Peptide arrays are an exciting and rapidly growing technology with a broad range of applications in the basic and applied life sciences. Peptide arrays have been under development for approximately 25 years, and commercial versions have been available for the past decade. The arrays typically comprise hundreds to thousands of distinct peptide sequences and have proven important for determining substrate specificities of enzymes, profiling antibodies, mapping epitopes, studying ligand–receptor interactions, and identifying ligands that mediate cell adhesion. Despite this, peptide arrays still find modest use, particularly when compared to oligonucleotide arrays, and have not yet realized their potential in becoming a standard method in laboratories and in the drug discovery process. In this review, we provide a discussion of the many approaches that have been developed to prepare and apply peptide arrays, including important advances in the last 3 years and remaining challenges to making these tools broadly useful.
Biomolecular arrays are planar substrates having large numbers of molecules immobilized in patterns, such that each region of the surface presents one specific molecule. The majority of work has been directed toward arrays prepared from either oligonucleotides, peptides, proteins, or small molecules. Of these, oligonucleotide arrays are the most developed; they are commercially available and widely used. Oligonucleotide array development began in the early 1990s through efforts led by Patrick Brown, Stephen Fodor, and Edwin Southern.1–4 These researchers used different approaches, based either on the immobilization of presynthesized DNA or the in situ synthesis of DNA directly on the substrate, and by 1995 were using the arrays to profile gene expression.4 In 1996, DNA arrays were used to profile the expression of as many as 1000 genes5 and soon after, the first whole-genome microarray of yeast was reported6 and a new high-density random array was presented.7 Now, 25 years later, commercially available arrays contain several million oligonucleotides and are routine tools in the laboratory.8,9 Oligonucleotide arrays have been used in clinical applications to better understand viruses and human disease, for genetic screening, and for the implementation of personalized medicine.10,11 The rapid pace of technical advancement of the early arrays, which performed rather poorly, has led to the robust and inexpensive arrays currently in use. By analogy, peptide arrays are still at the early stage of development and can be expected to see improvements that result in an important, if somewhat less ubiquitous, technology for the life sciences.
The development of peptide, protein, and carbohydrate arrays has been motivated in part by the incomplete knowledge provided by genomic information alone. This is because gene expression and mRNA levels do not correlate with protein activity, while several additional factors, including post-translation modifications, alternative splicing, allosteric ligands, colocalization, and degradation, serve to regulate the activities of proteins. A systematic understanding of the roles of the approximately 20 000 genes12 in the human genome, which code for ~100 000 transcriptomes13 and give rise to the expression of more than 1 million proteoforms,14,15 requires systems level information on proteomic activities. Peptide arrays offer one route to this end.
Early work by the Schreiber16 and Snyder17 groups has demonstrated protein arrays and their applications to identify protein–protein interactions, substrates for enzymes, and protein targets of small molecules, but in practice protein arrays have not yet had a broader impact. This can be attributed to the challenges inherent in making protein arrays, which includes expressing and purifying large numbers of proteins, immobilizing proteins with control over the orientation, increasing feature density, and maintaining the activities of the immobilized proteins (and preventing their denaturation at the interface). These challenges are avoided when working with arrays of peptides, which are relatively easy to synthesize, stable, and compatible with many immobilization chemistries. While many protein functions cannot be recapitulated at the peptide level due to lack of tertiary structure and sequence truncation, the use of peptides is still appropriate for many applications.
In comparison to DNA arrays, peptide arrays are more challenging to develop. First, the assays performed on DNA arrays are essentially all based on hybridization with fluorescent probes, whereas peptide arrays are used in a wide variety of assay and detection formats. Peptide arrays have been used to determine binding, enzyme activity, and cell adhesion and can be analyzed using fluorescence imaging, surface plasmon resonance spectroscopy, and mass spectrometry. These many applications make it difficult to identify materials and surface chemistries that are optimized for this broad range of applications. Moreover, peptide arrays are often used to assay samples that have high concentrations of protein, and nonspecific adsorption of proteins often leads to false positive and negative results. The use of “inert” surface chemistries can control these unwanted interactions but are still uncommon in peptide arrays. Further, peptides are more heterogeneous in their chemistry than are oligonucleotides. Peptides have greater functional group diversity in side chains, are synthesized in slightly lower yield than oligonucleotides, and have a wider distribution in properties including solubility, stability, and aggregation. Despite the impressive progress made in peptide array research and commercialization over the past 20 years, development has lagged behind that of DNA arrays but is expected to increase.
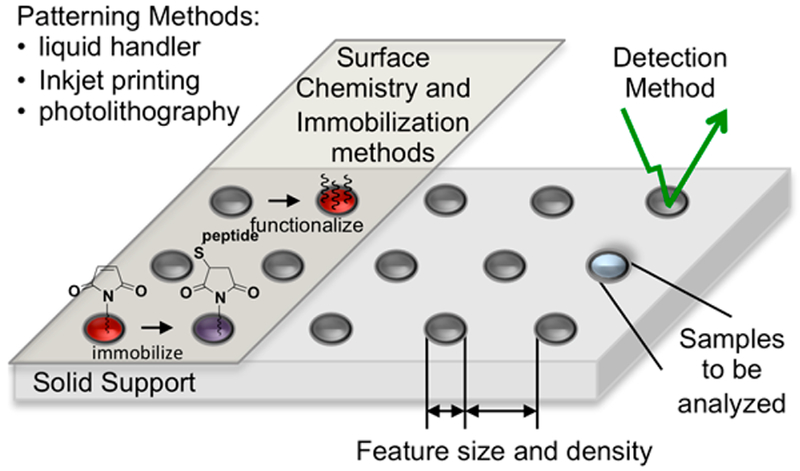
Several technical considerations are important in preparing peptide arrays, and among the various methods that have been reported, there are substantial differences in approach. We organize this review to address each of these technical areas, which include solid supports, surface and immobilization chemistries, patterning methods, and detection methods (Figure 1). We also describe the applications that have been addressed with peptide arrays and include a discussion of the currently available commercial arrays.18–23 Finally, we identify current limitations of peptide arrays including high costs of reagents, compatibility with complex samples, and high frequencies of false positive and negative data stemming from inconsistencies in immobilization methods, surface densities, and nonspecific interactions.
Figure 1.
Technical components of peptide arrays.
For our purposes, we consider arrays to have a minimum of 100 distinct peptide sequences, and we mostly do not address important work that has used lower density arrays. We also direct the reader to several previous reviews that describe applications of peptide arrays22,24–28 as well as the important approaches that have used solid-phase synthesis to prepare the arrays.29–34 Our goal is to add to this prior work by giving a technical description that applies to the many types of peptide arrays and to organize the current approaches, the commercialization, and applications of peptide arrays.
Finally, we do not include a discussion of the related protein chips,16,17,35,36 phage display array,37 or solution phase methods for peptide libraries,38–41 and instead give references where these methods are described.
TECHNICAL FACTORS
Peptide arrays are composed of a large number of peptides spatially arranged in an addressable format on a solid support. The array format has the benefit of allowing many experiments to be performed on a single sample. The specific applications will depend on the number of peptides in the array, the compatibility of the array material with samples (for example, those that are not inert to protein adsorption), the control over peptide attachment and density, and the compatibility with different detection methods. The reported approaches vary in their use of the different strategies, including the choice of peptide synthesis methodology, solid support and functionalization, immobilization method, patterning strategy, and detection method. We next describe the recent developments for each technical factor and how the approaches compare and offer our perspective on the advantages and limitations.
Peptide Synthesis
There are two widely used methods for synthesizing peptides present in arrays: Merrifield solid-phase synthesis and in situ synthesis. Merrifield solid-phase peptide synthesis (SPPS) is the traditional method for synthesizing peptides, assembling the peptides on a solid support, deprotection of the side chains, cleavage from the support, purification if necessary, and finally immobilization to the array support.42 Peptides prepared in this way are generally of high quality, having fewer impurities resulting from incomplete synthesis. This benefit of SPPS, however, comes with substantial expense and time associated with the synthesis of hundreds or thousands of distinct sequences.
This drawback in part motivated the development of in situ peptide synthesis, which now uses automated instrumentation for parallel synthesis of the peptides directly on the solid support of the array.21,43 In situ peptide synthesis has the benefits that it uses minimal amounts of reagents and necessarily eliminates the need (or possibility) for peptide purification. This yields substantial benefits in cost and time needed to prepare the arrays. This approach, however, makes it difficult to verify the purity and quality of peptides in the arrays.
The first important reports of the parallel, in situ synthesis of hundreds of peptides in an array format were introduced in the mid-1980s by Mario Geysen21 and Richard Houghten.43 Peptides were synthesized on polyethylene/poly(acrylic acid) solid supports and were used to identify viral antigens for antibody binding using the ELISA assay.21,43 The in situ method is the basis for three significant approaches to peptide arrays: SPOT, particle-based synthesis, and photolithograpic methods.
The SPOT method was introduced by Ronald Frank and utilizes Fmoc-protected amino acids to synthesize peptides in parallel directly on a membrane support.32 Solutions containing the amino acids and coupling reagents are dispensed onto specific locations of the membrane. After the coupling reaction have occurred, the entire membrane is washed and treated to remove the terminal amino protecting groups and therefore allow the next set of amino acid reagents to be dispensed onto the membrane for coupling. Many iterative cycles of coupling and washing are required to complete the synthesis of the peptide array. In the early version of this method, the amino acid solutions were manually pipetted onto the membranes, but more recent embodiments use automated liquid handling systems to replace the laborious preparations. A very significant benefit of this method lies in the minimized use of reagents, giving substantially lower costs than when peptides are synthesized prior to immobilization. This benefit and the ability to rapidly prepare custom arrays made SPOT synthesis a widely used technique.
However, the SPOT method has a few disadvantages. First, the porous membranes that were initially used led to diffusion of the amino acid solutions and therefore limited the density of spots that could be prepared; in practice, the region for each peptide had a minimal size of approximately 1 mm in diameter. An improvement to the SPOT methodology was introduced that increased array density by isolating the synthesized peptides and respotting on glass or other supports, though this format requires additional steps.44 The cellulose membrane that commonly serves as the support has the additional limitation that it is not inert to nonspecific protein adsorption. Hence, nonspecific adsorption of proteins in the assay sample often leads to a decrease in quantitative resolution and can be incompatible with the analysis of complex samples such as cell lysates. Finally, the cellulose support makes the SPOT peptide arrays incompatible with certain detection methods, including SPR and mass spectrometry.
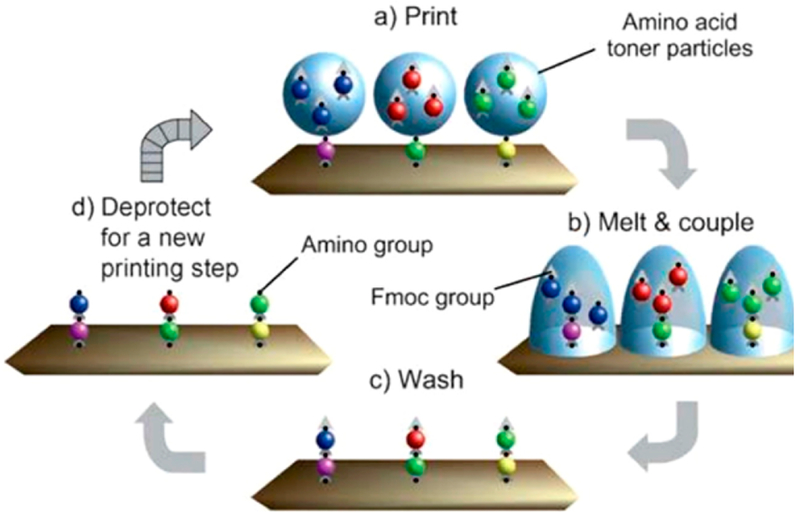
Particle-based synthesis was introduced by Stadler and co-workers in 2007 and, like the SPOT method, delivers Fmoc-protected amino acids to regions of a support to direct the in situ synthesis of peptides.45 However, instead of depositing solutions of reagents with a liquid handler, this method uses a 24-ink laser printer to transfer toner particles having Fmoc protected amino acids in a solid form. After transfer to the solid support, the particles are melted, which allows the coupling reactions to occur. Following coupling, the surface is washed of excess reagents, and the amino acids are deprotected to allow a next round of coupling reactions. This process is repeated until the desired peptide sequence is complete (Figure 2).45–47 This strategy harnesses the impressive laser printing technology, which avoids diffusion of the reagents and has the important benefit of giving smaller spot sizes and higher density arrays. The other steps in peptide synthesis, including washing and deprotection, are similar to those for the SPOT method, as are the benefits and limitations of the particle-based method.
Figure 2.
General methodology for particle-based synthesis. Amino acid toner particles are printed onto an electrostatically charged surface. Once printed, the toner particles are melted onto the surface to allow coupling. The surface is washed, and the coupled Fmoc-protected amino acids are deprotected.46 Reproduced from Combinatorial Synthesis of Peptide Arrays with a Laser Printer, Stadler, V.; Felgenhauer, T.; Beyer, M.; Fernandez, S.; Leibe, K.; Guttler, S.; Gröning, M.; König, K.; Torralba, G.; Hausmann, M.; Lindenstruth, V.; Nesterov, A.; Block, I.; Pipkorn, R.; Poustka, A.; Bischoff, F. R.; Breitling, F. Angewandte Chemie International Edition, Vol. 47, Issue 37 (ref 46). Copyright 2008 Wiley.
