Abstract
Long noncoding RNAs (lncRNAs) are a new class of regulators. LncRNAs are defined as endogenous transcribed RNA molecules with transcript length of >200 nt. Accumulating evidence has shown that lncRNAs are involved in many physiological processes such as cell cycle regulation, cell apoptosis and survival, cancer migration and metabolism. However, the biological and molecular mechanisms of lncRNAs in pancreatic cancer are still unclear. Recent studies have reported that many lncRNAs are dysregulated in pancreatic cancer and closely associated with tumorigenesis, diagnosis and prognosis. In this review, we described the regulation and functional role of lncRNAs and the potential underlying mechanism involved in pancreatic cancer, outlined the roles of lncRNA in pancreatic cancer, and discussed the potential possibility of lncRNAs as therapeutic targets in clinical practice. Moreover, the potential of lncRNAs used as sensitive biomarkers for diagnosis, prognosis and prediction of response to therapy in pancreatic cancer will also be discussed.
Keywords: Long noncoding RNAs, pancreatic cancer, tumorigenesis
INTRODUCTION
Previous studies over the past decades mostly focused on the protein-coding genes. However, only <2% of the genome sequences have protein-coding ability, and the remaining is transcribed into noncoding RNAs (ncRNAs), which were regarded as transcriptional “noise” or body “garbage” previously. Recently, there is an explosive expansion in the understanding of biological function of ncRNA transcripts, exemplified by the significant role of microRNAs (miRNAs) in various human diseases including cancer. Recently, other types of short or long ncRNAs, especially the long noncoding RNAs (lncRNAs), have attracted attention due to their large number and biological significance. Defined as being at least 200 nucleotides in length, lncRNAs are characterized by their abundant presence in the human genome, as well as their tissue-specific expression patterns. Moreover, lncRNAs can affect various physiological processes such as cell cycle regulation, cell apoptosis and survival, cancer migration and metabolism.[1,2,3] Although lncRNAs have an impact on various human diseases, the biological function and molecular mechanisms of lncRNA in human diseases remain largely unknown. Dysregulation of lncRNA in various cancers is considered as one of the leading forces during tumorigenesis. LncRNAs could interact specially with DNA and RNA, and alterations in lncRNA expression levels could affect a broad spectrum of genes and phenotypic changes.[4]
Pancreatic cancer is a malignant disease with poor prognosis, ranking the forth lethal malignancy worldwide. It is estimated that there are ~45,000 new cases diagnosed annually in United States, and 5-year overall survival rates are <5%.[5] Invasion and metastasis are the characteristics of pancreatic cancer. Metastasis-associated genes and proteins have been studied at the genetic level. It is reported that mutation of KRAS, p16, TP53 and SMAD4 are involved in the metastatic process. However, the underlying mechanism is still controversial. In spite of the progress in surgical techniques, chemotherapy and radiotherapy, the survival of patients with pancreatic cancer has not substantially improved. One of the main reasons for the low overall survival is poor understanding of the complicated molecular mechanism in the progress of pancreatic cancer, including altered cell physiology, dysfunction of cell growth and apoptosis signals, insensitivity to all cycle arrest signals, and evasion from programmed cell death (apoptosis).[6] Moreover, tumor microenvironment, inflammation, oxidative stress and hypoxia were also involved in promoting pancreatic cancer initiation, progression and metastasis. Recent studies found that many lncRNAs are dysregulated in pancreatic cancer. In this review, we discuss the current understanding of the lncRNAs regarding their biogenesis and functional characteristics as well as their roles in cancer development, outline the roles of lncRNA in pancreatic cancer, and discuss the potential possibility of lncRNAs as therapeutic targets and biomarkers in clinical practice.
OVERVIEW OF LNC-RNA IN CANCER
LncRNAs are defined as endogenous transcribed RNA molecules with transcript length of >200 nt and those that lack coding potential but possess regulatory roles in their primary or spliced form.[7] LncRNAs are poorly conserved and expressed at various levels, including chromatin modification, transcription and posttranscriptional processing. It is estimated that the numbers of lncRNA in the human genome range from 7,000 to 23,000.[8] According to the genomic location with respect to protein-coding genes, lncRNAs can be classified into subtypes that include intergenic, intronic, exonic, overlapping and antisense lncRNA. Based on the regulatory mechanisms, the lncRNAs can be divided into four categories: (1) regulators of gene expression, (2) act as miRNA decoys to free target mRNAs, (3) regulators of mRNA translation, and (4) regulator of protein activities.
