ABSTRACT
Macroautophagy is a process through which eukaryotic cells degrade large substrates including organelles, protein aggregates, and invading pathogens. Over 40 autophagy-related (ATG) genes have been identified through forward-genetic screens in yeast. Although homology-based analyses have identified conserved ATG genes in plants, only a few atg mutants have emerged from forward-genetic screens in Arabidopsis thaliana. We developed a screen that consistently recovers Arabidopsis atg mutations by exploiting mutants with defective LON2/At5g47040, a protease implicated in peroxisomal quality control. Arabidopsis lon2 mutants exhibit reduced responsiveness to the peroxisomally-metabolized auxin precursor indole-3-butyric acid (IBA), heightened degradation of several peroxisomal matrix proteins, and impaired processing of proteins harboring N-terminal peroxisomal targeting signals; these defects are ameliorated by preventing autophagy. We optimized a lon2 suppressor screen to expedite recovery of additional atg mutants. After screening mutagenized lon2-2 seedlings for restored IBA responsiveness, we evaluated stabilization and processing of peroxisomal proteins, levels of several ATG proteins, and levels of the selective autophagy receptor NBR1/At4g24690, which accumulates when autophagy is impaired. We recovered 21 alleles disrupting 6 ATG genes: ATG2/At3g19190, ATG3/At5g61500, ATG5/At5g17290, ATG7/At5g45900, ATG16/At5g50230, and ATG18a/At3g62770. Twenty alleles were novel, and 3 of the mutated genes lack T-DNA insertional alleles in publicly available repositories. We also demonstrate that an insertional atg11/At4g30790 allele incompletely suppresses lon2 defects. Finally, we show that NBR1 is not necessary for autophagy of lon2 peroxisomes and that NBR1 overexpression is not sufficient to trigger autophagy of seedling peroxisomes, indicating that Arabidopsis can use an NBR1-independent mechanism to target peroxisomes for autophagic degradation.
Abbreviations: ATG: autophagy-related; ATI: ATG8-interacting protein; Col-0: Columbia-0; DSK2: dominant suppressor of KAR2; EMS: ethyl methanesulfonate; GFP: green fluorescent protein; IAA: indole-3-acetic acid; IBA: indole-3-butyric acid; ICL: isocitrate lyase; MLS: malate synthase; NBR1: Next to BRCA1 gene 1; PEX: peroxin; PMDH: peroxisomal malate dehydrogenase; PTS: peroxisomal targeting signal; thiolase: 3-ketoacyl-CoA thiolase; UBA: ubiquitin-associated; WT: wild type
KEYWORDS: LON2 protease, organelle quality control, pexophagy, peroxisome turnover, suppressor genetics
Introduction
Eukaryotes use macroautophagy, hereafter referred to as autophagy, to maintain cellular homeostasis during development and in response to environmental changes. This degradation process targets protein aggregates, pathogens, damaged or superfluous organelles, and other cellular components [1–6]. During autophagy, a double-membrane structure known as a phagophore surrounds and engulfs the fated cargo. Once enclosed, the outer membrane of the resulting double-membrane-bound autophagosome fuses with a lysosome (in metazoans) or vacuole (in plants and fungi) to deliver the cargo into the lysosome or vacuole lumen. The cargo is broken down, and constituent amino acids, lipids, and carbohydrates are returned to the cytosol, where these molecules can be recycled to synthesize new organelles and proteins.
In plants, autophagy promotes survival during starvation and harsh environmental conditions. Autophagy is upregulated in plants during nutrient deprivation [7], and plants lacking autophagy are hypersensitive to carbon and nitrogen starvation [8], fungal infection [9], and various abiotic stresses, including heat stress [10], drought and salt stress [11], and oxidative stress [12]. Autophagy is also upregulated during seed maturation [13], suggesting a role during embryo development. Although autophagy is not required for plant survival in optimal growth conditions [8], plants lacking autophagy accumulate the selective autophagy receptor NBR1 [14] and certain peroxisomal proteins [15,16], suggesting that basal autophagy also contributes to general intracellular processes.
Nearly half of the over 40 identified autophagy-related (ATG) proteins are part of the core autophagy machinery that is conserved across kingdoms, including in the reference plant Arabidopsis thaliana [3–6]. These core ATG proteins can be grouped by the functions that they support. ATG1, ATG11, ATG13, and ATG101 are involved in initiating autophagosome formation [7,17,18]. ATG2, ATG6, ATG9, and ATG18 participate in phagophore expansion [18–24]. ATG8 is a ubiquitin-like protein that decorates the phagophore via conjugation to phosphatidylethanolamine [8,25,26], and this ATG8 lipidation requires ATG3, ATG4, ATG5, ATG7, ATG10, ATG12, and ATG16 [8,27–32].
A variety of receptors with differing cargo specificity mediate selective autophagy by controlling which substrates are engulfed by autophagosomes. Selective autophagy receptors link organelles, protein aggregates, or other cargo to the autophagy machinery by binding to both the fated cargo and ATG8 [33]. These receptors generally bind ATG8 via an ATG8-interacting motif with the consensus core sequence [W/Y/F]xx[L/I/V] with neighboring acidic (D or E) residues [33,34]. The ability to recognize not only specific types of organelles but also damaged or unnecessary organelles is paramount to maintaining cellular homeostasis, and the molecular mechanisms that enact various selective autophagy pathways continue to be unraveled.
Several selective autophagy receptors have been characterized [4,33]. For example, mammalian NBR1 and SQSTM1/p62 (sequestosome 1) bind both LC3/ATG8 and ubiquitin [33]. Plants encode NBR1 orthologs (known as Joka2 in tobacco), but plant orthologs of SQSTM1/p62 have not been identified [14,35]. Arabidopsis nbr1 mutants display sensitivity to only a subset of the stressors to which atg mutants display sensitivity [10,36], suggesting that NBR1 acts as a selective autophagy receptor for a subset of autophagic cargos. For example, Arabidopsis NBR1 is implicated in clearing ubiquitinated protein aggregates following heat stress [10] and in limiting cauliflower mosaic virus infection by targeting virus particle-forming capsid proteins for autophagic degradation [37] but appears not to participate in autophagy of proteasomes [38]. Several plant-specific ATI (ATG8-interacting) proteins [39,40] are implicated in autophagy. For example, ATI1/At2g45980 plays a role in autophagy of plastids [41], and ATI3A/At1g17780 is implicated in autophagy of the endoplasmic reticulum (ER) [40]. Besides ATI proteins, Arabidopsis uses a ubiquitin receptor to target a transcription factor for autophagic degradation [42] and uses a proteasome subunit as a receptor for autophagy of proteasomes [38].
One target of autophagy is the peroxisome, an organelle that sequesters oxidative reactions. Most notably, peroxisomes house fatty acid β-oxidation, which converts fatty acids into acetyl-CoA and produces hydrogen peroxide (H2O2) as a byproduct [43]. Peroxisomes use various enzymes to detoxify H2O2, including catalase [44], which is itself susceptible to oxidative damage by H2O2 [45]. Pexophagy, the selective autophagy of peroxisomes, contributes to peroxisome homeostasis in plants by degrading damaged or obsolete peroxisomes [46]. Pexophagy occurs at a higher basal rate than autophagy of other organelles in Arabidopsis seedlings, as evidenced by the higher relative accumulation of peroxisomal versus other organellar proteins in atg5 mutants [15,47]. Moreover, Arabidopsis atg mutants accumulate peroxisomes [16] and nonfunctional, oxidized catalase [15,47], suggesting a role for pexophagy in maintaining peroxisome function. However, the molecular mechanisms by which plant cells recognize peroxisomes in need of turnover remain unclear.
In addition to quality control via pexophagy, plant cells regulate peroxisome homeostasis through the peroxisomal matrix protease LON2 [48–50]. Like E. coli Lon [51,52] and yeast Pln/peroxisomal Lon [53,54], LON2 is an AAA ATPase thought to act as both a chaperone and a protease that recovers misfolded proteins and degrades proteins that cannot be refolded; the chaperone function of LON2 is implicated in preventing pexophagy [50]. Dysfunctional LON2 results in enlarged peroxisomes and heightened pexophagy [48,50,55]. Notably, the peroxisomal glyoxylate cycle enzymes ICL (isocitrate lyase/At3g21720) and MLS (malate synthase/At5g03860) are degraded at wild-type rates when LON2 is disrupted, weakly stabilized when autophagy is disrupted, and strongly stabilized when both LON2 and autophagy are disrupted [48,50], implicating both LON2 and autophagy in ICL and MLS turnover. In contrast, the peroxisomal β-oxidation enzyme thiolase (3-ketoacyl-CoA thiolase/At2g33150) is destabilized in lon2 mutants [49] but stabilized when autophagy or both LON2 and autophagy are disrupted [48,50], hinting that thiolase is primarily turned over via pexophagy.
We have previously recovered 1 atg3, 2 atg2, and 5 atg7 alleles by screening for suppressors of Arabidopsis lon2 mutants [48,56]. Peroxisomal β-oxidation converts the auxin precursor indole-3-butyric acid (IBA) to the active hormone indole-3-acetic acid (IAA) [57–59], and IBA treatment promotes abundant lateral root production [58,60] due to the IAA produced by metabolism of IBA. Unlike wild type, lon2 scarcely forms lateral roots in response to IBA [49] because lon2 peroxisomes are excessively degraded by pexophagy [48], decreasing peroxisome abundance and thereby presumably decreasing IBA-to-IAA conversion. Because screening for suppressors restoring lon2 IBA responsiveness affords a homology-independent method to uncover autophagy components in Arabidopsis [48], we expanded the lon2 suppressor screen. This effort yielded 21 alleles of 6 ATG genes, including 1 atg3, 2 atg2, 2 atg5, 2 atg18a, 3 atg16, and 11 atg7 mutants. We did not recover atg11 or nbr1 mutants in our screen, but using reverse genetics, we demonstrated that loss of ATG11 incompletely suppresses lon2 phenotypes and that the selective autophagy receptor NBR1 is neither necessary for pexophagy in the lon2 mutant nor sufficient for stimulating pexophagy when overexpressed in Arabidopsis seedlings.
Results
Forward-genetic screening strategy for Arabidopsis autophagy mutants
We used a 4-step strategy (Figure 1) to recover 21 mutants carrying mutations in 6 different Arabidopsis ATG genes (Table 1). First, to identify potential atg mutants, we screened approximately 500,000 8-day-old lon2-2 M2 seedlings from 104 mutagenized pools for the presence of lateral roots on medium containing 8 µM IBA (Figure 1(a)). Unlike wild-type seedlings, lon2 seedlings fail to form abundant lateral roots in response to IBA [49], and preventing autophagy in lon2 mutants suppresses this phenotype [48].
Figure 1.
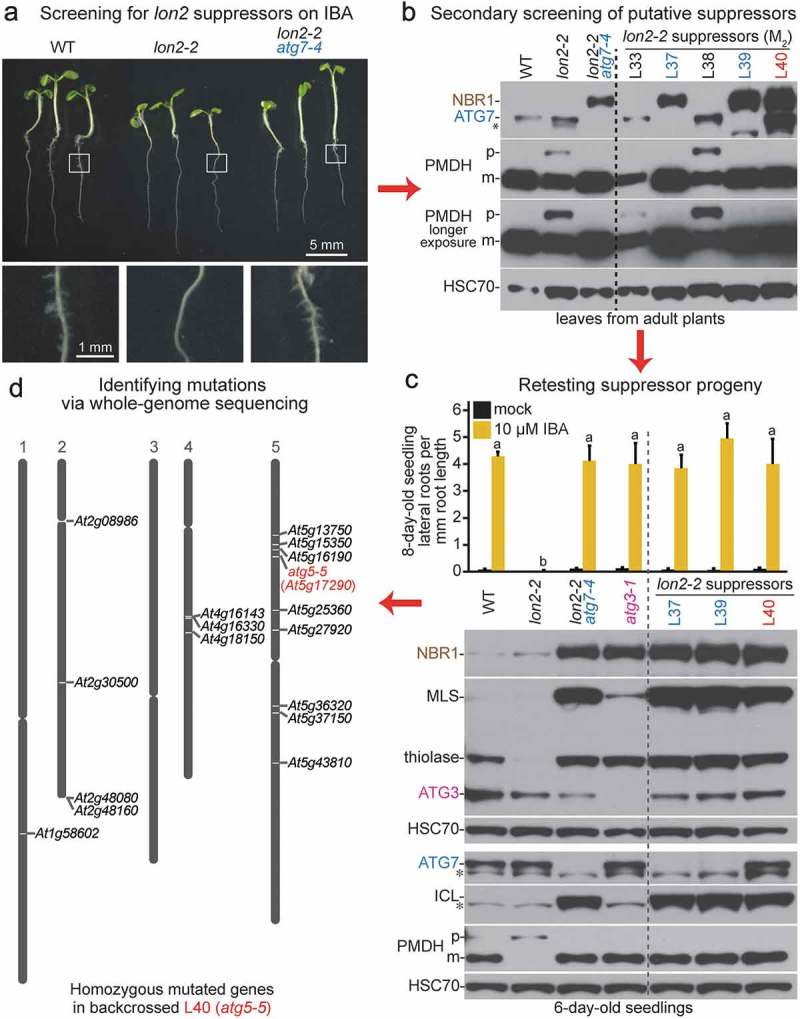
Four-step strategy for isolating Arabidopsis autophagy-defective mutants. (a) Initial screening for lon2 suppressors. lon2 forms few lateral roots in response to IBA; putative suppressors that formed lateral roots in the presence of IBA similar to wild-type and lon2-2 atg7-4 were moved to soil for propagation. Top panel: wild-type (WT), lon2-2, and lon2-2 atg7-4 seedlings were grown on media containing 8 µM IBA and imaged at 8 days; scale bar: 5 mm. Bottom row: magnified images of roots outlined in the top panel showing lateral roots, which are absent in lon2-2; scale bar: 1 mm. (b) Secondary screening of putative suppressors. Leaf extracts from approximately 30-day-old adult controls (left of dashed line) or M2 putative suppressor plants (right of dashed line) were processed for immunoblotting with antibodies to the indicated proteins. NBR1 is a selective autophagy receptor that accumulates in atg mutants [14]. PMDH is synthesized as a precursor (p) that is processed to a mature form (m) in the peroxisome. HSC70 is a loading control. The asterisk indicates a protein cross-reacting with the ATG7 antibody. (c) Retesting progeny of putative suppressors that displayed restored PTS2 processing in the M2 generation. Top: lateral root density of 8-day-old controls (left of dashed line) or M3 or M4 suppressor seedlings (right of dashed line) grown without or with IBA. Error bars show standard deviations (n = 8). Statistically significant (P < 0.0001) differences determined by one-way ANOVA are depicted by different letters above the bars. Bottom: extracts from 6-day-old controls (left of dashed line) or M3 or M4 suppressor seedlings (right of dashed line) were processed for immunoblotting. Membranes from duplicate gels were serially probed with antibodies to the indicated proteins to obtain the top 3 and bottom 4 panels. MLS, thiolase, and ICL are peroxisomal proteins that are stabilized when both LON2 and autophagy are defective [48]. Asterisks indicate proteins cross-reacting with the ATG7 or ICL antibodies. (d) Identifying mutations via whole-genome sequencing. The L40 suppressor was backcrossed to the original lon2-2 line, IBA-sensitive F2 seedlings were selected, and genomic DNA from pooled F3 seedlings was sequenced. Homozygous single-nucleotide polymorphisms consistent with EMS mutagenesis (G/C to A/T transitions) and causing nonsynonymous mutations in coding regions, altering splice sites, or occurring in introns or untranslated regions are indicated by locus identifiers and displayed to the right of the 5 Arabidopsis chromosomes using The Arabidopsis Information Resource Chromosome Map Tool.