The photolithographic method uses light to direct peptide synthesis on a solid support (typically glass) and was first implemented by Fodor and co-workers in 1991.3 In the photolithographic approach, the amino acid reagents are protected with a photolabile group, for example, the nitroveratryloxycarbonyl (NVOC) group, such that the next amino acid can be incorporated into a growing peptide only if the peptide is first irradiated to remove the protecting group. The original method used a set of photolithographic masks to direct light to a subset of features that each required the addition of the same amino acid in the synthesis. Fodor and co-workers introduced this method by preparing an array of 1024 peptides in only 10 steps (though this example used a subset of the amino acids).3 Photolithographic synthesis can be fully automated, and because the synthesis area is defined by the area that is illuminated, this method can prepare features that are small (on the order of 10 μm48) and therefore generate very high-density arrays. The significant drawbacks are the need for expensive mask sets and many cycles of synthesis, since only one of the 20 amino acids can be added in each round.
A more recent improvement to the photolithographic method introduced a maskless format and combines acid-labile Boc protected amino acids with photogenerated acid (PGA) precursors and digital photolithography. Solid supports are again functionalized with tert-butyloxycarbonyl (Boc)-protected amino groups. Peptide reaction sites are separated by a physical barrier, such as a hydrophobic fluorocarbon alkyl monolyer film,49 and are filled with a PGA. Digital light patterns are projected on the surface, and the photogenerated acid removes the Boc-protecting group.50 The main disadvantage to using a PGA precursor for this method is the need for physical barriers to isolate the solutions in contact with each peptide. However, the photolithographic methods have seen significant development and can now be used to synthesize ~10 million peptides on a single slide.48 One drawback that has limited the broader use of these methods stems from the need for specialized equipment, light sources, masks, and optics.
Solid Support and Surface Functionalization
The choice of material used as the solid support can be a critical determinant in how well the array performs, yet in many cases the substrate materials have not been optimized for their uses. For example, the availability of well-defined surface chemistries can lead to better control over the density of peptide so that measured activities can more directly be compared for different peptides in the array. Additionally, well-defined surface chemistries can ensure a regular microenvironment, which gives a more uniform activity for each peptide than is possible with polymeric or hydrogel layers. The solid support is also an important choice when considering the use of different detection methods that have special requirements for the properties of the support. For instance, surface plasmon resonance spectroscopy requires a noble metal film of a specific thickness and mass spectrometry requires a conductive support.
Polymeric materials are often used as the solid support for peptide arrays. In the pioneering work by Geysen and colleagues, peptide libraries were synthesized directly on a polyacrylate grafted polyethylene rods arranged in a microtiter plate array.21 Chemical modification of the polymer supports installs functional groups that can be used for either in situ synthesis or for immobilization of peptides. For example, as demonstrated in Merrifield’s early work, polystyrene can be chloromethylated and then subsequently reacted with an amino acid or peptide to form a benzyl ester.42 Another example is the use of a polymethylpentene support that is first oxidized with nitric acid to generate carboxylate groups that can then react with an amino group through amide coupling.51 Functional groups can be installed in the grafting process for subsequent immobilization. For example, Geysen and co-workers grafted acrylic acid onto polyethylene and utilized the resulting carboxylate for amide coupling.21 Similary, Dikmans and colleagues grafted amino poly(ethylene glycol) (PEG) functionality into a polypropylene support which allowed subsequent coupling of peptides onto the support.52
The majority of peptide arrays prepared by SPOT synthesis30 use a nitrocellulose or polyvinylidene difluoride (PVDF) membrane as the solid support. When SPOT arrays are prepared on cellulose membranes, the hydroxyl groups are converted into more reactive functional groups for in situ peptide synthesis or chemical immobilization of peptides.
Glass and other oxide-terminated materials are commonly used as the support and benefit from the availability of alkylsiloxane monolayer surface chemistries. Aminoalkoxysilanes and glycidyl alkoxysilanes, for example, are commonly used for functionalizing glass or silicon surfaces to anchor peptides through amide coupling3 or epoxide ring opening,17,53 respectively. Glass substrates that are modified with a layer of streptavidin54 or aldehyde groups55 are commercially available and find wide use. These functional handles allow the peptides to be immobilized through chemical reactions directly or through subsequent treatment with homo- or heterobifunctional linkers. Polylysine coated glass slides can be used as solid supports for immobilizing peptides and proteins through electrostatic interactions56 or through coupling with the lysine ε-amino groups.51
With all supports, it is important to recognize that the immobilized peptides may not be active for interaction with enzymes or proteins used in the assay. For example, steric interactions between the protein and the support may prevent access of the binding site on a protein with the immobilized peptide. To address this issue, spacer molecules can be introduced to extend the peptides further from the solid support and increase accessibility. Often, additional small amino acids such as glycine or alanine can be incorporated in the N- or C-terminus of the peptides to serve as a spacer.30,57 Likewise, a PEG linker54 or dextran coatings58 can be added to separate the peptide from the surface.
Many researchers prefer the use of cross-linked three-dimensional hydrogels as the surface functionalization because these materials can incorporate much higher densities of peptide per unit area.59,60 Printing peptides onto polyacrylamide or agarose gels also distances the peptide from the solid support, potentially reducing the steric limitations between peptides and analytes. The hydrophilic nature of these gels more closely mimics the biological conditions for the analyte–peptide interaction. However, it is likely that the peptides present within the gel are not all equal in their activity; because of a range of microenvironments, some will not be functionally available or will incur additional energetic costs for binding. Finally, the slower diffusion of macromolecules in the hydrogels can lead to perturbations in the kinetics of interaction and give skewed results.61
An often overlooked problem with the surfaces used in peptide arrays is that most proteins will adsorb to them nonspecifically. In fact, most proteins will adsorb rapidly and strongly to most artificial surfaces. The adsorbed proteins can then contribute to false positive results or obstruct interactions of the immobilized peptide and give false negative results. A common strategy to address this issue is to use blocking proteins that adsorb to the solid support in order to prevent further adsorption of proteins from the sample. However, blocking proteins can likewise obstruct interactions with the immobilized peptides to give false-negative results. A more effective solution is to use surfaces that are functionalized to prevent the nonspecific adsorption of protein. Among the most effective options are self-assembled monolayers (SAMs) terminated in oligo(ethylene glycol) groups.62,63 These surfaces have been important in realizing quantitative assays and are significant in allowing the use of otherwise challenging samples, including complex cell lysates and tissue extracts.18 Polyethylene glycol) methacrylate (PEGMA)-grafted glass slides have also been used to minimize the nonspecific adsorptions of proteins.64 While not yet applied to peptide arrays, Jiang and co-workers have developed several zwitterionic polymer biomaterials that prevent nonspecific protein adsorption. In 2009, they introduced poly(carboxybetaine acrylamide)-(polyCBAA)-grafted surfaces that prevent nonspecific protein interactions from human blood serum, plasma, and aged serum.65 More recently, they described poly(ectoine) hydrogels as a potential biomaterial due to its low nonspecific protein adsorption properties.66
The choice of support also limits the immobilization strategies, peptide densities, and the detection method available for analyzing the array in an experiment. These factors will be discussed in detail in the following sections.
Immobilization Methods
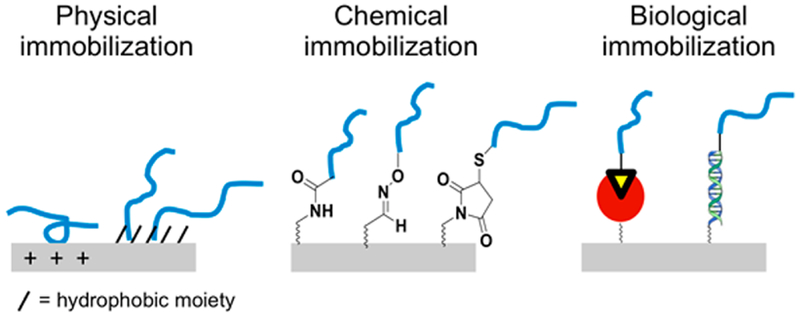
For the preparation of arrays from presynthesized peptides, there are three general methods used to immobilize peptides (Figure 3). We again emphasize that the cost of a peptide prepared by SPPS is substantial given the synthesis cost for a single 10-residue peptide is in the low tens of dollars. However, if the peptides are used to prepare many identical copies of the array, the amortized cost may be reasonable. A benefit of these methods, however, is that the peptides can be purified and yield higher quality arrays, with reduced batch-to-batch variations than may occur in arrays prepared by the in situ synthesis. Moreover, if arrays are prepared from post-translationally modified peptides, such as the use of glycopeptides, the in situ syntheses may not be feasible. In any event, the choice of immobilization strategy is an important consideration and is discussed next.
Figure 3.
Immobilization methods for peptide arrays. Peptides can be immobilized on to array surfaces through physical adsorption, chemical reaction, or biological interactions.
Physical Immobilization
The simplest way for attaching peptides to solid supports relies on physical adsorption. Most peptides of a suitable length or with a complementary property to the support will adsorb rapidly to the support. For example, hydrophobic amino acids will adsorb to a hydrophobic material and acidic amino acids will adsorb onto a positively charged surface. Peptide arrays made by adsorbing presynthesized peptides onto nitrocellulose or PVDF membranes have been demonstrated in SPOT arrays.32 This approach is analogous to the immobilization of proteins in ELISA and Western blotting applications that have been used for many years.
Fabricating peptide arrays by physical adsorptions is simple but presents several obvious drawbacks as discussed earlier, including the need for blocking nonspecific adsorption sites that can disrupt interactions between protein of interest and the immobilized peptides. Peptides attached through nonspecific adsorption may not be stable, in that the adsorbed peptide can be displaced by proteins in the sample, with the added concern that not all peptides will be subject to replacement at the same rate. It is also difficult to control the densities of the immobilized peptides, which will affect the level of signal that is measured in an assay. Finally, peptides presented on the solid support via physical adsorption are not oriented in a consistent way, and a fraction of the immobilized peptides are not bioactive in the intended assay, which can lead to substantial variation in otherwise identical arrays.
Chemical Immobilization
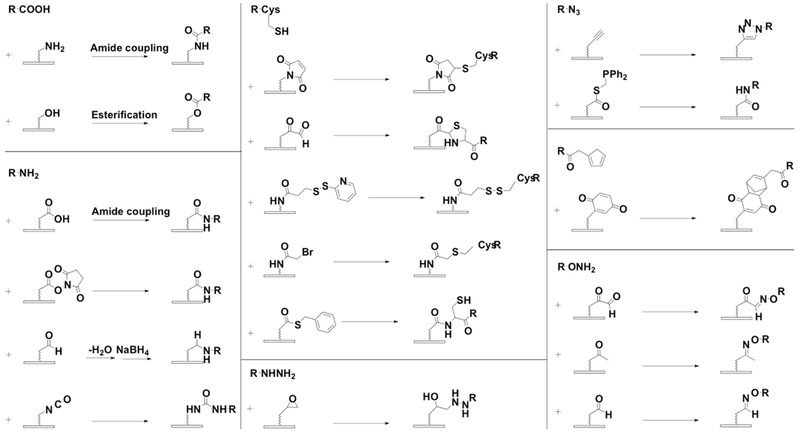
The most common strategies use selective or nonselective chemical reactions to covalently attach peptides to the support (Figure 4). These approaches usually require chemical modification of the surface to install the relevant functional groups for attachment, and they have the benefit that the peptides are covalently attached and have no risk of dissociating from the surface during an assay.
Figure 4.
Common chemical reactions used to immobilize peptides to materials.
A broad variety of reactions have been used to immobilize peptides, with many using the nucleophilic α-amino group that peptides naturally possess to condense with a carboxylate group on the support.21,51 On polylysine-coated surfaces, the side chain amino groups provide a functional handle for the coupling of peptides onto solid support through an amide coupling reaction. Likewise, the carboxylate groups on peptides can react through esterification reactions with the hydroxyl groups presented on cellulose membranes that are used in SPOT synthesis.
Aldehyde groups on a solid support provide an activated carbonyl that reacts with primary amines on peptides to form Schiff base linkages.55 Similarly, solid supports that are activated with succinimidyl ester or isocyanate groups react efficiently with peptide amine groups.67 These immobilization schemes do not require additional chemical modification of the peptide reagents, allowing simple routine implementation. However, when more than one of these nucleophilic side chains are present within a peptide, it is difficult to control the regiochemical point of attachment.
