Use of biomarkers to diagnose, predict the prognosis of, and guide the treatment of neuropsychiatric conditions is in its infancy. However, the era of biomarkers is upon us. This article provides an overview of biomarkers, their clinical uses, and challenges in their development and use.
Keywords: biomarker, bio-measures, psychiatry, treatment selection, prognostication. diagnosis
Abstract
Psychiatrists and mental health professionals regularly perform various clinical tasks (e.g., detection, differential diagnosis, prognostication, treatment selection and implementation). How well they perform each of these tasks has a direct impact on patient outcomes. Measurement-based care has brought greater precision to these tasks and has improved outcomes. This article provides an overview of the types of biomeasures and biomarkers, the clinical uses of biomarkers, and the challenges in their development and clinical use. Although still in their infancy, biomarkers hold the promise of bringing even greater precision and even better outcomes in mental health. Biomeasures that could become biomarkers include genetic, proteomic, metabolomic, and immunologic measures, as well as physiological, functional, and brain structural measures. Mechanistic markers reflect and are based on the specific pathobiological processes that are involved in the development of a clinically defined condition. Some clinically relevant biomarkers may rely on this mechanistic understanding while others may not. Clinical biomarkers serve three broadly defined goals. Diagnostic markers define what is wrong. Prognostic markers define what will happen in the natural course of the condition, although they may also predict the course of illness during treatment. Theranostic markers address issues pertinent to treatment by defining whether, when, whom, and how to treat. Other biomarkers may be used to monitor the overall effect of treatment regardless of the therapeutic effects or to monitor the specific therapeutic effects of the intervention on the disorder itself. Biomarkers can also be used to estimate susceptibility/risk of developing the condition or the biological consequences of having had the disorder.
“Medicine is the science of uncertainty and the art of probability,” Sir William Osler (1) A millennium ago, physicians often diagnosed diabetes on the basis of the sweetness of the patient’s urine (2). In ancient Greece, Hippocrates attributed melancholia to an excess of black bile in the body (3). As the influence of “spirits” and “the emotions and ambitions of the gods” as explanations for acts of nature and diseases gave way to chemistry and compounds, the first biomarkers or at least biomeasures were born. These measures often served dual purposes: to understand what was wrong and to define what to do for the patient. Today, we address the same problems but with new technologies. We are presently “measurement-rich” and “application-poor”—but maybe not for long!
Concepts and Terms
Biomeasures and Biomarkers
Biomarkers depend on biomeasures, i.e., any measurable characteristic of living tissue or fluids (e.g., bioassays of blood, urine, or cerebral spinal fluid; tissue biopsies of organs; and electrophysiological and functional measures of the brain at rest and in response to medications or selected tasks) (4). Biomarkers must be reliable by being objective and minimally affected by the will, behavior, or attitudes of both patients and evaluators or by transient environmental influences. For example, patients’ efforts affect reaction times but not visual evoked responses (4). Similarly, physiological responses to tasks that engage and thus depend on the patient’s willingness to attend to and conduct the task would be less suitable as biomarkers, whereas images reflecting receptor density or binding in the brain would qualify. Actigraphy would not be suitable as a biomarker because the subject can choose to be active or not, but body temperature over the 24-hour cycle would be more suitable.
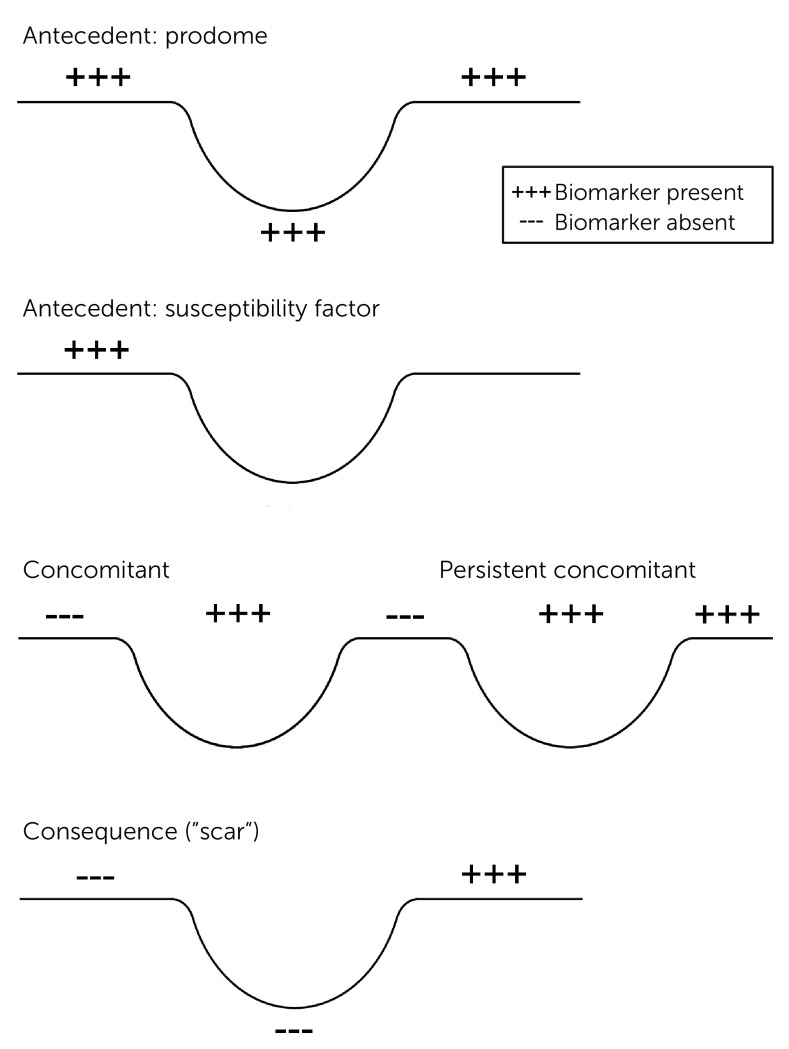
Biomarkers may be present before, during, or following the symptomatic expression of the illness (Figure 1). The antecedents are more likely to be mechanistically and potentially genetically relevant, whereas the consequences or “scars” might play a larger role in predicting the subsequent course of illness, the likelihood of complications, or residual disability. For example, some sleep EEG features appear to be trait-like (i.e., present before, during, and after the depressive episode), whereas others are largely episode concomitants (5).
FIGURE 1.
Antecedents, Concomitants, and Consequences of Illness Episodesa
aCopyright 2018 Curbstone Consultant LLC. Reprinted with permission
In addition, the types and nature of biomarkers may change over the course of the condition. Early biological indicators of bipolar illness may differ from those found among persons with an established multiyear history of the condition as the disease processes evolve over time. For example, left ventricular hypertrophy develops after years of chronic hypertension.
Furthermore, age and gender can affect the types and presence of biomarkers. For example, various sleep EEG features are affected by age, even among healthy persons. Women’s menstrual cycle phases, as well as smoking, diet, and exercise, can affect the presence, expression, and interpretation of biomarkers.
Whether one or a panel of biomeasures become clinically useful biomarkers depends on how well the test performs, costs and burdens to patients and care systems, practical limitations (e.g., invalid results for patients taking oral contraceptives or antihypertensive medications), comparative cost and performance vis-à-vis alternative competing tests, and clinical utility (i.e., what the biomarker adds to what we already know). In terms of acceptable test performance, Kraemer and colleagues (4) suggested a kappa of .6 for binary calls (e.g., a positive versus a negative diagnosis) and a Kendall or Spearman rank correlation coefficient of at least .6 for continuous variables. For example, a biomarker designed to reflect the severity of anxiety should correlate at least .6 with the anxiety symptom rating scale.