It was suspected for many years that lncRNAs play an important role in cancer, however this notion lacks critical supporting evidence. Recently, accumulating evidence demonstrates that lncRNAs, such as protein-coding genes, are central players in cancer development. Thus, they may open a new strategy for cancer therapy. The underlying mechanisms are proposed as followed: (1) mediators on signaling pathway - lncRNAs can be a part of molecular signals pathway in the process of gene regulation; (2) molecular guides - lncRNAs can serve as guides by binding specific proteins and then localizing them to specific targets; (3) scaffold function - some lncRNAs can take specific proteins altogether to form scaffolding complexes; these lncRNAs provide central platforms for molecular assemble; and (4) molecular decoys - lncRNAs can downregulate their targets or specific microRNAs via the decoy effect.[9] LncRNAs are classified oncogenic lncRNAs, tumor-suppressor lncRNAs and biomarker lncRNAs, according to their role in cancer development. Oncogenic lncRNAs contribute to initiate the inappropriate activation and cellular transformation process, thus resulting in progressive and uncontrolled cell growth. Several examples of oncogenic lncRNAs include MALAT1, H19 and HOTAIR. Conversely, tumor-suppressor lncRNAs such as GAS5 and p21 protect cells against deleterious mutations and delay cancer progress.[10] The types of oncogenic lncRNAs and tumor-suppressor lncRNAs are summarized in Table 1. The expression of lncRNA in cancer can be regulated by epigenetic alteration, genetic polymorphism and transcription factors, such as p53, Sox2, MYC and Oct4.[9] The following parts mainly focus on the potential role of lncRNA in pancreatic cancer progression and prognosis.
Table 1.
Examples of oncogenic and tumor-suppressor lncRNAs
| Types | LncRNA | Gemomic location | Role in pancreatic cancer | Molecular mechanism | Refs. |
|---|---|---|---|---|---|
| Oncogenic lncRNAs | HOTAIR | Intergenic, Hox Clocus | Enhance metastasis; increase cell proliferation; altered cell cycle progression; decrease apoptosis | Gene silencing by binding PRC2 and LSD1/CoREST complexes. Interacting with EZH2 | 17,18,19,20 |
| MALAT1 | Intergenic,Chr11q13 | Facilitates cell growth, migration and invasion; induces EMT | Contributory to nuclear paraspeckle function | 21,22,23,24,25,26 | |
| HOTTIP | 5’ tip of the HOXA locus | Increase proliferation, prevent apoptosis, and induce migration; chemo-sensitivity resistance | Interacting with HOXA13 | 29,30,31 | |
| PVT1 | human chromosome 8q24 | Increase proliferation, decrease apoptosis, and induce mobility and invasion; chemo-sensitivity resistance | Closed interaction between PVT1 and MYC in nucleus; binds to miR-200 family | 33,34,35 | |
| H19 | IGF2 locus in Chr11 | Promotes cell growth and proliferation; regulates EMT | Controls imprinting; Containing miR-675; activated by cMYC |
40,41,42,43 | |
| ATB | Promotes cell invasion; induces EMT | Binding miR-200 family members, upregulates ZEB1 and ZEB2 | 36 | ||
| Tumor-suppressor lncRNAs | GAS5 | Intergenic,Chr1q25.1 | Induces apoptosis and growth arrest; decreased in pancreatic cancer tissues | Binds GR and block transcriptional induction by GR | 12 |
| LincRNA-p21 | Intergenic,upstream of p21/cdkn1a | Induces apoptosis | Induced by p53 upon DNA damage; binds hnRNP-k | 12 |
lncRNA: Long noncoding RNA; HOTAIR: HOX transcript antisense RNA; MALAT1: Metastasis-associated lung adenocarcinoma transcript 1; HOTTIP: HOXA transcript at the distal tip; PVT1: Plasmacytoma variant translocation gene 1; ATB: Activated by transforming growth factor; GAS5: Growth arrest-specific 5; IGF2: Insulin-like growth factor 2; CDKN1A: Cyclin-dependent kinase inhibitor 1A; PRUNE2: Protein prune homolog 2; EMT: Epithelial-to-mesenchymal transition; PRC2: Polycomb repressive complex 2; LSD1: Lysine-specific demethylase 1; EZH2: Enhancer of zeste homolog 2; ZEB: Zinc finger E-box-binding homeobox; GR: Glucocorticoid receptor; hnRNP-k: Heterogeneous nuclear ribonucleoprotein K
ROLE OF LNC-RNA IN BIOLOGICAL ACTIVIVITY OF PANCREATIC CANCER
Recently, increasing studies have demonstrated that lncRNAs play crucial roles in tumorigenesis of pancreatic cancer, which are involved in a variety of biological processes by regulating cell proliferation, migration, invasion and epithelial–mesenchymal transition (EMT). LncRNAs expression is significantly different between pancreatic cancer tissues and noncancerous tissues by a complementary DNA (cDNA) microarray interrogating putative lncRNAs analysis. Moreover, the expression profiles of lncRNA between primary adenocarcinoma samples and metastatic samples collected from secondary tumors are also different. In total, 355 differentially expressed transcripts including 221 protein-coding mRNAs and 134 noncoding RNAs were found in a metastasis-associated signature.[11] The expressions of oncogenic lncRNAs, such as HOTAIR, MALAT1 and H19, are higher in pancreatic cancers than that in the adjacent normal pancreatic tissue and are associated with more aggressive tumors, whereas the tumor suppressor lncRNAs expression such as gas5 and ENST00000480739 was remarkably decreased in pancreatic cancer tissues compared with their corresponding nontumorous tissues. Abnormal expression level of lncRNA indicates the functional importance of lncRNA in pancreatic cancer tissue and their possible use as novel prognostic and diagnostic biomarkers.