The selective reaction of thiols with several electrophilic groups has made the use of cysteine-terminated peptides an important method. For example, cysteine-terminated peptides can react with glutaraldehyde or glyoxylyl on the support to form a thiazolidine ring23,68 or with maleimide groups.69 Likewise, peptide immobilization can occur via disulfide bond formation through sulfhydryl–disulfide exchange reactions70 or by nucleophilic substitution with bromoacetyl groups.71 Another elegant method uses native chemical ligation of cysteine-terminated peptides with a thioester-functionalized surface.72,73
Other nucleophilic functional groups incorporated into a peptide can also be utilized for chemo-selective immobilization. For example, alpha nucleophiles, such as hydroxylamine and hydrazine, react with aldehydes and ketones to form oximes and hydrazones, respectively. The Lam group has utilized this chemoselective ligation to covalently attach hydroxylamine-containing peptides onto glyoxylyl-derivatized glass surface23 and ketone-modified scaffolds.74 Chan and Yousaf introduced a method that uses SAMs presenting hydroquinone groups, which undergo electrochemical oxidation to the benzoquinone and then react with ligands containing oxyamine groups. This method allows for control over ligand density and also benefits from an inert background to prevent nonspecific protein adsorption.75 Ellman and co-workers also immobilized hydroxyl amine-substituted fluorogenic peptide substrates onto an aldehyde-derivatized surface for studies of protease substrate specificity.76 Similarly, peptides modified with a hydrazide group can be immobilized onto epoxide-coated glass via nucleophilic ring opening.53
Bioorthogonal functional groups can be incorporated into synthesized peptides and used for chemoselective immobilization. For example, azide-containing peptides can be chemically immobilized through click chemistry onto a cyclooctyne-modified surface77,78 or with Staudinger ligation onto a phosphinothioester-functionalized glass slide.79 The Diels–Alder reaction can also be utilized for peptide immobilization. We reported the use of a quinone-functionalized surface to capture peptides modified with a cyclopentadiene group.80,81 Finally, it should be obvious that any chemistries used in the immobilization of large numbers of peptides in an array should be robust, high yielding, and general with respect to the peptide sequence.
Biological Immobilization
Peptides can also be immobilized using biological strategies, either based on ligand–receptor interactions or enzyme-mediated reactions. The specific noncovalent complex between a biotin tag and avidin or streptavidin is a common example for capturing tagged peptides onto solid supports. Yao and co-workers printed biotinylated peptides onto an avidin-derivatized glass slide,72 and Lam and co-workers deposited biotinylated synthetic peptides onto a neutravidin-coated polystyrene microscope slide to form peptide microarrays.82 Streptavidin-coated membranes83 and glass slides54 are commercially available for capturing biotinylated peptides for fabricating peptide arrays.
The hybridization of two complementary oligonucleotides has also been used to pattern the immobilization of peptides. This allows for an array to be “self-assembled” by adding a collection of peptide–oligonucleotide conjugates to an oligonucleotide array and allowing specific hybridization to localize each peptide to its designated region. Harris and co-workers used an array of fluorogenic peptide substrates encoded with nucleic acids to profile protease activity84 and to screen for peptide-based protease inhibitors.85 Other biological strategies for presenting peptide arrays include the use of phage display,37,86 yeast surface display,87 ribosome display,88 and polysome display.89 Details of these methods are beyond the scope of our review.
Patterning Methods
The preparation of a peptide array necessarily requires patterning methods that can direct the immobilization or in situ synthesis of each peptide at its designated region of the support. The many different approaches offer contrasting benefits in terms of peptide spot density, speed, cost, compatibility with reagents, and array quality. The following discussion outlines the common methods and their strengths and weaknesses.
Presynthesized peptides are typically patterned onto a functionalized surface using a robotic liquid handling system. The density of peptide spots prepared using this method depends on the minimum dispensing capacity of the robotic liquid handler, the hydrophobicity of the surface (which will prevent spreading of the peptide solution), and solvent evaporation (rapid evaporation leads to incomplete immobilization). Even so, PEPSCAN has reported arrays having spot sizes of 200 μm and a spot density of ~300 spots/cm2.
Peptide arrays made with the in situ synthesis approaches are patterned by the method used to deposit reagents onto the support. Spot sizes are determined by the dispensed volume of reagent and the physical properties of both the membrane support and the solvents. The densities of peptide arrays prepared by SPOT are typically modest as reagents are transferred either with manual pipetting or mechanical microspotting with 96 or 384 pin automated liquid handlers. Commercial instruments are available for preparing the arrays, including the MultiPep synthesizer by Intavis that can synthesize up to 2400 peptides on four membranes (measuring 100 mm × 150 mm) in a few hours to give spots approximately 2–3 mm in diameter and at densities of 5 spots per cm2. There are also semiautomated liquid handler instruments that can be used for SPOT synthesis, including the SpotBot by Arrayit and Syro by MultiSynTech.
The SPOT arrays can also be fabricated using noncontact inkjet printers. The drop-on-demand (DOD) inkjet printing, for example, uses mechanical actuators to eject pico- to nanoliter volumes of liquid onto a solid support in a predefined pattern90,91. SPOT synthesis with inkjet printing was first described by Frank and co-workers in 2004.52 The automated synthesizer uses magnetically controlled DOD inkjet printing capable of dispensing 2500 spots of up to 24 different reagents onto a rotating disk in 3 min. While this technique allows for the preparation of large peptide arrays, it has not been widely adopted. However, it remains a popular approach for a variety of nonpeptide array applications such as printing paper documents, constructing biosensors,92 and synthesizing oligonucleotide arrays.93 Inkjet-printed microarrays are commercially available from Arrayjet. Their inkjet printers are capable of printing 640 features per second, with a minimum volume of 100 pL resulting in a feature size of 90 μm and a spot density of 4000 spots/cm2.
Laser printing has emerged as an important patterning approach for in situ synthesis and is used in particle-based synthesis, as described above.46,47 Breitling and co-workers reported the preparation of arrays with a density of 40 000 spots/cm2.45 A commercial service by PEPperPRINT can print up to 275 000 reagent spots within a minute and prepares standard arrays with 9 000 peptide spots on standard 75.4 mm × 25 mm slides. This company has also prepared larger arrays for discovery experiments having up to 60 000 spots with a spot size of approximately 250 μm.
Photolithography uses light to activate select regions of the solid support for coupling reactions, usually by removing a photoactive protecting group to allow further synthesis.3 The original processes used physical masks to allow light activation of specific regions, followed by exposure of the entire array to an amino acid reagent which only couples at the activated regions. This process is repeated to incorporate each of the amino acids at their intended positions and requires the fabrication of many masks. One drawback of the photolithographic method is that it requires many cycles of deprotection and synthesis as only a single amino acid can be added to the array in each step. If all 20 amino acids are used to synthesize the peptides, 20 cycles of deprotection and coupling will be required for each position in the peptide; 200 lithographic steps would be required to prepare an array of 10-mer peptides. Further, because of spatial registration loss introduced in the sequential replacement of masks, the synthesis is not in high yield near the edges of the spots.
A significant advance was reported by Cerrina and colleagues,94 who used digital micromirror arrays, the same devices that were found in common LCD projectors, to direct light to the desired regions of the surface. This “maskless” array synthesis strategy uses virtual masks that are designed on a computer and projected with the digital micromirror device to create any pattern on a solid support. This innovation avoids the need to prepare expensive mask sets and provides higher quality synthesized peptides throughout the spot as it avoids the physical realigning of masks during the synthesis. Affymetrix, now part of Thermo Fisher Scientific, commercializes gene chip arrays using maskless photolithography consisting of up to 1.3 million oligonucleotide features with diameters near 10 μm. This approach has since been applied to peptide arrays.95–97 For example, Schafer-N provides a commercial source of peptide arrays on 1 cm × 2 cm glass slides synthesized in this way. Peptides on these arrays are synthesized in a square pattern with each spot having a size between 10 and 100 μm and can give arrays with more than 2 million peptides at a density of 1 000 000 spots/cm2.95,96 While this method requires specialized instrumentation, maskless photolithography is now fully automated and well-suited to the preparation of high-density arrays.
While peptide arrays synthesized by in situ methods are prepared rapidly and inexpensively, the many steps involved can pose challenges for scaling to higher volume production. Following the demonstration of photolithographic synthesis of peptide arrays on 200 mm silicon wafers,98 Stafford and co-workers produced arrays using mask-based patterned synthesis on wafers with Boc-protected amino acids and a photoresist containing a photoacid generator. The synthesis involved 90 lithography cycles, each 20 min, and resulted in arrays having 8 μm spots with 12 μm center-to-center spacing (~694 000 spots/cm2). The wafers are cut into 13 75 mm × 25 mm slides that consist of 24 identical arrays of 330 000 unique peptides (~8 million peptides per slide and ~103 million peptides per wafer). These peptide arrays are high density, low cost, and scalable for multiple experiments.
Detection Methods
The vast majority of peptide arrays are analyzed with label-dependent assays, though more recent work has developed practical strategies for using “label-free” methods. The labeled assays are typically rapid and convenient to perform but always risk interference giving rise to false positive (and negative) results. To take one example, fluorescent labels were responsible for the incorrect finding that resveratrol was a sirtuin activator.99 The most popular assays used for peptide array studies are based on radioactivity, chemiluminescence, colorimetry, and fluorescence.
The use of radioactive labels is always discouraged for reasons of safety, complicated waste disposal, and expense. However, they remain important in assays that involve the transfer of molecules to the peptides, such as ATP ([γ-32/33P]ATP) used in kinase assays100,101 and SAM ([methyl-3H]-S-adenosyl-l-methionine) used in methyltransferase assays.102
Chemiluminescent, colorimetric, and fluorescent detection methods are preferred when antibodies are used to analyze arrays after an experiment. These methods can be tedious as they require multiple blocking, washing, and incubation steps before the array is treated with a primary antibody that recognizes and binds to the reacted peptides. For chemiluminescent and colorimetric detection, the arrays are often treated with a secondary antibody that is conjugated to a reporter enzyme, such as alkaline phosphatase or horseradish peroxidase, to catalyze a chemiluminescent57,103 or colorimetric104–106 reaction. The arrays are treated with substrates that result in colorimetric changes or chemiluminescence that can be measured using standard equipment. For fluorescent detection, the secondary antibody is fluorescently labeled and no further steps are required to measure peptide array reactivity.72 Secondary antibodies are not required if antibodies targeting the desired peptide product with the desired conjugated enzyme or fluorescent label are available. Antibody dependent methods usually give relative rather than absolute quantification of results and can be limited by the availability of antibodies and cross-reactivity.
Fluorescent methods are also important in assays that do not rely on antibodies. One example is chemical modification of products of enzyme activity to introduce a fluorophore.107 They are also important in assays of protease activity, where protease action can either release a fluorophore from the surface or can disrupt a FRET interaction.76
More recent work has introduced label-free methods to analyze biomolecular arrays, with surface plasmon resonance (SPR) spectroscopy and mass spectrometry (MS) emerging as the most significant. The label-free methods have several advantages: they avoid artifacts stemming from labels, they can be quite general in measuring a broad range of activities, and they have the possibility of identifying unanticipated activities that would not have been observed with a particular labeling strategy.
SPR is an electromagnetic technique used to measure the interaction of a soluble protein with an immobilized molecule, such as a peptide, in real-time. SPR effectively measures changes in the refractive index of a solution within approximately 100 nm of a metal interface.108 In SPR, polarized light is reflected from the backside of a metalized glass slide, where it excites a collective motion of electrons in the metal giving rise to an electric field that decays in the solution. The decay characteristics depend on the local refractive index and establish a resonant condition with a particular angle of incident light. This angle can be measured by observing a dip in the intensity of reflected light, and the angle at which this minimum reflectivity occurs shifts with the refractive index. In this way, when soluble proteins bind to ligands that are immobilized at the metal film, there is an increase in the local refractive index that is measured in real-time by monitoring shifts in the angle of minimum intensity of the reflected light. The “in situ” nature of the measurement makes SPR important for measuring rates of biomolecular interactions and gives both kinetic and thermodynamic information on binding. Many different methods can be used to immobilize or capture target substrates onto the sensor surface and to enhance binding signal.109,110
SPR has traditionally been used to monitor a single biomolecular complex in an experiment, though important work has translated SPR into an imaging technique that can simultaneously monitor many interactions between a soluble protein and ligands within an array. In 2002, Corn and co-workers introduced a technique using microfluidic devices to prepare an SPR-compatible peptide array and that serves as a small volume cell to direct the flow of a target molecule over the peptide array.70 This method was used to measure binding affinities of FLAG derived peptides to the anti-FLAG M2 antibody in parallel.
In 2010, Nomura and co-workers published an SPR method for studying kinase activity on a peptide array that can measure ~1000 samples per day.111 Biotinylated peptide libraries or recombinant proteins with a FLAG-GST tag were treated with a tyrosine kinase in solution. Following phosphorylation, the peptides or proteins were selectively captured onto the sensor surface through streptavidin or anti-FLAG antibodies, respectively. Antiphosphotyrosine antibody was flowed over the sensor surface and binding was monitored by SPR. After the binding affinity was measured for the substrate, the sensor surface was washed to remove biotinylated peptides or FLAG-tagged proteins and recycled, allowing fast turnover of sample analysis.
While SPR is a label-free, sensitive, and quantitative method that can incorporate many surface chemistries, it requires specialized instrumentation and still has a lower throughput than most label-dependent detection methods or mass spectrometry. In addition, SPR is compatible only with peptide arrays that can be prepared on gold (or other metallic films); SPOT peptide arrays on cellulose membranes cannot be analyzed using SPR.