Table 1.
ATG Mutant alleles recovered as lon2-2 suppressors in this work.
| Gene (accession number) | Protein function | Allele | Alias1 | Nucleotide change | Protein or transcript change |
|---|---|---|---|---|---|
| ATG2 (At3g19190) | Phagophore expansion | atg2-6 | L60 | g6271a | Intron 6 splice-acceptor site |
| atg2-7 | L78, L79 | g795a | W90Stop | ||
| ATG3 (At5g61500) | ATG8 lipidation (E2-like) |
atg3-2 | L13, L14, L15, L16 | g1613a | Intron 5 splice-acceptor site |
| ATG5 (At5g17290) | ATG8 lipidation (E3-like) |
atg5-5 | L40, L41 | g1387a | Intron 4 splice-acceptor site |
| atg5-6 | L43, L44, L45 | g2615a | E323K | ||
| ATG7 (At5g45900) | ATG8 lipidation (E1-like) |
atg7-62 | L61 | g1648a | W344Stop |
| atg7-10 | L21, L23, L24, L27 | g140a | Intron 1 splice-donor site | ||
| atg7-11 | L160 | g552a | G91E | ||
| atg7-12 | L37, L46 | g715a | Intron 2 splice-acceptor site | ||
| atg7-13 | L17, L18 | g721a | W119Stop | ||
| atg7-14 | L123 | g940a | W192Stop | ||
| atg7-15 | L39 | g1454a | W307Stop | ||
| atg7-16 | L66 | g2269a | Intron 7 splice-acceptor site | ||
| atg7-17 | L20, L25 | g2394a | G535D | ||
| atg7-19 | L71 | c2562t | T559I | ||
| atg7-21 | L92, L93, L96, L97 | c2848t | Q629Stop | ||
| ATG16 (At5g50230) | ATG8 lipidation (membrane tethering) | atg16-1 | L65 | g662a | W47Stop |
| atg16-2 | L68 | g650a; g760a |
Intron 2 splice-acceptor site; R80K | ||
| atg16-3 | L53 | c810t | Q97Stop | ||
| ATG18a (At3g62770) | Phagophore expansion | atg18a-3 | L1 | c463t | Q155Stop |
| atg18a-4 | L154 | g1921a | W364Stop |
1In all but one case, multiple isolates of allelic mutations were from the same M2 pool, indicating that the isolates were likely siblings. The exception was atg7-12, which was isolated from 2 independent pools.
2atg7-6 was independently isolated in a pilot lon2-2 suppressor screen [48].
Second, to eliminate false positives and mutants with elevated auxin levels or responsiveness, we moved seedlings with lateral roots to soil and collected leaf tissue from approximately 30-day-old plants for immunoblot analysis. We used antibodies recognizing NBR1, PMDH (peroxisomal malate dehydrogenase/At5g09660), and ATG7 to identify suppressors with impaired autophagy and improved peroxisome function (Figure 1(b)). The selective autophagy receptor NBR1 is degraded at a basal rate via autophagy and accumulates when autophagy is impaired [14]. PMDH is synthesized as precursor containing an N-terminal peroxisomal-targeting signal 2 (PTS2) that is removed following peroxisomal import. The incomplete PTS2 processing in lon2 plants [49] presumably stems from elevated pexophagy, which reduces the number of peroxisomes; preventing autophagy restores PMDH processing in lon2-2 by increasing the number of import-competent peroxisomes [48]. M2 plants with elevated NBR1 levels and restored PMDH processing were prioritized as likely atg mutants, and plants with decreased ATG7 levels were candidate atg7 mutants (Figure 1(b)).
Third, to confirm lon2-2 suppression, we collected M3 or M4 progeny of putative suppressors; retested IBA responsiveness; and examined thiolase, MLS or ICL, and ATG3 levels in 6-day-old seedlings (Figure 1(c)). Thiolase is a peroxisomal enzyme that is destabilized in lon2-2 seedlings [49] and stabilized in lon2-2 atg mutants [48]. Similarly, the peroxisomal enzymes ICL and MLS are stabilized in lon2-2 atg double mutants [48]. Mutants with elevated thiolase and ICL or MLS were prioritized as likely atg mutants, and plants with decreased ATG3 levels were candidate atg3 mutants.
Fourth, we used DNA sequencing to identify causal lesions. We sequenced ATG7 or ATG3 in suppressors with decreased levels of the respective proteins and prepared DNA for whole-genome sequencing for the remaining suppressors (Figure 1(d)).
New atg7 alleles recovered as lon2 suppressors
ATG7, which encodes an E1-like enzyme involved in ATG8 lipidation [8], was the most commonly mutated gene among our lon2 suppressors (Figure 2; Table 1). Most of these suppressors were initially identified as atg7 mutants due to low or undetectable ATG7 levels (Figure 2(c)). atg7-19 displayed near wild-type ATG7 levels and was identified by whole-genome sequencing (Fig. S1B). We identified 5 nonsense mutations (atg7-6, atg7-13, atg7-14, atg7-15, and atg7-21), 3 splice-site mutations (atg7-10, atg7-12, and atg7-16), and 3 missense mutations (atg7-11, atg7-17, and atg7-19) (Figure 2(a)). The 3 missense alleles alter residues that are conserved in ATG7 enzymes from Arabidopsis, humans, fruit flies, and budding yeast (Fig. S2). Interestingly, atg7-19 alters the conserved Thr559 residue immediately following the ATG7 active site Cys558 residue (Figure 2(a); Fig. S2). Although primarily novel atg7 alleles were recovered, a few duplicate alleles were isolated. The atg7-12 splice-site allele was found in 2 independent M2 pools, and an atg7-6 nonsense allele was previously recovered in our pilot lon2 suppressor screen [48]. Moreover, the nonsense allele atg7-13 harbors a mutation in a different nucleotide of the same codon as the previously published allele atg7-5 [48]; although non-allelic, both of these mutations result in the same protein alteration (W119Stop; Figure 2(a)). All of the atg7 mutants restored IBA responsiveness to lon2-2 (Figure 2(b)) and displayed the characteristic immunoblot phenotypes of 6-day-old lon2-2 atg double mutants: accumulated NBR1, stabilized MLS and thiolase, and restored PTS2 processing (Figure 2(c)). The early nonsense and splice-site mutations are expected to encode null alleles, and, indeed, we did not detect ATG7 protein in most of these mutants (Figure 2(c)). The 3 missense alleles (atg7-11, atg7-17, and atg7-19) accumulated detectable ATG7 protein (Figure 2(c)) and could therefore retain partial function, but these mutants nevertheless suppressed lon2-2 as well as the nonsense and splice-site alleles (Figure 2(b, c)).
Figure 2.
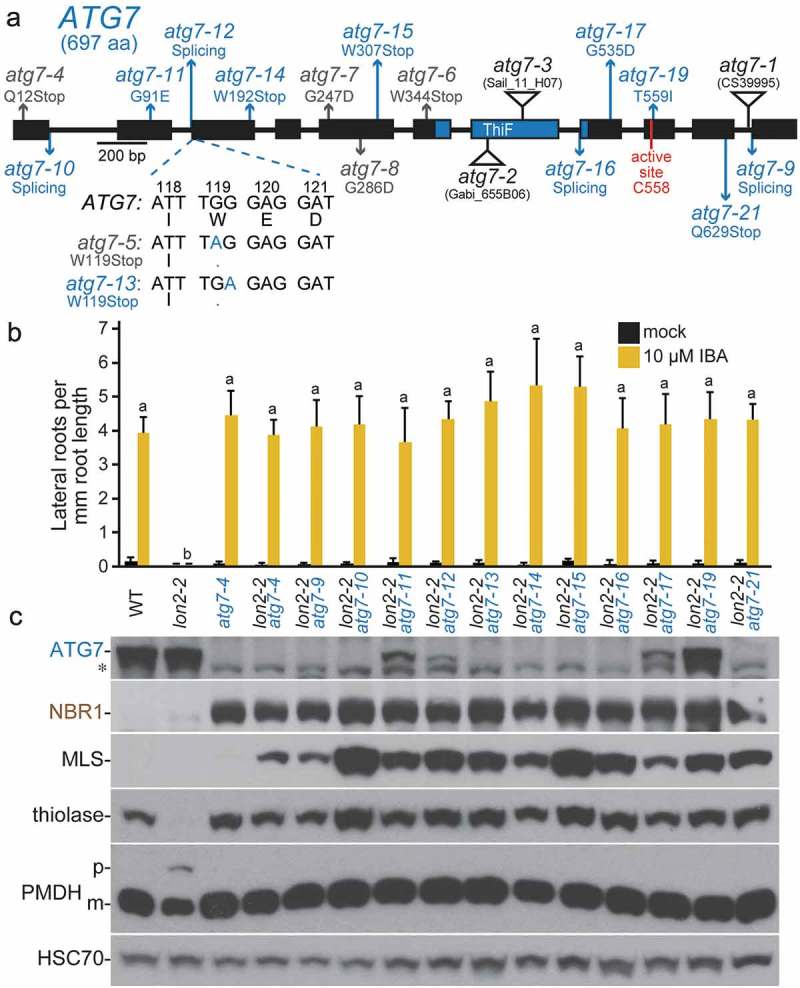
Numerous novel atg7 alleles recovered as lon2 suppressors. (a) Diagram of the ATG7 gene. Boxes and lines represent protein-coding regions and introns, respectively. The ThiF-like adenylation domain and active-site Cys residue are indicated. The positions of new atg7 mutations identified as lon2 suppressors are in blue; previously described EMS-derived lon2 suppressors [48] are in gray, and T-DNA insertion alleles [8,9,61] are indicated by triangles. atg7-9 is an unpublished allele (alias 8–30) from the pilot lon2-2 suppressor screen [48] that carries a g2959a mutation in the intron 10 splice acceptor site. The sequence of the atg7-13 nonsense allele compared to atg7-5 and wild-type ATG7 is shown below the gene diagram. aa, amino acids. (b) Lateral root density of 8-day-old wild type (WT), lon2-2, atg7-4, and lon2-2 atg7 seedlings grown without or with IBA. Error bars show standard deviations (n = 8). Statistically significant (P < 0.0001) differences determined by one-way ANOVA are depicted by different letters above the bars. (c) Extracts from 6-day-old seedlings were processed for immunoblotting with antibodies to the indicated proteins. The asterisk indicates a protein cross-reacting with the ATG7 antibody.
New atg3, atg5, and atg16 alleles recovered as lon2 suppressors
In addition to ATG7, we isolated novel alleles of 3 other genes involved in ATG8 lipidation. We identified the atg3-2 splice-site mutation by sequencing ATG3, which encodes an E2-like enzyme that conjugates ATG8 to phosphatidylethanolamine [32], from a mutant that lacked detectable ATG3 protein (Figure 3(c)).
Figure 3.