If the test is “accurate enough,” its clinical utility will depend on what the test adds to the clinical decision that it is designed to inform (6). For example, a specific threshold can be set with a particular measure that identifies individual patients who have a high certainty of doing poorly or well with a treatment. The degree of certainty—if high enough—can lead the clinician to strongly encourage or avoid the treatment (for examples, see Grieve and colleagues [7], South and colleagues [8], Kuk and colleagues [9, 10], and Li and colleagues [11]). More definitive biomarkers are more likely to be tightly tied to pathobiology or etiology (12). Often, several biomarkers are combined in a panel to increase the accuracy instead of relying on just one marker.
An important approach to dealing with the heterogeneous nature of psychiatric syndromes (complex disease phenotypes) has been to identify endophenotypes. In essence, this effort often combines clinical features with particular biological features that reflect the links between specific genes and the disease expression (13). The term—borrowed from insect biology—refers to the measurement of biological processes that lie between the genes and the clinical presentation. Fundamentally, the idea is that an endophenotype will be more biologically and etiologically homogeneous than the wider complex phenotype (syndrome) from which the endophenotype is drawn (14, 15). Because the endophenotype should be based on shared genes, it should be heritable, tend to cosegregate with the condition, and be found in some unaffected relatives in multiply affected families. Earlier work has been done on eye movement dysfunction in schizophrenia (see Levy and colleagues [16] for an overview), and other work has been done in alcoholism (17, 18).
Precision Medicine, Personalized Care, or Targeted Therapy
Personalized or precision medicine refers to the tailoring of medical treatment to the patient on the basis of individual patient characteristics (19). Fundamentally, treatment selection derives from our ability to sort patients into subgroups that differ in their biology, prognosis, or response to treatments (20). Biomarkers can aid us in selecting among multiple treatment options. This subgrouping enables the identification and selection of individual patients who are most suitable for (i.e., very likely to benefit from) or who are best advised to avoid specific therapies and the associated treatment costs and risks. This kind of targeted therapy is also referred to as “stratified medicine” (21). The notion is that through an analysis of biomarkers in the patient population, disorders can be stratified into subsets that exhibit differential outcomes and responses to specific therapeutics (22).
Beyond stratification, the term “precision medicine” is also used to include the creation of unique medical products for an individual patient, such as developing a cancer-fighting vaccine based on molecules derived from the patient’s own tumor.
From Biomarker Research to Clinical Applications: Please Mind the Gap!
Typically, when clinicians choose to use biomarkers or any laboratory test, they are often asking whether the patient does or does not have the condition or will or will not have a good outcome, etc. Unless the biomarker is almost uniquely associated with the condition in a highly sensitive and specific manner (as in the relation of hemoglobin SS to sickle cell disease), the actual performance of a biomarker test will depend substantially on the clinical context in which it is used.
When we ask, “How likely is my patient with a positive test to actually have the disease?” we are asking about the predictive value of a positive test. The answer depends on two test properties—sensitivity and specificity—and on the prevalence of the diagnosis or outcome in the population we are testing. Because prevalence affects the predictive value of any test, the same diagnostic test will have a different predictive accuracy according to the clinical context in which it is being applied.
Specifically, using the same test in a population with a higher prevalence of the positive outcome simultaneously increases the positive predictive value (PPV) and decreases the negative predictive value (NPV) of the test. Of course, if the clinical sample to which the biomarker is being applied has a comparable prevalence—in terms of the disorder or outcome being tested for—as the research populations in which the test was developed, the PPV, false discovery rate (FDR), and NPV will be comparable.
PPV depends on the number of true and false positives as shown in the following equations:
or
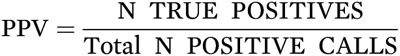 |
To calculate the actual numbers noted above, one needs the test performance characteristics of sensitivity and specificity and the prevalence of the condition in the population being tested. That is:
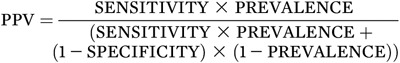 |
A concrete example is presented in Table 1, which illustrates the impact of prevalence changes on PPV for a test with 99% sensitivity and 95% specificity (23). Table 1 shows how the prevalence of diabetes—the outcome being sought—affects the performance of even a very good test. The first and last rows show that when the prevalence of diabetes rises with age from 1% (among 30-year-olds) to 20% (among 70-year-olds), the PPV rises from 17% to 83%—a huge difference in the clinical interpretation of the same test result. (The middle rows of the table show how this result is calculated.)
TABLE 1.
Impact on Positive Predictive Value (PPV) as Prevalence Changes, for a Test With 99% Sensitivity and 95% Specificitya
| Prevalence | |||
|---|---|---|---|
| Variable | 1% | 10% | 20% |
| a. N in population | 1,000 | 1,000 | 1,000 |
| b. N diseased | 10 | 100 | 200 |
| c. N not diseased | 990 | 900 | 800 |
| d. N of true positives on the test (b×.99) | 10 | 99 | 198 |
| e. N of false positives on the test [c×(1–.95)] | 50 | 45 | 40 |
| f. Total N positive on the test (d+e) | 60 | 144 | 238 |
| PPV (d/f) (%) | 17 | 69 | 83 |
Source: Shah (23)
Similarly, prevalence affects the FDR, which answers the question, “How often is a positive test wrong?” The FDR also depends on prevalence because it is calculated by using PPV as follows: FDR=1–PPV.
In brief, a biomarker or other lab test used in a very low-prevalence population will have a low PPV and a high FDR. Making biomarkers work well clinically depends on the timely and judicious use of the tests and on using the tests in a preferably target-rich environment.
Types of Biomeasurements
There are millions of variables that can be measured and that could become highly effective, clinically useful aids in understanding a patient’s condition or offer assistance in the conduct of our clinical tasks. It is beyond the scope of this article to detail all the available laboratory methods for performing these measures. The following sections highlight a few recent advances in the development of biomarkers in neuropsychiatric diseases. As our understanding of basic cellular biology, function, and communication in the normal and disordered central nervous system (CNS) grows, so will the number of potential biomarkers.
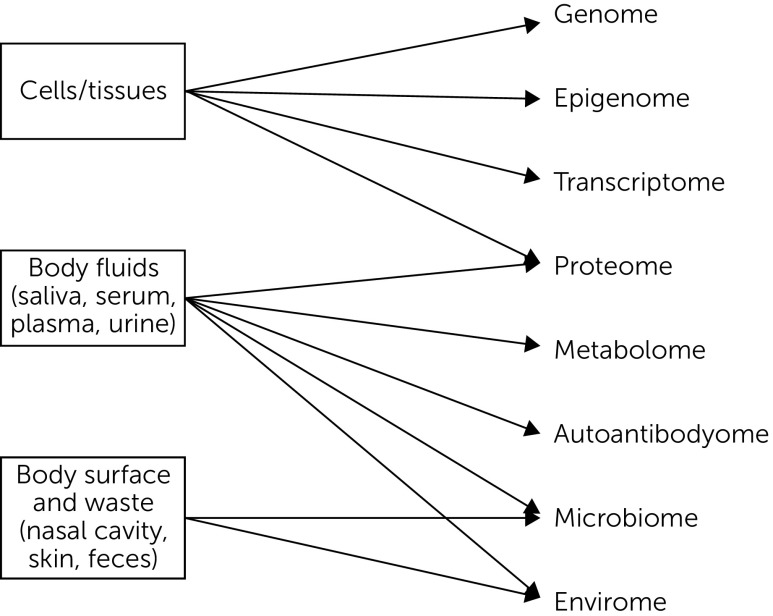
Broadly conceptualized, potential biomarkers can be based on the measurement of proteins, lipids, carbohydrates, electrolytes, nucleic acids, and so forth that regulate brain (and systemic) function. Promising systems that are being evaluated as sources of biomarkers include genomic, proteomic, metabolomic, and immunologic processes, among others. Gadad and colleagues (24) recently reviewed various approaches to developing biomarkers in mood disorders, with a focus on genetics, epigenetics, transcriptomics, proteomics, and metabolomics. Immunologic measurements and their potential relevance to neuropsychiatric diseases have also been reviewed (25, 26). Figure 2 summarizes these potential biomarker sources (27).