Other optical biosensors, including those that use resonant waveguide gratings (RWG),112,113 disk resonators,114 and biolayer interferometry,115,116 have also been introduced as methods for microarray analysis. While they will not be discussed in detail here, information about these techniques and their applications can be found elsewhere.117–119
Mass spectrometry (MS) is an important method for characterizing peptides and proteins and for identifying post-translational modifications that result in a change in mass.120 MS methods have the important benefit of providing molecular-level information on peptides and activities since an observed mass change is often consistent with one type of known modification. However, the sample preparation required in MS is usually tedious as it requires removal of salts and other species, often by HPLC or C18 spin columns. Further, these techniques are usually used to process soluble analytes and have not been compatible with arrays.
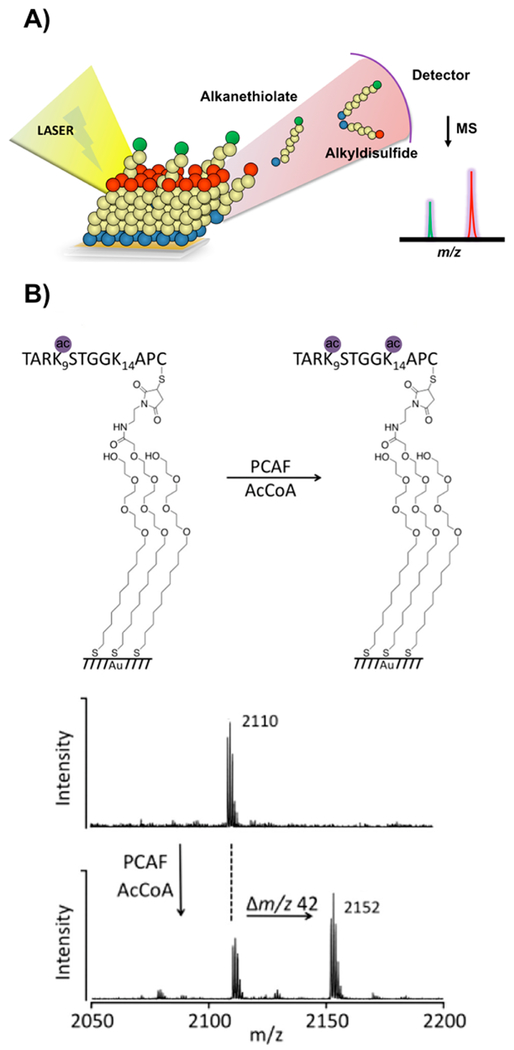
We introduced the use of matrix assisted laser-desorption ionization (MALDI) mass spectrometry to analyze self-assembled monolayers (SAMs) of alkanethiolates on gold (in a technique now known as SAMDI) MS.69,121–123 In SAMDI, laser irradiation of the monolayer results in release of the alkanethiolates (or corresponding disulfides) and provides the mass for each peptide-alkanethiolate conjugate in the monolayer (Figure 5). SAMDI is therefore able to identify mass changes, and the specific post-translational modification, of peptides modified by an enzyme. Most examples have used SAMs that present a maleimide group surrounded by SAMs that present a background of tri(ethylene glycol) (EG3) groups. The maleimide-presenting SAMs allow immobilization of cysteine-terminated peptides, while the EG3-presenting SAMs prevent nonspecific protein adsorption to the surface. We have demonstrated the use of SAMDI for a broad range of biochemical assays and have translated the method to operate in high throughput on 384 and 1536 spot plates, which can be analyzed in 15 or 30 min, respectively. Early examples used peptide arrays to study kinase activity,124 substrate specificity of deacetylase enzymes,125 and global deacetylase activities in cell lystes.18
Figure 5.
SAMDI mass spectrometry assays. (A) Laser irradiation of the monolayer releases alkanethiolates (or corresponding disulfides) and provides the mass of peptide-alkanethiolate conjugates.123 Readapted with permission from Mrksich, M. ACS Nano 2008, 2, 7–18 (ref 123). Copyright 2008 American Chemical Society. (B) Example of SAMDI MS detection of acetyltransferase activity.129 Readapted with permission from Kornacki, J. R.; Stuparu, A. D.; Mrksich, M. ACS Chemical Biology 2015, 10, 157–164 (ref 129). Copyright 2015 American Chemical Society.
CURRENT APPROACHES AND COMMERCIALIZATION
In the preceding section, we have discussed the many technical approaches that have contributed toward the development of peptide arrays. In practice, the current commercially available arrays can be grouped into five classes, for which we provide a comparative summary in Table 1. We discuss each of these commercial sources in the following section.
Table 1.
Current Methods for Preparing and Analyzing Peptide Arraysa
| Merrifield synthesis | SPOT | SAMDI | photolithography | particle-based | ||||
|---|---|---|---|---|---|---|---|---|
| technical components | peptide synthesis | SPPS | in situ | SPPS | in situ | in situ | ||
| solid support | glass | nitrocellulose or PVDF membranes | gold on glass/metal | functionalized glass or silicon | functionalized glass | |||
| surface chemistry | several methods available, e.g., polymer surface coating | amino derivitazation, typically with B-alanine or PEG | self-assembled mono-layersmaleimide and EG3 terminated | amino-derivatized polymer | amino-derivatized polymer (PEGMA) | |||
| inert layer | surface blocker with BSA or other blocking agents | membranes blocked with BSA or other blocking agents | self-assembled monolayers tri(ethylene) glycol | surfaces blocked with BSA or other blocking agents | surfaces blocked with BSA or other proteins | |||
| immobilization | many, e.g., cysteine-terminated peptide on glyoxylyl, bromomethylketone or disulfide surfaces | coupling of carboxyl group to aminofunctionalized surface | many, cysteine terminated peptides on maleimide surfaces | coupling of carboxyl group to amino functionalized surface | coupling of carboxyl group to amino functionalized surface | |||
| patterning method | liquid handler | liquid handler | inkjet printer | liquid handler | masks | mircomirrors/ digital patterning | laser printer | |
| maximum feature density | 318 spots/cm2 | 5 spots/cm2 | 4000 spots/cm2 | 16 spots/cm2 | 694 000 spots/cm2 | 1 000 000 spots/cm2 | 40 000 spots/cm2 | |
| minimum feature size | 250 μm | 2–3 mm | 90 μm | 1.25 mm | 8 μm | 10 μm × 10 μm | 50 μm | |
| detection methods | radioactivity | × | × | × | × | × | ||
| chemilluminescence | × | × | × | × | × | |||
| colorimetry | × | × | × | × | × | |||
| fluorescence | × | × | × | × | × | |||
| mass spectrometry | × | |||||||
| SPR | × | × | ||||||
| commercialization | PEPSCAN, New England Peptide, Biosynthesis | JPT, Intavis, Arrayit, MultiSynTech, Kinexus, Genscript | Arrayjet | SAMDI Tech | – | LC Sciences, Schafer-N | PEPperPRINT | |
References can be found in the text.
Merrifield synthesis peptide arrays are prepared through immobilization of presynthesized peptides onto functionalized glass surfaces.21,42,126,128 As described earlier, there are a variety of methods for functionalizing the glass support and immobilizing the peptides, but glass surfaces with a polymer coating functionalized to capture cysteine-terminated peptides, such as glyoxylyl, are among the most common.23 Merrifield synthesis peptide arrays synthesized by PEPSCAN have a maximum reported density of 318 spots/cm2 and can be analyzed with radioactive, chemiluminescent, colorimetric, and fluorescent labels. Merrifield synthesis peptide arrays are also available from New England Peptide and Bio-Synthesis.
SPOT peptide arrays are still among the most used.32,33,130–135 The peptides are typically synthesized directly on an amino-functionalized nitrocellulose membrane either using liquid handlers or inkjet printers. With liquid handlers, Intavis reported densities of 5 spots/cm2. To achieve higher density arrays, Intavis introduced the CelluSpot technology, which isolates the peptide-cellulose resin after SPOT synthesis and then spots solutions of the conjugated peptide onto a glass surface, increasing the density by nearly 10-fold with a spot size of ~1 mm. Inkjet printers from ArrayJet, however, can achieve densities of 4000 spots/cm2. The SPOT peptide arrays can be analyzed with several detection methods. Several sources of commercial arrays prepared by liquid handling are available, including those by Intavis, JPT, MultiSynTech, Kinexus, and Genscript. SPOT arrays prepared with an inkjet printer are available from Arrayjet.
SAMDI peptide arrays are prepared by immobilizing presynthesized peptides onto self-assembled monolayers of alkanethiolates on gold-coated glass or metal plates.136 Peptides are most commonly immobilized by reaction of a terminal cysteine residue with a maleimide group.69 Many other strategies, including biological immobilization through biotin-streptavidin, are also compatible with the use of monolayer surface chemistries.137–139 The monolayers are also terminated in a (EG3) group that serves to prevent nonspecific protein adsorption. This property is critical for reducing false positive and negative results and in allowing assays of cell lysates and other samples that contain high concentrations of protein. Another very significant benefit of the SAMDI arrays is that they can be analyzed by MALDI mass spectrometry to provide quantitative information on biochemical activities and can analyze a plate of 1536 peptides (corresponding to a density of 16 spots/cm2) in less than 1 h. The arrays are commercially available as a service from SAMDI Tech.
Photolithographic peptide arrays are prepared by using light to direct the synthesis of peptides directly onto functionalized glass or silicon supports3,48,95,96,98,140,141. The surfaces are usually modified with an amino-derivatized polymer for amino acid immobilization. Peptides can be synthesized either using photolabile-protecting groups on the amino acids or Boc-protected amino acids that are removed on excitation of a photoacid. Light directed patterning can be performed with a series of physical masks or digitally with a micromirror array. The maximum density reported for mask-based patterning is 694 000 spots/cm2 and required approximately 30 h for preparation. Digital patterning in the maskless format with micromirrors can achieve densities of 1 000 000 spots/cm2, and these arrays are available from Schafer-N. Digitally patterned photolithography peptide arrays have also been commercialized by LC Sciences.
The last type of peptide array commonly used is based on the SPOT arrays but uses laser printers to deliver reagents that are embedded in solid particles.45–47,142–145 The particle-based peptide arrays are usually prepared on amino-derivatized polymer-coated glass surfaces. In 2007, Breitling and co-workers reported a maximum density of 40 000 spots/cm2.45 The particle-based arrays can be analyzed with a variety of label-dependent detection methods. PEPperPRINT has commercialized particle-based arrays and can synthesize arrays having 60 000 peptides with a spot size of approximately 250 μm and densities of 1 033 spots/cm2.
There are further variations on these themes that continue to be reported. For example, in 2013, Nestrov-Mueller and co-workers introduced a microelectronic metal oxide semi-conductor (CMOS) chip printer that can synthesize 16 384 unique peptides onto a glass slide with a spot density of 10 000 spots/cm2.146 In 2016, this group introduced a combinatorial laser-induced forward transfer (cLIFT) method for peptide synthesis that can be used to prepare arrays with 17 000 spots/2cm2.143
APPLICATIONS
Peptide arrays offer an enormous opportunity to further understand the molecular pathways that underlie normal and pathological functions in cells, to guide the drug discovery process, and to diagnose and monitor treatment in disease. They also allow for a variety of other studies to understand sequence-dependent reactivity and properties of peptides. Among the early applications for peptide arrays are (1) epitope mapping for antibody binding, (2) identifying and characterizing binding interactions between protein and peptide ligands, (3) screening for active substrates of enzymes, (4) profiling enzyme activity in complex samples, and (5) identifying and studying peptides that mediate cell adhesion.
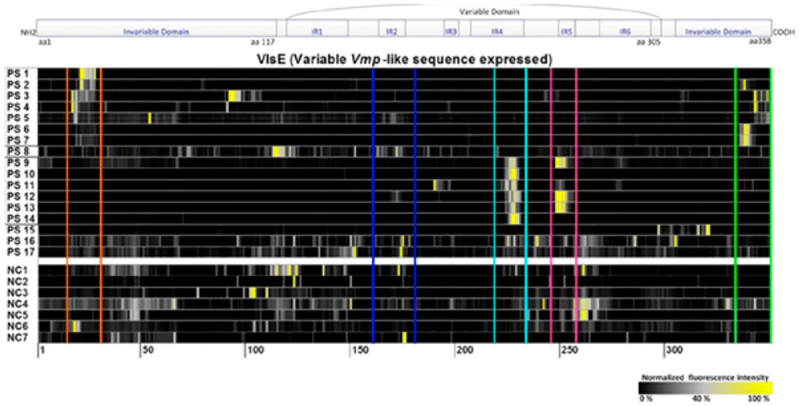
Perhaps the most common application of peptide arrays has been to map epitopes for antibody binding. Loeffler and co-workers designed a PEPperPRINT peptide array to map peptide antigens in VlsE, a surface lipoprotein that is responsible for the infectious properties of Lyme disease.144 The array had 335 peptides that were each 15 amino acids in length and tiled along the sequence of the VlsE antigen. The arrays were treated with sera from 17 patients infected with Lyme disease and seven uninfected patient sera to identify peptides that were recognized by antibodies (Figure 6). They found that the sera from infected patients fell into two main groups. The first group contained antibodies that preferably bound to the N- or C-terminal regions of the VlsE antigen (PS 1–7, highlighted in orange and green in Figure 6). The second group contained antibodies that recognized regions of the variable domain of VlsE (PS 9–14, highlighted in cyan and pink in Figure 6). A limitation of this approach is that the peptide arrays will not identify discontinuous epitopes that are present in the folded protein, including amino acids not proximal in the linear sequence. Reviews on methodology of epitope mapping and the importance of discontinuous epitopes can be found elsewhere.147,148
Figure 6.
