrivia et al., 1993), which is important for spatial and contextual learning and dopamine-specific LTP in the HPC (Wood et al., 2005, 2006). It appears, therefore, that CREB is necessary for late-phase or long-term memory consolidation, which is dependent on gene expression underlying de novo protein synthesis. Specifically, CREB may act as the transcriptional switch between protein synthesis-independent short-term memory and protein synthesis-dependent long-term memory.
CREB is regulated by multiple signaling cascades downstream of neuronal activity, including cAMP and Ca2+ (Figure 2) (Wheeler et al., 2012). Specifically, CREB phosphorylation occurs in conjunction with stimuli that induce LTP, and not simply with repeated neuronal firing (Deisseroth et al., 1996), indicating that signaling from the synapse is specifically required for CREB activation in the nucleus. Many molecules have been implicated in this signaling, including the cAMP-dependent protein kinase (PKA), extracellular signal-regulated kinase (ERK), and calmodulin-dependent protein kinases such as CaMKI, CaMKII, and CaMKIV (Bito et al., 1996; Benito and Barco, 2010). Many players in this signaling cascade leading to CREB phosphorylation are crucial for LTP (Malinow et al., 1989; Frey et al., 1993) and HPC transcription (Bading et al., 1993). Supporting this idea, constitutively active CREB (which contains mutations mimicking phosphorylation) primes hippocampal synapses for plasticity, thereby lowering the threshold for long-lasting, late-phase LTP (Barco et al., 2002) and leading to enhanced memory consolidation. Knockout (KO) of specific CREB isoforms impaired HPC LTP in mice (Bourtchuladze et al., 1994), although other studies have failed or only very weakly replicated this effect (Gass et al., 1998; Pittenger et al., 2002), which may be attributed to a different genetic background or that prior studies used a mutant that only knocked out specific isoforms and not other CREB related genes. A dominant negative-inhibitor of CREB (KREB) disrupts learning, and some Ca2+-dependent forms of LTP are impaired by this CREB KO (Pittenger et al., 2002). Additionally, constitutively active CREB overexpression in HPC leads to increases in NMDAR-dependent LTP and silent synapse formation (Marie et al., 2005) and also enhances LTP and memory consolidation (Suzuki et al., 2011), while dominant negative CREB expression impairs memory consolidation (Kathirvelu et al., 2013). CREB has also been shown to regulate intrinsic plasticity, such as cell excitability (Dong et al., 2006; Lopez de Armentia et al., 2007; Benito and Barco, 2010), which supports the premise that CREB activity may confer a competitive advantage allowing for the recruitment of CREB-induced cells during the formation of a memory trace (Han et al., 2007; Benito and Barco, 2010), a hypothesis corroborated experimentally in the amygdala (Kim et al., 2014; Yiu et al., 2014). Finally, a plethora of CREB target genes play a significant role in synaptic, structural, and intrinsic plasticity and signaling (Table 1), and newer studies are beginning to reveal the full complement of basal and memory-related genes that are targeted by CREB (Lakhina et al., 2015).
Figure 2: Common signaling cascades leading to activity-dependent TF activation.
Extracellular signals and changes in membrane potential can lead to increases in second-messenger (cAMP and Ca2+) or ERK signaling that converge on kinase activity within the nucleus resulting in phosphorylation and activation of CREB and SRF complexes. These bind to specific elements in promoter regions to regulate transcription of a variety of genes, including IEGs that encode other activity-dependent TFs, like c-fos and FosB.
Table 1:
Known gene targets of individual activity-dependent TFs.
A list of validated targets of some of the activity-dependent TFs is discussed here, along with references to supporting studies. Neither the gene list nor the references are comprehensive but are rather meant to illustrate (1) the existence of common targets and (2) the fact that many of the known gene targets are directly involved in the molecular mechanisms of synaptic plasticity (see Figure 1C).
SRF
SRF is a member of the MADS-box (MCM1, Agamous, Deficiens, SRF) family of TFs that share a conserved sequence motif and tend to recruit other TFs in multiregulatory complexes but have different DNA binding properties and heterodimer partners (Shore and Sharrocks, 1995). SRF binds the serum-response element (SRE) in many genes, including c-fos (Treisman, 1987), and is integral for IEG induction through Elk-1-dependent (see below) and -independent mechanisms (Xia et al., 1996). Binding to the SRE is dependent on SRF phosphorylation, which is downstream of some of the same signaling cascades that activate CREB (Figure 2), including ERK (Xia et al., 1996).
SRF is highly expressed in all subregions of HPC (Herdegen et al., 1997; Ramanan et al., 2005), where it plays an important role in plasticity and learning. SRF has many gene targets (Table 1), including IEGs and other genes associated with synaptic plasticity, and gene targeting appears to be regulated by various cofactors and phosphorylation states (Knöll and Nordheim, 2009). For example, the CCAAT-enhancer binding protein (C/EBP-β) gene requires SRF binding to SRE for transactivation of SRE-associated genes (Sealy et al., 1997) and Elk-1 contributes to SRF induction of c-fos (Marais et al., 1993), although SRF transcriptional activity may also be Elk-1-independent (Miranti et al., 1995).
SRF has been widely implicated in plasticity and learning. For example, SRF KO mice display impaired IEG induction, mild deficits in LTP (for discussion, see Etkin et al., 2006), and robust deficits in LTD in CA1 neurons, while displaying no change in basal excitatory signaling (Ramanan et al., 2005; Etkin et al., 2006). SRF appears especially important for structural plasticity that may underlie neuronal circuit remodeling; its expression closely follows neuronal development (Stringer et al., 2002), and its deletion produces profound structural changes in HPC (Knöll et al., 2006; Stritt and Knöll, 2010). In particular, SRF regulates neurite outgrowth and proper axonal guidance (Knöll et al., 2006; Li et al., 2014), in addition to dendritic spine development and neuronal migration (Stritt and Knöll, 2010). These functions appear to be mediated by SRF’s regulation of actin cytoskeletal genes (Table 1), and this regulation is associated with a host of outcomes including cell differentiation and development, intracellular trafficking, and synapse remodeling. SRF may also affect axonal development indirectly, as SRF deletion decreases myelination by reducing oligodendrocyte differentiation via paracrine signaling (Stritt et al., 2009). Despite SRF’s importance in actin cytoskeletal organization associated with circuitry remodeling and spine morphogenesis, it does not appear to be necessary for neuronal survival (Ramanan et al., 2005). These findings support the hypothesis that SRF translates synaptic activity into neuronal connectivity in HPC via actin cytoskeletal reorganization (Knöll and Nordheim, 2009).
Elk-1
Elk-1 is a member of the Ets family of oncogenes (Rao et al., 1989) and modulates HPC plasticity and memory consolidation through its interactions with other TFs, such as SRF. While Elk-1 is expressed in many cell types throughout the body, in the brain, it is exclusively expressed in neurons (Sgambato et al., 1998), with strong expression in both DG and CA regions of HPC. Elk-1 mRNA can be found in soma, dendrites, and axons, suggesting that it may play a role in local activity-dependent translation, in addition to its role in transcription. Elk-1 transcriptional activity is induced by growth factors (Marais et al., 1993) and glutamatergic signaling (Vanhoutte et al., 1999) through phosphorylation by the MAPK/ERK cascade (Figure 2). Elk-1 transcriptionally targets SRE to drive SRE-associated gene expression, such as IEGs (Hipskind et al., 1991; Janknecht and Nordheim, 1993). Elk-1 dimerizes with other cofactors and TFs, including SRF and CBP, to regulate gene expression (Hipskind et al., 1991; Janknecht and Nordheim, 1992, 1996). Unlike SRF, which can act in an Elk-1-independent manner (Miranti et al., 1995), there is scant evidence that Elk-1 can regulate gene expression without this dimerization.
