ABSTRACT
Our latest publication on the inhibition of Alzheimer disease (AD) through mitophagy consolidates the ‘defective mitophagy hypothesis of AD etiology’. Dementia (majorly AD) affects over 50 million people worldwide, and for AD there is no cure. AD leads to progressive loss of cognition, and pathological hallmarks of AD include aggregates of amyloid-β peptides extracellularly and MAPT (microtubule associated protein tau) intracellularly. However, there is no conclusive link between these pathological markers and cognitive symptoms. Anti-AD drug candidates have repeatedly failed, which led us to investigate other molecular etiologies to guide drug development. Mitochondria produce the majority of cellular ATP, affect Ca2+ and redox signaling, and promote developmental and synaptic plasticity. Mitochondrial dysfunction and accumulation of damaged mitochondria are common in brain tissues from AD patients and transgenic AD animal models, but the underlying molecular mechanisms are not fully understood. Damaged mitochondria are removed through multiple pathways, the major 2 being mitophagy and the ubiquitin proteasome pathway. Mitophagy is essential for clearance of damaged mitochondria to maintain mitochondrial homeostasis, ATP production, and neuronal activity and survival. These pieces of evidence converge on the ‘defective mitophagy hypothesis of AD etiology’, and the current cross-species study provides strong support for this hypothesis.
KEYWORDS: Mitophagy, mitochondria, Alzheimer’s disease, aging, memory
The takeaway of our study is that neuronal mitophagy is impaired in AD. Compared with normal controls, postmortem hippocampal brain tissues from AD patients show an accumulation of damaged mitochondria as well as reduced mitophagy. The mechanism underlying this may be an impaired initiation of the mitophagic machinery, evinced by lower phosphorylated (S172)-TBK1 and lower phosphorylated (S555)-ULK1 [1]. This mechanism is seen in both familial (APP) and sporadic (APOE/APOE4) AD patient iPSC-derived cortical neurons. For further mechanistic and interventional studies, we generated transgenic nematodes (C. elegans) expressing the DCT-1 mitophagy receptor fused with GFP together with the autophagosomal marker LGG-1 fused with DsRed in neurons of both the Aβ1-42(CL2355) and the TAU(BR5270) strains. Both Aβ1-42(CL2355) and TAU(BR5270) nematodes display defective energy metabolism and reduced mitophagy. Impaired mitochondrial function and reduced mitophagy are shown in the hippocampal tissues of an APP-PSEN1/PS1 AD mouse model. These cross-species results unambiguously indicate impaired mitophagic machinery in AD.
If defective mitophagy is a contributing factor to AD, then restoration of mitophagy should inhibit disease phenotypes. We established an in vivo drug-screening platform using nematodes to identify potent neuronal mitophagy inducers. We have identified 3 potent neuronal mitophagy-inducing agents, the NAD+ precursor nicotinamide mononucleotide (NMN), urolithin A (UA), and the antibiotic actinonin (AC). Our genetic mutation studies in the Aβ1-42(CL2355) nematodes show that UA- and AC-dependent memory improvement depends on PINK-1 and PDR-1 but not DCT-1. However, genetic mutation studies in the TAU(BR5270) AD nematodes indicate UA-induced memory improvement is dependent on both PINK-1 and PDR-1, whereas AC-induced memory improvement is only dependent on PINK-1. Reasons for the difference in AC-dependent molecular pathways of mitophagy in Aβ and TAU nematodes are unclear so far.
How does NAD+ induce mitophagy/autophagy? NAD+-dependent inhibition of cognitive loss in AD is indeed dependent on mitophagy, as knockout of any of 3 mitophagy genes (pink-1, pdr-1, and dct-1) eliminates cognitive benefit in both the Aβ1-42(CL2355) and the TAU(BR5270) AD nematodes1. NAD+ is a cofactor for other proteins, such as the sirtuins (SIRT1 to SIRT7), CD38, SARM1 (sterile alpha and TIR motif containing 1), and PARP (poly[ADP-ribose] polymerase) proteins, so there is a possibility of the involvement of different pathways (Figure 1). The NAD+-dependent deacetylase SIRT1 upregulates autophagy/mitophagy through multiple pathways: by deacetylating the autophagy machinery proteins ATG5, ATG7, and MAP1LC3/LC3; by increasing the expression of BECN1, a key member of the autophagy initiation class III phosphatidylinositol 3-kinase nucleation complex; by deacetylating LC3 at K49 and K51 in the nucleus, enabling its nucleocytoplasmic transportation and binding to ATG7 for autophagy; and by increasing the expression of autophagic proteins RAB7, LC3, ATG12, and BNIP3 through deacetylation and acetylation of the transcriptional factors FOXO1 and FOXO3. RAB7, a small GTPase, interacts with the UVRAG-VPS16 complex to enhance the maturation of autophagosomes and endosomes. The NAD+-SIRT1 pathway may also increase mitophagy through stimulation of autophosphorylation and activation of ATM (ATM serine/threonine kinase), which induces mitophagy via a STK11/LKB1-AMPK-TSC2 pathway. There are 3 mitochondrial sirtuins, SIRT3, SIRT4, and SIRT5. Recently, it has been shown that NAD+ replenishment induces mitochondrial SIRT3-PGAM5-FUNDC1-dependent mitophagy. Nuclear SIRT6 and SIRT7 induce autophagy through inhibition of MTOR. Whereas the NAD+-consuming enzyme CD38 plays a necessary role in autophagic fusion with lysosomes, the newly discovered NAD+-consuming enzyme SARM1 facilitates mitophagy via formation of a PINK1-SARM1-TRAF6 (TNF receptor associated factor 6) complex to stabilize PINK1 on depolarized mitochondria. Interestingly, SARM1 is required for activation of an injury-induced axon degeneration; thus, SARM1 may be involved in both neuroprotection and neuronal death. NAD+ also induces expression of the anti-inflammatory cytokine IL10 which induces mitophagy through MTOR inhibiton. NAD+ is involved with the TCA cycle, OXPHOS, and β-oxidation, so how these metabolic pathways interact with mitophagy is elusive. Noticeably, increased NAD+ may also inhibit autophagy/mitophagy through SIRT2, SIRT4, SIRT5, and PARPs. A possible balance of robust NAD+-dependent mitophagy induction and a mild NAD+-dependent mitophagy inhibition presents a still robust mitophagy induction (Figure 1).