Loeffler and co-workers mapped the antibody binding patterns in Lyme disease with peptide arrays that include tiled sequences from the antigen.144 Reproduced from Antibody fingerprints in lyme disease deciphered with high density peptide arrays, Weber, L. K.; Isse, A.; Rentschler, S.; Kneusel, R. E.; Palermo, A.; Hubbuch, J.; Nesterov-Mueller, A.; Breitling, F.; Loeffler, F. F. Engineering in Life Sciences Vol. 17, Issue 10 (ref 144). Copyright 2017 Wiley.
Schafer-Nielson and co-workers used a peptide array prepared by maskless photolithography to map peptide epitopes in human serum albumin (HSA) for an antibody reagent.96 The array had 595 peptides printed in five copies representing a complete tiling of the protein sequence. The array was treated with a polyclonal rabbit anti-HSA antibody and visualized with a Cy3-labeled goat antirabbit IgG. This study identified and characterized more than 20 linear epitopes for rabbit anti-HSA antibodies (Figure 7). In another example, Kaya and colleagues used a PEPperPRINT peptide array containing 26 364 different cardiovascular antigen-derived 15-mer peptides to compare antibody-binding profiles in patients with mycocarditis and dilated cardiomyopathy to those in healthy patients.149 They found three peptide sequences corresponding to three nonoverlapping antigens that are potentially cardiopathogenic. Ayogu and co-workers studied antigen binding specificity of autoantibodies in sera from patients with multiple sclerosis and flu-vaccine-associated narcoleptic patients using a peptide array prepared by maskless photolithography. The array had 2.2 million overlapping peptides that represented sequences found in all human protein-coding genes.150 They identified 14 082 peptides that corresponded to 1 588 proteins with differential reactivity for sera from the different patients. Lastly, Stafford and co-workers used peptide arrays prepared by Merrifield synthesis on functionalized glass to compare two peptide libraries, the first consisting of 96 random 20-mer peptides and the second consisting of 83 20-mer peptides designed from known Valley Fever epitopes, for use in monitoring antibody binding in sera of patients with coccidioidmycosis.151 Both arrays were treated with sera from infected and noninfected patients and tested for both IgG and IgM antibodies against Coccidioides. The arrays were then compared for sensitivity and specificity. The random peptide array was found to provide a more accurate diagnosis of the different stages of infection than the epitope peptide array.
Figure 7.
A small section of a peptide array prepared by maskless photolithography by Schafer-Nielson and co-workers for linear epitope mapping of HAS.96 Reprinted with permission from Hansen, L. B.; Buus, S.; Schafer-Nielsen, C. PLoS One 2013, 8, e68902 (ref 96). Copyright 2013 PLoS.
Peptide arrays have also found wide use in studying binding interactions between proteins and peptide ligands. For example, in order to better understand the consensus sequences for peptide binding to IgG and IL-2, Honda and co-workers used three SPOT peptide arrays, one random array of 512 4-mer peptides, one array of 234 8-mer peptides designed from the human IL-2 receptor, and one random 8-mer peptide array consisting of 640 peptides, to assay binding for IL-2 and IgG proteins. These experiments provide a substantial amount of data, particularly when the detection method is quantitative in resolving the amount of bound complex, and it can be difficult to predict the binding affinity for a new peptide sequence. These researchers used principle component analysis (PCA) to extrapolate and analyze binding properties of the 8-mer peptide arrays using data obtained from arrays having 4-mer peptides.132 In another example, Feller and co-workers used a SPOT peptide array to study the binding of SH3 domains in the CD2AP scaffolding protein and identified 40 candidate SH3 binding proteins.152
When using peptide arrays to identify ligands for receptor binding, it is necessary that the peptide bind with sufficient affinity, or more precisely, with a sufficiently slow dissociation rate constant so the complex is stable to the washing steps required before imaging of the array. In practice, it is challenging to characterize binding interactions with association constants in the low micromolar range. Many important regulatory interactions in the cell fall in this range and therefore have not been addressable with peptide arrays. We recently described a variation of the SAMDI method, termed PI-SAMDI (for Protein Interaction SAMDI), that can characterize weak protein–peptide interactions in an array format.153 This technique uses SAMs having both an immobilized peptide ligand and a peptide substrate for an enzyme. The receptor protein of interest is prepared as a fusion to a reporter enzyme. As the receptor binds to the immobilized ligand, the enzyme is brought to the interface where it can then modify its substrate peptide with up to 20-fold greater rate. This results in a “covalent record” of the binding interaction, and the affinity of the complex corresponds to the amount of product on the monolayer, which can be quantified with SAMDI mass spectrometry. PI-SAMDI retains the advantages of a SAMDI assay, quantitative, label-free, and high throughput with inert surfaces to prevent nonspecific protein adsorption and also the ability to study low-affinity interactions. PI-SAMDI was applied to study the binding of chromodomain proteins to methylated peptides found in the histone amino-terminal tails.
Li and co-workers demonstrated the use of imaging SPR to analyze binding of eight histone reader proteins to peptides derived from post-translationally modified histone amino terminal tails. The array contained 125 modified histone peptides and was prepared using Merrifield synthesis.154 The use of SPR allowed kinetic profiles of each binding protein to be measured. This study confirmed the strong interaction of the transcription initiation factor, TAF3, to the modified histone, H3K4me3, as well as the weak interaction between TAF3 and H3K4ac. Additionally, the DNA mismatch repair protein MSH6 was found to recognize H3K4me3.
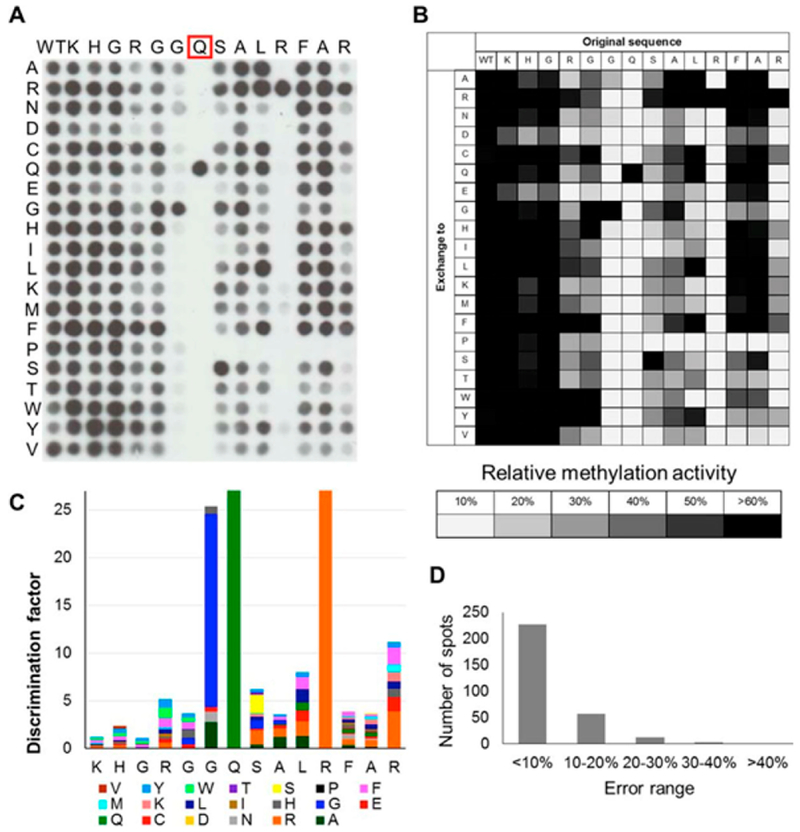
Another important application of peptide arrays is to characterize the specificities of enzymes for catalyzing modifications to peptides. Jeltsch and co-workers used a SPOT peptide array having 300 peptides to study the substrate specificity of the glutamine methyltransferase, HEMK2. The peptides were designed from a known HEMK2 substrate (residues 179–192 of eRF1) where each amino acid position was separately substituted with one of the 20 amino acids. Arrays were treated with HEMK2 and 3H-labeled S-adenosylmethionine and methylation was detected using autoradiography. The study found an unexpected minimal recognition motif based on the GQ sequence with an arginine four residues away was required for HEMK2 activity (Figure 8).155 In another example, Xu and co-workers mapped substrates for the protein arginine methyltransferase, CARM1, using an Intavis CelluSpot array.156 The array consisted of 192 15-mer peptides with a central arginine residue and was treated with purified CARM1 and 3H-labeled S-adenosylmethionine; CARM1 substrates were again detected using autoradiography. This work found that substrates containing proline-rich motifs were preferentially methylated by CARM1. Finally, Cesareni and colleagues used a SPOT peptide array consisting of ~6 000 phosphorylated peptides to profile protein tyrosine phosphatase (PTP) activity.157 The array was treated with mutants of the PTP domains that could bind the peptide substrates and that were fused to a GST tag. PTP substrates were then recognized using an anti-GST antibody conjugated to a fluorophore. The activity profiles of 16 PTP domains were obtained, and the results helped identify an important residue for tyrosine phosphatase specificity in the substrate binding pockets of PTP domains. However, the challenges in directly detecting phosphatase activity made the assay format more complicated (as shown below, SAMDI mass spectrometry gives a more direct assay of these activities).
Figure 8.
SPOT peptide array to study substrate specificity of the methyl transferase HEMK2.155 Reprinted with permission from Kusevic, D.; Kudithipudi, S.; Jeltsch, A. The Journal of Biological Chemistry 2016, 291, 6124–6133 (ref 155). Copyright 2016 American Society for Biochemistry and Molecular Biology.
Our group has used SAMDI peptide arrays to study substrate specificity of enzymes. One example used a SAMDI peptide array to identify active substrates of the sirtuin deacetylase, SIRT3.158 The active substrate was used to screen SIRT3 inhibitors, and a total of 306 inhibitors were identified from a library of 100 000 molecules. In another example, we used SAMDI peptide arrays to discover an example whereby one post-translational modification of a histone-derived peptide regulates a second post-translational modification, also known as histone crosstalk.129 We found that acetylation of Histone 3 at lysine 14 by the acetyltransferase PCAF is inhibited when the arginine residue at position 8 is first enzymatically methylated or deaminated.
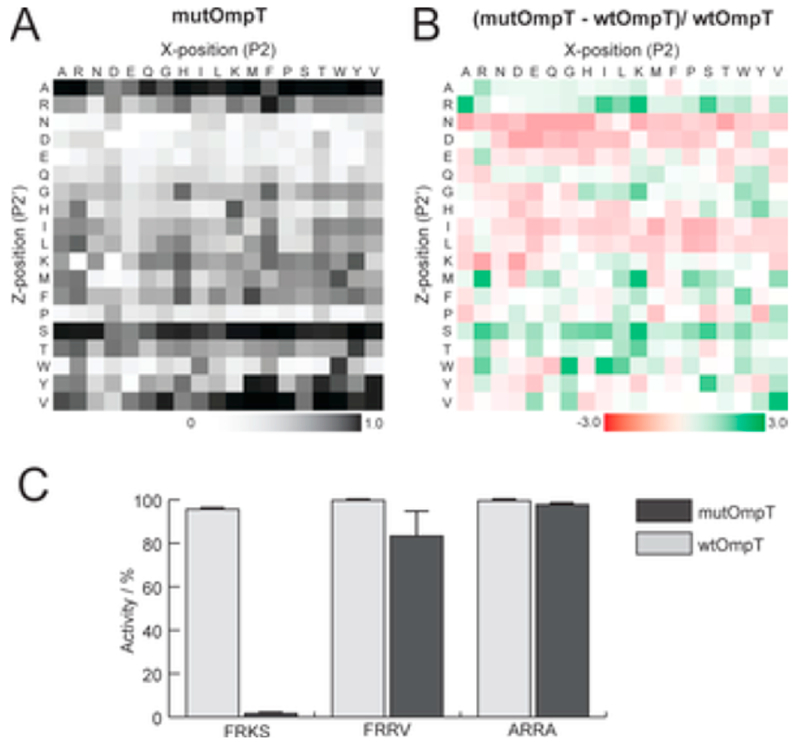
We recently reported an example that used peptide arrays to compare the substrate specificities of the OmpT protease with a mutant form that has been used because of its greater stability. Contrary to the current understanding, we found that the mutant does not share the same specificity as does the wildtype (Figure 9).159 Finally, in a study of the lysine deacetylase KDAC8 we found, with Fierke, that the specificity of the enzyme depends on the identity of the active site divalent metal ion, demonstrating a potential mechanism for regulating the activity of an enzyme for its substrates.160 These examples show that peptide arrays are critical for characterizing the specificity of an enzyme, as it is important to identify sequences that are both active and inactive for a given enzyme.