Both LTP and LTD in HPC neurons induce MAPK/ERK-dependent Elk-1 phosphorylation (Davis et al., 2000; Thiels et al., 2002), and mGluR-induced LTD requires SRF/Elk-1 for induction of IEGs (Lindecke et al., 2006). Elk-1 KO mice, while displaying normal development and few abnormal phenotypes, show no change in basal c-fos expression but a decrease in kainite-induced seizure induction of c-fos in HPC (Cesari et al., 2004). However, early growth response protein 1 (egr1)/zinc finger protein 268 (zif268) induction by seizures was normal in Elk-1 KO mice, suggesting that induction of IEGs may be compensated by other TFs, e.g. SRF, in Elk-1-deficient mice. This is in line with evidence indicating that activity-dependent induction of SRF-mediated transcription of IEGs acts in both an Elk-1-dependent and independent manner, with Elk-1 enhancing SRF-SRE transcriptional activation (Xia et al., 1996).
Although Elk-1 is induced by learning (Cammarota et al., 2000; Sananbenesi et al., 2002), to date, no study has identified a specific and critical role for HPC Elk-1 in memory formation. Indeed, SRF but not Elk-1 inhibition impairs spatial learning (Dash et al., 2005). Similarly, no impairments in learning have been observed in Elk-1 KO mice (Cesari et al., 2004). Delineating the function of Elk-1 in plasticity and learning is further complicated by its close association with SRF, which is uniquely important for HPC plasticity and learning (see above). This makes it difficult to accurately identify Elk-1 gene targets that are not directly transcribed by SRF (Table 1). However, given the critical role of MAPK/ERK signaling in synaptic plasticity and learning (Davis et al., 2000; Sweatt, 2004), it is likely Elk-1 is involved in some aspect of HPC plasticity and learning that remains to be determined.
IEG TFs associated with hippo-campal plasticity and learning
Egr1/Zif268/NGFI-A/Krox-24
Egr1, which is also known as zif268, nerve growth factor-induced A, and Krox-24, is an IEG encoded by the EGR1 gene. It shows rapid, robust induction following neuronal activation that return to basal levels within 24 h (Knapska and Kaczmarek, 2004). Egr1 expression is induced by NMDAR activation (Worley et al., 1993) but is also widely regulated by a number of different signaling cascades, including trophic factors, membrane-depolarization-induced glutamate signaling, and Ca2+ signaling (Condorelli et al., 1994; Ghosh et al., 1994; Knapska and Kaczmarek, 2004). The promoter region of EGR1 includes six SRE sites, a CRE site, and an activator protein 1 (AP1) binding site (Veyrac et al., 2014), indicating that its induction is downstream of CREB, SRF, Elk-1, and Fos TFs, and it is induced by MAPK/ERK signaling via Elk-1, SRF, and CREB (Davis et al., 2000), suggesting that egr1 is a nexus for activity-dependent gene expression.
EGR1 KO mice show decreased long-term in vivo LTP and impairment in long-term memory formation (Jones et al., 2001), as well as destabilized place cell representations (Renaudineau et al., 2009). Conversely, inducible overexpression of egr1 enhances long-term spatial memory and LTP in DG neurons (Penke et al., 2014), although egr1 may play a larger role in reconsolidation of a memory, rather than its initial consolidation (Lee et al., 2004). Although egr1 appears to be necessary for plasticity, few gene targets supporting this have thus far been uncovered. Nevertheless, NMDAR-dependent induction of egr1 leads to the subsequent repression of the synaptic structure protein PSD-95; this is followed by endocytosis of AMPA receptors and robust LTD (Qin et al., 2015). The majority of gene targets of egr1 have been identified in cancer studies and include genes involved in cell proliferation, differentiation, and survival, as well as apoptosis (Veyrac et al., 2014); however, preliminary microarray studies have further implicated additional target genes of egr1 in the brain (James et al., 2005). Notably, this included genes previously associated with plasticity, such as synapsin II and gephyrin, as well as serum/glucocorticoid regulated kinase and c-Jun (Table 1). Thus, although it is clear that egr1 is critical for learning, the mechanisms by which it controls synaptic function remain to be fully revealed.
AP1
AP1 is a TF complex composed of heterodimers between Fos family proteins, Jun family proteins, Jun dimerization proteins, and/or ATF proteins. A typical AP1 complex consists of Fos-Jun heterodimers that utilize leucine zippers present in both proteins for dimerization and a basic region that interacts with DNA. Because of the variety of AP1 complexes that can form, they are associated with diverse induction stimuli, a number of coactivators/corepressors, and a wide variety of gene targets. For the sake of brevity, we will focus mainly on the Fos family of TFs due to their well-known role in HPC plasticity and learning; however, the remaining members are also important for HPC function (Minatohara et al., 2015).
c-fos
The Fos family of TFs is comprised of c-fos, FosB (and its splice variants, ΔFosB and Δ2ΔFosB), Fra1, and Fra2. While not completely reliant upon neuronal activity (for example, see Wisden et al., 1990), the induction of these TFs is directly associated with neuronal activity, to the point that c-fos staining is commonly used as a marker of neuronal activity (Sagar et al., 1988). The list of environmental and cellular stimuli, signaling pathways, proteins, induction protocols, growth factors, etc., that have been implicated in the regulation of c-fos are too broad a subject for the current review, but it is clear that neurons produce c-fos in response to any activity-inducing stimulus. However, there is scant evidence investigating the gene targets of c-fos and their role in cellular function, specifically in memory consolidation and plasticity. The majority of its known gene targets come from studies in cancer, inflammation, and bone development (Grigoriadis et al., 1993; Matsuo et al., 2000, 2004; Matthews et al., 2007), while its targets in neurons are largely unknown. Many have made the claim that c-fos is merely induced by neuronal activity and does not contribute to long-term cellular changes. We propose here that c-fos is particularly important for plasticity and initial memory consolidation. This is based on preliminary studies described in more detail below showing a correlation between c-fos induction and plasticity, newer studies implicating a role for c-fos in plasticity and learning, and studies showing c-fos may represent aspects of activity associated with a memory engram.
c-fos plays a key role in downstream gene expression underlying neuronal activity (Dragunow and Robertson, 1987; Kaczmarek et al., 1988; Sheng et al., 1990); however, it may also underlie plasticity and learning (Kaczmarek, 1993) by regulating gene expression, including the expression of other TFs associated with plasticity (Sheng and Greenberg, 1990). c-fos is regulated by a variety of factors including SRF/Elk-1 and CREB (Sheng and Green-berg, 1990; Sheng et al., 1990; Hipskind et al., 1991) and is repressed by other factors, such as ΔFosB (Nakabeppu and Nathans, 1991; Renthal et al., 2008). c-fos is transiently and robustly induced, with a half-life ranging from minutes up to a couple hours (Sheng and Greenberg, 1990; Kovács, 1998; Ferrara et al., 2003). It is hypothesized to target a wide variety of genes associated with cell differentiation, cell and synapse development, synaptic plasticity, and learning (Alberini, 2009; West and Greenberg, 2011); however, conclusive evidence for c-fos-specific gene targets, especially those involved in HPC plasticity, has not yet been provided.
Correlative and some preliminary causal evidence suggest that c-fos is important for memory consolidation. In particular, c-fos is induced in the HPC by memory tasks, including spatial learning and a brightness discrimination form of associative learning (Tischmeyer et al., 1990; Guzowski et al., 2001), and inhibition of c-fos impairs memory for the brightness discrimination (Grimm et al., 1997). c-fos is also induced in HPC by LTP (Dragunow et al., 1989; Fleischmann et al., 2003) and has been found to be necessary for induction of LTP (Fleischmann et al., 2003) and LTD (Kemp et al., 2013). Furthermore, brain-specific KO of c-fos produces marked impairments in learning and memory and HPC plasticity (Fleischmann et al., 2003). Recent evidence using manipulations of c-fos to inactivate neuronal ensembles, reactivate consolidated memories, or generate false memory traces suggests a role for c-fos in more than just neuronal activation (Garner et al., 2012; Liu et al., 2012; Cruz et al., 2015). Thus, c-fos is required for transcription-dependent changes in HPC neurons underlying memory consolidation. Future studies examining c-fos gene targets and any link between c-fos and synaptic plasticity will reveal the full extent of its functions underling HPC memory consolidation.