FIGURE 2.
Molecular Biomeasures: The Personal Omics Profilea
aAdapted from Chen and Snyder (27). Reprinted from Current Opinion in Psychiatry. Copyright 2012. Reprinted with permission
Genomics (Genetics, Epigenetics, and Transcriptomics)
Our ability to evaluate the genomics, epigenetics, and transcriptomics entailed in any condition is truly remarkable. For example, in a recent review, Gadad and colleagues (24) noted that we can now genotype 2.5 million single-nucleotide polymorphisms across the genome in a single pass (28). Furthermore, they note that genotype imputation is now an essential tool to predict genotypes not directly assayed in genome-wide association scans. These imputed genotype data increase analytic power, and they provide greater coverage with higher resolution of the genome. In addition, next-generation sequencing, or massively parallel sequencing, technology allows for the sequencing of the whole exome or genome and the potential identification of causal genetic mutations (29).
Furthermore, we can now evaluate the processes involved in gene expression, such as DNA methylation, histone modifications, or microRNA expression. These seem to be promising approaches to understanding the role of genetics in the pathobiology of psychiatric conditions such as major depressive disorder (30, 31). Finally, RNA sequencing, also known as whole-transcriptome shotgun sequencing (24), may help identify the pathobiological pathways that underlie psychiatric conditions by examining gene expression, alternative mRNA splicing, and posttranscriptional modifications (32–35).
Proteomics
Mass spectrometry (MS), two-dimensional gel electrophoresis, or various other methods that rely on the use of antibodies can identify a host of proteins. Two-dimensional gel electrophoresis and MS are commonly used in neuropsychiatric studies (36–39), but their detection abilities depend on abundance, size, and other properties of proteins (40). Shotgun proteomics, also called liquid chromatography–tandem mass spectrometry, is able to separate and identify thousands of proteins by using a combination of chromatographic steps in a high-throughput manner prior to MS analyses (37, 39).
More recently, multiplex proteomics has revolutionized proteomic analyses (24). It employs multiplexed dye-coded microspheres, coated with specific capture reagents (antibodies or DNA-based aptamers) that may bind numerous analytes within the same biological sample. This technology enables the measurement of over 1,000 analytes to be assayed in a single sample and minimizes sampling errors, sample volume requirements, and reagent costs (24, 40, 41). Multiplex-based technologies have already been used to screen for putative biomarkers in several neuropsychiatric disorders, including Alzheimer’s disease, Parkinson’s disease, schizophrenia, major depressive disorder, and bipolar disorder (41). Finally, well-established antibody-based assays for proteins, such as western blots or enzyme-linked immunosorbent assays, have significant quantification advantages over MS but are limited by the availability of protein-specific antibodies, low throughput, and the cost per protein measured (24).
Metabolomics
Metabolomics entail the measurement of small molecules in biological samples (42). The sources of the metabolites may be endogenous (i.e., naturally produced), such as amino acids or vitamins, or exogenous (i.e., not naturally produced) drugs, food additives, environmental contaminants, or toxins (43, 44). Every metabolite in a specific metabolic pathway, such as the tyrosine, purine, or tryptophan pathway, can be quantified. Their baseline values or changes over time may serve diagnostic, theranostic, or prognostic purposes, as well as shed light on the pathobiology of the condition.
Various technologies have been used to characterize the metabolome, including nuclear magnetic resonance spectroscopy, MS, and multiplex-based metabolite screening. High-resolution proton nuclear magnetic resonance spectroscopy can analyze several thousand metabolites with high throughput (45, 46). MS has been used for targeted or large-scale metabolome analyses. High-performance liquid chromatography, gas chromatography, and targeted electrochemistry-based MS platforms have also been employed to quantify abundant metabolic biomarkers in serum, plasma, and cerebral spinal fluid (47). Furthermore, multiplex-based metabolite screening is an advanced and quantitative method that can quickly screen for metabolic biomarker signatures (48).
More recently, exosomes are being evaluated as potential biomarkers in neuropsychiatric conditions (49). Exosomes are small extracellular vesicles that are released from cells such as neurons. They include various RNA species and proteins that modulate normal and pathological functions in the CNS. Exosomes are particularly promising for diagnostic and mechanistic biomarker discovery purposes because they contain markers that can be tracked to the cell of origin (50).
Other Biomeasurements
The immunologic systems are characterized largely with the measurement of proteins. Recent advances in our understanding of these systems and their relations to CNS disorders suggest that these systems play a key role in depressive disorders, schizophrenia, Parkinson’s disease (for example, see Goldstein and Young [51]), Alzheimer’s disease, and multiple sclerosis (52). These disorders, or a subset of them, may be directly due to immunologic dysfunctions, or the disorders may lead to immunologic abnormalities as a consequence, or both. That is, the biological derangements may be antecedents, concomitants, or consequences of these conditions.
In addition, over the past three decades, our ability to image the structure and function of the CNS at various levels, including gross anatomical structures and specific functional brain circuits, has advanced remarkably (53–55). These advances have enabled us to challenge the brain with highly selective therapeutic medications, stimulation technologies, or neuropsychological tasks to probe and thereby define the CNS substrates that distinguish various psychiatric conditions. The brain at rest or under various challenge conditions provides a vast array of biomeasures that are potential mechanistic or clinical biomarkers.
The combination of imaging, genetic, and other blood markers has begun to reveal the mechanistic relationships between genetic diatheses and brain response patterns (56) and between clinical capacities and brain circuits (57, 58). It is entirely plausible that blood markers, genotype findings, and brain-imaging parameters taken together could become a panel of biomarkers to address many of the clinical decision challenges that we currently face (for example, see Zethelius and colleagues [59]). Furthermore, it is logical to expect that a combination of already available clinical indicators of susceptibility or risk (60), when combined with biomeasures that are associated with the syndrome (for example, see Goldstein and Young [51]), could provide a simple clinical-laboratory battery for very early detection. This would seem feasible for bipolar disorder, for example.
Uses of Biomarkers
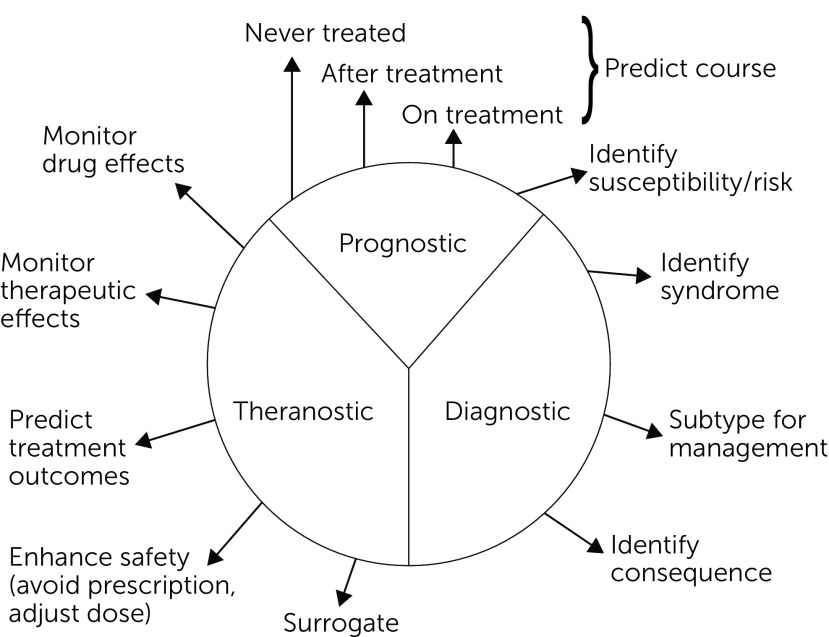
From a clinical perspective, biomarkers can be roughly grouped into four domains based on contexts of use: mechanistic, diagnostic, prognostic, and theranostic (61), with the latter three encompassing clinical aims or functions. Alternative categorizations of biomarkers have been proposed. For example, Bough and colleagues (62) suggested the following biomarker categories: mechanistic, toxicity, diagnostic, prognostic, pharmacodynamic, and predictive.