Impact of lncRNA on the biological capacities of pancreatic cancer
Recently, the analysis of lncRNA expression has found the crucial role of lncRNAs on pancreatic cancer proliferation, apoptosis, invasion and metastasis, and angiogenesis [Figure 1]. Here, we focus on characterized lncRNAs which can impact biological process of pancreatic cancer through targeting various key regulators.
Figure 1.
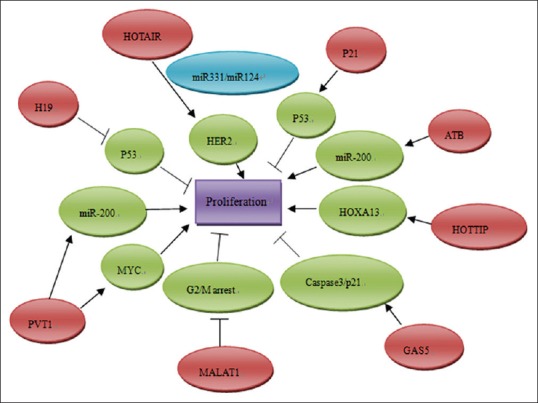
The functions and regulatory mechanisms of dysregulated long noncoding RNAs (lncRNAs) in regulating the proliferation of pancreatic cancer. The red ellipse diagrams imply the dysregulated lncRNAs in pancreatic cancer, and the green ellipse diagrams are their downstream proteins
HOTAIR is a 2158-bp lncRNA localized to a boundary in the HOXC gene cluster. HOTAIR binds to polycomb repressive complex 2 (PRC2) and is required for histone H3 lysine-27 trimethylation of the HOD locus.[12] PRC2 is a histone methyltransferase that affects epigenetic regulation during different biological processes. Studies have shown that increasing HOTAIR expression in epithelial cancer cells could increase invasive and metastatic abilities, whereas decreasing HOTAIR expression in highly metastatic, mesenchymal cancer cells, inhibited cell invasiveness and metastasis. Moreover, gene expression analysis showed that hundreds of genes were regulated by HOTAIR. Knocking down HOTAIR expression decreased expression of 552 genes and increased expression of 454 genes in pancreatic cancer cell line.[13] HOTAIR is a negative prognostic factor and shows pro-oncogenic activity in pancreatic cancer progression. The expression of HOTAIR was higher in pancreatic tumors compared with the normal pancreas and was more highly expressed in advanced tumors.[14] HOTAIR knockdown decreased pancreatic cancer cell proliferation, altered cell cycle progression and induced apoptosis. HOTAIR includes EZH2, SUZ12 and EED major subunits, which play a key role in the methyltransfer process. HOTAIR interacts with EZH2 in a region that could be bound by BRCA1. Overexpression of BRCA1 could inhibit the binding of HOTAIR to EZH2 and decrease HOTAIR-induced recruitment of PRC2 to its target genes. Other mechanisms of HOTAIR involved in cancer progression include recruiting LSD1, H3K27 methylation and H3K4 demethylation, and ultimately gene regulation. However, the precise mechanism of how HOTAIR overexpression enhances pancreatic cancer progression is still controversial and needs further investigation.[15]
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is an evolutionarily highly conserved and ubiquitously expressed lncRNA. MALAT-1 expression levels were upregulated in cancer tissue compared with adjacent noncancerous controls. It is not only overexpressed in early-stage metastasizing nonsmall cell lung cancer but also in breast, pancreas, colon, prostate and liver cancers. MALAT-1 serves as an oncogenic lncRNA that promotes malignancy phenotypes of pancreatic cancer. Elevated expression level of lncRNA MALAT-1 facilitates cell growth, migration and invasion in pancreatic cancer.