FosB
FosB is encoded by the FosB gene and shares many characteristics and gene targets with c-fos. Like c-fos, FosB has low basal expression in HPC (Herdegen et al., 1995) and is transiently and robustly induced by neuronal activity (Nestler et al., 1999), with a similar half-life in cells (Dobrazanski et al., 1991; Ferrara et al., 2003; Ulery et al., 2006). FosB KO mice show decreased neurogenesis in DG (Yutsudo et al., 2013) and impairment in rearing pups but have normal spatial learning and olfactory discrimination (Brown et al., 1996). Nevertheless, FosB gene targets and its role in plasticity and learning remain unknown; however, other FosB gene products (detailed below) may provide a novel mechanism for chronic activity-dependent gene expression associated with plasticity and learning.
ΔFosB
Splice variation of FosB gene transcripts produces a premature stop codon resulting in the truncated ΔFosB protein. The splice variant lacks two c-terminal degron domains lending it increased stability (Carle et al., 2007); most other Fos TFs have a half-life of a few hours, while ΔFosB has an unusually long-half life, up to 7 days in vivo (Hope et al., 1994; Andersson et al., 2003; Ulery-Reynolds et al., 2009). In addition, Ca2+-signaling may contribute to this stability, as CaMKII has been shown to phosphorylate ΔFosB at Ser27, thereby enhancing its stability (Ulery-Reynolds et al., 2009; Robison et al., 2013). While ΔFosB has a well-characterized role in the nucleus accumbens (NAc) in stress-and reward-related behavior (Robison and Nestler, 2011; Nestler, 2015), more recent evidence suggests that it is induced by a variety of stimuli in HPC, including ischemia (McGahan et al., 1998), drugs of abuse (Perrotti et al., 2008), stress (Perrotti et al., 2004; Vialou et al., 2015), antidepressants (Vialou et al., 2015), and spatial learning (Eagle et al., 2015). While the mechanism for the induction of HPC ΔFosB is unknown, ΔFosB is induced by SRF and CREB in NAc (Vialou et al., 2012). Engram-specific induction of ΔFosB may under-lie memory consolidation, as specific ΔFosB inhibition in HPC impairs multiple forms of learning (Eagle et al., 2015). Interestingly, ΔFosB overexpression in HPC also impaired memory consolidation, perhaps due to dysregulation of preferential connectivity underlying memory engrams (Eagle et al., 2015). While its gene targets are not well known in HPC, studies in other brain regions show that ΔFosB regulates a number of genes associated with plasticity, including c-fos, cdk5, CaMKII, and glutamate receptors, such as GluA2 (Nakabeppu and Nathans, 1991; Kelz et al., 1999; Chen et al., 2000; Robison et al., 2014). Moreover, ΔFosB overexpression regulates syn-apses in both NAc (Grueter et al., 2013) and HPC (Yutsudo et al., 2013) and induces immature spine formation in HPC CA1 pyramidal neurons (Eagle et al., 2015) and D1R-expressing medium spiny neurons of the NAc (Grueter et al., 2013).
Because of its unusual stability, ΔFosB is a unique target for studies examining long-term plasticity or learning in response to chronic environmental changes. This is especially relevant considering that other activity-dependent TFs, such as c-fos, homeostatically decrease expression in HPC after repeated daily trials of learning (Nikolaev et al., 1992; Hess et al., 1995; Bertaina-Anglade et al., 2000), which, interestingly, may even be regulated by ΔFosB (Renthal et al., 2008). Therefore, ΔFosB is an attractive target for further investigation into chronic HPC activity-dependent gene expression driving long-term behavioral adaptations or disease.
C/EBP-B, ATF, others
C/EBP-βb is expressed in adult, but not fetal, brain, primarily in HPC, cerebellum, and cortex (Kuo et al., 1990), where it binds the CCAAT box motif to regulate gene expression. C/EBP is induced in HPC by learning and is required for long-term memory consolidation (Taubenfeld et al., 2001a,b). It has been shown to colocalize with phosphorylated CREB, and CRE-mediated gene expression is required for its induction during memory formation (Athos et al., 2002). C/EBP also appears to be regulated by cAMP and mediates long-term facilitation in Aplysia (Alberini et al., 1994), but its role in HPC plasticity has yet to be established. C/EBP acts as a positive feedback mechanism for brain-derived neurotrophic factor (BDNF) by potentiating BDNF upregulation leading to further C/EBP induction (Bekinschtein et al., 2007; Bambah-Mukku et al., 2014), and BDNF strongly induces HPC LTP (Ying et al., 2002).
ATF4/CREB2, or ATF4, regulates CRE-mediated gene expression and has been hypothesized to act as a transcriptional repressor of plasticity and memory consolidation (Guan et al., 2002). Supporting this, ATF4 inhibition was shown to enhance HPC-dependent memory consolidation and LTP (Chen et al., 2003). Conversely, new evidence demonstrates that ATF4 knockdown decreases dendritic spines, glutamatergic neurotransmission, and expression of synaptic GluA1 and PSD95. Moreover, this decreases LTP and LTD in HPC CA1, impairs long-term memory formation, and is rescued by ATF4 overexpression (Liu et al., 2014; Pasini et al., 2015). Thus, ATF4 appears critical for memory formation, but its precise role in HPC function remains debatable.
More recently, a host of additional activity-dependent TFs important for plasticity and memory consolidation have begun to emerge. For example, the myocyte enhancer factor-2 family of TFs has been shown to act as a critical negative regulator of synaptic plasticity and learning (Rashid et al., 2014). Interestingly, stress may also be associated with HPC plasticity, with glucocorticoid and mineralocorticoid receptors having diverse roles in HPC plasticity in response to stress (Kim and Diamond, 2002; Avital et al., 2006; Berger et al., 2006; Gray et al., 2013). Finally, NF-kB, or nuclear factor kappa-B, which has a wide variety of functions in the brain and other tissues, may play a significant role in HPC plasticity and learning (Albensi and Mattson, 2000; Meffert et al., 2003; Kaltschmidt et al., 2006) through changes in synapse function and synaptogenesis (Boersma et al., 2011). The identification of these and other activity-dependent TFs and their associated gene targets illustrates the complexity of HPC transcription and its role in synaptic, structural, and intrinsic plasticity underlying learning, a knot that must be untangled in order to advance the science of learning and the treatment of diseases involving HPC dysfunction.
Pushing the field forward: uncovering novel pathways
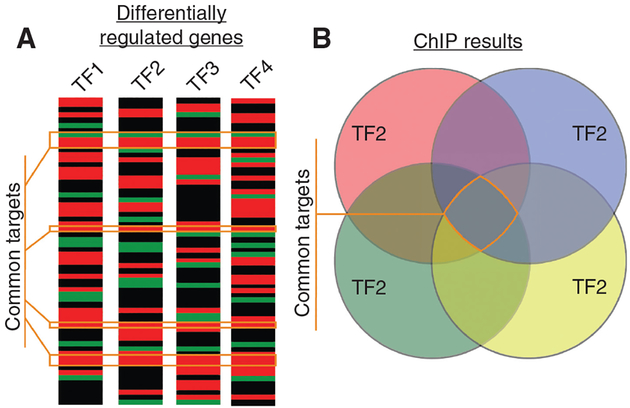
As catalogued in brief above, the role of activity-dependent TFs in HPC function has been extensively studied, and this has greatly advanced the learning and memory field. However, in order for future studies of these TFs to directly impact the treatment of neurological and psychiatric disorders involving dysfunction of the HPC, it will be necessary to determine novel HPC genes and pathways that are common targets of multiple TFs during learning and/or in models of disease. Such advances may arise from comparing data sets generated by mRNA sequencing (RNAseq) experiments on HPC tissue after learning or in a disease model. Overlaying such data sets generated from animals in which select TFs have been overexpressed or silenced could reveal common genes regulated in HPC by multiple activity-dependent TFs under relevant conditions (Figure 3A), many of which may become pharmacologically feasible therapeutic targets. Similarly, using chromatin immunoprecipitation (ChIP) for specific TFs followed by unbiased sequencing approaches will reveal direct binding targets (Figure 3B), and meta-analyses combining all of these approaches should uncover common path-ways whose dysfunction drives disease. Such ‘big data’ approaches are already underway in many labs, and the coming decade should give rise to a host of advances in the field of HPC gene expression that will impact both the science of learning and the treatment of disease.