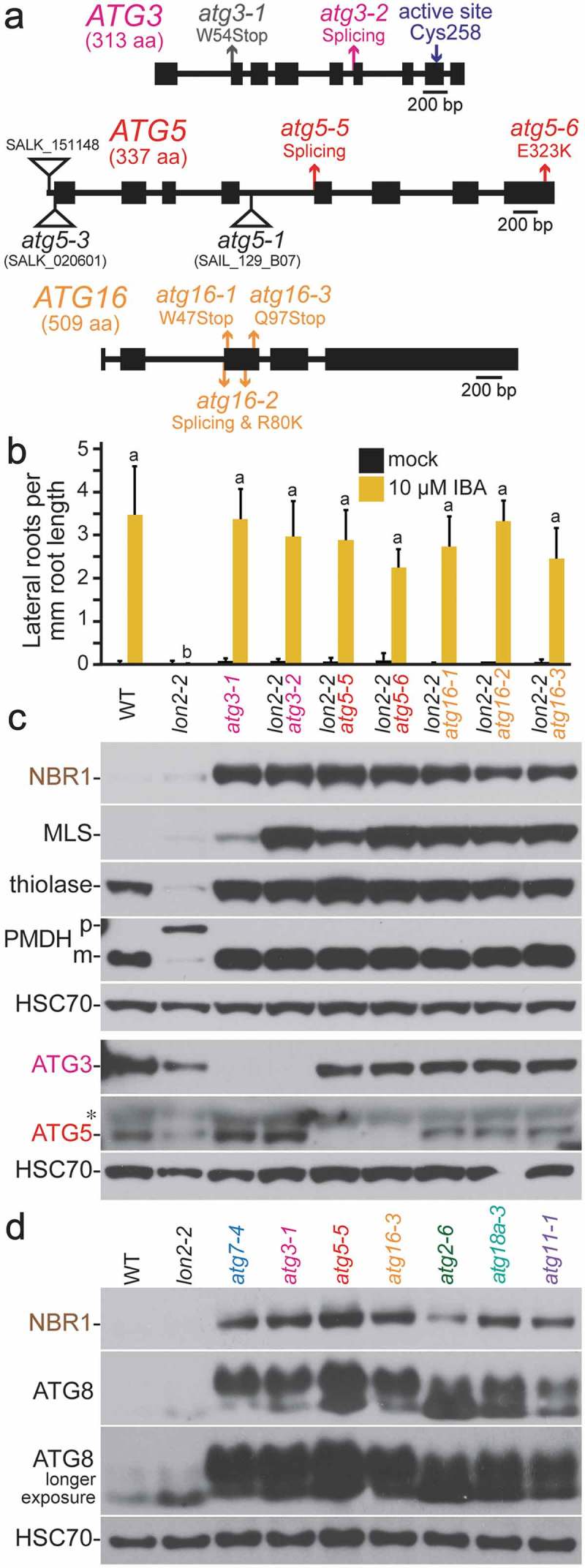
Novel atg3, atg5, and atg16 alleles recovered as lon2 suppressors. (a) Diagrams of the ATG3, ATG5, and ATG16 genes. Boxes and lines represent protein-coding regions and introns, respectively. The positions of new atg3, atg5, and atg16 mutations identified as lon2 suppressors are shown in pink, red, and orange, respectively; the previously described atg3-1 EMS-derived lon2 suppressor [48] is in gray, and T-DNA insertion alleles [29,72,73] are indicated by triangles. aa, amino acids. (b) Lateral root density of 8-day-old wild type (WT), lon2-2, atg3-1, lon2-2 atg3-2, lon2-2 atg5, and lon2-2 atg16 seedlings grown without or with IBA. Error bars show standard deviations (n = 8). Statistically significant (P < 0.0001) differences determined by one-way ANOVA are depicted by different letters above the bars. (c) Extracts from 6-day-old seedlings were processed for immunoblotting. Membranes from duplicate gels were serially probed with antibodies to the indicated proteins to obtain the top 5 and bottom 3 panels. The asterisk indicates a protein cross-reacting with the ATG5 antibody. (d) Extracts from 6-day-old seedlings were processed for immunoblotting and serially probed with antibodies to NBR1, ATG8A, and HSC70. ATG8 is lipidated and encoded by multiple genes in Arabidopsis.
We recovered 2 mutations in ATG5, which encodes a protein with 2 ubiquitin-like domains that is conjugated by the ubiquitin-like protein ATG12 to act as an E3-like enzyme in ATG8 lipidation [29,31,62]. We used whole-genome sequencing to identify the atg5-5 splice-site mutation (Figure 1(d), 3(a)) and the atg5-6 missense mutation (Figure 3(a), Fig. S1A), which altered a conserved acidic residue (Glu323) in the second ubiquitin-like domain to a Lys residue (Fig. S3). We did not detect ATG5 protein in either of these mutants (Figure 3(c)).
We recovered 3 mutations in ATG16, which in yeast encodes a protein that tethers the ATG12–ATG5 conjugate to the membrane to allow ATG8 lipidation and autophagosome formation [31]. Whole-genome sequencing revealed that atg16-1 and atg16-3 harbored nonsense mutations (W47Stop and Q97Stop, respectively) and that atg16-2 harbored both a splice-site mutation and a nearby missense mutation (R80K) (Figure 3(a), Fig. S1C to E). We surmise that the atg16-2 splice site mutation is causal because it is expected to prevent full-length ATG16 accumulation and is 5ʹ of the missense mutation.
All of the atg3, atg5, and atg16 alleles similarly restored lon2-2 IBA responsiveness (Figure 3(b)) and displayed the characteristic immunoblot phenotypes (Figure 3(c)) of other lon2-2 atg double mutant seedlings (e.g., Figure 1(c)).
Because the roles of ATG3 and ATG16 have not been confirmed in plants, we began this analysis by comparing ATG8 levels in an allele of each atg mutant in a wild-type LON2 background. As previously reported [29], ATG8 levels were elevated in atg5 and atg7 mutants (Figure 3(d)). We found similarly elevated ATG8 levels in our atg3 and atg16 alleles (Figure 3(d)), which is consistent with the possibility that autophagy is impaired in these mutants.
New atg2 alleles recovered as lon2 suppressors
We used whole-genome sequencing to identify 2 mutations in ATG2 (Figure 4(a, b), Fig. S4), which encodes a protein involved in phagophore expansion. Both atg2 mutants fully restored IBA responsiveness to lon2-2 (Figure 4(d)) and displayed the characteristic immunoblot phenotypes (Figure 4(e)) of other lon2-2 atg seedlings (Figure 1(c), 2(c), 3(c)). Moreover, atg2-6 displayed elevated ATG8 levels (Figure 3(d)). The atg2-7 early nonsense mutation (W90Stop, Figure 4(b)) is expected to prevent ATG2 accumulation. The atg2-6 mutation appeared in the middle of a cluster of mutations on chromosome 3 (Figure 4(a)) identified by sequencing backcrossed seedlings. However, the mutation was in intron 6 according to the primary prediction of Araport 11, the latest A. thaliana genome annotation. Araport 11 also predicts 2 ATG2 splice variants that would result in atg2-6 interrupting a splice-acceptor site. Indeed, the region of the intron following the mutated nucleotide can be translated in frame with exon 7 and lacks stop codons (Figure 4(b)). Moreover, aligning predicted ATG2 proteins of other Brassicaceae family members revealed that this region of the predicted Arabidopsis intron 6 is conserved and exonic in other Brassicaceae ATG2 orthologs (Figure 4(b)), supporting the conclusion that the mutation in atg2-6 disrupts a splice site in A. thaliana and suggesting that the ‘alternative’ splice variant encodes an ATG2 isoform that is essential for autophagy. Although the predicted splicing of A. lyrata ATG2 matched that of the A. thaliana ATG2 predicted by Araport 11, the A. lyrata genome was assembled based in part on alignment with A. thaliana [63], meaning that annotation errors in the A. thaliana genome may have been propagated to A. lyrata. Based on the whole-genome sequencing and Brassicaceae ATG2 alignment, we considered atg2-6 as a splice-site mutation and the causal suppressor lesion.
Figure 4.
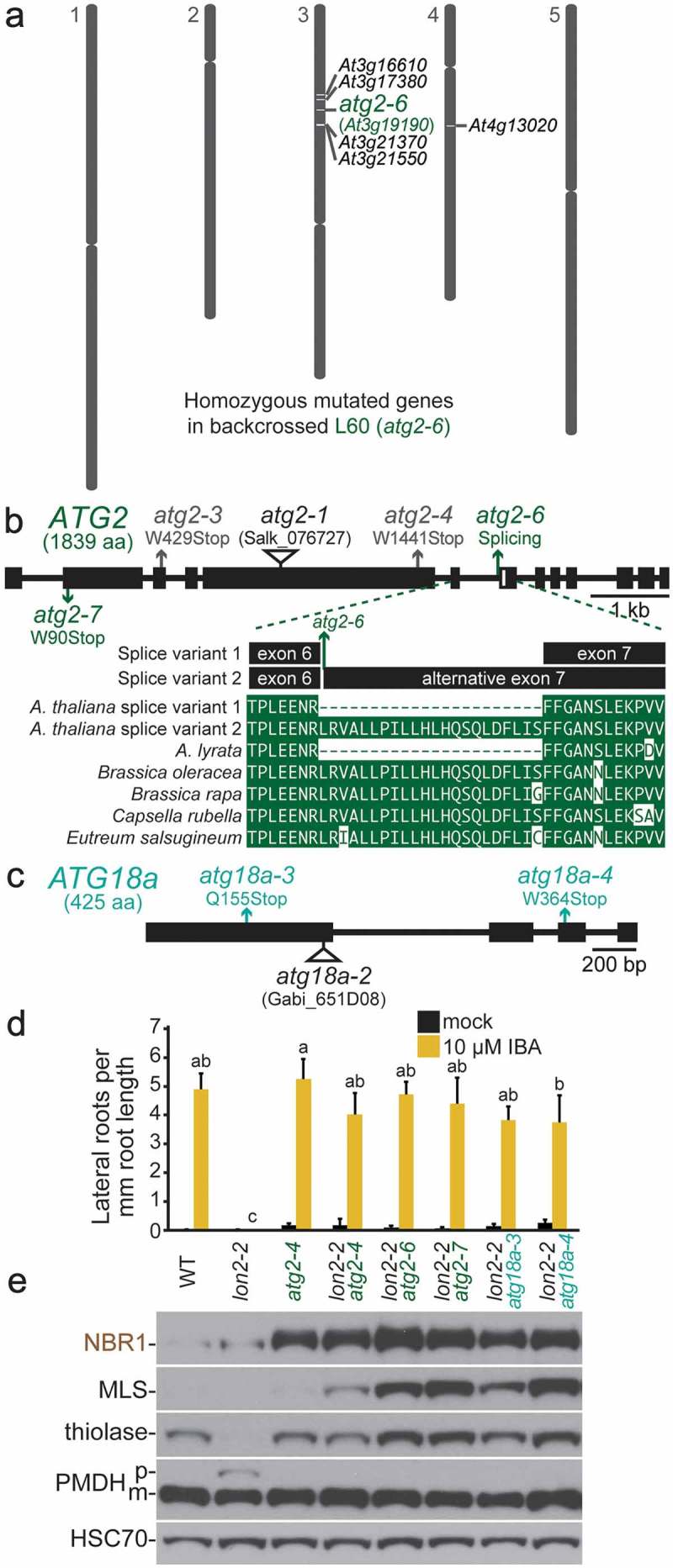
Novel atg2 and atg18a alleles recovered as lon2 suppressors. (a) The L60 suppressor was backcrossed to the original lon2-2 line, and genomic DNA from pooled IBA-sensitive F2 seedlings was sequenced and analyzed as in the legend to Figure 1(d). (b) ATG2 gene diagram with boxes and lines representing protein-coding regions and introns, respectively. The positions of new atg2 mutations identified as lon2 suppressors are shown in green, previously described EMS-derived lon2 suppressors [48] are in gray, and a T-DNA insertion allele [64] is indicated by a triangle. The partial alignment shows predicted Brassicaceae ATG2 proteins, including 2 A. thaliana ATG2 splice variants predicted by the latest genome annotation (Araport 11); the alternative ATG2 splice variant is interrupted by the atg2-6 mutation. aa, amino acids. (c) ATG18a gene diagram showing atg18a mutations identified as lon2 suppressors (teal) and a previously described T-DNA insertion allele [65] (triangle). (d) Lateral root density of 8-day-old wild type (WT), lon2-2, atg2-4, lon2-2 atg2, and lon2-2 atg18a seedlings grown without or with IBA. Error bars show standard deviations (n = 8). Statistically significant (P < 0.0001) differences determined by one-way ANOVA are depicted by different letters above the bars. (e) Extracts from 6-day-old seedlings were processed for immunoblotting with antibodies to the indicated proteins.
New atg18a alleles recovered as lon2 suppressors
Like ATG2, ATG18a is involved in phagophore expansion. We used whole-genome sequencing to identify the atg18a-3 and atg18a-4 nonsense mutations (Q155Stop, W364Stop, Figure 4(c), Fig. S5). atg18a-3 and atg18a-4 fully restored IBA responsiveness to lon2-2 (Figure 4(d)) and displayed the characteristic immunoblot phenotypes of other lon2-2 atg seedlings (Figure 4(e)). Moreover, atg18a-3 seedlings displayed similarly elevated ATG8 levels as other atg null alleles (Figure 3(d)).
To further probe the importance of ATG18a in pexophagy, we backcrossed lon2-2 atg18a-3 to remove lon2-2 and other unlinked mutations and examined atg18a-3 seedlings constitutively expressing GFP carrying a peroxisomal targeting signal (GFP-PTS1) [66] using confocal microscopy. atg7 and atg5 mutants accumulate peroxisomes in hypocotyls of developing seedlings [16], and atg18a mutants display clusters of oxidized peroxisomes in mesophyll cells [15]. We observed increased peroxisome abundance relative to wild type in atg18a-3 cotyledon epidermal cells, cotyledon mesophyll cells, and hypocotyl cells in 4- to 7-day-old seedlings (Figure 5), confirming that ATG18a promotes pexophagy during seedling development.
Figure 5.