Figure 3 highlights the clinical tasks or aims that can be addressed by clinical biomarkers. Not every mechanistic measure or marker will necessarily serve any particular clinical aim. Furthermore, some clinically valuable biomarkers may not be mechanistically important. Moreover, a specific biomarker may be germane to one or more of these clinical aims. Thus the same biomarker could have diagnostic and prognostic value clinically.
FIGURE 3.
Biomarker Categoriesa
aCopyright 2018 Curbstone Consultant LLC. Reprinted with permission
Mechanistic Biomarkers
Mechanistic biomarkers identify the key molecular pathophysiological processes that underpin the disorder. They can be used to molecularly classify a heterogeneous syndrome or phenotype, which describes most mental conditions. A mechanistic biomarker does not involve any therapeutic intervention (62). However, mechanistic biomarkers may serve one or another clinical function as noted above.
Susceptibility or Risk Biomarkers
A susceptibility/risk biomeasure or biomarker is associated with an increased, or in some instances reduced, likelihood of developing a disorder by an individual who does not yet have that condition (for example, see LaGasse and colleagues [63]). A high level of low-density lipoprotein cholesterol is an example of a susceptibility/risk biomarker that indicates an increased risk of coronary artery disease. Clinically, susceptibility/risk biomarkers can be used in clinical practice to guide preventive strategies (64).
Diagnostic Biomarkers
Ideally, diagnostic biomarkers are mechanistic measures that reflect one or more biological processes that underlie the disease phenotype. Diagnostic biomarkers might stratify patient populations into appropriate treatment cohorts, but not necessarily so. They may detect the onset of the disease process before the full clinical manifestations appear. In such cases, the diagnostic biomarker can also serve as a prognostic biomarker (see below) (62).
Diagnostic biomarkers can also aid in limiting the range of potential diagnoses to be included in the differential diagnosis or in characterizing particular aspects of the syndrome to focus attention on a specific group of treatments, without being sufficiently accurate to recommend one treatment over another. For example, Lamers and colleagues (65) highlighted the biological differences between melancholic and atypical depressions, which suggest different potential treatment considerations but without sufficient specificity as to recommend one treatment over another. Furthermore, diagnostic biomarkers may differentiate among some but not other mental conditions, because two “distinct” syndromes may share some common mechanisms along the multistep cascade of processes that underpin both conditions.
Conversely, diagnostic biomarkers could be developed to assist our understanding of mechanisms that underpin specific aspects of the syndrome of interest, without necessarily being diagnostic of the overall condition (66). For example, both hypersomnia and insomnia qualify as a criterion symptom in defining a major depressive episode, but the mechanisms that underlie each type of sleep disturbance could be quite different. A mechanistic understanding of each sleep disturbance could lead to different prognostic and theranostic biomarkers.
A recent report illustrates the development of a nine-biomarker diagnostic blood sample panel for differentiating patients with major depressive disorder from nondepressed patients in a control group (67). The results of the nine-biomarker panel—expressed as an “MDDScore” (range 1–9)—distinguished sharply between patients with major depressive disorder and the control group patients (area under the receiver operating characteristic [ROC] curve=.963, 95% confidence interval=.93–.99) when the training and validation sets were combined.
The ROC curve is created by plotting the false-positive rate on the x axis and the true-positive rate on the y axis at the optimal test threshold. The ROC curve is used to estimate the accuracy of the test in differentiating between persons with and without the designated condition. When the test is highly accurate, it has an area under the curve of >90%; an area of 80%−90% is accepted as good (accurate roughly eight times in ten). That is, a good test will detect 80%−90% of persons with the condition, and a positive test result will be correct (i.e., be a true positive) eight to nine times out of ten. Therefore, the .963 area under the curve noted above is clinically very promising. ROC curves, however, have several limitations, one of which is that they cannot estimate the net benefit achieved in particular contexts by using or not using the test (68). In addition, there are many additional important steps to determine whether and when any promising biomarker is appropriate for clinical use (69).
A toxicity biomarker is not “diagnostic” in the sense of identifying a recognized clinical phenotype, but it is relevant to the identification and explanation of selected aspects of a specific condition. Bough and colleagues (62) defined a toxicity biomarker as a nonmechanistic but important measure of the biological effect(s) incurred as a consequence of having the condition. For example, chronic depression has been associated with the development of hypertension (70, 71) or elevated inflammatory cytokines (72) over time. The development of these biological consequences (i.e., toxicity biomarkers) could have prognostic value, as illustrated by the prognostic implications of developing mitral valve regurgitation or left ventricular hypertrophy because of long-standing hypertension.
Prognostic Biomarkers
Prognostic biomarkers indicate the likely progression of the disorder—often with a specific clinical outcome in mind—such as the onset, full syndromal expression, response, remission, or relapse. Ideally, prognostic biomarkers will predict the disease course without treatment or once treatment has stopped, as suggested in the U.S. Food and Drug Administration’s [FDA’s] definition of a prognostic marker for oncology (73). A variant on a prognostic marker could also predict the likelihood or the timing of the onset of complications (e.g., development of a substance use disorder during the course of bipolar disorder, emergence of an opioid use disorder amid narcotic use for postsurgical pain control, or development of vascular dementia in the course of hypertension), especially if the complication was derived directly from the causal cascade that led to the original condition. For example, multiple episodes of depression with hypothalamic-pituitary-adrenal activation might lead to metabolic or immunologic complications. Prognostic markers may or may not be based on the direct measurement of specific causal pathobiological processes.
Theranostic Biomarkers
Theranostic (treatment-related) biomarkers address several different clinical tasks or aims, including treatment selection, avoidance or orchestration (predictive), treatment implementation or tailoring (pharmacodynamics or pharmacogenetics), treatment effects (disease progression monitoring), treatment outcomes evaluation (e.g., surrogates), and development of a personalized allocation of health care resources (patient-level care indicators) (74).
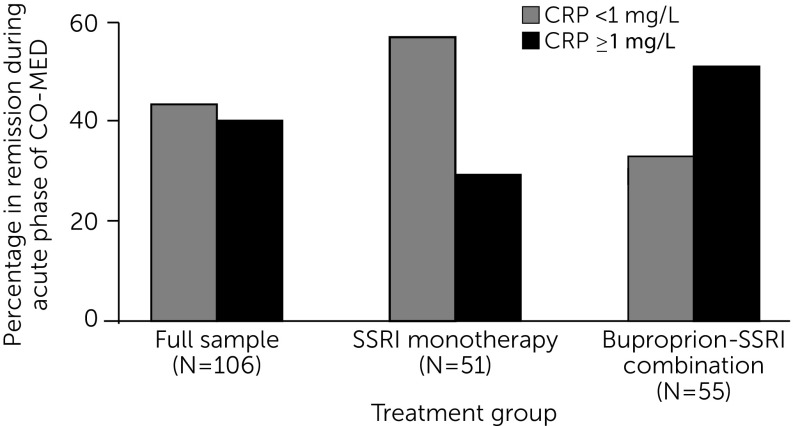
Predictive biomarkers are measures that identify specific patients who will fare well or poorly with an individual pharmacologic, somatic, or psychotherapeutic treatment (75). A predictive biomarker can also act as a moderator (76), a pretreatment measure that can recommend—at a specific threshold—one treatment over another because of either differential therapeutic effects or side effects. For example, Jha and colleagues (77) found the pretreatment level of C-reactive protein to be differentially associated with response to escitalopram versus escitalopram plus bupropion among depressed outpatients (Figure 4).
FIGURE 4.