Figure 3: Identifying novel genes and pathways regulated by activity-dependent HPC gene transcription.
(A) Hypothetical heat plots of RNAseq and proteomic experiments in which individual activity-dependent TFs are genetically or pharmacologically manipulated to reveal genes whose expression is directly or indirectly regulated by each TF. Comparing the unbiased output of such experiments will reveal common potential gene targets that may underlie learning or disease (orange boxes). (B) Similarly, ChIPseq experiments using antibodies against individual activity-dependent TFs to precipitate bound chromatin from HPC of mice undergoing learning or models of disease will reveal common gene targets for activity-dependent TF binding (orange box). Combining the data from such experiments will produce new molecular models of learning and disease.
References
- Albensi BC and Mattson MP (2000). Evidence for the involvement of TNF and NF-kB in hippocampal synaptic plasticity. Synapse 35, 151–159. [DOI] [PubMed] [Google Scholar]
- Alberini CM (2009). Transcription factors in long-term memory and synaptic plasticity. Physiol. Rev 89, 121–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberini CM and Kandel ER (2014). The regulation of transcription in memory consolidation. Cold Spring Harbor Perspect. Biol 7, a021741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberini CM, Ghirardl M, Metz R, and Kandel ER (1994). C/EBP is an immediate-early gene required for the consolidation of long-term facilitation in aplysia. Cell 76, 1099–1114. [DOI] [PubMed] [Google Scholar]
- Andersson M, Westin JE, and Cenci MA (2003). Time course of striatal ΔFosB-like immunoreactivity and prodynorphin mRNA levels after discontinuation of chronic dopaminomimetic treatment. Eur. J. Neurosci 17, 661–666. [DOI] [PubMed] [Google Scholar]
- Athos J, Impey S, Pineda VV, Chen X, and Storm DR (2002). Hippocampal CRE-mediated gene expression is required for contextual memory formation. Nat. Neurosci 5, 1119–1120. [DOI] [PubMed] [Google Scholar]
- Avital A, Segal M, and Richter-Levin G (2006). Contrasting roles of corticosteroid receptors in hippocampal plasticity. J. Neuro-sci 26, 9130–9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bading H, Ginty D, and Greenberg M (1993). Regulation of gene expression in hippocampal neurons by distinct calcium signaling pathways. Science 260, 181–186. [DOI] [PubMed] [Google Scholar]
- Bambah-Mukku D, Travaglia A, Chen DY, Pollonini G, and Alberini CM (2014). A positive autoregulatory BDNF feedback loop via C/EBPb mediates hippocampal memory consolidation. J. Neurosci 34, 12547–12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barco A, Alarcon JM, and Kandel ER (2002). Expression of constitutively active CREB protein facilitates the late phase of long-term potentiation by enhancing synaptic capture. Cell 108, 689–703. [DOI] [PubMed] [Google Scholar]
- Bekinschtein P, Cammarota M, Igaz LM, Bevilaqua LRM, Izquierdo I, and Medina JH (2007). Persistence of long-term memory storage requires a late protein synthesis- and BDNF-dependent phase in the hippocampus. Neuron 53, 261–277. [DOI] [PubMed] [Google Scholar]
- Benito E and Barco A (2010). CREB’s control of intrinsic and synaptic plasticity: implications for CREB-dependent memory models. Trends Neurosci. 33, 230–240. [DOI] [PubMed] [Google Scholar]
- Berger S, Wolfer DP, Selbach O, Alter H, Erdmann G, Reichardt HM, Chepkova AN, Welzl H, Haas HL, Lipp H-P, et al. (2006). Loss of the limbic mineralocorticoid receptor impairs behavioral plasticity. Proc. Natl. Acad. Sci. USA 103, 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertaina-Anglade V, Tramu G, and Destrade C (2000). Differential learning-stage dependent patterns of c-Fos protein expression in brain regions during the acquisition and memory consolidation of an operant task in mice. Eur. J. Neurosci 12, 3803–3812. [DOI] [PubMed] [Google Scholar]
- Bito H, Deisseroth K, and Tsien RW (1996). CREB phosphorylation and dephosphorylation: a Ca2+- and stimulus duration-dependent switch for hippocampal gene expression. Cell 87, 1203–1214. [DOI] [PubMed] [Google Scholar]
- Boersma MCH, Dresselhaus EC, De Biase LM, Mihalas AB, Bergles DE, and Meffert MK (2011). A requirement for nuclear factor-kB in developmental and plasticity-associated synaptogenesis. J. Neurosci 31, 5414–5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourtchuladze R, Frenguelli B, Blendy J, Cioffi D, Schutz G, and Silva AJ (1994). Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell 79, 59–68. [DOI] [PubMed] [Google Scholar]
- Brown JR, Ye H, Bronson RT, Dikkes P, and Greenberg ME (1996). A defect in nurturing in mice lacking the immediate early gene fosB. Cell 86, 297–309. [DOI] [PubMed] [Google Scholar]
- Cammarota M, Bevilaqua LRM, Ardenghi P, Paratcha G, Levi de Stein M, Izquierdo I, and Medina JH (2000). Learning-associated activation of nuclear MAPK, CREB and Elk-1, along with Fos production, in the rat hippocampus after a one-trial avoidance learning: abolition by NMDA receptor blockade. Mol. Brain. Res 76, 36–46. [DOI] [PubMed] [Google Scholar]
- Carle TL, Ohnishi YN, Ohnishi YH, Alibhai IN, Wilkinson MB, Kumar A, and Nestler EJ (2007). Proteasome-dependent and -independent mechanisms for FosB destabilization: identification of FosB degron domains and implications for ΔFosB stability. Eur. J. Neurosci 25, 3009–3019. [DOI] [PubMed] [Google Scholar]
- Cesari F, Brecht S, Vintersten K, Vuong LG, Hofmann M, Klingel K, Schnorr J-J, Arsenian S, Schild H, Herdegen T, et al. (2004). Mice deficient for the Ets transcription factor Elk-1 show normal immune responses and mildly impaired neuronal gene activation. Mol. Cell. Biol 24, 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhang Y, Kelz MB, Steffen C, Ang ES, Zeng L, Nestler EJ (2000). Induction of cyclin-dependent kinase 5 in the hippocampus by chronic electroconvulsive seizures: role of ΔFosB. J. Neurosci 20, 8965–8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Muzzio IA, Malleret G, Bartsch D, Verbitsky M, Pavlidis P, Yonan AL, Vronskaya S, Grody MB, Cepeda I, et al. (2003). Inducible enhancement of memory storage and synaptic plasticity in transgenic mice expressing an inhibitor of ATF4 (CREB-2). and C/EBP proteins. Neuron 39, 655–669. [DOI] [PubMed] [Google Scholar]
- Chrivia JC, Kwok RPS, Lamb N, Hagiwara M, Montminy MR, Goodman RH (1993). Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365, 855–859. [DOI] [PubMed] [Google Scholar]
- Condorelli DF, Albani PD, Amico C, Lukasiuk K, Kaczmarek L, Giuffrida-Stella AM (1994). Glutamate receptor-driven activation of transcription factors in primary neuronal cultures. Neurochem. Res 19, 489–499. [DOI] [PubMed] [Google Scholar]
- Conway AM, James AB, Zang J, and Morris BJ (2007). Regulation of neuronal cdc20 (p55cdc) expression by the plasticity-related transcription factor zif268. Synapse 61, 463–468. [DOI] [PubMed] [Google Scholar]
- Cruz FC, Javier Rubio F, and Hope BT (2015). Using c-fos to study neuronal ensembles in corticostriatal circuitry of addiction. Brain Res. 1628, 157–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash PK, Orsi SA, and Moore AN (2005). Sequestration of serum response factor in the hippocampus impairs long-term spatial memory. J. Neurochem 93, 269–278. [DOI] [PubMed] [Google Scholar]
- Davis S, Vanhoutte P, Pagès C, Caboche J, and Laroche S (2000). The MAPK/ERK cascade targets both Elk-1 and cAMP response element-binding protein to control long-term potentiation-dependent gene expression in the dentate gyrus in vivo. J. Neurosci 20, 4563–4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K, Bito H, and Tsien RW (1996). Signaling from synapse to nucleus: postsynaptic CREB phosphorylation during multiple forms of hippocampal synaptic plasticity. Neuron 16, 89–101. [DOI] [PubMed] [Google Scholar]