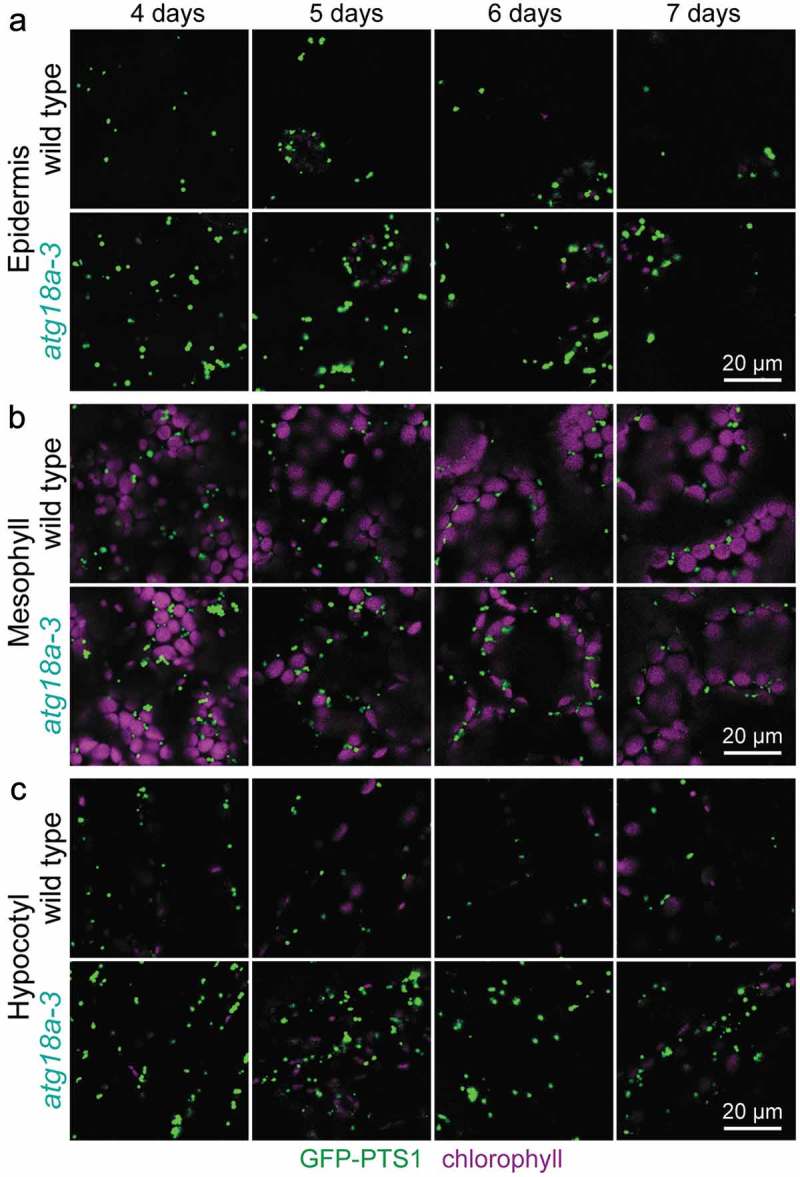
Increased peroxisome abundance in atg18a-3 seedlings. (a) Cotyledon epidermal cells, (b) cotyledon mesophyll cells, and (c) hypocotyl cells in light-grown 4-, 5-, 6-, and 7-day-old wild-type and twice backcrossed atg18a-3 seedlings expressing the peroxisomal matrix marker GFP-PTS1 were imaged for GFP fluorescence (green) and chlorophyll autofluorescence (magenta) using confocal microscopy. Scale bars: 20 µm.
Loss of ATG11 partially suppresses lon2 defects
Although ATG11 functions in Arabidopsis autophagy [17,18], no atg11 mutants emerged from our lon2-2 suppressor screen. To test the importance of ATG11 for lon2-related pexophagy, we crossed the atg11-1 T-DNA insertional allele (Figure 6(a)) [17] to lon2-2. Although the lon2-2 PTS2-processing defect was fully suppressed in lon2-2 atg11-1 (Figure 6(c)), atg11-1 only partially suppressed lon2-2 IBA resistance (Figure 6(b)), thiolase was only partially stabilized in lon2-2 atg11-1 (Figure 6(c)), and MLS was not stabilized (Figure 6(c)). This incomplete suppression suggested that some pexophagy still occurs in atg11-1. Additionally, ATG8 was only moderately elevated in atg11-1 (Figure 3(d)), and NBR1 accumulated to intermediate levels in both atg11-1 and lon2-2 atg11-1 (Figure 6(c)), consistent with only partially disrupted autophagy.
Figure 6.
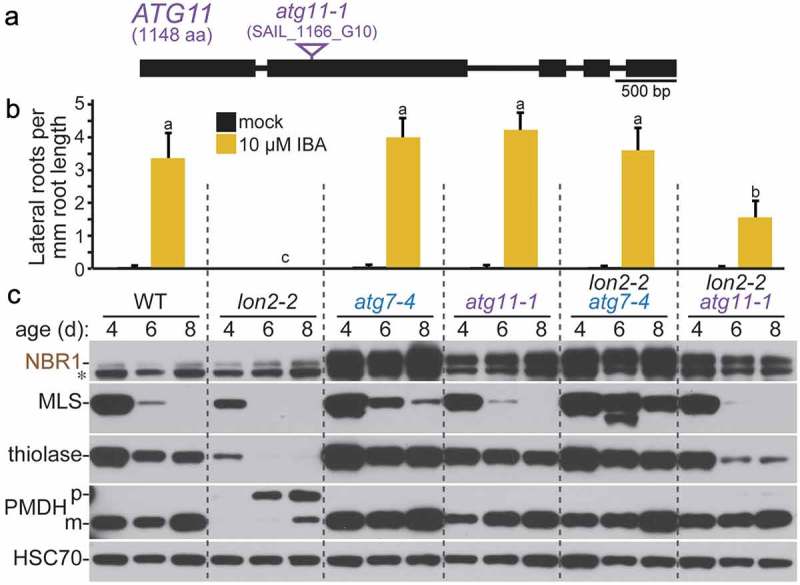
Loss of ATG11 partially suppresses lon2 defects. (a) Diagram of the ATG11 gene. Boxes and lines represent protein-coding regions and introns, respectively. A triangle marks the location of the atg11-1 T-DNA insertion allele [17]. aa, amino acids. (b) Lateral root density of 8-day-old wild type (WT), lon2-2, atg7-4, atg11-1, lon2-2 atg7-4, and lon2-2 atg11-1 seedlings grown without or with IBA. Error bars show standard deviations (n = 8). Statistically significant (P < 0.0001) differences determined by one-way ANOVA are depicted by different letters above the bars. (c) Extracts from 4-, 6-, and 8-day-old seedlings were processed for immunoblotting with antibodies to the indicated proteins. An asterisk indicates a protein cross-reacting with the NBR1 antibody.
NBR1 is not necessary for pexophagy of lon2 peroxisomes
NBR1 is a selective autophagy receptor in Arabidopsis [10,14,36] and is necessary and sufficient for pexophagy in mammals [67], but no nbr1 mutants emerged from our screen for lon2-2 suppressors. To test the necessity of NBR1 for Arabidopsis pexophagy, we obtained T-DNA insertional alleles of nbr1 (Figure 7(a)). These T-DNA insertions dramatically decreased full-length NBR1 protein accumulation (Figure 7(c)); however, the anti-NBR1 antibody was generated to the C-terminal UBA domain [14] and would not detect any N-terminal fragments of the protein that might accumulate in the mutants. Because intronic T-DNAs are sometimes removed by splicing [68,69], we characterized both intronic (nbr1-1) and exonic (nbr1-4) insertional mutants of NBR1 in combination with lon2-2. Both lon2 nbr1 double mutants resembled lon2-2 and did not form lateral roots in response to IBA (Figure 7(b)). Moreover, neither nbr1 allele stabilized MLS or thiolase or ameliorated the PTS2-processing defects of lon2 (Figure 7(c)). This lack of suppression suggested that NBR1 is dispensable for pexophagy in lon2. Because atg18a-3 accumulated peroxisomes (Figure 5), we also visualized nbr1-4 expressing the peroxisomal marker GFP-PTS1 to evaluate peroxisome abundance. Unlike atg18a-3, peroxisome abundance in hypocotyls of 6-day-old nbr1-4 seedlings resembled wild type (Figure 7(d)). The failure of nbr1-4 mutants to accumulate excess peroxisomes suggested that NBR1 is not necessary for peroxisome turnover under our standard growth conditions.
Figure 7.
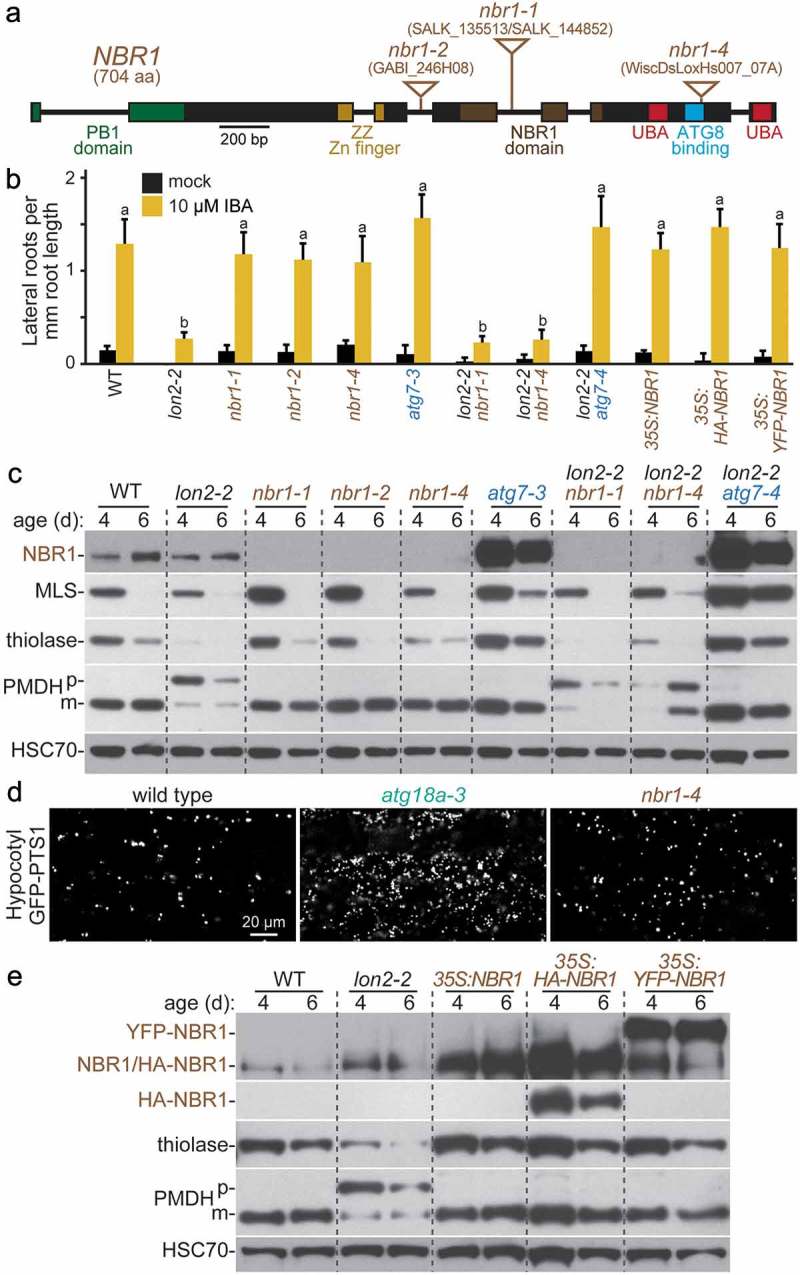
NBR1 is not necessary for pexophagy of lon2 peroxisomes, and excess NBR1 is not sufficient to induce pexophagy. (a) Diagram of the NBR1 gene. Boxes and lines represent protein-coding regions and introns, respectively. The PB1 domain, zinc finger, NBR1 domain, ATG8 binding site, and ubiquitin-associated (UBA) domains are indicated. Triangles mark the locations of T-DNA insertion alleles. aa, amino acids. (b) Lateral root density of 8-day-old wild type (WT), lon2-2, nbr1, atg7-3, lon2-2 nbr1, lon2-2 atg7-4, 35S:NBR1, 35S:HA-NBR1, and 35S:YFP-NBR1 seedlings grown without or with IBA. Error bars show standard deviations (n = 8). Statistically significant (P < 0.0001) differences determined by one-way ANOVA are depicted by different letters above the bars. (c) Extracts prepared from 4- and 6-day-old seedlings were processed for immunoblotting with antibodies to the indicated proteins. (d) Hypocotyl cells in 6-day-old wild-type, atg18a-3, and nbr1-4 light-grown seedlings expressing the peroxisomal matrix marker GFP-PTS1 were imaged for GFP fluorescence using confocal microscopy. Scale bar: 20 µm. (e) Extracts prepared from 4- and 6-day-old seedlings were processed for immunoblotting with antibodies to the indicated proteins.
If excess NBR1 was sufficient to induce pexophagy, then we would expect that overexpressing NBR1 would stimulate pexophagy, as is observed in mammalian cells [67], and phenocopy lon2. We drove untagged, HA-tagged, and YFP-tagged NBR1 from the constitutive cauliflower mosaic virus 35S promoter. Despite elevated NBR1 levels (Figure 7(e)), these lines resembled wild type in lateral root formation (Figure 7(b)), PTS2 processing (Figure 7(e)), and thiolase stability (Figure 7(e)). This failure of excess NBR1 to potentiate seedling pexophagy indicates that NBR1 is not limiting for pexophagy in Arabidopsis, which is consistent with the possibility that NBR1 is not a pexophagy receptor in Arabidopsis seedlings.