Figure 9.
Wood et al. used SAMDI peptide arrays to study and compare the substrate specificities of mutant and wild type OmpT.159 Reproduced from A Bottom-Up Proteomic Approach to Identify Substrate Specificity of Outer-Membrane Protease OmpT, Wood, S. E.; Sinsinbar, G.; Gudlur, S.; Nallani, M.; Huang, C.-F.; Liedberg, B.; Mrksich, M. Angew. Chem. Int. Ed., DOI: 10.1002/ange.201707535 (ref 159). Copyright 2017 Wiley.
The monolayer surface chemistries of the SAMDI arrays are among the best available for preventing the unwanted adsorption of protein; therefore, the arrays can be used to profile enzyme activities in complex samples derived from cell lysates and tissue extracts. Our group used SAMDI peptide arrays to profile the sirtuin and KDAC deacetylase families in cell lysates.18 These studies found that while KDAC activity remained fairly constant, sirtuin activity was significantly decreased during differentiation of CHRF megakaryocytic cells.
Another example of using peptide arrays to profile complex samples was described by Pieters and co-workers to study the activity of O-GlcNAcase in purified form as well as in cell lysates. This work used a Merrifield peptide array having six 15-mer peptides on a porous membrane of aluminum oxide.161 The arrays were first blocked with BSA to reduce nonspecific adsorption to the surface. They were then treated with the enzyme or lysate and O-GlcNAcase activity was measured by observing the decrease in binding of a fluorescently labeled anti-GlcNAc antibody. They found that different cancer cell lines had varying amounts of O-GlcNAcase activity, though the assay was not quantitative.
There are challenges in experiments that profile activies in cell lysates, however. For instance, enzyme activities can rapidly degrade once the lysate is generated and a substantial number of cells (~100 000) can be required to prepare suitable quantities of lysate. To begin to address these limitations, we recently introduced the Tandem Culture and Lysis-SAMDI (TCAL-SAMDI) assay,162 where SAMs are engineered to present a peptide substrate for the relevant enzyme as well as a peptide that mediates cell adhesion. Cells can be cultured on the monolayers and then upon lysis the released enzymes can directly, and without delay, modify the peptide substrate. After removal of the lysate, the array is analyzed with SAMDI mass spectrometry to quantify enzyme activities. We showed that this method could measure phosphatase activities from as few as 5 cells per spot. More recent work has used nanopatterned surfaces to present extracellular matrix proteins (as opposed to short peptides) to mediate the cell adhesion and make this method applicable to a broader range of cell types.163
The final general application of peptide arrays has been directed toward studying and identifying ligands for cell adhesion. Honda and co-workers used a SPOT peptide array to identify novel cell-adhesive peptides.164 The array had 180 peptides derived from fibronectin domains, and 18 of these peptides were identified as adhesion ligands in an anchorage-dependent cell assay. In addition, Kaur and co-workers used a SPOT peptide array to screen for peptides that mediate the adhesion of cancer cells.165 Previous work showed that the 12-mer p160 peptide bound to several tumor cell lines.166,167 The array had 70 peptides derived from the p160 peptide (VPWMEPAYQRFL) and were tested for adhesion of MDA-MB-435 and MCF-7 tumor cells. The bound cells were stained with CyQUANT dye, revealing two peptides that had selective adhesion for the tumor cells. These methods are important, and representative of the experiments that have resulted in the discovery of the RGD adhesion ligand by Ruoslahti,168 but must be interpreted with caution because of the propensity for nonspecific adhesion of protein and attachment of cells. In separate work, we used the monolayers that present peptides against inert backgrounds to decipher the sequence specificity of adhesion receptors and discover short peptide adhesion ligands.169,170 Peptide arrays have expanded the capabilities of protein research, allowing for a broad range of applications.
It is clear that peptide arrays will continue to play a central role in the applications described above and for new purposes as well. Some examples include identifying and optimizing reactions for the site-specific modification of peptide tags171, providing patterns of enzyme activities to understand cell function at a systems level and identifying peptides having nonbiological properties, including affinity for nanoparticle surfaces.172 It is already becoming difficult to fully understand the large data sets that are generated form peptide arrays, and as arrays increase in size, it will be more important to develop and apply complex analytical methods. A recent example from our group demonstrates the role of machine learning for more efficient peptide array design.173
SUMMARY
This review identifies the important approaches to peptide arrays and discusses the various technical options that must be considered. Of the five important approaches that are now practiced and commercially available, it is clear that no single approach offers advantages for all applications. Instead, the variations of peptide arrays enable the study of a wide range of applications. For example, the approaches based on in situ synthesis can prepare the highest density arrays and do so at relatively low reagent costs (though with varying expense in the synthesis hardware). The approaches that prepare arrays by immobilizing presynthesized peptides have the primary benefits of compatibility with a broader range of surface chemistries (that may not be sufficiently stable toward the conditions for in situ synthesis). The use of self-assembled monolayers, for example, brings the important benefits of chemistries that prevent the nonspecific adsorption of protein, give excellent control over the densities of peptides in the array, and are uniquely compatible with the use of SAMDI mass spectrometry to perform a broad range of label-free assays.
ACKNOWLEDGMENTS
Our recent work with peptide arrays has been supported by the National Cancer Institute of the National Institute of Health under Award Number U54CA199091, the Department of Defense, Defense Threat Reduction Agency under Award Number HDTRA1-15-1-0052, the Air Force Research Laboratory under Award Number FA8650-15-2-5518, and the NU-NTU Institute for Nano Medicine located at the International Institute for Nanotechnology, Northwestern University, USA, and the Nanyang Technological University, Singapore, under Award Number Agmt10/20/14.
Biographies
Biographies
Lindsey Szymczak received her B.A. in Biology and Chemistry from Cornell University in 2012. She is currently a Ph.D. candidate in the Chemistry Department at Northwestern University. Her research focuses on using peptide arrays to study enzyme activities in complex systems.
Hsin-Yu Kuo received her Ph.D. in Organic Chemistry from the University of Chicago in 2015. Her doctoral work focused on developing and applying biochemical assays for studying how enzyme activities are regulated in cellular processes. As a postdoctoral fellow at Northwestern University, she worked on building protein-based nanostructures for controlling receptor clustering at the cell membrane. Her research interests include bioanalytical tools development and bioinspired nanostructured materials.
Milan Mrksich received his Ph.D. in Chemistry at Caltech and was an American Cancer Society Postdoctoral Fellow at Harvard University. He was on the faculty in Chemistry at the University of Chicago for 16 years before moving to Northwestern as the Henry Wade Rogers Professor in the Departments of Chemistry, Biomedical Engineering and Cell & Molecular Biology. He also serves as a co-Director of the Institute of Chemical Biology and Nanomedicine at Hunan University. His research is concerned with the development of surface chemistries for applications in the life sciences.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- (1).Maskos U; Southern EM Nucleic Acids Res. 1992, 20, 1679–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Southern E Apparatus and method for analyzing polynucleotide sequences and method of generating oligonucleotide arrays. U.S. Patent 5,700,637, December 23, 1997.
- (3).Fodor S; Read J; Pirrung M; Stryer L; Lu A; Solas D Science 1991, 251, 767–773. [DOI] [PubMed] [Google Scholar]
- (4).Schena M; Shalon D; Davis RW; Brown PO Science 1995, 270, 467–470. [DOI] [PubMed] [Google Scholar]
- (5).Schena M; Shalon D; Heller R; Chai A; Brown PO; Davis RW Proc. Natl. Acad. Sci. U. S. A 1996, 93, 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Lashkari DA; DeRisi JL; McCusker JH; Namath AF; Gentile C; Hwang SY; Brown PO; Davis RW Proc. Natl. Acad. Sci. U. S. A 1997, 94, 13057–13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Michael KL; Taylor LC; Schultz SL; Walt DR Anal. Chem 1998, 70, 1242–1248. [DOI] [PubMed] [Google Scholar]
- (8).Davies RW; Wells GA; Stewart AFR; Erdmann J; Shah SH; Ferguson JF; Hall AS; Anand SS; Burnett MS; Epstein SE; Dandona S; Chen L; Nahrstaedt J; Loley C; König IR; Kraus WE; Granger CB; Engert JC; Hengstenberg C; Wichmann HE; et al. Circ.: Cardiovasc. Genet 2012, 5, 217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Telenti A; Pierce LCT; Biggs WH; di Iulio J; Wong EHM; Fabani MM; Kirkness EF; Moustafa A; Shah N; Xie C; Brewerton SC; Bulsara N; Garner C; Metzker G; Sandoval E; Perkins BA; Och FJ; Turpaz Y; Venter JC Proc. Natl. Acad. Sci. U. S. A 2016, 113, 11901–11906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Abegglen LM; Caulin AF; Chan A; Lee K; Robinson R; Campbell MS; Kiso WK; Schmitt DL; Waddell PJ; Bhaskara S; Jensen ST; Maley CC; Schiffman JD JAMA 2015, 314, 1850–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Jabara HH; Boyden SE; Chou J; Ramesh N; Massaad MJ; Benson H; Bainter W; Fraulino D; Rahimov F; Sieff C; Liu ZJ; Alshemmari SH; Al-Ramadi BK; Al-Dhekri H; Arnaout R; Abu-Shukair M; Vatsayan A; Silver E; Ahuja S; Davies EG; et al. Nat. Genet 2016, 48, 74–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).International Human Genome Sequencing Consortium. Nature 2004, 431, 931–945. [DOI] [PubMed] [Google Scholar]
- (13).Pan Q; Shai O; Lee LJ; Frey BJ; Blencowe BJ Nat. Genet 2008, 40, 1413–1415. [DOI] [PubMed] [Google Scholar]
- (14).Harper JW; Bennett EJ Nature 2016, 537, 328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Jensen ON Curr. Opin. Chem. Biol 2004, 8, 33–41. [DOI] [PubMed] [Google Scholar]
- (16).MacBeath G; Schreiber SL Science 2000, 289, 1760–1763. [DOI] [PubMed] [Google Scholar]
- (17).Zhu H; Klemic JF; Chang S; Bertone P; Casamayor A; Klemic KG; Smith D; Gerstein M; Reed MA; Snyder M Nat. Genet 2000, 26, 283–289. [DOI] [PubMed] [Google Scholar]