- Dobrazanski P, Noguchi T, Kovary K, Rizzo CA, Lazo PS, and Bravo R (1991). Both products of the fosB gene, FosB and its short form, FosB/SF, are transcriptional activators in fibroblasts. Mol. Cell Biol. 11, 5470–5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Green T, Saal D, Marie H, Neve R, Nestler EJ, and Malenka RC (2006). CREB modulates excitability of nucleus accumbens neurons. Nat. Neurosci 9, 475–477. [DOI] [PubMed] [Google Scholar]
- Dragunow M and Robertson HA (1987). Kindling stimulation induces c-fos protein(s) in granule cells of the rat dentate gyrus. Nature 329, 441–442. [DOI] [PubMed] [Google Scholar]
- Dragunow M, Abraham WC, Goulding M, Mason SE, Robertson HA, Faull RLM (1989). Long-term potentiation and the induction of c-fos mRNA and proteins in the dentate gyrus of unanesthetized rats. Neurosci. Lett 101, 274–280. [DOI] [PubMed] [Google Scholar]
- Eagle AL, Gajewski PA, Yang M, Kechner ME, Al Masraf BS, Kennedy PJ, Wang H, Mazei-Robison MS, and Robison AJ (2015). Experience-dependent induction of hippocampal ΔFosB controls learning. J. Neurosci 35, 13773–13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etkin A, Alarcón JM, Weisberg SP, Touzani K, Huang YY, Nordheim A, and Kandel ER (2006). A role in learning for SRF: deletion in the adult forebrain disrupts LTD and the formation of an immediate memory of a novel context. Neuron 50, 127–143. [DOI] [PubMed] [Google Scholar]
- Ferrara P, Andermarcher E, Bossis G, Acquaviva C, Brockly F, Jariel-Encontre I, and Piechaczyk M (2003). The structural determinants responsible for c-Fos protein proteasomal degradation differ according to the conditions of expression. Oncogene 22, 1461–1474. [DOI] [PubMed] [Google Scholar]
- Fleischmann A, Hvalby O, Jensen V, Strekalova T, Zacher C, Layer LE, Kvello A, Reschke M, Spanagel R, Sprengel R, et al. (2003). Impaired long-term memory and NR2A-type NMDA receptor-dependent synaptic plasticity in mice lacking c-Fos in the CNS. J. Neurosci 23, 9116–9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey U, Huang YY, and Kandel ER (1993). Effects of cAMP simulate a late stage of LTP in hippocampal CA1 neurons. Science 260, 1661–1664. [DOI] [PubMed] [Google Scholar]
- Garner AR, Rowland DC, Hwang SY, Baumgaertel K, Roth BL, Kentros C, and Mayford M (2012). Generation of a synthetic memory trace. Science 335, 1513–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gass P, Wolfer DP, Balschun D, Rudolph D, Frey U, Lipp H-P, and Schütz G (1998). Deficits in memory tasks of mice with CREB mutations depend on gene dosage. Learn Memory 5, 274–288. [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Ginty DD, Bading H, and Greenberg ME (1994). Calcium regulation of gene expression in neuronal cells. J. Neurobiol 25, 294–303. [DOI] [PubMed] [Google Scholar]
- Govindarajan A, Kelleher RJ, and Tonegawa S (2006). A clustered plasticity model of long-term memory engrams. Nat. Rev. Neurosci 7, 575–583. [DOI] [PubMed] [Google Scholar]
- Gray JD, Milner TA, and McEwen BS (2013). Dynamic plasticity: the role of glucocorticoids, brain-derived neurotrophic factor and other trophic factors. Neuroscience 239, 214–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriadis A, Schellander K, Wang Z, and Wagner E (1993). Osteoblasts are target cells for transformation in c-fos transgenic mice. J. Cell. Biol 122, 685–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm R, Schicknick H, Riede I, Gundelfinger ED, Herdegen T, Zuschratter W, and Tischmeyer W (1997). Suppression of c-fos induction in rat brain impairs retention of a brightness discrimination reaction. Learn Memory 3, 402–413. [DOI] [PubMed] [Google Scholar]
- Grueter BA, Robison AJ, Neve RL, Nestler EJ, and Malenka RC (2013). FosB differentially modulates nucleus accumbens direct and indirect pathway function. Proc. Natl. Acad. Sci. USA 110, 1923–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Giustetto M, Lomvardas S, Kim J-H, Miniaci MC, Schwartz JH, Thanos D, and Kandel ER (2002). Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell 111, 483–493. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Setlow B, Wagner EK, and McGaugh JL (2001). Experience-dependent gene expression in the rat hippocampus after spatial learning: a comparison of the immediate-early genes Arc, c-fos, and zif268. J. Neurosci 21, 5089–5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J-H, Kushner SA, Yiu AP, Cole CJ, Matynia A, Brown RA, Neve RL, Guzowski JF, Silva AJ, and Josselyn SA (2007). Neuronal competition and selection during memory formation. Science 316, 457–460. [DOI] [PubMed] [Google Scholar]
- Herdegen T, Kovary K, Buhl A, Bravo R, Zimmermann M, and Gass P (1995). Basal expression of the inducible transcription factors c-Jun, JunB, JunD, c-Fos, FosB, and Krox-24 in the adult rat brain. J. Compar. Neurol 354, 39–56. [DOI] [PubMed] [Google Scholar]
- Herdegen T, Blume A, Buschmann T, Georgakopoulos E, Winter C, Schmid W, Hsieh TF, Zimmermann M, and Gass P (1997). Expression of activating transcription factor-2, serum response factor and cAMP/Ca response element binding protein in the adult rat brain following generalized seizures, nerve fibre lesion and ultraviolet irradiation. Neuroscience 81, 199–212. [DOI] [PubMed] [Google Scholar]
- Hess U, Lynch G, and Gall C (1995). Changes in c-fos mRNA expression in rat brain during odor discrimination learning: differential involvement of hippocampal subfields CA1 and CA3. J. Neurosci 15, 4786–4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipskind RA, Roa VN, Muller CGF, Raddy ESP, and Nordheim A (1991). Ets-related protein Elk-1 is homologous to the c-fos regulatory factor p62TCF. Nature 354, 531–534. [DOI] [PubMed] [Google Scholar]
- Hope BT, Nye HE, Kelz MB, Self DW, Iadarola MJ, Nakabeppu Y, Duman RS, and Nestler EJ (1994). Induction of a long-lasting AP-1 complex composed of altered Fos-like proteins in brain by chronic cocaine and other chronic treatments. Neuron 13, 1235–1244. [DOI] [PubMed] [Google Scholar]
- Huber KM, Kayser MS, and Bear MF (2000). Role for rapid dendritic protein synthesis in hippocampal mGluR-dependent long-term depression. Science 288, 1254–1257. [DOI] [PubMed] [Google Scholar]
- James AB, Conway AM, and Morris BJ (2005). Genomic profiling of the neuronal target genes of the plasticity-related transcription factor – Zif268. J. Neurochem 95, 796–810. [DOI] [PubMed] [Google Scholar]
- Janknecht R and Nordheim A (1992). Elk-1 protein domains required for direct and SRF-assisted DNA-binding. Nucleic. Acids Res 20, 3317–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janknecht R and Nordheim A (1993). Gene regulation by Ets proteins. Biochim Biophys Acta 1155, 346–356. [DOI] [PubMed] [Google Scholar]
- Janknecht R and Nordheim A (1996). MAP kinase-dependent transcriptional coactivation by Elk-1 and its cofactor CBP. Biochem. Biophys Res. Commun 228, 831–837. [DOI] [PubMed] [Google Scholar]
- Jones MW, Errington ML, French PJ, Fine A, Bliss TVP, Garel S, Charnay P, Bozon B, Laroche S, and Davis S (2001). A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nat. Neurosci 4, 289–296. [DOI] [PubMed] [Google Scholar]
- Kaczmarek L (1993). Molecular biology of vertebrate learning: is c-fos a new beginning? J. Neurosci. Res 34, 377–381. [DOI] [PubMed] [Google Scholar]
- Kaczmarek L, Siedlecki JA, and Danysz W (1988). Proto-onco-gene c-fos induction in rat hippocampus. Mol. Brain Res 3, 183–186. [DOI] [PubMed] [Google Scholar]