Discussion
A facile screen for Arabidopsis atg mutants
Screening for lon2-2 suppressors revealed 21 alleles (20 novel alleles) in 6 ATG genes (Table 1), providing new mutants for investigating autophagy in Arabidopsis. A pilot lon2-2 suppressor screen has uncovered atg2, atg3, and atg7 mutants [48], and the expanded screen described here uncovered additional alleles of these genes plus atg5, atg16, and atg18a mutants. In contrast to these lon2 suppressor screens, a single atg2 allele emerged from a forward-genetic screen for plants with enhanced cell death in response to fungal infection [22], a single atg2 allele has emerged from a hydroxyurea-resistance screen [70] that primarily recovers cat2 (catalase/At4g35090) mutants [71], and 2 atg2 alleles along with single atg7 and atg18a mutants have been recovered from a microscopy-based screen for aggregated peroxisomes [15]. The lon2-2 suppressor screen combines high throughput with specificity and is the only forward-genetic screen to report atg3, atg5, or atg16 mutants. Although atg5 T-DNA insertional mutants have been characterized [29,72,73], no Arabidopsis atg3 or atg16 mutants have been reported aside from those arising from lon2-2 suppressor screens described here and previously [48], and these alleles will be useful for future studies to determine whether ATG3 and ATG16 function similarly in Arabidopsis as in other organisms. Moreover, our atg7 missense alleles (Figure 2(a), Fig. S2) may inform ATG7 structure-function studies.
Roughly half (11 of 21) of the lon2-2 suppressors harbored a mutation in ATG7 (Table 1); the reason for this abundance of atg7 mutants is mysterious. When adding the atg7 alleles identified in our pilot screen [48], 16 of 29 lon2-2 suppressors were atg7 mutants, and atg7-6 was independently isolated in both iterations of the screen (Table 1) [48]. Although ATG7 encodes a large protein (697 amino acids), ATG2 and ATG11 encode larger proteins (1861 and 1148 amino acids, respectively) and thus would seem to be more likely mutagenic targets. However, our combined screening recovered only 4 atg2 and no atg11 mutants. Moreover, ATG2 has 12 introns compared to 10 introns in ATG7, suggesting that ATG2 would present more opportunities than ATG7 for mutations in splice sites in addition to coding regions. The frequency of atg7 mutants recovered in this screen might be explained by the structure and function of the ATG7 protein. As an E1-like enzyme, ATG7 activates both ATG8 and ATG12 and coordinates transfer to their respective E2-like enzymes, ATG3 and ATG10 [32,74–76]. ATG7 is the interaction hub among these proteins, requiring precise spatial arrangement [32,74–77], so it is tempting to speculate that ATG7 might be more susceptible to functional perturbation via missense mutations than other ATG proteins. Indeed, 5 of the 16 atg7 mutants identified as lon2-2 suppressors (atg7-7, atg7-8, atg7-11, atg7-17, and atg7-19; Figure 2(a), Fig. S2) harbor missense mutations whereas our only non-atg7 lon2-2 suppressor to harbor an atg missense mutation is atg5-6 (Figure 3(a), Fig. S3). However, fewer mutations in other ATG genes were recovered, hindering comparison of the relative proportions of null versus missense alleles; further screening might recover missense alleles of other ATG genes. Moreover, even discounting missense mutations, 11 of our atg7 mutants harbor nonsense or splice-site mutations, which still exceeds the number of alleles recovered for any other ATG gene (Table 1), indicating that the explanation for the apparent bias towards atg7 mutants may exist beyond the structure or function of ATG7. Perhaps the ATG7 locus is more accessible to EMS mutagenesis than other ATG genes.
ATG18a appears to act non-redundantly with the other 7 ATG18 isoforms [23] in Arabidopsis seedling pexophagy. The atg18a-3 and atg18a-4 nonsense alleles fully restored lon2-2 IBA responsiveness and PTS2 processing, and lon2-2 atg18a seedlings displayed similarly stabilized NBR1, MLS, and thiolase as other lon2-2 atg double mutants (Figure 4(d, e)). In addition, ATG8 elevation in atg18a-3 was similar to the elevation in atg2-6 (Figure 3(d)). Moreover, the increased peroxisome abundance in atg18a-3 seedlings (Figure 5) and the isolation of an atg18a mutant from a microscopy-based screen for aggregated peroxisomes [15] are consistent with a requirement for ATG18a in pexophagy. ATG18a functions beyond pexophagy; RNAi-based ATG18a downregulation confers hypersensitivity to starvation [23] and various abiotic stresses [11,12]. Although most of the ATG18 isoforms are upregulated during seed maturation [13], whether the remaining 7 Arabidopsis ATG18 isoforms [23] function in autophagy remains an open question.
Because no atg11 mutants emerged from our screen for lon2-2 suppressors, we crossed the atg11-1 T-DNA insertional allele to lon2-2 and observed only partial suppression of lon2-2 phenotypes (Figure 6), suggesting that pexophagy is incompletely blocked in atg11-1. This conclusion is supported by the observation that atg11-1 accumulates less catalase than other atg mutants [18]. Moreover, ATG8 and NBR1 are only partially stabilized in atg11-1 relative to atg7-4 and other null alleles in core ATG genes (Figure 3(d), 6(c)), consistent with previous reports that autophagy is incompletely blocked in atg11-1 [17,18]. As ATG11 is thought to act as a scaffold for ATG1a and ATG13 within the phagophore assembly site [17], these findings imply that autophagy can still occur, albeit less efficiently, in atg11-1 seedlings. Despite the large size of ATG11 (1148 amino acids), its role as a scaffold may allow some tolerance for missense mutations, which, combined with the observation that atg11-1 only partially restored lon2-2 IBA responsiveness (Figure 6(b)), may explain why atg11 mutants did not emerge from lon2-2 suppressor screens. In Pichia pastoris, Atg11 binds both Atg8 and the pexophagy receptor Atg30 [78], so it will be interesting to determine whether ATG11 plays a similar role in Arabidopsis pexophagy. Arabidopsis ATG11 also interacts with ATG8 [17], and additional studies are needed to decipher the role of this protein in processes upstream and downstream of ATG8 lipidation.
Besides ATG11, mutants of 5 other single-copy Arabidopsis ATG orthologs did not emerge from our lon2-2 suppressor screen: ATG6/At3g61710, ATG9/At2g31260, ATG10/At3g07525, ATG20/At5g0614, and ATG101/At5g66930. Of these 5, only the putative ATG20 ortholog SNX1 (Sorting Nexin 10), which acts in endosomal sorting [79], has not been directly implicated in autophagy in plants. Conversely, although Arabidopsis ATG6 is important for autophagy [21], atg6 defects in pollen germination confer male sterility [80,81] that would preclude recovery from our lon2-2 suppressor screen. Arabidopsis ATG9 is a transmembrane protein critical for autophagosome formation [24,82–84], but autophagic flux is only partially compromised in atg9 mutants [18,82]. It would be interesting to determine whether atg9 mutants partially suppress lon2-2 similar to atg11 mutants (Figure 6). ATG10 is an E2-like enzyme that conjugates ATG12 during autophagosome formation and is essential for Arabidopsis autophagy [30]. ATG101 interacts with ATG11 and ATG13a [17], but atg101 mutants have not been described. The genes encoding ATG10 (226 amino acids) and ATG101 (215 amino acids) are relatively small targets for EMS mutagenesis, so further screening might yield mutants of these genes.
In addition to ATG18, several ATG genes are present as gene families in Arabidopsis: ATG1 (3 genes), ATG4 (2 genes), ATG8 (9 genes), ATG12 (2 genes), and ATG13 (2 genes). The roles of these genes in Arabidopsis autophagy have been experimentally validated using reverse-genetic and biochemical approaches [7,8,27,84,85]. The failure of these genes to emerge from forward-genetic screens for lon2-2 suppressors is consistent with the possibility that, unlike ATG18a (Figure 4), the genes in these families function redundantly in pexophagy.
Arabidopsis pexophagy receptors remain to be identified
In spite of the conservation of autophagy across kingdoms, identified pexophagy receptors differ among species, and a pexophagy receptor has not been reported in plants. Pichia pastoris Atg30 [86] and Saccharomyces cerevisiae Atg36 [87] act as pexophagy receptors, but plants lack Atg30 or Atg36 orthologs. We did not recover mutants of candidate pexophagy receptors in our screen for lon2-2 suppressors. Because NBR1 is a pexophagy receptor in mammalian cells [67] and a selective autophagy receptor in Arabidopsis [10,14,36,37], we used reverse genetics to investigate the role of Arabidopsis NBR1 in pexophagy. Two T-DNA insertional alleles of nbr1 failed to suppress lon2-2 defects (Figure 7(b, c)), and NBR1 overexpression failed to phenocopy lon2 seedling defects (Figure 7(b, e)), which presumably result from heightened pexophagy. Moreover, peroxisome abundance in nbr1-4 seedlings resembled wild type rather than the increased abundance of peroxisomes observed in atg18a-3 (Figure 7(d)). These data indicate that NBR1 is not the sole pexophagy receptor in lon2 seedlings but do not eliminate the possibility that NBR1 acts redundantly in pexophagy or in other contexts. Intriguingly, peroxisomal catalase accumulates in nbr1 mutants during heat stress [36], suggesting that NBR1 might be important for LON2-independent pexophagy in Arabidopsis. Further studies are needed to fully elucidate the roles of NBR1 in Arabidopsis selective autophagy.
Perhaps plants employ a pexophagy receptor that is distinct from those identified in yeast and mammalian cells. The plant-specific ATI (ATG8-interacting) proteins [39,40] are pexophagy receptor candidates but did not emerge from our screen for lon2-2 suppressors. The ubiquitin-binding protein DSK2 [88] is another pexophagy receptor candidate in Arabidopsis. Arabidopsis DSK2 binds to and targets the transcription factor BES1 for autophagic degradation [42]. DSK2 also interacts with 2 peroxisomal membrane proteins [89]. Because Arabidopsis has 2 DSK2 paralogs (At2g17190 and At2g17200) [90], recovering dsk2 mutants as lon2 suppressors is unlikely, and directed approaches will be needed to explore the potential roles of DSK2 and ATI proteins in pexophagy.
Potential roles for peroxins in pexophagy
PEX (peroxin) proteins are necessary for peroxisome biogenesis and matrix protein import [91]; many peroxins are found on the peroxisome membrane where they would be accessible to the autophagy machinery. Several peroxins have been implicated in pexophagy in Arabidopsis as well as yeast and mammalian cells. In general, peroxins or other peroxisomal membrane proteins might mediate pexophagy in 3 ways.
First, post-translationally modified peroxins could interact with a selective autophagy receptor. In mammalian cells, PEX5 phosphorylation by ataxia-telangiectasia mutated (ATM) leads to pexophagy [92]. Selective autophagy receptors, including NBR1, often recognize ubiquitinated cargo [4,33]. Because the matrix protein import peroxin PEX5 is ubiquitinated during its typical function [93], ubiquitinated PEX5 is a candidate pexophagy signal.
Second, peroxisomal membrane proteins could interact directly with a selective autophagy receptor. For example, Pichia pastoris Atg37 is a peroxisomal membrane protein that binds to the pexophagy receptor Atg30 and modulates pexophagy [94,95]. Moreover, the membrane peroxins Pex3 and Pex14 interact with the P. pastoris pexophagy receptor Atg30 [86,94,96]. Pex3 also binds to the Saccharomyces cerevisiae pexophagy receptor Atg36 [87], and PEX3 plays a pivotal role in pexophagy in mammalian cells [97]. Decreasing PEX14 levels in mammalian cells reduces pexophagy, perhaps as a result of reduced recruitment of PEX5 [67]. In Arabidopsis, the membrane peroxins PEX2/At1g79810 and PEX12/At3g04460 interact with DSK2 [89], making these peroxins candidates for pexophagy regulators.
Third, peroxins could interact directly with ATG8, bypassing the need for a bridging receptor. Although the relative simplicity of ATG8-interacting motifs has confounded bioinformatic approaches to identifying ATG8-interacting proteins, a recent refinement considering acidic residue positioning reveals that Arabidopsis PEX10/At2g26350 and PEX6/At1g03000 interact with ATG8 [34]. Interestingly, pexophagy contributes to defects observed in pex1 and pex6 mutants in Arabidopsis [98,99], yeast [100], and humans [101], suggesting that PEX6 prevents rather than promotes pexophagy, perhaps by competing with the pexophagy signal for interaction with ATG8 or by removing ubiquitinated substrates, such as PEX5, from the peroxisomal surface.
Despite the potential importance of peroxins in pexophagy, pex mutants are unlikely to emerge as lon2-2 suppressors because pex mutants are generally IBA resistant [91] and thus not expected to restore IBA-responsive lateral rooting to lon2 (Figure 1(a)). Alternative approaches will be needed to dissect the roles of peroxins in Arabidopsis pexophagy.
Conclusions
We used forward genetics to recover 21 alleles of 6 ATG genes. All 6 genes are core ATG genes, underscoring the functional conservation of these components. These mutant alleles will be useful in future studies interrogating the role of these genes in Arabidopsis autophagy, especially the atg3 and atg16 alleles, which disrupt genes without available T-DNA insertional alleles. Using the lon2 mutant as a system for studying pexophagy in Arabidopsis, we also investigated the role of ATG11 and NBR1 in this process and found that ATG11 plays a limited role in pexophagy whereas NBR1 does not appear to be involved in lon2-related pexophagy. lon2 mutants will undoubtedly serve as a useful platform for future studies elucidating the regulation of pexophagy in Arabidopsis.