Baseline C-Reactive Protein (CRP) Levels Moderate Remission in Depressiona
aCO-MED, Combining Medications to Enhance Depression Outcomes; SSRI, selective serotonin reuptake inhibitor. Source, Jha et al. (77). Reprinted from Psychoneuroendocrinology. Copyright 2017. Reprinted with permission
Importantly, other predictive biomarkers may be especially useful in identifying patients for whom a treatment is to be avoided because of the high risk of unacceptable side effects. For example, a genetic test has been found to be very useful in identifying specific patients who should avoid carbamazepine because of their high risk of developing Stevens-Johnson syndrome (78).
Response-indicator biomarkers (also called pharmacodynamic biomarkers) are used posttreatment to show that a change has occurred following treatment. These measures are markers of a pharmacological or physiological response to treatment regardless of treatment benefit (62, 79). For example, the treatment may affect specific metabolic pathways with or without affecting the person’s depression (46).
On the other hand, efficacy-response biomarkers are used as an indicator of a clinical end point and the effects of the treatment on this end point. For example, an efficacy-response biomarker for depression would reflect biological change that accompanies the therapeutic effect (e.g., symptomatic change) in the course of treatment. Such changes suggest that the biological measure is assessing part of the disease process that is accounting for some of the clinical symptom expression (73).
Finally, surrogate biomarkers fall within the category of theranostic markers. Surrogate biomarkers are obtained before and during treatment to reflect the therapeutic and biological effects of the treatment after it has been initiated (62, 79, 80). A surrogate biomarker is intended to be a reliable early indicator of the full effect of the treatment (81). Serum nicotine levels would be a surrogate end point biomarker for an antismoking agent that does not provide nicotine substitution, such as varenicline.
Surrogate markers can play an important role in clinical trials by shortening the length of the trial if the outcome prediction made by the surrogate marker is accurate. Surrogate biomarkers can be used alone or combined with clinical parameters to inform clinicians early in the course of treatment whether it will eventually produce the desired outcome for a particular patient, which enables timelier treatment plan revisions by reducing the time needed to decide that the treatment is not going to work well enough to continue it.
Clinical Findings and Their Relation to Biomarkers
Clinical findings have been widely evaluated as potential contributors to a variety of clinical tasks, including prognostication, treatment selection, and, of course, treatment monitoring. Indeed, psychiatric diagnoses are currently based nearly exclusively on clinical findings and history.
Clinical findings may include personality or temperament, as well as demographic features, such as education and social class. By and large, clinical features have been useful in providing overall estimates of outcomes for large groups, thereby helping to stratify or categorize patients into groups by features. For example, risk factors such as being Caucasian, male, or a substance abuser are associated with a higher risk of suicide attempts (82).
On the other hand, individual features may also inform clinical decisions. Consider a report by Uher and colleagues (83), who identified the interest-anhedonia symptom dimension, or by Fava and colleagues (84), who identified an anxiety symptom dimension as predictive of responses to antidepressant medications. These are simple symptom ratings that are easily obtained from patients. Biomarkers can be added to these clinical assessments, but to earn a place in practice the biomarker must provide a meaningful improvement over these easily obtained, less burdensome clinical measures. Presently, we have yet to arrive at such a clinically useful convergence.
Another potential role for clinical measures is in the development of person-level clinical indicators (74)—measures that can be used to allocate care resources to individuals whose needs can be predicted by using clinical features alone or with a combination of clinical and biological indicators. These person-level clinical indicators typically entail patient self-reports or clinical assessments that predict an individual patient’s course of illness, which in turn informs the allocation of care system resources.
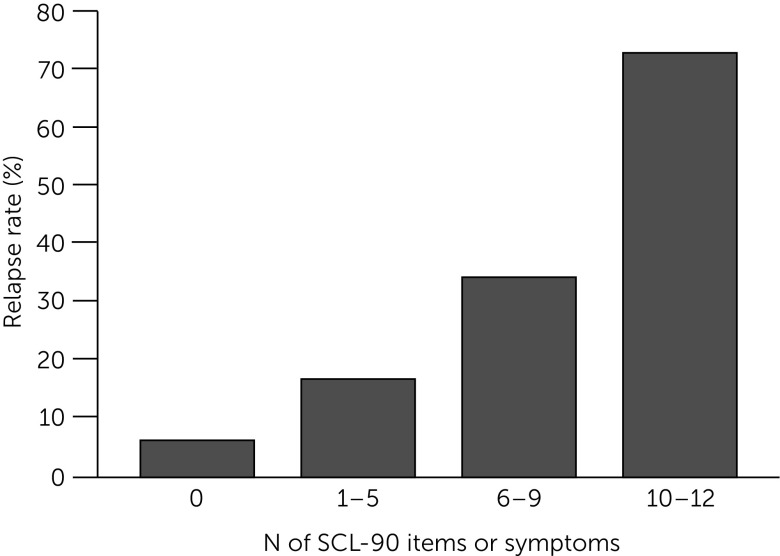
For example, the presence of depressive symptom remission compared with symptom response portends a more favorable prognosis (85, 86), which allows for less frequent follow-up visits for those in remission. We recently found that a brief 12-item symptom checklist accurately predicted the risk of relapse over the six months following the onset of remission from a major depressive episode (Figure 5) (87).
FIGURE 5.
Personalized Care Indicatora
aSCL-90, Symptom Checklist–90. Source, Judd et al. (86). Copyright 2016 American Psychiatric Association. Reprinted with permission
This type of clinical care indicator could be readily supplemented or even replaced by biomarkers that inform decisions about care planning and resource allocation. Demonstrating the feasibility of this same notion, Zeisler and colleagues (88) found that a biomarker based on the ratio of two serum markers known to change with preeclampsia could reliably predict preeclampsia over the ensuing several weeks in a large cohort of pregnant women.
Challenges in the Development of Biomarkers
Steps in Biomarker Development
Biomarker development consists of two components: analytical validation and clinical validation (73, 89, 90). Analytical validation requires generating data that describe the test performance characteristics of the biomarker measurement itself. As Scher and colleagues (73) stated, those characteristics “include the device, imaging modality, or assay, and the range of conditions under which the patient gives reproducible and accurate results.” Once analytical validity is established, clinical validity is addressed by studies to demonstrate that the biomarker is suitable for its stated purpose—and thereby to establish that the test results will inform the medical decision (73).
The specific steps needed to develop and evaluate a biomarker depend on the designated aim of the biomarker (73). Commonly, the first step entails evaluating a biomarker during a clinical trial or in a large sample of patients, some of whom will demonstrate the outcome of interest (e.g., response or relapse) These retrospective hypothesis-generating data lead to a prospective evaluation of the idea in a proof-of-concept study. The American Heart Association criteria for the evaluation of novel biomarkers of cardiovascular risk suggest that biomarker development steps should be clinically informed and use common sense (91). A similar approach could be adopted to develop biomarkers in mental health and other specialty areas.
Table 2 shows the case of a novel risk marker for cardiovascular disease (91). The first step–a proof of concept—entails determining whether the novel marker levels differ between participants who have and do not have the outcome to be predicted. Subsequently, a prospective validation is required. Having established that the marker performs the task for which it was identified or designed, the next few steps focus on clinical validation. Incremental value refers to the additional information provided over what is already available with established markers. Clinical utility evaluates the effect of this information on clinical behavior and ability to generate treatment recommendations. Clinical outcomes and cost ask about the “return on investment.” In other words, biomarker development and evaluation ultimately ask whether the additional information obtained with the biomarker is of clinical value and worth the added cost. This is a multistep journey (69).
TABLE 2.