[16] Knockdown of MALAT-1 inhibited tumor cell growth. Moreover, MALAT1 associated with clinical parameters and promoted tumor cell invasion and metastasis. Liu et al. study showed that the relative level of MALAT1 was significantly higher in pancreatic cancer compared with the adjacent normal pancreatic tissues. The expression of MALAT1 was significantly associated with tumor size, tumor stage and depth of invasion in normal pancreatic tissues. Overexpression of MALAT1 was an independent predictor of disease-specific survival. Patients with higher MALAT1 expression had a poorer disease-free survival.[17,18] The hypothesis of underlying mechanisms includes (1) MALAT-1 enhances stem-like phenotypes in pancreatic cancer cells and regulate sstem-like cell marker expression; (2) MALAT-1 induces EMT by activating Wnt-signaling pathway; (3) Downregulation of MALAT-1 could induce the G2/M cell cycle arrest, thus promoting cell apoptosis.[19,20] Therefore, MALAT-1 may be used as a potential therapeutic target of pancreatic cancer in clinic.
Role of lncRNAs in regulating chemotherapy sensitivity
The optimized treatment of pancreatic adenocarcinoma is to remove the entire tumor. However, curative surgery can be performed in <20% of patients, because of the absence of specific symptoms and the lack of early detection. Chemotherapy plays a critical role in prolonging overall survival of patients with pancreatic cancer. Unfortunately, the biological features of pancreatic cancer result in resistance to chemotherapy. Recent studies suggest that lncRNAs are significantly involved in chemotherapy sensitivity. Hou et al. identified differentially expressed lncRNAs and mRNA using microarray profiling of lung squamous cell carcinoma patients treated with cisplatin. The results showed that compared with the progressive disease samples, 953 lncRNAs were consistently upregulated and 749 lncRNAs were downregulated in partial response patients.[21] To uncover the precise role played by lncRNAs in cancer chemotherapy, sensitivity has potential to overcome chemotherapy resistance.
HOXA transcript at the distal tip (HOTTIP), located at the 5′ tip of the HOXA locus, is associated with the polycomb repressor complex 2 (PRC2) and WD repeat containing protein 5 (WDR5)/mixed lineage leukemia 1 (MLL1) chromatin modifying complexes. It directly controls the HOXA locus gene expression via interaction with the WDR5/MLL1 complex. A recent paper showed a regulatory loop between HOTTIP and its target, Hoxa13 in hepatocellular carcinomas (HCCs).[22] The expression of HOTTIP is increased in pancreatic cancer tissues and cell lines. Knockdown of HOTTIP in pancreatic cancer cells decreased proliferation, induced apoptosis, and decreased migration. Moreover, HOTTIP knockdown enhances the chemosensitivity of human pancreatic cancer cells to gemcitabine in vitro and in vivo. HOTTIP exerts its function in pancreatic cancer at least partly by controlling Hoxa13. There is positive correlation between HOTTIP and Hoxa13 expression. Decreased expression of Hoxa13 led to a significant reduction of HOTTIP expression in pancreatic cancer. HOTTIP/Hoxa13 axis plays a critical role in pancreatic cancer tumorigenesis as well as chemotherapy sensitivity.[23] However, Cheng et al. showed that HOTTIP regulates not Hoxa13 but several other HOX genes such as Hoxa10, Hoxb2, Hoxa11, Hoxa9 and Hoxa1 in Panc1 cells,[24] indicating that regulation of HOTTIP expression and the agents that target this lncRNA is complicated.