Materials and methods
Plant material and growth conditions
The Columbia-0 (Col-0) accession of Arabidopsis thaliana transformed with 35S:GFP-PTS1 [66] was used as the wild-type control. lon2-2 (SALK_043857) [49] crossed to 35S:GFP-PTS1 was mutagenized to isolate suppressors (see Mutant Isolation below) and used as the lon2-2 control for subsequent experiments. lon2-2 atg2-4 35S:PTS2-GFP and lon2-2 atg7-4 35S:PTS2-GFP were previously described [48]. atg7-3 (SAIL_11_H07) [9,22], atg11-1 (SAIL_1166_G10) [17], nbr1-1 (SALK_135513) [10], and nbr1-2 (GABI_246H08) [10] were previously described and were obtained from the Arabidopsis Biological Resource Center (ABRC) at Ohio State University. We found that the T-DNA interrupting NBR1 in SALK_144852 (Figure 7(a)) was at the identical position as the T-DNA in nbr1-1 (SALK_135513). lon2-2 atg7-9 35S:PTS2-GFP is an unpublished allele originating from a screen for lon2-2 35S:PTS2-GFP suppressors [48]. atg2-4 35S:PTS2-GFP, atg3-1 35S:PTS2-GFP, and atg7-4 35S:PTS2-GFP were obtained by crossing the respective lon2-2 atg 35S:PTS2-GFP double mutant [48] to 35S:PTS2-GFP [102] to remove the lon2-2 mutation. atg2-6 35S:GFP-PTS1, atg5-5 35S:GFP-PTS1, atg16-3 35S:GFP-PTS1, and atg18a-3 35S:GFP-PTS1 were obtained by crossing the respective lon2-2 atg 35S:GFP-PTS1 double mutant to 35S:GFP-PTS1 to remove the lon2-2 mutation. The presence of the T-DNA insertion in atg11-1, lon2-2, nbr1-1, and nbr1-2 was confirmed by PCR of genomic DNA using a gene-specific primer and a T-DNA left-border specific primer (Table S1).
nbr1-4 (WiscDsLoxHs007_07A) was obtained from the ABRC. The position of the T-DNA insertion was confirmed by PCR of genomic DNA using a gene-specific primer and a T-DNA left-border specific primer (Table S1), and the exact position of the insertion was determined by sequencing the PCR product (Lone Star Labs, Houston, TX, USA). nbr1-4 35S:GFP-PTS1 was obtained by crossing.
To generate NBR1-overexpressing lines, an NBR1 cDNA in pENTR223 (stock G25119) from the ABRC was cloned using LR Clonase II (Invitrogen, 11791020) into pEarleyGate destination vectors pEG100, pEG104, and pEG201 [103] from the ABRC. The resultant 35S:NBR1, 35S:YFP-NBR1, and 35S:HA-NBR1 plasmids were used to transform Agrobacterium tumefaciens GV3101 (pMP90) [104] by electroporation. The resultant A. tumefaciens strains were used to transform Col-0 plants using the floral dip method [105]. Transformed plants were selected for glufosinate ammonium (Basta, Gold Biotechnology, P-165–1) resistance, and overexpression was confirmed by immunoblot analysis using anti-NBR1 [14]. Homozygous progeny were used for phenotypic analysis.
Seeds were surface-sterilized using 3% (w:v) sodium hypochlorite solution containing 0.01% (v:v) Triton X-100 (Sigma-Aldrich, T9284), resuspended in 0.1% agar (Fisher Scientific, BD-214030), and stratified at 4°C overnight. Seedlings were grown at 22°C on plant nutrient (PN) medium [106] supplemented with 0.5% (w:v) sucrose (VWR, JT4097-6) (PNS) and solidified with 0.6% (w:v) agar. To quantify IBA sensitivity, seedlings were grown on PNS plates under continuous white light for 4 days, transferred to new PNS plates with either ethanol (mock) or 10 µM IBA (Sigma-Aldrich, I5386; made from a 100-mM stock in ethanol), and grown under light filtered with yellow long-pass filters (to slow photochemical breakdown of indolic compounds) [107] for an additional 4 days. Lateral roots emerged from the primary root were counted using a stereomicroscope (S6E, Leica Microsystems, Buffalo Grove, IL, USA), and primary root lengths were measured using a ruler. Measurements were completed at least twice with similar results.
Mutant isolation
lon2-2 35S:GFP-PTS1 seeds (1.3 g, ~65,000 seeds) were mutagenized for 16 h at room temperature with 0.24% to 0.25% (v:v) ethylmethane sulfonate (Sigma-Aldrich, M0880), rinsed extensively with water, and grown in 104 M1 pools. For screening, 0.1 g of M2 seeds (~5000 seeds) per pool were surface-sterilized, stratified, and plated on PNS medium supplemented with 8 µM IBA and solidified with 1.0% (w:v) agar in square Petri dishes (89 x 89 mm). Plates were positioned vertically, and seedlings were grown under yellow-filtered light for 8 days, when M2 seedlings with 3 or more lateral roots were moved to soil. Leaf tissue was collected from approximately 30-day-old M2 plants for immunoblot analysis using anti-PMDH, anti-NBR1, anti-ATG7, and anti-HSC70 antibodies (as described below). M3 seeds from M2 plants with multiple lateral roots, complete PTS2 processing, and elevated NBR1 protein levels were collected and re-tested for IBA resistance (as described above in Plant Materials and Growth Conditions), and 6-day-old tissue was tested by immunoblot analysis using anti-thiolase, anti-ATG3, anti-HSC70, and anti-ICL or anti-MLS (as described below). Lines exhibiting IBA sensitivity and thiolase and ICL or MLS stabilization were retained as lon2 suppressors. All suppressors were genotyped to confirm lon2-2 homozygosity (Table S1). After suppressor mutation identification, PCR-based genotyping markers designed using dCAPS Finder 2.0 [108] were used to confirm the presence of the identified lesions (Table S2) and follow mutations in the progeny of crosses.
Whole-genome sequencing
Genomic DNA from 10 suppressors was prepared for whole-genome sequencing. Genomic DNA from 6 suppressors (atg2-6, atg5-6, atg7-19, atg16-1, atg16-2, and atg16-3) was prepared from F2 seedlings from lines backcrossed to lon2-2 35S:GFP-PTS1. Surface-sterilized F2 seeds were plated on PNS medium supplemented with 8 µM IBA and solidified with 0.6% (w:v) agar and grown at 22°C under light filtered with yellow long-pass filers. At 10 days, between 50 and 100 F2 seedlings with lateral roots were moved to sterile filter paper atop PNS medium and grown at 22°C under white light for approximately 15 more days before collecting tissue. Genomic DNA from one suppressor (atg5-5) was prepared from F3 seedlings pooled from 3 F3 lines from a backcross to lon2-2 35S:GFP-PTS1, and genomic DNA from 3 suppressors (atg2-7, atg18a-3, and atg18a-4) was prepared from un-backcrossed lines.
Genomic DNA for whole-genome sequencing was prepared as previously described [109]. DNA was sequenced with HiSequation 2000 sequencers (Illumina, San Diego, CA, USA) at the Genome Technology Access Center at Washington University in St. Louis and aligned with the TAIR 10 build of the A. thaliana Col-0 genome using Novoalign (Novocraft; http://novocraft.com). SNPs were identified using SAMtools [110] and annotated with snpEFF [111]. Mutations were filtered using a script that prioritized homozygous canonical EMS-derived mutations (G-to-A and C-to-T) causing non-synonymous amino acid changes or altered splice sites but also retained mutations in introns and heterozygous EMS-consistent mutations. We disregarded mutations that were present in our lab stock of Col-0 or in multiple suppressors from different pools, indicating an origin in the starting line. Chromosomal nucleotide positions and gene identifiers of genes with homozygous EMS-consistent mutations are listed in Data S1.
Sequencing genomic DNA from the L1 isolate revealed mutations in both ATG18a (c463t causing Q155Stop) and ATG18f (g708a causing A198T). To determine which of these lesions was linked to lon2 suppression, we crossed L1 to lon2-2 35S:GFP-PTS1 and used genotyping markers (Table S2) to isolate F3 plants that were homozygous for the atg18a lesion but homozygous for wild-type ATG18f and vice versa. The resultant F4 progeny were tested for IBA responsive lateral rooting and 6-day-old immunoblot phenotypes to reveal that the atg18a mutation suppressed lon2-2 whereas the atg18f mutation did not alter lon2-2 phenotypes. We used the backcrossed lon2-2 atg18a-3 mutant in subsequent analyses.
Individual gene sequencing
ATG7 or ATG3 was sequenced directly in suppressors with reduced ATG7 or ATG3 protein levels, respectively. ATG7 was PCR amplified using the primers indicated in Table S3, and ATG3 was PCR amplified using previously described primers [48]. Amplicons were purified using the DNA Clean & Concentrator kit (Zymo Research, D4004) and sequenced directly (Lone Star Labs, Houston, TX, USA or Genewiz, Houston, TX, USA) with the primers used for amplification.
Immunoblot analysis
For most immunoblots, extracts were prepared from seedlings or adult leaves by homogenizing frozen tissue in 2 volumes of 2X NuPAGE LDS sample buffer (212 mM Tris HCl [Fisher Scientific, BP1531], 282 mM Tris base [Fisher Scientific, BP1525], 4% [w:v] lithium dodecyl sulfate [Worldwide Medical Products, 0782], 1.02 mM EDTA [Genesee Scientific, 20–148], 20% [w:v] glycerol [Worldwide Medical Products, 0854], 0.44 mM Coomassie Brilliant Blue G250 [Bio-Rad 161–0406], 0.332 mM Phenol Red [Sigma-Aldrich P5530], pH 8.5) containing 50 mM dithiothreitol (Gold Biotechnology, DTT10) and heating at 100°C for 5 min. Samples were electrophoresed on Bolt 10% Bis-Tris Plus gels (Invitrogen, NW00100BOX/NW00102BOX/NW00107BOX) or NuPAGE 10% Bis-Tris Midi gels (Invitrogen, WG1202BOX) using 1X MOPS running buffer (50 mM MOPS [Gold Biotechnology, M-790–1], 50 mM Tris base, 0.1% SDS [Dot Scientific, DSL22010], 1 mM EDTA, pH 7.7) and transferred to Amersham Protran Premium nitrocellulose membrane (VWR, 10120–006) for 40 min at 24 V using NuPAGE transfer buffer (25 mM Bicine [Gold Biotechnology, B-785–1], 25 mM Bis-Tris [Gold Biotechnology, B-020–1], 1 mM EDTA, 0.05 mM Chlorobutanol [Sigma-Aldrich 11,205–4], 10% [v:v] methanol, pH 7.2).
For ATG8 immunoblot analysis (Figure 3(d)), extracts were prepared by homogenizing frozen seedlings in 2 volumes of 2X Tricine SDS sample buffer (900 mM Tris HCl, 24% glycerol, 8% SDS, 0.005% Coomassie Brilliant Blue G250, 0.005% Phenol Red, pH 8.45) containing 6 M urea (JT Baker, 4204–01) and 50 mM dithiothreitol. Samples were electrophoresed on 10% Tricine gels (Invitrogen, EC66752BOX) using 1X Tricine SDS running buffer (100 mM Tris base, 100 mM Tricine [Sigma-Aldrich T0377], 0.1% SDS, pH 8.3) and transferred to Amersham Protran Premium nitrocellulose membrane for 1 h at 24 V using Tris-glycine transfer buffer (12 mM Tris base, 96 mM glycine, 20% [v:v] methanol, pH 8.3).
Membranes were blocked in 8% (w:v) Carnation non-fat dry milk solution (or 5% [w:v] bovine serum albumin [Gibco-BRL, 11018–017] for the anti-ATG3 and anti-ATG5 antibodies) in 20 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Tween 20 (Promega, H5151) and then incubated overnight at 4°C with primary antibodies in blocking solution. Rabbit antibodies against ATG3 [30] (1:10,000), ATG5 (1:2000; Agrisera, AS15 3060), ATG7 [8] (1:1000), ATG8A (1:2500; Abcam, ab77003; previously known as APG8A), ICL [112] (1:1000), MLS [113] (1:25,000), NBR1 [14] (1:2000), the PED1 isoform of thiolase [114] (1:20,000), and PMDH2 [115] (1:5000) were diluted as indicated. Mouse antibodies against HSC70 (1:100,000; StressGen Bioreagents, SPA-817) were used. Primary antibodies were visualized with goat anti-rabbit (1:5000; Santa Cruz Biotechnology, SC-2030 or GenScript, A00098) or anti-mouse (1:5000; Santa Cruz Biotechnology, SC-516102) horseradish peroxidase-conjugated secondary antibodies diluted in blocking buffer. Horseradish peroxidase was visualized with ProSignal Pico ECL reagent (Genesee Scientific, 20-300B) and autoradiography film (Genesee Scientific, 30-810C). Membranes were reblocked and sequentially probed with the indicated antibodies without stripping the membrane between incubations.
Confocal fluorescence microscopy
Wild-type 35S:GFP-PTS1, atg18a-3 35S:GFP-PTS1, and nbr1-4 35S:GFP-PTS1 seedlings grown on PNS under continuous white light were mounted in water, and fluorescence was visualized using a LSM 710 laser scanning confocal microscope (Carl Zeiss Microscopy, Thornwood, NY, USA) equipped with a meta detector. Samples were imaged using a 63X (Figure 5) or 40X (Figure 7(d)) oil immersion objective. GFP and chlorophyll were excited with a 488-nm argon laser. GFP emission was collected between 493 nm and 572 nm, and chlorophyll autofluorescence was collected between 620 nm and 719 nm. Each image in Figure 5 is an average of 2 exposures using a 47-µm pinhole, corresponding to a 0.8-µm optical section. Each image in Figure 7(d) is an average of 4 exposures using a 47-µm pinhole, corresponding to a 1.2-µm optical section.