Phases of Evaluation of a Novel Risk Markera
| Phase | Description |
|---|---|
| 1. Proof of concept | Do novel marker levels differ between subjects with and without outcome? |
| 2. Prospective validation | Does the novel marker predict development of future outcomes in a prospective cohort or nested case-cohort/case-cohort study? |
| 3. Incremental value | Does the novel marker add predictive information to established, standard risk markers? |
| 4. Clinical utility | Does the novel risk marker change predicted risk sufficiently to change recommended therapy? |
| 5. Clinical outcomes | Does use of the novel risk marker improve clinical outcomes, especially when tested in a randomized clinical trial? |
| 6. Cost-effectiveness | Does use of the marker improve clinical outcomes sufficiently to justify the additional costs of testing and treatment? |
From Hlatky et al. (91)
Specification of the Intended Use Is Required for FDA Biomarker Approval
For FDA approval, the clinical aim or goal of the biomarker must be specified. Table 3 summarizes the indications from the FDA for biomarker development in oncology (73). These aims are largely applicable to the development of biomarkers for mental illnesses. For example, the prognostic biomarker in oncology must estimate the likelihood of a specific clinical outcome (e.g., recurrence, progression, or death) for a patient who has completed anticancer therapy. Such a test enables determination as to which patients should be considered for more treatment. Similarly, in mental illnesses, a prognostic indicator could be developed to estimate the risk of relapse after continuation phase treatment for recurrent major depression to aid in the clinical decision as to which patients should and should not receive maintenance phase treatment.
TABLE 3.
FDA Biomarker Classification Based on Contexts of Usea
| Biomarker Type | When Biomarker Is Measured | What Biomarker Indicates |
|---|---|---|
| Prognostic | Prior to treatment | Indicates (estimates) the risk or likelihood that a patient who receives no further cancer-directed therapy will experience a specified clinical outcome, such as recurrence, progression, or death |
| Predictive | Prior to treatment | Interpret with defined criteria to identify patients who are likely to benefit from a specific treatment compared with patients who do not meet the specified criteria |
| Response indicator | During or after treatment | Demonstrates a pharmacological or physiological response to the treatment but does not necessarily signify patient benefit. Examples are declines in prostate-specific antigen, measures of tumor shrinkage, or pharmacodynamic changes in a parameter to show the on-target effect of a drug as proof of mechanism or to optimize dosing. |
| Efficiency response (surrogate) | After treatment | Provides an early and accurate prediction of both a clinical end point and the effects of the treatment on that end point |
Source, Scher et al. (73). Copyright 2013 Macmillan Publishers Ltd. Reprinted with permission
Note that in this scheme for oncology, a predictive indicator need not be a moderator that distinguishes between two active treatments. However, there must be evidence that the biomarker is not simply predicting responses to supportive, nonspecific placebo effects. For mental illnesses, demonstration that the biomarker is a moderator between drug and placebo would be very important given the probability of placebo response.
A predictive biomarker is usually and ideally obtained prior to treatment initiation. However, if the ultimate treatment outcomes are not seen for weeks or even months after starting therapy, a predictive biomarker of clinical value could include the baseline and early postbaseline biomarkers alone or in combination with clinical parameters.
Conclusions
The development and use of biomarkers with neuropsychiatric conditions is in its infancy. Our ability to specify the detailed pathobiology is remarkably limited compared with the advances in oncology or cardiology. Furthermore, the field has historically regarded biomarkers as methods to rule out and refer out any patients with a positive marker. Patients with epilepsy, cognitive decline or dementia, encephalopathies, and many other medical-neurological conditions with comorbid psychopathology are often referred out for treatment. With the development of biomarkers for “mental illnesses,” a much-needed bridge to medicine and neurology will be strengthened.
To prepare ourselves for biomarkers as “rule-in” indicators, we need to be familiar with the methods used to develop and evaluate these biomarkers, along with their strengths and limitations in clinical practice. Because the clinical value of a biomarker depends substantially on how and when it is used, we need to understand the conditions and contexts that form the basis for obtaining valid and useful biomarker results.
Finally, biomarkers in psychiatry are certain to present ethical and social challenges, especially if they have prognostic, diagnostic, or early susceptibility features. Although we may eagerly want to detect heart disease early, we are likely less eager to seek early detection of dementia or schizophrenia, conditions for which our treatments are only modestly effective.
Despite these challenges, the era of biomarkers is upon us. To optimally serve our patients, they and we must prepare to participate in this new, exciting, and hopeful journey.
Acknowledgments
The authors appreciate the editorial assistance of Jon Kilner, M.S., M.A.
Footnotes
Dr. Rush has received consulting or speaking fees from Akili Inc., Brain Resource Ltd., Compass Inc., Curbstone Consultant LLC., Emmes Corp., Liva-Nova, Mind-Linc, National Institute of Drug Abuse, Santium Inc., Sunovion, Taj Medical, and Takeda USA and royalties from Guilford Press and the University of Texas Southwestern Medical Center. The National Institutes of Health (NIH) has filed two patents in which Dr. Rush is named as a co-inventor. Under federal law, the NIH is required to pay the authors a portion of any royalties the NIH receives. The author does not endorse any commercial use of the patents. The patents are Methods to Predict the Outcome of Treatment With Antidepressant Medication; inventors (U.S. Patent No. 7,795,033) and Methods to Identify Patients at Risk of Developing Adverse Events During Treatment With Antidepressant Medication (U.S. Patent No. 7,906,283). Dr. Ibrahim reports no financial relationships with commercial interests.
References
- 1.Bean RB, Bean WB: Sir William Osler: Aphorisms From His Bedside Teachings and Writings. New York, Henry Schumann, 1950 [Google Scholar]
- 2.Swidorski D: Diabetes History. Madeira Beach, FL, Defeat Diabetes Foundation, 2014. https://www.defeatdiabetes.org/diabetes-history/. Accessed Aug 14, 2017
- 3.Telles-Correia D, Marques JG: Melancholia before the twentieth century: fear and sorrow or partial insanity? Front Psychol 2015; 6:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kraemer HC, Schultz SK, Arndt S: Biomarkers in psychiatry: methodological issues. Am J Geriatr Psychiatry 2002; 10:653–659 [PubMed] [Google Scholar]
- 5.Thase ME, Fasiczka AL, Berman SR, et al. : Electroencephalographic sleep profiles before and after cognitive behavior therapy of depression. Arch Gen Psychiatry 1998; 55:138–144 [DOI] [PubMed] [Google Scholar]
- 6.Rush AJ: Narrowing the gaps between what we know and what we do in psychiatry. J Clin Psychiatry 2015; 76:1366–1372 [DOI] [PubMed] [Google Scholar]
- 7.Grieve SM, Korgaonkar MS, Gordon E, et al. : Prediction of nonremission to antidepressant therapy using diffusion tensor imaging. J Clin Psychiatry 2016; 77:e436–e443 [DOI] [PubMed] [Google Scholar]
- 8.South C, Rush AJ, Carmody TJ, et al. : Accurately identifying patients who are excellent candidates or unsuitable for a medication: a novel approach. Neuropsychiatr Dis Treat 2017; 13:3001–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuk AY, Li J, Rush AJ: Recursive subsetting to identify patients in the STAR*D: a method to enhance the accuracy of early prediction of treatment outcome and to inform personalized care. J Clin Psychiatry 2010; 71:1502–1508 [DOI] [PubMed] [Google Scholar]