PVT1 is a copy number amplification-associated lncRN, which maps to band 4 of region 2 on the long arm of chromosome 8 (8q24). The location of PVT1 near Myc at human chromosome 8q24 forms a complexity of chromosomal translocation breakpoints. Co-amplification of human Myc and PVT1 has been shown to contribute to tumorigenesis. There is close interaction between PVT1 and Myc in the nucleus, and depletion of PVT1 results in the reduction of MYC protein levels. Moreover, PVT1 demonstrates a net-binding preference toward the mir-200 family. PVT1 are involved in many biological processes, including proliferation, apoptosis, mobility and invasion. It is a predictor of tumor progression and patient survival in various types of cancers, such as ovarian, breast and colorectal cancer.[25,26] You et al. identified PVT1 as a novel regulator of gemcitabine sensitivity using random homozygous knockout technology.[27] The study showed that overexpression of antisense PVT1 leads to increased sensitivity to gemcitabine, whereas overexpression of sense PVT1 results in decreased sensitivity of pancreatic cancer to gemcitabine. However, the regulation networks within the Myc-PVT1 locus and with other gene networks are complex. Further characterization of PVT1 gene and its regulation may reveal their roles in regulation of gemcitabine sensitivity.
Role of lncRNAs in EMT
EMT is a crucial process in tumor progression. It enables the tumor cells to acquire fibroblast-like properties and show reduced intercellular adhesion. EMTs are involved in many biological processes of carcinoma progression, such as induced proliferation, increased resistance to apoptosis and chemo sensitivity, the increased ability of invasion and migration, and the acquisition of stem cell–like properties.[28] During the process of EMT, expression of epithelial markers such as E-cadherin is decreased, and mesenchymal markers such as vimentin and N-cadherin can be acquired. It is reported that EMT is induced by several growth factors involved in tumor progression, such as VEGF and TGF-β.[29,30] Several molecular factors have been reported to involve in the process of EMT, including Snail, Slug, Twist, ZEB1/2, SIP1 and E12/E47. Accumulating data from high-throughput genomic approaches demonstrated the existence of lncRNAs potentially implicated in EMT [Figure 2].
Figure 2.
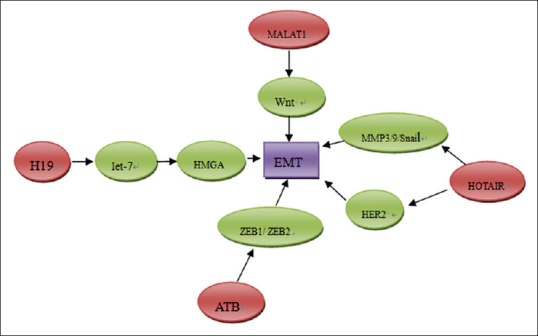
The role of long noncoding RNA in promoting epithelial mesenchymal transition of pancreatic cancer
H19 is a paternally imprinted gene located close to the telomeric region of chromosome 11p15.5. Whether H19 serves as an oncogene or has tumor-suppressor activity is still controversial. H19 was initially proposed to possess tumor-suppressive properties based on its ability to inhibit tumorigenicity.[31] However, recent evidence showed that H19 was overexpressed in many types of cancers, including bladder, hepatocellular, colon and breast cancer.[32,33,34] Ma et al. showed that H19 was significantly highly expressed in pancreatic cancer tissues than corresponding adjacent nontumorous tissues. Moreover, H19 levels are remarkably increased in primary tumors that subsequently metastasized compared with those that did not metastasis, indicating that H19 might play a role in pancreatic cancer metastasis. In vitro study showed that H19 promoted migration and invasion of pancreatic cancer cells, and H19 regulated the pancreatic cancer cell migration and invasion partly via regulating let-7. H19 antagonized let-7 and affected its target Hmga2, which induced EMT in pancreatic cancer cells. H19-siRNA treatment decreased HMGA2 protein levels, as well as EMT marker vimentin, but increased E-cadherin. The results demonstrated that H19/let-7/HMGA2/EMT-signaling axis played important roles in pancreatic cancer progression via EMT.[35]
LncRNA activated by TGF-b (lncRNA-ATB) is a novel lncRNA, which could be activated by TGF-β. The expression of lncRNA-ATB was increased in HCC, gastric cancer and breast cancer. It is reported that lncRNA-ATB promotes cell invasion by competitively binding miR-200 family members, upregulates ZEB1 and ZEB2, and inducing EMT in HCC.[36] Lnc-ATB mediated the process of EMT to promote invasion and metastasis via TGF-β/miR-200s/ZEB-signaling network. However, there is no report on the role of lncRNA-ATB in pancreatic cancer progression at present. The agitated effects of lncRNA-ATB on the invasion-metastasis cascade suggest that lncRNA-ATB could be an effective target for antimetastasis therapies.
LNC-RNA AS BIOMARKERS AND THERAPEUTIC TARGET IN CANCER
Recently, technological advance in high-throughput sequencing has been applied to identify the dysregulated expression of lncRNAs in various types of cancer. LncRNAs may be used as sensitive biomarkers for diagnosis, prognosis and prediction of response to therapy.