- Kaltschmidt B, Ndiaye D, Korte M, Pothion S, Arbibe L, Prüllage M, Pfeiffer J, Lindecke A, Staiger V, Israël A, et al. (2006). NF-kB regulates spatial memory formation and synaptic plasticity through protein kinase A/CREB signaling. Mol. Cell Biol 26, 2936–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathirvelu B, East BS, Hill AR, Smith CA, and Colombo PJ (2013). Lentivirus-mediated chronic expression of dominant-negative CREB in the dorsal hippocampus impairs memory for place learning and contextual fear conditioning. Neurobiol. Learn Mem. 99, 10–16. [DOI] [PubMed] [Google Scholar]
- Kelz MB, Chen J, Carlezon WA, Whisler K, Gilden L, Beckmann AM, Steffen C, Zhang Y-J, Marotti L, Self DW, et al. (1999). Expression of the transcription factor ΔFosB in the brain controls sensitivity to cocaine. Nature 401, 272–276. [DOI] [PubMed] [Google Scholar]
- Kemp A, Tischmeyer W, and Manahan-Vaughan D (2013). Learning-facilitated long-term depression requires activation of the immediate early gene, c-fos, and is transcription dependent. Behav. Brain. Res 254, 83–91. [DOI] [PubMed] [Google Scholar]
- Kida S, Josselyn SA, de Ortiz SP, Kogan JH, Chevere I, Masushige S, and Silva AJ (2002). CREB required for the stability of new and reactivated fear memories. Nat. Neurosci 5, 348–355. [DOI] [PubMed] [Google Scholar]
- Kim JJ and Diamond DM (2002). The stressed hippocampus, synaptic plasticity and lost memories. Nat. Rev. Neurosci 3, 453–462. [DOI] [PubMed] [Google Scholar]
- Kim J, Kwon JT, Kim HS, Josselyn SA, and Han JH (2014). Memory recall and modifications by activating neurons with elevated CREB. Nat. Neurosci 17, 65–72. [DOI] [PubMed] [Google Scholar]
- Knapska E and Kaczmarek L (2004). A gene for neuronal plasticity in the mammalian brain: Zif268/Egr-1/NGFI-A/Krox-24/TIS8/ZENK? Prog. Neurobiol 74, 183–211. [DOI] [PubMed] [Google Scholar]
- Knöll B and Nordheim A (2009). Functional versatility of transcription factors in the nervous system: the SRF paradigm. Trends Neurosci. 32, 432–442. [DOI] [PubMed] [Google Scholar]
- Knöll B, Kretz O, Fiedler C, Alberti S, Schutz G, Frotscher M, Nordheim A (2006). Serum response factor controls neuronal circuit assembly in the hippocampus. Nat. Neurosci 9, 195–204. [DOI] [PubMed] [Google Scholar]
- Kovács KJ (1998). Invited review c-Fos as a transcription factor: a stressful (re)view from a functional map. Neurochem. Int 33, 287–297. [DOI] [PubMed] [Google Scholar]
- Kuo CF, Xanthopoulos KG, and Darnell JE Jr, (1990). Fetal and adult localization of C/EBP: evidence for combinatorial action of transcription factors in cell-specific gene expression. Development 109, 473–481. [DOI] [PubMed] [Google Scholar]
- Lakhina V, Arey Rachel N, Kaletsky R, Kauffman A, Stein G, Keyes W, Xu D, and Murphy Coleen T (2015). Genome-wide functional analysis of CREB/long-term memory-dependent transcription reveals distinct basal and memory gene expression programs. Neuron 85, 330–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latinkic BV, Zeremski M, and Lau LF (1996). Elk-1 can recruit SRF to form a ternary complex upon the serum response element. Nucleic. Acids Res. 24, 1345–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JLC, Everitt BJ, and Thomas KL (2004). Independent cellular processes for hippocampal memory consolidation and reconsolidation. Science 304, 839–843. [DOI] [PubMed] [Google Scholar]
- Li CL, Sathyamurthy A, Oldenborg A, Tank D, and Ramanan N (2014). SRF phosphorylation by glycogen synthase kinase-3 promotes axon growth in hippocampal neurons. J. Neurosci 34, 4027–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindecke A, Korte M, Zagrebelsky M, Horejschi V, Elvers M, Widera D, Prüllage M, Pfeiffer J, Kaltschmidt B, and Kaltschmidt C (2006). Long-term depression activates transcription of immediate early transcription factor genes: involvement of serum response factor/Elk-1. Eur. J. Neurosci 24, 555–563. [DOI] [PubMed] [Google Scholar]
- Liu X, Ramirez S, Pang PT, Puryear CB, Govindarajan A, Deisseroth K, and Tonegawa S (2012). Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature 484, 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Pasini S, Shelanski ML, and Greene LA (2014). Activating transcription factor 4 (ATF4). modulates post-synaptic development and dendritic spine morphology. Front Cell Neurosci. 8, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonze BE and Ginty DD (2002). Function and regulation of CREB family transcription factors in the nervous system. Neuron 35, 605–623. [DOI] [PubMed] [Google Scholar]
- Lopez de Armentia M, Jancic D, Olivares R, Alarcon JM, Kandel ER, Barco A (2007). cAMP response element-binding protein-mediated gene expression increases the intrinsic excitability of CA1 pyramidal neurons. J. Neurosci 27, 13909–13918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinow R, Schulman H, and Tsien R (1989). Inhibition of postsynaptic PKC or CaMKII blocks induction but not expression of LTP. Science 245, 862–866. [DOI] [PubMed] [Google Scholar]
- Manahan-Vaughan D, Kulla A, and Frey JU (2000). Requirement of translation but not transcription for the maintenance of long-term depression in the CA1 region of freely moving rats. J. Neurosci 20, 8572–8576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais R, Wynne J, and Treisman R (1993). The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell 73, 381–393. [DOI] [PubMed] [Google Scholar]
- Marie H, Morishita W, Yu X, Calakos N, and Malenka RC (2005). Generation of silent synapses by acute in vivo expression of CaMKIV and CREB. Neuron 45, 741–752. [DOI] [PubMed] [Google Scholar]
- Matsuo K, Owens JM, Tonko M, Elliott C, Chambers TJ, and Wagner EF (2000). Fosl1 is a transcriptional target of c-Fos during osteoclast differentiation. Nat. Genet 24, 184–187. [DOI] [PubMed] [Google Scholar]
- Matsuo K, Galson DL, Zhao C, Peng L, Laplace C, Wang KZQ, Bachler MA, Amano H, Aburatani H, Ishikawa H, et al. (2004). Nuclear factor of activated T-cells (NFAT). rescues osteoclastogenesis in precursors lacking c-Fos. J. Biol. Chem 279, 26475–26480. [DOI] [PubMed] [Google Scholar]
- Matthews CP, Colburn NH, and Young MR (2007). AP-1 a target for cancer prevention. Curr Cancer Drug Targets 7, 317–324. [DOI] [PubMed] [Google Scholar]
- Mayr B and Montminy M (2001). Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2, 599–609. [DOI] [PubMed] [Google Scholar]
- McGahan L, Hakim AM, Nakabeppu Y, and Robertson GS (1998). Ischemia-induced CA1 neuronal death is preceded by elevated FosB and Jun expression and reduced NGFI-A and JunB levels. Mol. Brain Res, 56, 146–161. [DOI] [PubMed] [Google Scholar]
- McGaugh JL (2000). Memory – a century of consolidation. Science 287, 248–251. [DOI] [PubMed] [Google Scholar]
- Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS, and Baltimore D (2003). NF-kB functions in synaptic signaling and behavior. Nat. Neurosci, 6, 1072–1078. [DOI] [PubMed] [Google Scholar]
- Miano JM, Long X, and Fujiwara K (2007). Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am. J. Physiol. Cell Physiol 292, C70–C81. [DOI] [PubMed] [Google Scholar]
- Minatohara K, Akiyoshi M, and Okuno H (2015). Role of immediate–early genes in synaptic plasticity and neuronal ensembles underlying the memory trace. Front Mol. Neurosci 8, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranti CK, Ginty DD, Huang G, Chatila T, and Greenberg ME (1995). Calcium activates serum response factor-dependent transcription by a Ras- and Elk-1-independent mechanism that involves a Ca2+/calmodulin-dependent kinase. Mol. Cell Biol. 15, 3672–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno M, Yamada K, Maekawa N, Saito K, Seishima M, and Nabeshima T (2002). CREB phosphorylation as a molecular marker of memory processing in the hippocampus for spatial learning. Behav. Brain Res. 133, 135–141. [DOI] [PubMed] [Google Scholar]