Statistical analysis
One-way ANOVA with the Duncan post-hoc test was used to assess statistical significance (SPSS Statistics software, Version 24.0.0.0).
Accession numbers
Sequence data can be found in the Arabidopsis Genome Initiative under the following accession numbers: At3g19190 (ATG2), At5g61500 (ATG3), At5g17290 (ATG5), At5g45900 (ATG7), At4g30790 (ATG11), At5g50230 (ATG16), At3g62770 (ATG18a), At5g47040 (LON2), and At4g24690 (NBR1). The Brassicaceae ATG2 orthologs used for alignment have the following accession numbers: EFH59425.1 (Arabidopsis lyrata), XP_013585639.1 (Brassica oleracea), XP_009145821.1 (Brassica rapa), XP_006296813.1 (Capsella rubella), and XP_006406527.1 (Eutreum salsugineum).
Funding Statement
This research was supported by the National Science Foundation (MCB-1516966) and the Robert A. Welch Foundation (C-1309). RJL was supported in part by the National Institutes of Health (F31GM125367). Genomic sequencing at the Genome Technology Access Center at Washington University School of Medicine was supported by the National Institutes of Health (P30CA91842 and UL1RR024992). Confocal microscopy equipment was obtained through a Shared Instrumentation Grant from the National Institutes of Health (S10RR026399).
Acknowledgments
We thank Hiba Zafar for assistance with screening and Charles Danan for assistance with identifying lon2-2 atg7-9 35S:PTS2-GFP. We thank Yun-Ting Kao and Joseph Faust for the script for facilitating analysis of whole-genome sequencing data and Lucia Strader (Washington University in St. Louis) for assistance with whole-genome sequencing. We thank Richard Vierstra (Washington University in St. Louis) for the anti-ATG3 and anti-ATG7 antibodies, Masayoshi Maeshima (Nagoya University) for the anti-ICL antibody, John Harada (University of California, Davis) for the anti-MLS antibody, Terje Johansen (University of Tromsø) for the anti-NBR1 antibody, and Steven Smith (University of Western Australia) for the anti-PMDH2 antibody. We thank Janet Braam, Kathryn Smith, Melissa Traver, Andrew Woodward, and Zachary Wright for critical comments on the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental material
Supplemental data for this article can be accessed here.
References
- [1].Reggiori F, Klionsky DJ.. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics. 2013. June;194(2):341–361. PMID: 23733851; PMCPMC3664846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Mizushima N, Levine B.. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010. September;12(9):823–830. PMID: 20811354; PMCPMC3127249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Li F, Vierstra RD. Autophagy: A multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 2012. September;17(9):526–537. PMID: 22694835 [DOI] [PubMed] [Google Scholar]
- [4].Michaeli S, Galili G, Genschik P, et al. Autophagy in plants–what’s new on the menu? Trends Plant Sci. 2016;21(2):134–144. [DOI] [PubMed] [Google Scholar]
- [5].Meijer WH, van der Klei IJ, Veenhuis M, et al. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy. 2007. Mar-Apr;3(2):106–116. PMID: 17204848 [DOI] [PubMed] [Google Scholar]
- [6].Yoshimoto K. Beginning to understand autophagy, an intracellular self-degradation system in plants. Plant Cell Physiol. 2012. August;53(8):1355–1365. PMID: 22764279 [DOI] [PubMed] [Google Scholar]
- [7].Suttangkakul A, Li F, Chung T, et al. The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell. 2011. October;23(10):3761–3779. PMID: 21984698; PMC3229148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Doelling JH, Walker JM, Friedman EM, et al. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem. 2002. September 6;277(36):33105–33114. PMID: 12070171. [DOI] [PubMed] [Google Scholar]
- [9].Lai Z, Wang F, Zheng Z, et al. A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 2011. June;66(6):953–968. PMID: 21395886. [DOI] [PubMed] [Google Scholar]
- [10].Zhou J, Wang J, Cheng Y, et al. NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 2013;9(1):e1003196 PMID: 23341779; PMCPMC3547818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Liu Y, Xiong Y, Bassham DC. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy. 2009. October;5(7):954–963. PMID: 19587533 [DOI] [PubMed] [Google Scholar]
- [12].Xiong Y, Contento AL, Nguyen PQ, et al. Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 2007. January;143(1):291–299. PMID: 17098847; PMC1761971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Di Berardino J, Marmagne A, Berger A, et al. Autophagy controls resource allocations and protein storage accumulation in Arabidopsis seeds. J Exp Bot. 2018;69(6):1403–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Svenning S, Lamark T, Krause K, et al. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy. 2011. September;7(9):993–1010. PMID: WOS:000294477100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Shibata M, Oikawa K, Yoshimoto K, et al. Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. Plant Cell. 2013. December;25(12):4967–4983. PMID: 24368788; PMC3903999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kim J, Lee H, Lee HN, et al. Autophagy-related proteins are required for degradation of peroxisomes in Arabidopsis hypocotyls during seedling growth. Plant Cell. 2013. December;25(12):4956–4966. PMID: 24368791; PMC3903998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Li F, Chung T, Vierstra RD. AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell. 2014. February;26(2):788–807. PMID: 24563201; PMC3967041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kang S, Shin KD, Kim JH, et al. Autophagy-related (ATG) 11, ATG9 and the phosphatidylinositol 3-kinase control ATG2-mediated formation of autophagosomes in Arabidopsis. Plant Cell Rep. 2018. April;37(4):653–664. PMID: 29350244. [DOI] [PubMed] [Google Scholar]
- [19].Suzuki K, Kubota Y, Sekito T, et al. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007. February;12(2):209–218. PMID: 17295840. [DOI] [PubMed] [Google Scholar]
- [20].Kihara A, Noda T, Ishihara N, et al. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001. February 5;152(3):519–530. PMID: 11157979; PMCPMC2196002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Patel S, Dinesh-Kumar SP. Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy. 2008. January 1;4(1):20–27. PMID: WOS:000252211800003. [DOI] [PubMed] [Google Scholar]
- [22].Wang Y, Nishimura MT, Zhao T, et al. ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J. 2011. October;68(1):74–87. PMID: 21645148. [DOI] [PubMed] [Google Scholar]
- [23].Xiong Y, Contento AL, Bassham DC. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 2005. May;42(4):535–546. PMID: WOS:000228746000008 [DOI] [PubMed] [Google Scholar]
- [24].Zhuang X, Chung KP, Cui Y, et al. ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc Natl Acad Sci USA. 2017. January 17;114(3):E426–E425. PMID: 28053229; PMCPMC5255614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sláviková S, Shy G, Yao Y, et al. The autophagy-associated Atg8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. J Exp Bot. 2005;56(421):2839–2849. [DOI] [PubMed] [Google Scholar]
- [26].Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001. March;2(3):211–216. PMID: 11265251. [DOI] [PubMed] [Google Scholar]
- [27].Chung T, Phillips AR, Vierstra RD. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A and ATG12B loci. Plant J. 2010. May;62(3):483–493. PMID: 20136727 [DOI] [PubMed] [Google Scholar]
- [28].Yoshimoto K, Hanaoka H, Sato S, et al. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 2004. November;16(11):2967–2983. PMID: WOS:000225228500010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Thompson AR, Doelling JH, Suttangkakul A, et al. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 2005. August;138(4):2097–2110. PMID: 16040659; PMCPMC1183398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Phillips AR, Suttangkakul A, Vierstra RD. The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics. 2008. March;178(3):1339–1353. PMID: WOS:000254921600022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Romanov J, Walczak M, Ibiricu I, et al. Mechanism and functions of membrane binding by the Atg5–atg12/Atg16 complex during autophagosome formation. Embo J. 2012;31(22):4304–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yamaguchi M, Matoba K, Sawada R, et al. Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7. Nat Struct Mol Biol. 2012. December;19(12):1250–1256. PMID: 23142983. [DOI] [PubMed] [Google Scholar]
- [33].Rogov V, Dotsch V, Johansen T, et al. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell. 2014. January 23;53(2):167–178. PMID: 24462201. [DOI] [PubMed] [Google Scholar]
- [34].Xie Q, Tzfadia O, Levy M, et al. hfAIM: A reliable bioinformatics approach for in silico genome-wide identification of autophagy-associated Atg8-interacting motifs in various organisms. Autophagy. 2016;12(5):876–887. PMID: WOS:000375330100010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zientara-Rytter K, Lukomska J, Moniuszko G, et al. Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy. 2011. October;7(10):1145–1158. PMID: 21670587; PMC3242614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhou J, Zhang Y, Qi J, et al. E3 ubiquitin ligase CHIP and NBR1-mediated selective autophagy protect additively against proteotoxicity in plant stress responses. PLoS Genet. 2014. January;10(1):e1004116 PMID: 24497840; PMC3907298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hafrén A, Macia J-L, Love AJ, et al. Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc Natl Acad Sci USA. 2017. March 7;114(10):E2026–E2035. PMID: 28223514; PMCPMC5347569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Marshall RS, Li F, Gemperline DC, et al. Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in Arabidopsis. Mol Cell. 2015. June 18;58(6):1053–1066. PMID: WOS:000360986700017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Honig A, Avin-Wittenberg T, Ufaz S, et al. A new type of compartment, defined by plant-specific Atg8-interacting proteins, is induced upon exposure of Arabidopsis plants to carbon starvation. Plant Cell. 2012. January;24(1):288–303. PMID: WOS:000300881800025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhou J, Wang Z, Wang X, et al. Dicot-specific ATG8-interacting ATI3 proteins interact with conserved UBAC2 proteins and play critical roles in plant stress responses. Autophagy. 2018;14(3):487–504. PMID: 29313416; PMCPMC5915045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Michaeli S, Honig A, Levanony H, et al. Arabidopsis ATG8-INTERACTING PROTEIN1 is involved in autophagy-dependent vesicular trafficking of plastid proteins to the vacuole. Plant Cell. 2014. October;26(10):4084–4101. PMID: WOS:000345920900023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Nolan TM, Brennan B, Yang M, et al. Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev Cell. 2017. April 10;41(1):33–46. PMID: 28399398; PMCPMC5720862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].van Den Bosch H, Schutgens RB, Wanders RJ, et al. Biochemistry of peroxisomes. Annu Rev Biochem. 1992;61:157–197. PMID: 1353950. [DOI] [PubMed] [Google Scholar]
- [44].Willekens H, Chamnongpol S, Davey M, et al. Catalase is a sink for H2O2 and is indispensable for stress defence in C3 plants. Embo J. 1997. August 15;16(16):4806–4816. PMID: 9305623; PMC1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Anand P, Kwak Y, Simha R, et al. Hydrogen peroxide induced oxidation of peroxisomal malate synthase and catalase. Arch Biochem Biophys. 2009. November;491(1–2):25–31. PMID: 19800310. [DOI] [PubMed] [Google Scholar]
- [46].Young PG, Bartel B. Pexophagy and peroxisomal protein turnover in plants. BBA - Mol Cell Res. 2016. May;1863(5):999–1005. PMID: 26348128; PMCPMC4779433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Yoshimoto K, Shibata M, Kondo M, et al. Organ-specific quality control of plant peroxisomes is mediated by autophagy. J Cell Sci. 2014. March 15;127(Pt 6):1161–1168. PMID: 24463818. [DOI] [PubMed] [Google Scholar]
- [48].Farmer LM, Rinaldi MA, Young PG, et al. Disrupting autophagy restores peroxisome function to an Arabidopsis lon2 mutant and reveals a role for the LON2 protease in peroxisomal matrix protein degradation. Plant Cell. 2013. October;25(10):4085–4100. PMID: 24179123; PMC3877801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lingard MJ, Bartel B. Arabidopsis LON2 is necessary for peroxisomal function and sustained matrix protein import. Plant Physiol. 2009. November;151(3):1354–1365. PMID: WOS:000271430500036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Goto-Yamada S, Mano S, Nakamori C, et al. Chaperone and protease functions of LON protease 2 modulate the peroxisomal transition and degradation with autophagy. Plant Cell Physiol. 2014. March;55(3):482–496. PMID: 24492254. [DOI] [PubMed] [Google Scholar]
- [51].Gur E, Sauer RT. Recognition of misfolded proteins by Lon, a AAA+ protease. Genes Dev. 2008. August 15;22(16):2267–2277. PMID: 18708584; PMCPMC2518814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wohlever ML, Baker TA, Sauer RT. Roles of the N domain of the AAA+ Lon protease in substrate recognition, allosteric regulation and chaperone activity. Mol Microbiol. 2014. January;91(1):66–78. PMID: 24205897; PMC3877180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Aksam EB, Koek A, Jourdan S, et al. A peroxisomal Lon protease and peroxisome degradation by autophagy play key roles in vitality of Hansenula polymorpha cells. Autophagy. 2007. Mar-Apr;3(2):96–105. PMID: 17172804 [DOI] [PubMed] [Google Scholar]