LncRNA diagnostic biomarkers
Some lncRNAs commonly show restricted tissue-specific and cancer-specific expression patterns. For example, prostate cancer antigen 3 (PCA3) is a prostate-specific gene and highly overexpressed in prostate cancer. A meta-analysis including 13 clinical trials about urine PCA3 test in the diagnosis of prostate cancer showed that the pooled sensitivity and specificity were 0.62 (95% confidence interval [CI]: 0.59–0.65) and 0.75 (95% CI: 0.73–0.76), respectively. Urine PCA3 test has acceptable sensitivity and specificity in the diagnosis of prostate cancer.[37] The Progensa® PCA3 test has been approved by FDA for the detection of prostate cancer.[38] Using PCA3 in conjunction with PSA can improve the performance of PSA-based diagnostic accuracy. Combination PSA-PCA3 strategies were able to substantially reduce the false-positive tests.[39,40,41] The PCA3 test for diagnosis of prostate cancer is one of the most effective clinical translations of a cancer-associated lncRNA gene. Other lncRNA-based diagnoses for cancer, such as pancreatic cancer, are underway and have not been widely used. It is anticipated that advances in the study of lncRNAs will yield new sensitive diagnostic biomarkers for early diagnosis of pancreatic cancer.
LncRNA prognostic biomarkers
Abnormally expressed lncRNAs may have either oncogenic or tumor suppressor functions during cancer initiation, development and progression. A complementary DNA (cDNA) microarray interrogating putative lncRNAs showed that lncRNAs expression is significantly altered between pancreatic cancer tissues and noncancerous tissues.[11] Many lncRNAs have been not only been implied to be involved in the development and progression of pancreatic cancer but are also the clinical prognostic indicators. LncRNA H19, HOTAIR, MALAT1, etc., are elevated in pancreatic cancer in comparison to adjacent normal tissues and correlate with more aggressive status of pancreatic cancer. These lncRNAs were independent unfavorable prognostic factors and positively correlated with overall survival in patients with cancer. These lncRNAs could become desirable candidates for monitoring high-risk populations, predicting prognosis and monitoring recurrence.
LncRNA-based therapies
LncRNA are interesting targets in cancer therapy and have multiple advantages as novel-specific therapeutic targets. The role that lncRNA played in apoptosis, proliferation of cancer is of great relevance for the design of new drugs. Selectively, silencing oncogenic lncRNAs provides a new strategy for cancer therapy. LncRNAs can be easily manipulated through RNAi or LNA GapmeRs. Systemic administration of siRNA to patients with solid cancers has been conducted by Davis et al. early in 2010 and achieved great success.[42] As another strategy for knockdown of lncRNAs, LNA GapmeRs consist of a DNA stretch flanked by LNA nucleotides, which interact selectively with the lncRNA. LNA GapmeRs degrade the targeted lncRNA via an RNAse H-dependent mechanism.[43] However, more functional and structural studies on lncRNA biology are necessary before the new therapeutic options are applied.
CONCLUSIONS
In conclusion, there are many studies showing that lncRNAs fulfill important functions in cancer initiation, progression, invasion and metastasis, as well as prognosis. In this review, we have discussed some examples of lncRNAs and the potential underlying mechanism involved in pancreatic cancer. LncRNAs may be used as diagnostic, prognostic biomarkers of cancer and provide a new strategy for pancreatic cancer treatment. However, there are still many challenges that remain to be solved. For example, the biological and molecular characteristics of most lncRNAs remain unknown and the mechanism of lncRNA in pancreatic cancer regulation is still not understood. More studies are needed to elucidate the association of lncRNAs and pancreatic cancer.
Financial support and sponsorship
The present study was supported by the National Natural Science Foundation of China (Grant nos. 81802888) and the Key Research and Development Project of Shandong Province (Grant no. 2018GSF118088).