- Mo J, Kim C-H, Lee D, Sun W, Lee HW, and Kim H (2015). Early growth response 1 (Egr-1). directly regulates GABAA receptor a2, a4, and θ subunits in the hippocampus. J. Neuro-chem 133, 489–500. [DOI] [PubMed] [Google Scholar]
- Montminy MR, Bilezikjian LM (1987). Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature 328, 175–178. [DOI] [PubMed] [Google Scholar]
- Mouravlev A, Young D, and During MJ (2007). Phosphorylation-dependent degradation of transgenic CREB protein initiated by heterodimerization. Brain Res. 1130, 31–37. [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y and Nathans D (1991). A naturally occurring truncated form of FosB that inhibits Fos/Jun transcriptional activity. Cell 64, 751–759. [DOI] [PubMed] [Google Scholar]
- Nestler EJ (2015). ΔFosB: a transcriptional regulator of stress and antidepressant responses. Eur. J. Pharmacol 753, 66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ, Kelz MB, and Chen J (1999). ΔFosB: a molecular mediator of long-term neural and behavioral plasticity. Brain Res. 835, 10–17. [DOI] [PubMed] [Google Scholar]
- Neves G, Cooke SF, and Bliss TVP (2008). Synaptic plasticity, memory and the hippocampus: a neural network approach to causality. Nat. Rev. Neurosci 9, 65–75. [DOI] [PubMed] [Google Scholar]
- Nguyen PV, Abel T, and Kandel ER (1994). Requirement of a critical period of transcription for induction of a late phase of LTP. Science 265, 1104–1107. [DOI] [PubMed] [Google Scholar]
- Nikolaev E, Kaminska B, Tischmeyer W, Matthies H, and Kaczmarek L (1992). Induction of expression of genes encoding transcription factors in the rat brain elicited by behavioral training. Brain Res. Bull 28, 479–484. [DOI] [PubMed] [Google Scholar]
- Pasini S, Corona C, Liu J, Greene Lloyd A, and Shelanski Michael L (2015). Specific downregulation of hippocampal ATF4 reveals a necessary role in synaptic plasticity and memory. Cell Rep. 11, 183–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penke Z, Chagneau C, and Laroche S (2011). Contribution of Egr1/zif268 to activity-dependent Arc/Arg3.1 transcription in the dentate gyrus and area CA1 of the hippocampus. Front Behav. Neurosci, 5, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penke Z, Morice E, Veyrac A, Gros A, Chagneau C, LeBlanc P, Samson N, Baumgärtel K, Mansuy IM, Davis S, et al. (2014). Zif268/Egr1 gain of function facilitates hippocampal synaptic plasticity and long-term spatial recognition memory. Phil. Trans. R Soc. Lond B: Biol. Sci 369, 20130159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotti LI, Hadeishi Y, Ulery PG, Barrot M, Monteggia L, Duman RS, Nestler EJ (2004). Induction of ΔFosB in reward-related brain structures after chronic stress. J. Neurosci 24, 10594–10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotti LI, Weaver RR, Robison B, Renthal W, Maze I, Yazdani S, Elmore RG, Knapp DJ, Selley DE, Martin BR, et al. (2008). Distinct patterns of ΔFosB induction in brain by drugs of abuse. Synapse 62, 358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittenger C, Huang YY, Paletzki RF, Bourtchouladze R, Scanlin H, Vronskaya S, and Kandel ER (2002). Reversible inhibition of CREB/ATF transcription factors in region CA1 of the dorsal hippocampus disrupts hippocampus-dependent spatial memory. Neuron 34, 447–462. [DOI] [PubMed] [Google Scholar]
- Qin X, Jiang Y, Tse YC, Wang Y, Wong TP, and Paudel HK (2015). Early growth response 1 (Egr-1). regulates N-Methyl-d-aspartate receptor (NMDAR)-dependent transcription of PSD-95 and a-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor (AMPAR) trafficking in hippocampal primary neurons. J. Biol. Chem 290, 29603–29616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan N, Shen Y, Sarsfield S, Lemberger T, Schutz G, Linden DJ, and Ginty DD (2005). SRF mediates activity-induced gene expression and synaptic plasticity but not neuronal viability. Nat. Neurosci 8, 759–767. [DOI] [PubMed] [Google Scholar]
- Rao V, Huebner K, Isobe M, ar-Rushdi A, Croce C, and Reddy E (1989). elk, tissue-specific ets-related genes on chromo-somes X and 14 near translocation breakpoints. Science 244, 66–70. [DOI] [PubMed] [Google Scholar]
- Rashid AJ, Cole CJ, and Josselyn SA (2014). Emerging roles for MEF2 transcription factors in memory. Genes. Brain. Behav, 13, 118–125. [DOI] [PubMed] [Google Scholar]
- Renaudineau S, Poucet B, Laroche S, Davis S, and Save E (2009). Impaired long-term stability of CA1 place cell representation in mice lacking the transcription factor zif268/egr1. Proc. Natl. Acad. Sci. USA 106, 11771–11775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Carle TL, Maze I, Covington HE, Truong H-T, Alibhai I, Kumar A, Montgomery RL, Olson EN, and Nestler EJ (2008). ΔFosB mediates epigenetic desensitization of the c-fos gene after chronic amphetamine exposure. J. Neurosci 28, 7344–7349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restivo L, Tafi E, Ammassari-Teule M, and Marie H (2009). Viral-mediated expression of a constitutively active form of CREB in hippocampal neurons increases memory. Hippocampus 19, 228–234. [DOI] [PubMed] [Google Scholar]
- Robison AJ and Nestler EJ (2011). Transcriptional and epigenetic mechanisms of addiction. Nat. Rev. Neurosci 12, 623–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison AJ, Vialou V, Mazei-Robison M, Feng J, Kourrich S, Collins M, Wee S, Koob G, Turecki G, Neve R, et al. (2013). Behavioral and structural responses to chronic cocaine require a feedforward loop involving ΔFosB and calcium/calm-odulin-dependent protein kinase II in the nucleus accumbens shell. J. Neurosci 33, 4295–4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison AJ, Vialou V, Sun HS, Labonte B, Golden S, Dias C, Turecki G, Tamminga C, Russo S, Mazei-Robison M, et al. (2014). Fluoxetine epigenetically alters the CaMKIIa promoter in nucleus accumbens to regulate ΔFosB binding and antidepressant effects. Neuropsychopharmacology 39, 1178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar S, Sharp F, and Curran T (1988). Expression of c-fos protein in brain: metabolic mapping at the cellular level. Science 240, 1328–1331. [DOI] [PubMed] [Google Scholar]
- Sananbenesi F, Fischer A, Schrick C, Spiess J, and Radulovic J (2002). Phosphorylation of hippocampal Erk-1/2, Elk-1, and p90-Rsk-1 during contextual fear conditioning: interactions between Erk-1/2 and Elk-1. Mol. Cell Neurosci 21, 463–476. [DOI] [PubMed] [Google Scholar]
- Sealy L, Malone D, and Pawlak M (1997). Regulation of the cfos serum response element by C/EBPb. Mol. Cell Biol. 17, 1744–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgambato V, Vanhoutte P, Pagès C, Rogard M, Hipskind R, Besson M-J, Caboche J (1998). In vivo expression and regulation of Elk-1, a target of the extracellular-regulated kinase signaling pathway, in the adult rat brain. J. Neurosci 18, 214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M and Greenberg ME (1990). The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron 4, 477–485. [DOI] [PubMed] [Google Scholar]
- Sheng M, McFadden G, and Greenberg ME (1990). Membrane depolarization and calcium induce c-fos transcription via phosphorylation of transcription factor CREB. Neuron 4, 571–582. [DOI] [PubMed] [Google Scholar]
- Shore P and Sharrocks AD (1995). The MADS-box family of transcription factors. Eur. J. Biochem 229, 1–13. [DOI] [PubMed] [Google Scholar]