- (18).Kuo H-Y; DeLuca TA; Miller WM; Mrksich M Anal. Chem 2013, 85, 10635–10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Adler S; Frank R; Lanzavecchia A; Weiss S FEBS Lett. 1994, 352, 167–170. [DOI] [PubMed] [Google Scholar]
- (20).Jellis CL; Cradick TJ; Rennert P; Salinas P; Boyd J; Amirault T; Gray GS Gene 1993, 137, 63–68. [DOI] [PubMed] [Google Scholar]
- (21).Geysen HM; Meloen RH; Barteling SJ Proc. Natl. Acad. Sci. U. S. A 1984, 81, 3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Katz C; Levy-Beladev L; Rotem-Bamberger S; Rito T; Rudiger SGD; Friedler A Chem. Soc. Rev 2011, 40, 2131–2145. [DOI] [PubMed] [Google Scholar]
- (23).Falsey JR; Renil M; Park S; Li S; Lam KS Bioconjugate Chem. 2001, 12, 346–353. [DOI] [PubMed] [Google Scholar]
- (24).Arsenault R; Griebel P; Napper S Proteomics 2011, 11, 4595–4609. [DOI] [PubMed] [Google Scholar]
- (25).Thiele A; Stangl GI; Schutkowski M Mol. Biotechnol 2011, 49, 283–305. [DOI] [PubMed] [Google Scholar]
- (26).Foong YM; Fu J; Yao SQ; Uttamchandani M Curr. Opin. Chem. Biol 2012, 16, 234–242. [DOI] [PubMed] [Google Scholar]
- (27).Chiari M; Cretich M; Corti A; Damin F; Pirri G; Longhi R Proteomics 2005, 5, 3600–3603. [DOI] [PubMed] [Google Scholar]
- (28).Cretich M; Damin F; Pirri G; Chiari M Biomol. Eng 2006, 23, 77–88. [DOI] [PubMed] [Google Scholar]
- (29).Hilpert K; Winkler DF; Hancock RE Biotechnol. Genet. Eng. Rev 2007, 24, 31–106. [DOI] [PubMed] [Google Scholar]
- (30).Frank R Tetrahedron 1992, 48, 9217–9232. [Google Scholar]
- (31).Frank R; Dübel S Cold Spring Harb. Protoc. 2006, DOI: 10.1101/pdb.prot4566. [DOI] [Google Scholar]
- (32).Frank R J. Immunol. Methods 2002, 267, 13–26. [DOI] [PubMed] [Google Scholar]
- (33).Reineke U; Volkmer-Engert R; Schneider-Mergener J Curr. Opin. Biotechnol 2001, 12, 59–64. [DOI] [PubMed] [Google Scholar]
- (34).Volkmer R; Tapia V; Landgraf C FEBS Lett. 2012, 586, 2780–2786. [DOI] [PubMed] [Google Scholar]
- (35).Arenkov P; Kukhtin A; Gemmell A; Voloshchuk S; Chupeeva V; Mirzabekov A Anal. Biochem 2000, 278, 123–131. [DOI] [PubMed] [Google Scholar]
- (36).Zhu H; Bilgin M; Bangham R; Hall D; Casamayor A; Bertone P; Lan N; Jansen R; Bidlingmaier S; Houfek T; Mitchell T Miller P; Dean RA; Gerstein M; Snyder M Science 2001, 293, 2101–2105. [DOI] [PubMed] [Google Scholar]
- (37).Scott J; Smith G Science 1990, 249, 386–390. [DOI] [PubMed] [Google Scholar]
- (38).Houghten RA; Pinilla C; Blondelle SE; Appel JR; Dooley CT; Cuervo JH Nature 1991, 354, 84–86. [DOI] [PubMed] [Google Scholar]
- (39).Liu R; Enstrom AM; Lam KS Exp. Hematol 2003, 31, 11. [DOI] [PubMed] [Google Scholar]
- (40).Songyang Z; Blechner S; Hoagland N; Hoekstra MF; Piwnica-Worms H; Cantley LC Curr. Biol 1994, 4, 973–982. [DOI] [PubMed] [Google Scholar]
- (41).Lam KS; Lebl M; Krchňák V Chem. Rev 1997, 97, 411–448. [DOI] [PubMed] [Google Scholar]
- (42).Merrifield RB J. Am. Chem. Soc 1963, 85, 2149–2154. [Google Scholar]
- (43).Houghten RA Proc. Natl. Acad. Sci. U. S. A 1985, 82, 5131–5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Dikmans A; Beutling U; Schmeisser E; Thiele S; Frank R QSAR Comb. Sci. 2006, 25, 1069–1080. [Google Scholar]
- (45).Beyer M; Nesterov A; Block I; König K; Felgenhauer T; Fernandez S; Leibe K; Torralba G; Hausmann M; Trunk U; Lindenstruth V; Bischoff FR; Stadler V; Breitling F Science 2007, 318, 1888–1888. [DOI] [PubMed] [Google Scholar]
- (46).Stadler V; Felgenhauer T; Beyer M; Fernandez S; Leibe K; Güttler S; Groning M; König K; Torralba G; Hausmann M; Lindenstruth V; Nesterov A; Block I; Pipkorn R; Poustka A; Bischoff FR; Breitling F Angew. Chem., Int. Ed 2008, 47, 7132–7135. [DOI] [PubMed] [Google Scholar]
- (47).Breitling F; Felgenhauer T; Nesterov A; Lindenstruth V; Stadler V; Bischoff FR ChemBioChem 2009, 10, 803–808. [DOI] [PubMed] [Google Scholar]
- (48).Legutki JB; Zhao Z-G; Greving M; Woodbury N; Johnston SA; Stafford P Nat. Commun 2014, 5, 4785. [DOI] [PubMed] [Google Scholar]
- (49).Gao X; LeProust E; Zhang H; Srivannavit O; Gulari E; Yu P; Nishiguchi C; Xiang Q; Zhou X Nucleic Acids Res. 2001, 29, 4744–4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Pellois JP Nat. Biotechnol 2002, 20, 922. [DOI] [PubMed] [Google Scholar]
- (51).Saxinger C; Conrads TP; Goldstein DJ; Veenstra TD BMC Immunol. 2005, 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Dikmans AJ; Morr M; Zander N; Adler F; Türk G; Frank R Mol. Diversity 2004, 8, 197–207. [DOI] [PubMed] [Google Scholar]
- (53).Pokholok DK; Harbison CT; Levine S; Cole M; Hannett NM; Lee TI; Bell GW; Walker K; Rolfe PA; Herbolsheimer E; Zeitlinger J; Lewitter F; Gifford DK; Young RA Cell 2005, 122, 517–527. [DOI] [PubMed] [Google Scholar]
- (54).Rothbart SB; Krajewski K; Strahl BD; Fuchs SM Methods Enzymol 2012, 512, 107–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Schutkowski M; Reimer U; Panse S; Dong L; Lizcano JM; Alessi DR; Schneider-Mergener J Angew. Chem., Int. Ed 2004, 43, 2671–2674. [DOI] [PubMed] [Google Scholar]
- (56).Haab BB; Dunham MJ; Brown PO Genome Biology 2000, 1, preprint0001.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Dürauer A; Kopecky E; Berger E; Seifert M; Hahn R; Jungbauer A J. Biochem. Biophys. Methods 2006, 66, 45–57. [DOI] [PubMed] [Google Scholar]
- (58).Gregorius K; Mouritsen S; Elsner HI J. Immunol. Methods 1995, 181, 65–73. [DOI] [PubMed] [Google Scholar]
- (59).Kiyonaka S; Sada K; Yoshimura I; Shinkai S; Kato N; Hamachi I Nat. Mater 2004, 3, 58–64. [DOI] [PubMed] [Google Scholar]
- (60).Parker LL; Brueggemeier SB; Rhee WJ; Wu D; Kent SBH; Kron SJ; Palecek SP Analyst 2006, 131, 1097–1104. [DOI] [PubMed] [Google Scholar]
- (61).Schuck P; Zhao H Methods Mol. Biol. (N. Y. NY, U. S.) 2010, 627, 15–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Love JC; Estroff LA; Kriebel JK; Nuzzo RG; Whitesides GM Chem. Rev 2005, 105, 1103–1170. [DOI] [PubMed] [Google Scholar]
- (63).Prime KL; Whitesides GM J. Am. Chem. Soc 1993, 115, 10714–10721. [Google Scholar]
- (64).Beyer M; Felgenhauer T; Ralf Bischoff F; Breitling F; Stadler V Biomaterials 2006, 27, 3505–3514. [DOI] [PubMed] [Google Scholar]
- (65).Yang W; Xue H; Li W; Zhang J; Jiang S Langmuir 2009, 25, 11911–11916. [DOI] [PubMed] [Google Scholar]
- (66).Jain P; Hung H-C; Lin X; Ma J; Zhang P; Sun F; Wu K; Jiang S Langmuir 2017, 33, 11264. [DOI] [PubMed] [Google Scholar]
- (67).Benters R; Niemeyer CM; Wöhrle D ChemBioChem 2001, 2, 686–694. [DOI] [PubMed] [Google Scholar]
- (68).Han X; Shigaki S; Yamaji T; Yamanouchi G; Mori T; Niidome T; Katayama Y Anal. Biochem 2008, 372, 106–115.18028866 [Google Scholar]
- (69).Houseman BT; Gawalt ES; Mrksich M Langmuir 2003, 19, 1522–1531. [Google Scholar]
- (70).Wegner GJ; Lee HJ; Corn RM Anal. Chem 2002, 74, 5161–5168. [DOI] [PubMed] [Google Scholar]
- (71).Takahashi M; Nokihara K; Mihara H Chem. Biol 2003, 10, 53–60. [DOI] [PubMed] [Google Scholar]
- (72).Lesaicherre M-L; Uttamchandani M; Chen GYJ; Yao SQ Bioorg. Med. Chem. Lett 2002, 12, 2079–2083. [DOI] [PubMed] [Google Scholar]
- (73).Camarero JA; Kwon Y; Coleman MA J. Am. Chem. Soc 2004, 126, 14730–14731. [DOI] [PubMed] [Google Scholar]
- (74).Xu Q; Miyamoto S; Lam KS Mol. Diversity 2004, 8, 301–310. [DOI] [PubMed] [Google Scholar]
- (75).Chan EWL; Yousaf MN J. Am. Chem. Soc 2006, 128, 15542–15546. [DOI] [PubMed] [Google Scholar]
- (76).Salisbury CM; Maly DJ; Ellman JA J. Am. Chem. Soc 2002, 124, 14868–14870. [DOI] [PubMed] [Google Scholar]
- (77).Wammes AEM; Fischer MJE; de Mol NJ; van Eldijk MB; Rutjes FPJT; van Hest JCM; van Delft FL Lab Chip 2013, 13, 1863–1867. [DOI] [PubMed] [Google Scholar]
- (78).Shamsi F; Coster H; Jolliffe KA Surf. Sci 2011, 605, 1763–1770. [Google Scholar]
- (79).Soellner MB; Dickson KA; Nilsson BL; Raines RT J. Am. Chem. Soc 2003, 125, 11790–11791. [DOI] [PubMed] [Google Scholar]
- (80).Houseman BT; Huh JH; Kron SJ; Mrksich M Nat. Biotechnol 2002, 20, 270–274. [DOI] [PubMed] [Google Scholar]
- (81).Yousaf MN; Mrksich M J. Am. Chem. Soc 1999, 121, 4286–4287. [Google Scholar]
- (82).Aina OH; Sroka TC; Chen M-L; Lam KS Biopolymers 2002, 66, 184–199. [DOI] [PubMed] [Google Scholar]
- (83).De Keersmaecker K; Versele M; Cools J; Superti-Furga G; Hantschel O Leukemia 2008, 22, 2208–2216. [DOI] [PubMed] [Google Scholar]
- (84).Winssinger N; Damoiseaux R; Tully DC; Geierstanger BH; Burdick K; Harris JL Chem. Biol 2004, 11, 1351–1360. [DOI] [PubMed] [Google Scholar]
- (85).Winssinger N; Ficarro S; Schultz PG; Harris JL Proc. Natl. Acad. Sci. U. S. A 2002, 99, 11139–11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Smith GP; Petrenko VA Chem. Rev 1997, 97, 391–410. [DOI] [PubMed] [Google Scholar]
- (87).Gai SA; Wittrup KD Curr. Opin. Struct. Biol 2007, 17, 467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Zahnd C; Amstutz P; Pluckthun A Nat. Methods 2007, 4, 269–279. [DOI] [PubMed] [Google Scholar]
- (89).Mattheakis LC; Bhatt RR; Dower WJ Proc. Natl. Acad. Sci. U. S. A 1994, 91, 9022–9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Delaney JT; Smith PJ; Schubert US Soft Matter 2009, 5, 4866–4877. [Google Scholar]
- (91).Saunders RE; Derby B. Int. Mater. Rev 2014, 59, 430–448. [Google Scholar]
- (92).Setti L; Fraleoni-Morgera A; Ballarin B; Filippini A; Frascaro D; Piana C Biosens. Bioelectron 2005, 20, 2019–2026. [DOI] [PubMed] [Google Scholar]
- (93).Hughes TR; Mao M; Jones AR; Burchard J; Marton MJ; Shannon KW; Lefkowitz SM; Ziman M; Schelter JM; Meyer MR; Kobayashi S; Davis C; Dai HY; He YDD; Stephaniants SB; Cavet G; Walker WL; West A; Coffey E; Shoemaker DD; et al. Nat. Biotechnol 2001, 19, 342–347. [DOI] [PubMed] [Google Scholar]
- (94).Singh-Gasson S; Green RD; Yue Y; Nelson C; Blattner F; Sussman MR; Cerrina F Nat. Biotechnol 1999, 17, 974–978. [DOI] [PubMed] [Google Scholar]
- (95).Buus S; Rockberg J; Forsström B; Nilsson P; Uhlen M; Schafer-Nielsen C Mol. Cell. Proteomics 2012, 11, 1790–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Hansen LB; Buus S; Schafer-Nielsen C PLoS One 2013, 8, e68902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Shin D-S; Lee K-N; Yoo B-W; Kim J; Kim M; Kim Y-K; Lee Y-S J. Comb. Chem 2010, 12, 463–471. [DOI] [PubMed] [Google Scholar]
- (98).Price JV; Tangsombatvisit S; Xu G; Levy D; Baechler EC; Gozani O; Varma M; Utz PJ; Liu CL Nat. Med 2012, 18, 1434–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Kaeberlein M; McDonagh T; Heltweg B; Hixon J; Westman EA; Caldwell SD; Napper A; Curtis R; DiStefano PS; Fields S; Bedalov A; Kennedy BK J. Biol. Chem 2005, 280, 17038–17045. [DOI] [PubMed] [Google Scholar]