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Kitagawa M, Kitagawa K, Kotake Y, Niida H, Ohhata T. Cell cycle regulation by long non-coding RNAs. Cell Mol Life Sci. 2013;70:4785–94. doi: 10.1007/s00018-013-1423-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–9. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gutschner T, Diederichs S. The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 2012;9:703–19. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 6.Aghdassi A, Phillips P, Dudeja V, Dhaulakhandi D, Sharif R, Dawra R, et al. Heat shock protein 70 increases tumorigenicity and inhibits apoptosis in pancreatic adenocarcinoma. Cancer Res. 2007;67:616–25. doi: 10.1158/0008-5472.CAN-06-1567. [DOI] [PubMed] [Google Scholar]
- 7.Bánfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE, Jr, et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012;22:1646–57. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jia H, Osak M, Bogu GK, Stanton LW, Johnson R, Lipovich L. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA. 2010;16:1478–87. doi: 10.1261/rna.1951310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng K, Guo X, Wang H, Xia J. The lncRNA-MYC regulatory network in cancer. Tumour Biol. 2014;35:9497–503. doi: 10.1007/s13277-014-2511-y. [DOI] [PubMed] [Google Scholar]
- 10.Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol. 2013;26:155–65. doi: 10.1038/modpathol.2012.160. [DOI] [PubMed] [Google Scholar]
- 11.Tahira AC, Kubrusly MS, Faria MF, Dazzani B, Fonseca RS, Maracaja-Coutinho V, et al. Long noncoding intronic RNAs are differentially expressed in primary and metastatic pancreatic cancer. Mol Cancer. 2011;10:141. doi: 10.1186/1476-4598-10-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–25. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stratford JK, Bentrem DJ, Anderson JM, Fan C, Volmar KA, Marron JS, et al. A six-gene signature predicts survival of patients with localized pancreatic ductal adenocarcinoma. PLoS Med. 2010;7:e1000307. doi: 10.1371/journal.pmed.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai B, Song XQ, Cai JP, Zhang S. HOTAIR: A cancer-related long non-coding RNA. Neoplasma. 2014;61:379–91. doi: 10.4149/neo_2014_075. [DOI] [PubMed] [Google Scholar]
- 16.Jiao F, Hu H, Yuan C, Wang L, Jiang W, Jin Z, et al. Elevated expression level of long noncoding RNA MALAT-1 facilitates cell growth, migration and invasion in pancreatic cancer. Oncol Rep. 2014;32:2485–92. doi: 10.3892/or.2014.3518. [DOI] [PubMed] [Google Scholar]
- 17.Liu JH, Chen G, Dang YW, Li CJ, Luo DZ. Expression and prognostic significance of lncRNA MALAT1 in pancreatic cancer tissues. Asian Pac J Cancer Prev. 2014;15:2971–7. doi: 10.7314/apjcp.2014.15.7.2971. [DOI] [PubMed] [Google Scholar]
- 18.Pang EJ, Yang R, Fu XB, Liu YF. Overexpression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour Biol. 2015;36:2403–7. doi: 10.1007/s13277-014-2850-8. [DOI] [PubMed] [Google Scholar]
- 19.Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Upregulated MALAT-1contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst. 2012;8:2289–94. doi: 10.1039/c2mb25070e. [DOI] [PubMed] [Google Scholar]
- 20.Jiao F, Hu H, Han T, Yuan C, Wang L, Jin Z, et al. Long noncoding RNA MALAT-1 enhances stem cell-like phenotypes in pancreatic cancer cells. Int J Mol Sci. 2015;16:6677–93. doi: 10.3390/ijms16046677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou Z, Xu C, Xie H, Xu H, Zhan P, Yu L, et al. Long noncoding RNAs expression patterns associated with chemo response to cisplatin based chemotherapy in lung squamous cell carcinoma patients. PLoS One. 2014;9:e108133. doi: 10.1371/journal.pone.0108133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, Procino A, et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59:911–23. doi: 10.1002/hep.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Z, Zhao X, Zhou Y, Liu Y, Zhou Q, Ye H, et al. The long non-coding RNA HOTTIP promotes progression and gemcitabine resistance by regulating Hoxa13 in pancreatic cancer. J Transl Med. 2015;13:84. doi: 10.1186/s12967-015-0442-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng Y, Jutooru I, Chadalapaka G, Corton JC, Safe S. The long non-coding RNA HOTTIP enhances pancreatic cancer cell proliferation, survival and migration. Oncotarget. 2015;6:10840–52. doi: 10.18632/oncotarget.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahashi Y, Sawada G, Kurashige J, Uchi R, Matsumura T, Ueo H, et al. Amplification of PVT-1 is involved in poor prognosis via apoptosis inhibition in colorectal cancers. Br J Cancer. 2014;110:164–71. doi: 10.1038/bjc.2013.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guan Y, Kuo WL, Stilwell JL, Takano H, Lapuk AV, Fridlyand J, et al. Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin Cancer Res. 2007;13:5745–55. doi: 10.1158/1078-0432.CCR-06-2882. [DOI] [PubMed] [Google Scholar]