- Silva AJ, Zhou Y, Rogerson T, Shobe J, and Balaji J (2009). Molecular and cellular approaches to memory allocation in neural circuits. Science 326, 391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer JL, Belaguli NS, Iyer D, Schwartz RJ, and Balasubramanyam A (2002). Developmental expression of serum response factor in the rat central nervous system. Dev. Brain Res 138, 81–86. [DOI] [PubMed] [Google Scholar]
- Stritt C and Knöll B (2010). Serum response factor regulates hippocampal lamination and dendrite development and Is connected with reelin signaling. Mol. Cell Biol 30, 1828–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stritt C, Stern S, Harting K, Manke T, Sinske D, Schwarz H, Vingron M, Nordheim A, and Knoll B (2009). Paracrine control of oligodendrocyte differentiation by SRF-directed neuronal gene expression. Nat. Neurosci 12, 418–427. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Fukushima H, Mukawa T, Toyoda H, Wu L-J, Zhao M-G, Xu H, Shang Y, Endoh K, Iwamoto T, et al. (2011). Upregulation of CREB-mediated transcription enhances both short- and long-term memory. J. Neurosci 31, 8786–8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt JD (2004). Mitogen-activated protein kinases in synaptic plasticity and memory. Curr. Opin. Neurobiol 14, 311–317. [DOI] [PubMed] [Google Scholar]
- Tao X, Finkbeiner S, Arnold DB, Shaywitz AJ, and Greenberg ME (1998). Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron 20, 709–726. [DOI] [PubMed] [Google Scholar]
- Taubenfeld SM, Milekic MH, Monti B, and Alberini CM (2001a). The consolidation of new but not reactivated memory requires hippocampal C/EBPb. Nat. Neurosci 4, 813–818. [DOI] [PubMed] [Google Scholar]
- Taubenfeld SM, Wiig KA, Monti B, Dolan B, Pollonini G, and Alberini CM (2001b). Fornix-dependent induction of hippocampal CCAAT enhancer-binding protein β and δ co-localizes with phosphorylated cAMP response element-binding protein and accompanies long-term memory consolidation. J. Neurosci 21, 84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiels E, Kanterewicz BI, Norman ED, Trzaskos JM, and Klann E (2002). Long-term depression in the adult hippocampus in vivo involves activation of extracellular signal-regulated kinase and phosphorylation of Elk-1. J. Neurosci 22, 2054–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischmeyer W, Kaczmarek L, Strauss M, Jork R, and Matthies H (1990). Accumulation of c-fos mRNA in rat hippocampus during acquisition of a brightness discrimination. Behav. Neural. Biol 54, 165–171. [DOI] [PubMed] [Google Scholar]
- Treisman R (1987). Identification and purification of a polypeptide that binds to the c-fos serum response element. EMBO J. 6, 2711–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulery PG, Rudenko G, and Nestler EJ (2006). Regulation of ΔFosB stability by phosphorylation. J. Neurosci 26, 5131–5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulery-Reynolds PG, Castillo MA, Vialou V, Russo SJ, and Nestler EJ (2009). Phosphorylation of ΔFosB mediates its stability in vivo. Neuroscience 158, 369–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhoutte P, Barnier J-V, Guibert B, Pagès C, Besson M-J, Hipskind RA, and Caboche J (1999). Glutamate induces phosphorylation of Elk-1 and CREB, along with c-fos activation, via an extracellular signal-regulated kinase-dependent pathway in brain slices. Mol. Cell Biol, 19, 136–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrac A, Besnard A, Caboche J, Davis S, and Laroche S (2014). The transcription factor Zif268/Egr1, brain plasticity, and memory In: Molecular Basis of Memory: Progress in Molecular Biology and Translational Science. (New York, USA: Academic Press; ), pp. 89–129. [DOI] [PubMed] [Google Scholar]
- Vialou V, Robison AJ, Laplant QC, Covington HE 3rd, Dietz DM, Ohnishi YN, Mouzon E, Rush AJ 3rd, Watts EL, Wallace DL, et al. (2010). DeltaFosB in brain reward circuits mediates resilience to stress and antidepressant responses. Nat. Neurosci 13, 745–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vialou V, Feng J, Robison AJ, Ku SM, Ferguson D, Scobie KN, Mazei-Robison MS, Mouzon E, Nestler EJ (2012). Serum response factor and cAMP response element binding protein are both required for cocaine induction of ΔFosB. J. Neurosci 32, 7577–7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vialou V, Thibault M, Kaska S, Cooper S, Gajewski P, Eagle A, Mazei-Robison M, Nestler EJ, and Robison AJ (2015). Differential induction of FosB isoforms throughout the brain by fluoxetine and chronic stress. Neuropharmacology 99, 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AE and Greenberg ME (2011). Neuronal activity–regulated gene transcription in synapse development and cognitive function. Cold Spring Harbor Perspect. Biol 3, a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DG, Groth RD, Ma H, Barrett CF, Owen SF, Safa P, and Tsien RW (2012). Ca(V)1 and Ca(V)2 channels engage distinct modes of Ca2+ signaling to control CREB-dependent gene expression. Cell 149, 1112–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisden W, Errington ML, Williams S, Dunnett SB, Waters C, Hitchcock D, Evan G, Bliss TVP, and Hunt SP (1990). Differential expression of immediate early genes in the hippocampus and spinal cord. Neuron 4, 603–614. [DOI] [PubMed] [Google Scholar]
- Wood MA, Kaplan MP, Park A, Blanchard EJ, Oliveira AMM, Lombardi TL, and Abel T (2005). Transgenic mice expressing a truncated form of CREB-binding protein (CBP) exhibit deficits in hippocampal synaptic plasticity and memory storage. Learn Memory 12, 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MA, Attner MA, Oliveira AMM, Brindle PK, and Abel T (2006). A transcription factor-binding domain of the coactivator CBP is essential for long-term memory and the expression of specific target genes. Learn Memory 13, 609–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley PF, Bhat RV, Baraban JM, Erickson CA, McNaughton BL, and Barnes CA (1993). Thresholds for synaptic activation of transcription factors in hippocampus: correlation with long-term enhancement. J. Neurosci 13, 4776–4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z, Dudek H, Miranti CK, Greenberg ME (1996). Calcium influx via the NMDA receptor induces immediate early gene transcription by a MAP kinase/ERK-dependent mechanism. J. Neurosci 16, 5425–5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin JCP, Wallach JS, Del Vecchio M, Wilder EL, Zhou H, Quinn WG, and Tully T (1994). Induction of a dominant negative CREB transgene specifically blocks long-term memory in Drosophila. Cell 79, 49–58. [DOI] [PubMed] [Google Scholar]
- Ying SW, Futter M, Rosenblum K, Webber MJ, Hunt SP, Bliss TV, and Bramham CR (2002). Brain-derived neurotrophic factor induces long-term potentiation in intact adult hippocampus: requirement for ERK activation coupled to CREB and upregulation of Arc synthesis. J. Neurosci 22, 1532–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiu AP, Mercaldo V, Yan C, Richards B, Rashid AJ, Hsiang HL, Pressey J, Mahadevan V, Tran MM, Kushner SA, et al. (2014). Neurons are recruited to a memory trace based on relative neuronal excitability immediately before training. Neuron 83, 722–735. [DOI] [PubMed] [Google Scholar]
- Yutsudo N, Kamada T, Kajitani K, Nomaru H, Katogi A, Ohnishi YH, Ohnishi YN, Takase K. i., Sakumi K, Shigeto H, et al. (2013). fosB-null mice display impaired adult hippocampal neurogenesis and spontaneous epilepsy with depressive behavior. Neuropsychopharmacology 38, 895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]