- (100).Dostmann WRG; Taylor MS; Nickl CK; Brayden JE; Frank R; Tegge WJ Proc. Natl. Acad. Sci. U. S. A 2000, 97, 14772–14777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Smith FD; Samelson BK; Scott JD Biochem. J 2011, 438, 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Rathert P; Dhayalan A; Murakami M; Zhang X; Tamas R; Jurkowska R; Komatsu Y; Shinkai Y; Xiaodong C; Jeltsch A Nat. Chem. Biol 2008, 4, 344–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Landgraf C; Panni S; Montecchi-Palazzi L; Castagnoli L; Schneider-Mergener J; Volkmer-Engert R; Cesareni G PLoS Biol. 2004, 2, e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Blake MS; Johnston KH; Russell-Jones GJ; Gotschlich EC Anal. Biochem 1984, 136, 175–179. [DOI] [PubMed] [Google Scholar]
- (105).Hawkes R; Niday E; Gordon J Anal. Biochem 1982, 119, 142–147. [DOI] [PubMed] [Google Scholar]
- (106).Verastegui M; Moro P; Guevara A; Rodriguez T; Miranda E; Gilman RH J. Clin. Microbiol 1992, 30, 1557–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Akita S; Umezawa N; Kato N; Higuchi T Bioorg. Med. Chem 2008, 16, 7788–7794. [DOI] [PubMed] [Google Scholar]
- (108).Schuck P Annu. Rev. Biophys. Biomol. Struct 1997, 26, 541–566. [DOI] [PubMed] [Google Scholar]
- (109).Inoue Y; Mori T; Yamanouchi G; Han X; Sonoda T; Niidome T; Katayama Y Anal. Biochem 2008, 375, 147–149. [DOI] [PubMed] [Google Scholar]
- (110).Inamori K; Kyo M; Nishiya Y; Inoue Y; Sonoda T; Kinoshita E; Koike T; Katayama Y Anal. Chem 2005, 77, 3979–3985. [DOI] [PubMed] [Google Scholar]
- (111).Takeda H; Goshima N; Nomura N In Surface Plasmon Resonance: Methods and Protocols; Mol NJ; Fischer MJE, Eds.; Humana Press: Totowa, NJ, 2010; pp 131–145. [DOI] [PubMed] [Google Scholar]
- (112).Fang Y In Optical Guided-wave Chemical and Biosensors II; Springer, 2010; pp 27–42. [Google Scholar]
- (113).Li G; Ferrie AM; Fang Y JALA 2006, 11, 181–187. [Google Scholar]
- (114).Grist SM; Schmidt SA; Flueckiger J; Donzella V; Shi W; Talebi Fard S; Kirk JT; Ratner DM; Cheung KC; Chrostowski L Opt. Express 2013, 21, 7994–8006. [DOI] [PubMed] [Google Scholar]
- (115).Picaud S; Filippakopoulos P Microarrays 2015, 4, 370–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (116).Hundsberger H; Önder K; Schuller-Götzburg P; Virok DP; Herzog J; Rid R BMC Genomics 2017, 18, 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (117).Fernández Gavela A; Grajales García D; Ramirez JC; Lechuga LM Sensors 2016, 16, 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (118).Damborský P; Švitel J; Katrlík J Essays Biochem. 2016, 60, 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (119).Schmidt S; Flueckiger J; Wu W; Grist SM; Fard ST; Donzella V; Khumwan P; Thompson ER; Wang Q; Kulik P; Wang X; Sherwali A; Kirk J; Cheung KC; Chrostowski L; Ratner D In SPIE NanoScience + Engineering; SPIE, 2014; p 38. [Google Scholar]
- (120).de Rond T; Danielewicz M; Northen T Curr. Opin. Biotechnol 2015, 31, 1–9. [DOI] [PubMed] [Google Scholar]
- (121).Su J; Mrksich M Angew. Chem., Int. Ed 2002, 41, 4715–4718. [DOI] [PubMed] [Google Scholar]
- (122).Min D-H; Yeo W-S; Mrksich M Anal. Chem 2004, 76, 3923–3929. [DOI] [PubMed] [Google Scholar]
- (123).Mrksich M ACS Nano 2008, 2, 7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (124).Min D-H; Su J; Mrksich M Angew. Chem. Int. Ed 2004, 43, 5973–5977. [DOI] [PubMed] [Google Scholar]
- (125).Gurard-Levin ZA; Kim J; Mrksich M ChemBioChem 2009, 10, 2159–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (126).Gemini-Piperni S; Milani R; Bertazzo S; Peppelenbosch M; Takamori ER; Granjeiro JM; Ferreira CV; Teti A; Zambuzzi W Biotechnol. Bioeng 2014, 111, 1900–1905. [DOI] [PubMed] [Google Scholar]
- (127).de Bourayne M; Gallais Y; El Ali Z; Rousseau P; Damiens M-H; Cochet C; Filhol O; Chollet-Martin S; Pallardy M; Kerdine-Römer S J. Leukocyte Biol. 2017, 101, 703–715. [DOI] [PubMed] [Google Scholar]
- (128).Tsang SH; Chan L; Tsai Y-T; Wu W-H; Hsu C-W; Yang J; Tosi J; Wert KJ; Davis RJ; Mahajan VB Trans. Am. Ophthamol Soc. 2014, 112, 103–115. [PMC free article] [PubMed] [Google Scholar]
- (129).Kornacki JR; Stuparu AD; Mrksich M ACS Chem. Biol 2015, 10, 157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (130).Kramer A; Schneider-Mergener J In Combinatorial Peptide Library Protocols; Cabilly S, Ed.; Humana Press: Totowa, NJ, 1998; pp 25–39. [Google Scholar]
- (131).Deschamps T; Bazot Q; Leske DM; MacLeod R; Mompelat D; Tafforeau L; Lotteau V; Maréchal V; Baillie GS; Gruffat H; Wilson JB; Manet E J. Gen. Virol 2017, 98, 251–265. [DOI] [PubMed] [Google Scholar]
- (132).Kume A; Kawai S; Kato R; Iwata S; Shimizu K; Honda H J. Biosci. Bioeng 2017, 123, 230–238. [DOI] [PubMed] [Google Scholar]
- (133).López-Pérez PM; Grimsey E; Bourne L; Mikut R; Hilpert K Front. Chem 2017, 5, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (134).Tanaka M; Hikiba S; Yamashita K; Muto M; Okochi M Acta Biomater. 2017, 49, 495–506. [DOI] [PubMed] [Google Scholar]
- (135).Webster CG; Thillier M; Pirolles E; Cayrol B; Blanc S; Uzest M Insect Science 2017, DOI: 10.1111/1744-7917.12469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (136).Gurard-Levin ZA; Mrksich M Annu. Rev. Anal. Chem 2008, 1 767–800. [DOI] [PubMed] [Google Scholar]
- (137).Anderson LL; Berns EJ; Bugga P; George AL; Mrksich M Anal. Chem 2016, 88, 8604–8609. [DOI] [PubMed] [Google Scholar]
- (138).Diagne AB; Li S; Perkowski GA; Mrksich M; Thomson RJ ACS Comb. Sci 2015, 17, 658–662. [DOI] [PubMed] [Google Scholar]
- (139).Ban L; Pettit N; Li L; Stuparu AD; Cai L; Chen W; Guan W; Han W; Wang PG; Mrksich M Nat. Chem. Biol 2012, 8, 769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (140).Mortensen R; Nissen TN; Fredslund S; Rosenkrands I; Christensen JP; Andersen P; Dietrich J Sci. Rep 2016, 6, 22030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (141).Forsström B; Axnäs BB; Stengele K-P; Bühler J; Albert TJ; Richmond TA; Hu FJ; Nilsson P; Hudson EP; Rockberg H ; Uhlen M Mol. Cell. Proteomics 2014, 13, 1585–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (142).Vernet T; Choulier L; Nominé Y; Bellard L; Baltzinger M; Travé G; Altschuh D J. Mol. Recognit 2015, 28, 635–644. [DOI] [PubMed] [Google Scholar]
- (143).Loeffler FF; Foertsch TC; Popov R; Mattes DS; Schlageter M; Sedlmayr M; Ridder B; Dang F-X; von Bojničić-Kninski C; Weber LK; Fischer A; Greifenstein J; Bykovskaya V; Buliev I; Bischoff FR; Hahn L; Meier MAR; Brase S; Powell AK; Balaban TS; et al. Nat. Commun 2016, 7, 11844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (144).Weber LK; Isse A; Rentschler S; Kneusel RE; Palermo A; Hubbuch J; Nesterov-Mueller A; Breitling F; Loeffler FF Engineering in Life Sciences 2017, 17, 1078–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (145).Nesterov-Mueller A; Maerkle F; Hahn L; Foertsch T; Schillo S; Bykovskaya V; Sedlmayr M; Weber LK; Ridder B; Soehindrijo M; Muenster B; Striffler J; Bischoff FR; Breitling F; Loeffler FF Microarrays 2014, 3, 245–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (146).Loeffler FF; Cheng Y-C; Muenster B; Striffler J; Liu FC; Ralf Bischoff F; Doersam E; Breitling F; Nesterov-Mueller A In Fundamentals and Application of New Bioproduction Systems; Zeng A-P, Ed.; Springer Berlin Heidelberg: Berlin, Heidelberg, Germany, 2013; pp 1–23. [Google Scholar]
- (147).Abbott WM; Damschroder MM; Lowe DC Immunology 2014, 142, 526–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (148).Ahmad TA; Eweida AE; Sheweita SA Trials in Vaccinology 2016, 5, 71–83. [Google Scholar]
- (149).Müller A-M; Bockstahler M; Hristov G; Weiß C; Fischer A; Korkmaz-Icöz S; Giannitsis E; Poller W; Schultheiss H-P; Katus HA; Kaya Z Clin. Immunol 2016, 173, 64–75. [DOI] [PubMed] [Google Scholar]
- (150).Zandian A; Forsström B; Häggmark-Månberg A; Schwenk JM; Uhlén M; Nilsson P; Ayoglu B J. Proteome Res. 2017, 16, 1300–1314. [DOI] [PubMed] [Google Scholar]
- (151).Navalkar KA; Johnston SA; Woodbury N; Galgiani JN; Magee DM; Chicacz Z; Stafford P Clin. Vaccine Immunol. 2014, 21, 1169–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (152).Rouka E; Simister PC; Janning M; Kumbrink J; Konstantinou T; Muniz JRC; Joshi D.; O’Reilly N; Volkmer R; Ritter B; Knapp S; von Delft F; Kirsch KH; Feller SM J. Biol. Chem 2015, 290, 25275–25292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (153).O’Kane PT; Mrksich M J. Am. Chem. Soc 2017, 139, 10320–10327. [DOI] [PubMed] [Google Scholar]
- (154).Zhao S; Yang M; Zhou W; Zhang B; Cheng Z; Huang J; Zhang M; Wang Z; Wang R; Chen Z; Zhu J; Li H Proc. Natl. Acad. Sci. U. S. A 2017, 114, E7245–E7254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (155).Kusevic D; Kudithipudi S; Jeltsch A J. Biol. Chem 2016, 291, 6124–6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (156).Shishkova E; Zeng H; Liu F; Kwiecien NW; Hebert AS; Coon JJ; Xu W Nat. Commun 2017, 8, 15571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (157).Palma A; Tinti M; Paoluzi S; Santonico E; Brandt BW; Hooft van Huijsduijnen R; Masch A; Heringa J; Schutkowski M; Castagnoli L; Cesareni G J. Biol. Chem 2017, 292, 4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (158).Patel K; Sherrill J; Mrksich M; Scholle MD J. Biomol. Screening 2015, 20, 842–848. [DOI] [PubMed] [Google Scholar]
- (159).Wood SE; Sinsinbar G; Gudlur S; Nallani M; Huang CF; Liedberg B; Mrksich M Angew. Chem 2017, DOI: 10.1002/ange.201707535. [DOI] [PubMed] [Google Scholar]
- (160).Castaneda CA; Lopez JE; Joseph CG; Scholle MD; Mrksich M; Fierke CA Biochemistry 2017, 56, 5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (161).Sharif S; Shi J; Bourakba M; Ruijtenbeek R; Pieters RJ Anal. Biochem 2017, 532, 12–18. [DOI] [PubMed] [Google Scholar]
- (162).Berns EJ; Cabezas MD; Mrksich M Small 2016, 12, 3811–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (163).Cabezas MD; Mirkin CA; Mrksich M Nano Lett. 2017, 17, 1373–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (164).Kato R; Kaga C; Kunimatsu M; Kobayashi T; Honda H J. Biosci. Bioeng 2006, 101, 485–495. [DOI] [PubMed] [Google Scholar]
- (165).Ahmed S; Mathews AS; Byeon N; Lavasanifar A; Kaur K Anal. Chem 2010, 82, 7533–7541. [DOI] [PubMed] [Google Scholar]
- (166).Askoxylakis V; Mier W; Zitzmann S; Ehemann V; Zhang J; Krämer S; Beck C; Schwab M; Eisenhut M; Haberkorn U J. Nucl. Med 2006, 47, 981–988. [PubMed] [Google Scholar]
- (167).Askoxylakis V; Zitzmann S; Mier W; Graham K; Krämer S; von Wegner F; Fink RHA; Schwab M; Eisenhut M; Haberkorn U Clin. Cancer Res. 2005, 11, 6705–6712. [DOI] [PubMed] [Google Scholar]
- (168).Ruoslahti E; Pierschbacher M Science 1987, 238, 491–497. [DOI] [PubMed] [Google Scholar]
- (169).Sánchez-Cortés J; Mrksich M Chem. Biol 2009, 16, 990–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (170).Sánchez-Cortés J; Mrksich M ACS Chem. Biol 2011, 6, 1078–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (171).Ramil CP; An P; Yu Z; Lin Q J. Am. Chem. Soc 2016, 138, 5499–5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (172).Shao Q; Hall CK Langmuir 2016, 32, 7888–7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (173).Xue AY; Szymczak LC; Mrksich M; Bagheri N Anal. Chem 2017, 89, 9039–9047. [DOI] [PMC free article] [PubMed] [Google Scholar]