- 27.You L, Chang D, Du HZ, Zhao YP. Genome-wide screen identifies PVT1 as a regulator of Gemcitabine sensitivity in human pancreatic cancer cells. Biochem Biophys Res Commun. 2011;407:1–6. doi: 10.1016/j.bbrc.2011.02.027. [DOI] [PubMed] [Google Scholar]
- 28.Fan F, Samuel S, Evans KW, Lu J, Xia L, Zhou Y, et al. Overexpression of snail induces epithelial-mesenchymal transition and a cancer stem cell-like phenotype in human colorectal cancer cells. Cancer Med. 2012;1:5–16. doi: 10.1002/cam4.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang AD, Camp ER, Fan F, Shen L, Gray MJ, Liu W, et al. Vascular endothelial growth factor receptor-1 activation mediates epithelial to mesenchymal transition in human pancreatic carcinoma cells. Cancer Res. 2006;66:46–51. doi: 10.1158/0008-5472.CAN-05-3086. [DOI] [PubMed] [Google Scholar]
- 30.Miyazono K. Transforming growth factor-beta signaling in epithelial-mesenchymal transition and progression of cancer. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:314–23. doi: 10.2183/pjab.85.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hao Y, Crenshaw T, Moulton T, Newcomb E, Tycko B. Tumour-suppressor activity of H19 RNA. Nature. 1993;365:764–7. doi: 10.1038/365764a0. [DOI] [PubMed] [Google Scholar]
- 32.Byun HM, Wong HL, Birnstein EA, Wolff EM, Liang G, Yang AS. Examination of IGF2 and H19 loss of imprinting in bladder cancer. Cancer Res. 2007;67:10753–8. doi: 10.1158/0008-5472.CAN-07-0329. [DOI] [PubMed] [Google Scholar]
- 33.Lottin S, Adriaenssens E, Dupressoir T, Berteaux N, Montpellier C, Coll J, et al. Overexpression of an ectopic H19 gene enhances the tumorigenic properties of breast cancer cells. Carcinogenesis. 2002;23:1885–95. doi: 10.1093/carcin/23.11.1885. [DOI] [PubMed] [Google Scholar]
- 34.Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abu-lail R, Hochberg A, et al. The H19 non-coding RNA is essential for human tumor growth. PLoS One. 2007;2:e845. doi: 10.1371/journal.pone.0000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma C, Nong K, Zhu H, Wang W, Huang X, Yuan Z, et al. H19 promotes pancreatic cancer metastasis by derepressing let-7's suppression on its target HMGA2-mediated EMT. Tumour Biol. 2014;35:9163–9. doi: 10.1007/s13277-014-2185-5. [DOI] [PubMed] [Google Scholar]
- 36.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–81. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 37.Xue WJ, Ying XL, Jiang JH, Xu YH. Prostate cancer antigen 3 as a biomarker in the urine for prostate cancer diagnosis: A meta-analysis. J Cancer Res Ther. 2014;10(Suppl):C218–21. doi: 10.4103/0973-1482.145881. [DOI] [PubMed] [Google Scholar]
- 38.Groskopf J, Aubin SM, Deras IL, Blase A, Bodrug S, Clark C, et al. APTIMA PCA3 molecular urine test: Development of a method to aid in the diagnosis of prostate cancer. Clin Chem. 2006;52:1089–95. doi: 10.1373/clinchem.2005.063289. [DOI] [PubMed] [Google Scholar]
- 39.Wei JT, Feng Z, Partin AW, Brown E, Thompson I, Sokoll L, et al. Can urinary PCA3 supplement PSA in the early detection of prostate cancer? J Clin Oncol. 2014;32:4066–72. doi: 10.1200/JCO.2013.52.8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Birnbaum JK, Feng Z, Gulati R, Fan J, Lotan Y, Wei JT, et al. Projecting benefits and harms of novel cancer screening biomarkers: A study of PCA3 and prostate cancer. Cancer Epidemiol Biomarkers Prev. 2015;24:677–82. doi: 10.1158/1055-9965.EPI-14-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salameh A, Lee AK, Cardó-Vila M, Nunes DN, Efstathiou E, Staquicini FI, et al. PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3. Proc Natl Acad Sci U S A. 2015;112:8403–8. doi: 10.1073/pnas.1507882112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–70. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang G, Lu X, Yuan L. LncRNA: A link between RNA and cancer. Biochim Biophys Acta. 2014;1839:1097–109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]