- 10.Kuk A, Li J, Rush AJ: Variable and threshold selection to control predictive accuracy in logistic regression. Appl Stat 2014; 63:657–672 [Google Scholar]
- 11.Li J, Kuk AY, Rush AJ: A practical approach to the early identification of antidepressant medication non-responders. Psychol Med 2012; 42:309–316 [DOI] [PubMed] [Google Scholar]
- 12.Lindstrom TM, Robinson WH: Biomarkers for rheumatoid arthritis: making it personal. Scand J Clin Lab Invest Suppl 2010; 242:79–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shields J, Gottesman II: Cross-national diagnosis of schizophrenia in twins: the heritability and specificity of schizophrenia. Arch Gen Psychiatry 1972; 27:725–730 [DOI] [PubMed] [Google Scholar]
- 14.Gottesman II, Gould TD: The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry 2003; 160:636–645 [DOI] [PubMed] [Google Scholar]
- 15.Walters JT, Owen MJ: Endophenotypes in psychiatric genetics. Mol Psychiatry 2007; 12:886–890 [DOI] [PubMed] [Google Scholar]
- 16.Levy DL, Sereno AB, Gooding DC, et al. : Eye tracking dysfunction in schizophrenia: characterization and pathophysiology. Curr Top Behav Neurosci 2010; 4:311–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dick DM, Jones K, Saccone N, et al. : Endophenotypes successfully lead to gene identification: results from the collaborative study on the genetics of alcoholism. Behav Genet 2006; 36:112–126 [DOI] [PubMed] [Google Scholar]
- 18.Hinrichs AL, Wang JC, Bufe B, et al. : Functional variant in a bitter-taste receptor (hTAS2R16) influences risk of alcohol dependence. Am J Hum Genet 2006; 78:103–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu YF, Goldstein DB, Angrist M, et al. : Personalized medicine and human genetic diversity. Cold Spring Harb Perspect Med 2014; 4:a008581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Timmerman L: What’s in a name? A lot, when it comes to “precision medicine.” Xconomy, Feb 4, 2013. http://www.xconomy.com/national/2013/02/04/whats-in-a-name-a-lot-when-it-comes-to-precision-medicine/#
- 21.Trusheim MR, Berndt ER, Douglas FL: Stratified medicine: strategic and economic implications of combining drugs and clinical biomarkers. Nat Rev Drug Discov 2007; 6:287–293 [DOI] [PubMed] [Google Scholar]
- 22.Lyman GH, Moses HL: Biomarker tests for molecularly targeted therapies: the key to unlocking precision medicine. N Engl J Med 2016; 375:4–6 [DOI] [PubMed] [Google Scholar]
- 23.Shah C: Public Health and Preventative Medicine in Canada, 5th ed. Collingwood, Ontario, Saunders Canada, 2003 [Google Scholar]
- 24.Gadad BS, Jha MK, Czysz A, et al. : Peripheral biomarkers of major depression and antidepressant treatment response: current knowledge and future outlooks. J Affect Disord 2017 (Epub ahead of print), doi: 10.1016/j.jad.2017.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wohleb ES, Franklin T, Iwata M, et al. : Integrating neuroimmune systems in the neurobiology of depression. Nat Rev Neurosci 2016; 17:497–511 [DOI] [PubMed] [Google Scholar]
- 26.Jones KA, Thomsen C: The role of the innate immune system in psychiatric disorders. Mol Cell Neurosci 2013; 53:52–62 [DOI] [PubMed] [Google Scholar]
- 27.Chen R, Snyder M: Systems biology: personalized medicine for the future? Curr Opin Pharmacol 2012; 12:623–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lohoff FW: Overview of the genetics of major depressive disorder. Curr Psychiatry Rep 2010; 12:539–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato T: Whole genome/exome sequencing in mood and psychotic disorders. Psychiatry Clin Neurosci 2015; 69:65–76. doi: 10.1111/pcn.12247 [DOI] [PubMed] [Google Scholar]
- 30.Maze I, Shen L, Zhang B, et al. : Analytical tools and current challenges in the modern era of neuroepigenomics. Nat Neurosci 2014; 17:1476–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oh G, Wang SC, Pal M, et al. : DNA modification study of major depressive disorder: beyond locus-by-locus comparisons. Biol Psychiatry 2015; 77:246–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morin R, Bainbridge M, Fejes A, et al. : Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. Biotechniques 2008; 45:81–94. doi: 10.2144/000112900 [DOI] [PubMed] [Google Scholar]
- 33.Chu Y, Corey DR: RNA sequencing: platform selection, experimental design, and data interpretation. Nucleic Acid Ther 2012; 22:271–274. doi: 10.1089/nat.2012.0367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z, Gerstein M, Snyder M: RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009; 10:57–63. doi: 10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jansen R, Penninx BW, Madar V, et al. : Gene expression in major depressive disorder. Mol Psychiatry 2016; 21:339–347 [DOI] [PubMed] [Google Scholar]
- 36.Lista S, Faltraco F, Prvulovic D, et al. : Blood and plasma-based proteomic biomarker research in Alzheimer’s disease. Prog Neurobiol 2013; 101–102:1–17 [DOI] [PubMed] [Google Scholar]
- 37.Martins-de-Souza D, Harris LW, Guest PC, et al. : The role of proteomics in depression research. Eur Arch Psychiatry Clin Neurosci 2010; 260:499–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martins-de-Souza D: Proteomics as a tool for understanding schizophrenia. Clin Psychopharmacol Neurosci 2011; 9:95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schirle M, Bantscheff M, Kuster B: Mass spectrometry-based proteomics in preclinical drug discovery. Chem Biol 2012; 19:72–84 [DOI] [PubMed] [Google Scholar]
- 40.Xu HB, Zhang RF, Luo D, et al. : Comparative proteomic analysis of plasma from major depressive patients: identification of proteins associated with lipid metabolism and immunoregulation. Int J Neuropsychopharmacol 2012; 15:1413–1425 [DOI] [PubMed] [Google Scholar]
- 41.Chen CS, Zhu H: Protein microarrays. Biotechniques 2006; 40:423. [DOI] [PubMed] [Google Scholar]
- 42.Oliver SG, Winson MK, Kell DB, et al. : Systematic functional analysis of the yeast genome. Trends Biotechnol 1998; 16:373–378. doi: 10.1016/S0167-7799(98)01214-1 [DOI] [PubMed] [Google Scholar]
- 43.Wishart DS: Current progress in computational metabolomics. Brief Bioinform 2007; 8:279–293. doi: 10.1093/bib/bbm030 [DOI] [PubMed] [Google Scholar]
- 44.Nordström A, O’Maille G, Qin C, et al. : Nonlinear data alignment for UPLC-MS and HPLC-MS based metabolomics: quantitative analysis of endogenous and exogenous metabolites in human serum. Anal Chem 2006; 78:3289–3295. doi: 10.1021/ac060245f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abo R, Hebbring S, Ji Y, et al. : Merging pharmacometabolomics with pharmacogenomics using “1000 Genomes” single-nucleotide polymorphism imputation: selective serotonin reuptake inhibitor response pharmacogenomics. Pharmacogenet Genomics 2012; 22:247–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaddurah-Daouk R, Krishnan KR: Metabolomics: a global biochemical approach to the study of central nervous system diseases. Neuropsychopharmacology 2009; 34:173–186 [DOI] [PubMed] [Google Scholar]
- 47.Martins-de-Souza D: Proteomics, metabolomics, and protein interactomics in the characterization of the molecular features of major depressive disorder. Dialogues Clin Neurosci 2014; 16:63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rotroff DM, Corum DG, Motsinger-Reif A, et al. : Metabolomic signatures of drug response phenotypes for ketamine and esketamine in subjects with refractory major depressive disorder: new mechanistic insights for rapid acting antidepressants. Transl Psychiatry 2016; 6:e894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanninen K, Bister N, Koistnaho J, et al. : Exomes as new diagnostic tools in CNS diseases. Biochimica et Biophysica Acta 2016; 1862:403–410 [DOI] [PubMed] [Google Scholar]
- 50.Vries HE, Schwaninger M: Neuroinflammation: a common denominator for stroke, multiple sclerosis and Alzheimer’s disease. Biochim Biophys Acta 2016; 1862:297–298 [DOI] [PubMed] [Google Scholar]