Figure 1.
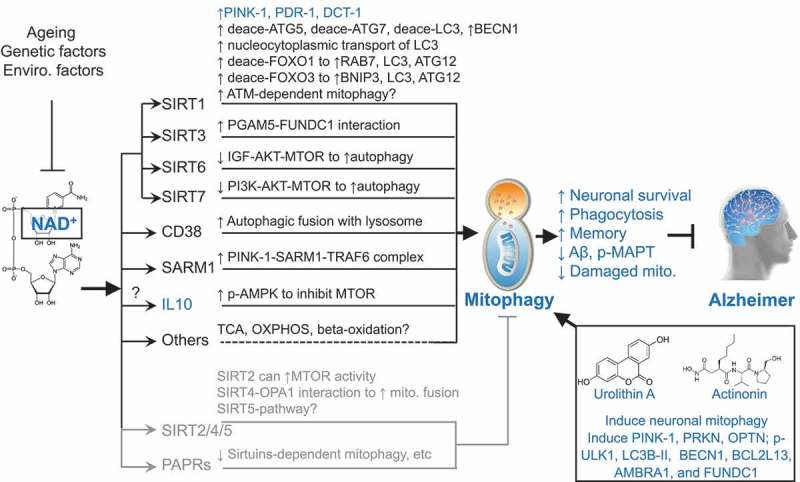
Schematic representation of how NAD+, urolithin A, and actinonin induce mitophagy, and inhibit Alzheimer disease (AD). Cellular NAD+ levels are reduced with ageing as well as affected by genetic and environmental (enviro.) factors. NAD+ is a cofactor of sirtuins (SIRT1 to SIRT7), CD38, SARM1, and PARPs. The nuclear SIRT1, SIRT6, SIRT7, mitochondrial SIRT3, CD38, and SARM1 induce mitophagy/autophagy. NAD+ may induce mitophagy through other pathways such as through the induction of the mitophagy inducer IL10 and fundamental metabolic pathways. Increased NAD+ may also inhibit autophagy/mitophagy through cytoplasmic SIRT2, mitochondrial SIRT4, mitochondrial SIRT5, and the DNA damage sensor PARPs. One reasonable explanation is that a robust NAD+-dependent mitophagy induction and a mild NAD+-dependent mitophagy inhibition give an outcome of a remaining robust induced mitophagy. UA and AC are robust mitophagy inducers, both inducing expression of mitophagy/autophagy proteins such as PINK1, PRKN, OPTN, p-ULK1, LC3B-II, BECN1, BCL2L13, AMBRA1, and FUNDC1. Results marked in blue are from the current study. Results marked in dark (induction of mitophagy) and gray (inhibition of mitophagy) are from previous publications. See manuscript for details and references. deace, deacetylated.
Collectively, our study strongly indicates defective mitophagy in AD with mitophagy reduction as a possible major driver of AD pathology. It is important to point out that defective mitophagy solely may not suffice to induce AD; genetic factors (e.g., mutations of APP and PSEN1, APOE variant, and MAPT) and environmental challenges may work together to induce and exacerbate AD. Many questions need to be addressed regarding this hypothesis: What are the connections between mitophagy, Aβ, and p-MAPT, and which occurs first? Nonetheless, manipulation of mitophagy might therefore be an attractive therapeutic target for AD patients. NAD+ precursors, including NMN and nicotinamide riboside (NR), are promising drug candidates in view of their natural existence in the human body and their safety and efficiency in preclinical trials. Large randomized, double-blind, placebo-controlled studies of the effects of neuronal mitophagy inducers on AD patients are necessary.
Funding Statement
This work was supported by the Helse Sør-Øst RHF [017056]; Norges Forskningsråd [#262175 and #277813].
Acknowledgments
I acknowledge the invaluable collaborations and continued supports from Drs. Vilhelm Bohr, Mark P. Mattson, Nektarios Tavernarakis, M. Zameel Cader, and Hilde Nilsen. I appreciate Dr. Yujun Hou, Dr. Konstantinos Palikaras, and all the other co-authors for all their contributions in the current study. I thank Jesse Keer for reading the manuscript. I am grateful for the generous support from the HELSE SøR-ØST (E.F.F., #2017056), the Research Council of Norway (E.F.F., #262175 and #277813), and an Akershus University Hospital Strategic grant (E.F.F., #269901). The Fang group has CRODA with Chromadex. This ´autophagic puncta´ was written based on our recent publication and several major findings from other research papers. I was unable to cite these references due to the one citation limitation of this ´autophagic puncta´.
Disclosure statement
E.F.F. has Cooperative Research and Development Agreement (CRADA) arrangement with ChromaDex.
Reference
- [1].Fang EF, Huo Y, Palikaras K, et al. Mitophagy inhibits amyloid-β and tau pathology and reverses cognitive deficits in models of Alzheimer’s disease. Nat Neurosci. 2019;22(3):401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]