- [54].Bartoszewska M, Williams C, Kikhney A, et al. Peroxisomal proteostasis involves a Lon family protein that functions as protease and chaperone. J Biol Chem. 2012. August 10;287(33):27380–27395. PMID: 22733816; PMC3431691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Rinaldi MA, Patel AB, Park J, et al. The roles of β-oxidation and cofactor homeostasis in peroxisome distribution and function in Arabidopsis thaliana. Genetics. 2016. November;204(3):1089–1115. PMID: 27605050; PMCPMC5105844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Bartel B, Farmer LM, Rinaldi MA, et al. Mutation of the Arabidopsis LON2 peroxisomal protease enhances pexophagy. Autophagy. 2014. March;10(3):518–519. PMID: 24413187; PMCPMC4077889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Strader LC, Bartel B. Transport and metabolism of the endogenous auxin precursor indole-3-butyric acid. Mol Plant. 2011. February 28;2011(4):477–486. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Zolman BK, Yoder A, Bartel B. Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics. 2000. November;156(3):1323–1337. PMID: 11063705; PMCPMC1461311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Strader LC, Culler AH, Cohen JD, et al. Conversion of endogenous indole-3-butyric acid to indole-3-acetic acid drives cell expansion in Arabidopsis seedlings. Plant Physiol. 2010. August;153(4):1577–1586. PMID: 20562230; PMCPMC2923913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Zolman BK, Silva ID, Bartel B. The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid β-oxidation. Plant Physiol. 2001;127(3):1266–1278. [PMC free article] [PubMed] [Google Scholar]
- [61].Hofius D, Schultz-Larsen T, Joensen J, et al. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell. 2009. May 15;137(4):773–783. PMID: 19450522. [DOI] [PubMed] [Google Scholar]
- [62].Matsushita M, Suzuki NN, Obara K, et al. Structure of Atg5•Atg16, a complex essential for autophagy. J Biol Chem. 2007. March 2;282(9):6763–6772. PMID: 17192262. [DOI] [PubMed] [Google Scholar]
- [63].Hu TT, Pattyn P, Bakker EG, et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet. 2011. May;43(5):476–481. PMID: 21478890; PMCPMC3083492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Yoshimoto K, Jikumaru Y, Kamiya Y, et al. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell. 2009. September;21(9):2914–2927. PMID: 19773385; PMC2768913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Lenz HD, Haller E, Melzer E, et al. Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J. 2011. June;66(5):818–830. PMID: 21332848. [DOI] [PubMed] [Google Scholar]
- [66].Zolman BK, Bartel B. An Arabidopsis indole-3-butyric acid-response mutant defective in PEROXIN6, an apparent ATPase implicated in peroxisomal function. Proc Natl Acad Sci USA. 2004. February 10;101(6):1786–1791. PMID: 14745029; PMCPMC341854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Deosaran E, Larsen KB, Hua R, et al. NBR1 acts as an autophagy receptor for peroxisomes. J Cell Sci. 2013. February 15;126(Pt 4):939–952. PMID: WOS:000317585300008. [DOI] [PubMed] [Google Scholar]
- [68].Rodríguez MC, Wawrzynska A, Sirko A. Intronic T-DNA insertion in Arabidopsis NBR1 conditionally affects wild-type transcript level. Plant Signal Behav. 2014;9(12):e975659 PMID: 25482782; PMCPMC4622546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Chehab EW, Kim S, Savchenko T, et al. Intronic T-DNA insertion renders Arabidopsis opr3 a conditional jasmonic acid-producing mutant. Plant Physiol. 2011. June;156(2):770–778. PMID: 21487047; PMCPMC3177274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Hackenberg T, Juul T, Auzina A, et al. Catalase and NO CATALASE ACTIVITY1 promote autophagy-dependent cell death in Arabidopsis. Plant Cell. 2013. November;25(11):4616–4626. PMID: WOS:000329174400025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Juul T, Malolepszy A, Dybkaer K, et al. The in vivo toxicity of hydroxyurea depends on its direct target catalase. J Biol Chem. 2010. July 9;285(28):21411–21415. PMID: 20452979; PMCPMC2898382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kulich I, Pečenková T, Sekereš J, et al. Arabidopsis exocyst subcomplex containing subunit EXO70B1 is involved in autophagy-related transport to the vacuole. Traffic. 2013;14(11):1155–1165. [DOI] [PubMed] [Google Scholar]
- [73].Ono Y, Wada S, Izumi M, et al. Evidence for contribution of autophagy to Rubisco degradation during leaf senescence in Arabidopsis thaliana. Plant Cell Environ. 2013. June;36(6):1147–1159. PMID: 23215962. [DOI] [PubMed] [Google Scholar]
- [74].Hong SB, Kim B-W, Lee K-E, et al. Insights into noncanonical E1 enzyme activation from the structure of autophagic E1 Atg7 with Atg8. Nat Struct Mol Biol. 2011. December;18(12):1323–1330. PMID: WOS:000298011600017. [DOI] [PubMed] [Google Scholar]
- [75].Yamazaki-Sato H, Tanida I, Ueno T, et al. The carboxyl terminal 17 amino acids within Apg7 are essential for Apg8 lipidation, but not for Apg12 conjugation. FEBS Lett. 2003;551(1–3):71–77. [DOI] [PubMed] [Google Scholar]
- [76].Noda NN, Satoo K, Fujioka Y, et al. Structural basis of Atg8 activation by a homodimeric E1, Atg7. Mol Cell. 2011. November 4;44(3):462–475. PMID: 22055191. [DOI] [PubMed] [Google Scholar]
- [77].Taherbhoy AM, Tait SW, Kaiser SE, et al. Atg8 transfer from Atg7 to Atg3: a distinctive E1-E2 architecture and mechanism in the autophagy pathway. Mol Cell. 2011. November 4;44(3):451–461. PMID: 22055190; PMCPMC3277881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Farré JC, Burkenroad A, Burnett SF, et al. Phosphorylation of mitophagy and pexophagy receptors coordinates their interaction with Atg8 and Atg11. EMBO Rep. 2013;14(5):441–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Jaillais Y, Fobis-Loisy I, Miege C, et al. AtSNX1 defines an endosome for auxin-carrier trafficking in Arabidopsis. Nature. 2006. September 7;443(7107):106–109. PMID: 16936718. [DOI] [PubMed] [Google Scholar]
- [80].Qin G, Ma Z, Zhang L, et al. Arabidopsis AtBECLIN 1/AtAtg6/AtVps30 is essential for pollen germination and plant development. Cell Res. 2007. March;17(3):249–263. PMID: WOS:000246128100007. [DOI] [PubMed] [Google Scholar]
- [81].Fujiki Y, Yoshimoto K, Ohsumi Y. An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol. 2007. March;143(3):1132–1139. PMID: WOS:000244757700007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Shin KD, Lee HN, Chung T. A revised assay for monitoring autophagic flux in Arabidopsis thaliana reveals involvement of AUTOPHAGY-RELATED9 in autophagy. Mol Cells. 2014. May;37(5):399 PMID: 24805779; PMCPMC4044311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Floyd BE, Morriss SC, MacIntosh GC, et al. Evidence for autophagy-dependent pathways of rRNA turnover in Arabidopsis. Autophagy. 2015;11(12):2199–2212. PMID: 26735434; PMCPMC4835196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Hanaoka H, Noda T, Shirano Y, et al. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 2002. July;129(3):1181–1193. PMID: 12114572; PMCPMC166512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Woo J, Park E, Dinesh-Kumar S. Differential processing of Arabidopsis ubiquitin-like Atg8 autophagy proteins by Atg4 cysteine proteases. Proc Natl Acad Sci USA. 2014. January 14;111(2):863–868. PMID: 24379391; PMCPMC3896200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Farré J-C, Manjithaya R, Mathewson RD, et al. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell. 2008. March;14(3):365–376. PMID: 18331717; PMCPMC3763908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Motley AM, Nuttall JM, Hettema EH. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. Embo J. 2012. June 29;31(13):2852–2868. PMID: 22643220; PMCPMC3395097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Lin Y-L, Sung S-C, Tsai H-L, et al. The defective proteasome but not substrate recognition function is responsible for the null phenotypes of the Arabidopsis proteasome subunit RPN10. Plant Cell. 2011;23(7):2754–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Kaur N, Zhao Q, Xie Q, et al. Arabidopsis RING peroxins are E3 ubiquitin ligases that interact with two homologous ubiquitin receptor proteins. J Integr Plant Biol. 2013. January;55(1):108–120. PMID: WOS:000318600300001. [DOI] [PubMed] [Google Scholar]
- [90].Farmer LM, Book AJ, Lee K-H, et al. The RAD23 family provides an essential connection between the 26S proteasome and ubiquitylated proteins in Arabidopsis. Plant Cell. 2010. January;22(1):124–142. PMID: WOS:000275926100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Kao Y-T, Gonzalez KL, Bartel B. Peroxisome function, biogenesis, and dynamics in plants. Plant Physiol. 2018. January;176(1):162–177. PMID: 29021223; PMCPMC5761812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Zhang J, Tripathi DN, Jing J, et al. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat Cell Biol. 2015. October;17(10):1259–1269. PMID: WOS:000362213500006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Platta HW, El Magraoui F, Bäumer BE, et al. Pex2 and Pex12 function as protein-ubiquitin ligases in peroxisomal protein import. Mol Cell Biol. 2009. October 15;29(20):5505–5516. PMID: WOS:000270271100010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Zientara-Rytter K, Ozeki K, Nazarko TY, et al. Pex3 and Atg37 compete to regulate the interaction between the pexophagy receptor, Atg30, and the Hrr25 kinase. Autophagy. 2018;14(3):368–384. PMID: 29260977; PMC5915033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Nazarko TY, Ozeki K, Till A, et al. Peroxisomal Atg37 binds Atg30 or palmitoyl-CoA to regulate phagophore formation during pexophagy. J Cell Biol. 2014. February 17;204(4):541–557. PMID: 24535825; PMC3926955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Burnett SF, Farré J-C, Nazarko TY, et al. Peroxisomal Pex3 activates selective autophagy of peroxisomes via interaction with the pexophagy receptor Atg30. J Biol Chem. 2015. March 27;290(13):8623–8631. PMID: 25694426; PMCPMC4375511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Yamashita S-I, Abe K, Tatemichi Y, et al. The membrane peroxin PEX3 induces peroxisome-ubiquitination-linked pexophagy. Autophagy. 2014. September;10(9):1549–1564. PMID: 25007327; PMCPMC4206534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Rinaldi MA, Fleming WA, Gonzalez KL, et al. The PEX1 ATPase stabilizes PEX6 and plays essential roles in Arabidopsis peroxisome biology. Plant Physiol. 2017;174:2231–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Gonzalez KL, Ratzel SE, Burks KH, et al. A pex1 missense mutation improves peroxisome function in a subset of Arabidopsis pex6 mutants without restoring PEX5 recycling. Proc Natl Acad Sci USA. 2018. April 3;115(14):E3163–E3172. PMID: WOS:000429012500014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Nuttall JM, Motley AM, Hettema EH. Deficiency of the exportomer components Pex1, Pex6, and Pex15 causes enhanced pexophagy in Saccharomyces cerevisiae. Autophagy. 2014. May;10(5):835–845. PMID: 24657987; PMCPMC5119063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Law KB, Bronte-Tinkew D, Di Pietro E, et al. The peroxisomal AAA ATPase complex prevents pexophagy and development of peroxisome biogenesis disorders. Autophagy. 2017. May 4;13(5):868–884. PMID: 28521612; PMCPMC5446072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Woodward AW, Bartel B. The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol Biol Cell. 2005. February;16(2):573–583. PMID: 15548601; PMC545895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Earley KW, Haag JR, Pontes O, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006. February;45(4):616–629. PMID: 16441352. [DOI] [PubMed] [Google Scholar]
- [104].Koncz C, Schell J. The promoter of T L-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet. 1986;204(3):383–396. [Google Scholar]
- [105].Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998. December;16(6):735–743. PMID: 10069079 [DOI] [PubMed] [Google Scholar]
- [106].Haughn GW, Somerville C. Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet. 1986;204(3):430–434. PMID: WOS:A1986D929100010 [Google Scholar]
- [107].Stasinopoulos TC, Hangarter RP. Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol. 1990. August;93(4):1365–1369. PMID: 16667626; PMCPMC1062681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Neff MM, Turk E, Kalishman M. Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 2002. December;18(12):613–615. PMID: 12446140 [DOI] [PubMed] [Google Scholar]
- [109].Thole JM, Beisner ER, Liu J, et al. Abscisic acid regulates root elongation through the activities of auxin and ethylene in Arabidopsis thaliana. G3: Genes Genomes Genetics. 2014. July 1;4(7):1259–1274. PMID: WOS:000339326600008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009. August 15;25(16):2078–2079. PMID: 19505943; PMCPMC2723002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Cingolani P, Platts A, Wang LL, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: sNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012. Apr-Jun;6(2):80–92. PMID: 22728672; PMCPMC3679285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Maeshima M, Yokoi H, Asahi T. Evidence for no proteolytic processing during transport of isocitrate lyase into glyoxysomes in castor bean endosperm. Plant Cell Physiol. 1988. March;29(2):381–384. PMID: WOS:A1988M630000025 [Google Scholar]
- [113].Olsen LJ, Ettinger WF, Damsz B, et al. Targeting of glyoxysomal proteins to peroxisomes in leaves and roots of a higher plant. Plant Cell. 1993. August;5(8):941–952. PMID: 8400872; PMCPMC160329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Lingard MJ, Monroe-Augustus M, Bartel B. Peroxisome-associated matrix protein degradation in Arabidopsis. Proc Natl Acad Sci USA. 2009. March 17;106(11):4561–4566. PMID: 19246395; PMC2657447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Pracharoenwattana I, Cornah JE, Smith SM. Arabidopsis peroxisomal malate dehydrogenase functions in β-oxidation but not in the glyoxylate cycle. Plant J. 2007;50(3):381–390. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