- 51.Goldstein BI, Young LT: Toward clinically applicable biomarkers in bipolar disorder: focus on BDNF, inflammatory markers, and endothelial function. Curr Psychiatry Rep 2013; 15:425. doi: 10.1007/S11920-013-0425-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dantzer R, O’Connor JC, Freund GG, et al. : From inflammation to sickness and depression: when the immune system subjugates the brain. Nat Rev Neurosci 2008; 9:46–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wager TD, Woo CW: Imaging biomarkers and biotypes for depression. Nat Med 2017; 23:16–17 [DOI] [PubMed] [Google Scholar]
- 54.Drysdale AT, Grosenick L, Downar J, et al. : Resting-state connectivity biomarkers define neurophysiological subtypes of depression. Nat Med 2017; 23:28–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williams LM: Precision psychiatry: a neural circuit taxonomy for depression and anxiety. Lancet Psychiatry 2016; 3:472–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hariri AR, Drabant EM, Weinberger DR: Imaging genetics: perspectives from studies of genetically driven variation in serotonin function and corticolimbic affective processing. Biol Psychiatry 2006; 59:888–897 [DOI] [PubMed] [Google Scholar]
- 57.Etkin A, Gyurak A, O’Hara R: A neurobiological approach to the cognitive deficits of psychiatric disorders. Dialogues Clin Neurosci 2013; 15:419–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Etkin A, Patenaude B, Song YJ, et al. : A cognitive-emotional biomarker for predicting remission with antidepressant medications: a report from the iSPOT-D trial. Neuropsychopharmacology 2015; 40:1332–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zethelius B, Berglund L, Sundström J, et al. : Use of multiple biomarkers to improve the prediction of death from cardiovascular causes. N Engl J Med 2008; 358:2107–2116. doi: 10.1056/NEJMoa0707064 [DOI] [PubMed] [Google Scholar]
- 60.Axelson D, Goldstein B, Goldstein T, et al. : Diagnostic precursors to bipolar disorder in offspring of parents with bipolar disorder: a longitudinal study. Am J Psychiatry 2015; 172:638–646. doi: 10.1176/appi.ajp.2014.14010035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weickert CS, Weickert TW, Pillai A, et al. : Biomarkers in schizophrenia: a brief conceptual consideration. Dis Markers 2013; 35:3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bough KJ, Amur S, Lao G, et al. : Biomarkers for the development of new medications for cocaine dependence. Neuropsychopharmacology 2014; 39:202–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.LaGasse JM, Brantley MS, Leech NJ, et al. : Successful prospective prediction of type 1 diabetes in schoolchildren through multiple defined autoantibodies: an 8-year follow-up of the Washington State Diabetes Prediction Study. Diabetes Care 2002; 25:505–511 [DOI] [PubMed] [Google Scholar]
- 64.FDA-NIH Biomarker Working Group: BEST (Biomarkers, EndpointS, and Other Tools) Resource. Silver Spring, MD, US Food and Drug Administration, and Bethesda, MD, National Institutes of Health, 2016 [PubMed]
- 65.Lamers F, Vogelzangs N, Merikangas KR, et al. : Evidence for a differential role of HPA-axis function, inflammation and metabolic syndrome in melancholic versus atypical depression. Mol Psychiatry 2013; 18:692–699 [DOI] [PubMed] [Google Scholar]
- 66.Cuthbert BN, Insel TR: Toward the future of psychiatric diagnosis: the seven pillars of RDoC. BMC Med 2013; 11:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bilello JA, Thurmond LM, Smith KM, et al. : MDDScore: confirmation of a blood test to aid in the diagnosis of major depressive disorder. J Clin Psychiatry 2015; 76:e199–e206 [DOI] [PubMed] [Google Scholar]
- 68.Halligan S, Altman DG, Mallett S: Disadvantages of using the area under the receiver operating characteristic curve to assess imaging tests: a discussion and proposal for an alternative approach. Eur Radiol 2015; 25:932–939. doi: 10.1007/s00330-014-3487-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rush AJ: Medical test development and implementation: a multistep journey. J Clin Psychiatry 2015; 76:e220–e221 [DOI] [PubMed] [Google Scholar]
- 70.Rubio-Guerra AF, Rodriguez-Lopez L, Vargas-Ayala G, et al. : Depression increases the risk for uncontrolled hypertension. Exp Clin Cardiol 2013; 18:10–12 [PMC free article] [PubMed] [Google Scholar]
- 71.Meng L, Chen D, Yang Y, et al. : Depression increases the risk of hypertension incidence: a meta-analysis of prospective cohort studies. J Hypertens 2012; 30:842–851 [DOI] [PubMed] [Google Scholar]
- 72.Raison CL, Miller AH: Is depression an inflammatory disorder? Curr Psychiatry Rep 2011; 13:467–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Scher HI, Morris MJ, Larson S, et al. : Validation and clinical utility of prostate cancer biomarkers. Nat Rev Clin Oncol 2013; 10:225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perlman CM, Hirdes JH, Vigod S: Psychiatric rehospitalization: development of a personal-level indicator for care planning and quality assurance. Prim Care Companion CNS Disord; 17(4). (Epub ahead of print, July 23, 2015). doi: 10.4088/PCC.15m01784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kennedy SH, Downar J, Evans KR, et al. : The Canadian Biomarker Integration Network in Depression (CAN-BIND): advances in response prediction. Curr Pharm Des 2012; 18:5976–5989 [DOI] [PubMed] [Google Scholar]
- 76.Papakostas GI, Fava M: Predictors, moderators, and mediators (correlates) of treatment outcome in major depressive disorder. Dialogues Clin Neurosci 2008; 10:439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jha MK, Minhajuddin A, Gadad BS, et al. : Can C-reactive protein inform antidepressant medication selection in depressed outpatients? Findings from the CO-MED trial. Psychoneuroendocrinology 2017; 78:105–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hung SI, Chung WH, Liu ZS, et al. : Common risk allele in aromatic antiepileptic-drug induced Stevens-Johnson syndrome and toxic epidermal necrolysis in Han Chinese. Pharmacogenomics 2010; 11:349–356 [DOI] [PubMed] [Google Scholar]
- 79.Schmidt HD, Shelton RC, Duman RS: Functional biomarkers of depression: diagnosis, treatment, and pathophysiology. Neuropsychopharmacology 2011; 36:2375–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Biomarkers Definitions Working Group : Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther 2001; 69:89–95 [DOI] [PubMed] [Google Scholar]
- 81.Gispen-de Wied CC, Kritsidima M, Elferink AJ: The validity of biomarkers as surrogate endpoints in Alzheimer’s disease by means of the Quantitative Surrogate Validation Level of Evidence Scheme (QSVLES). J Nutr Health Aging 2009; 13:376–387 [DOI] [PubMed] [Google Scholar]
- 82.Depression Guideline Panel: Clinical Practice Guideline, Number 5. Depression in Primary Care: Vol 1, Detection and Diagnosis. AHCPR pub no 93-0550, Rockville, MD, Agency for Health Care Policy and Research, 1993
- 83.Uher R, Perlis RH, Henigsberg N, et al. : Depression symptom dimensions as predictors of antidepressant treatment outcome: replicable evidence for interest-activity symptoms. Psychol Med 2012; 42:967–980. doi: 10.1017/S0033291711001905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fava M, Rush AJ, Alpert JE, et al. : Difference in treatment outcome in outpatients with anxious versus nonanxious depression: a STAR*D report. Am J Psychiatry 2008; 165:342–351 [DOI] [PubMed] [Google Scholar]
- 85.Paykel ES, Ramana R, Cooper Z, et al. : Residual symptoms after partial remission: an important outcome in depression. Psychol Med 1995; 25:1171–1180 [DOI] [PubMed] [Google Scholar]
- 86.Judd LL, Schettler PJ, Rush AJ, et al. : A new empirical definition of major depressive episode recovery and its positive impact on future course of illness. J Clin Psychiatry 2016; 77:1065–1073 [DOI] [PubMed] [Google Scholar]
- 87.Judd LL, Schettler PJ, Rush AJ: A brief clinical tool to estimate individual patients’ risk of depressive relapse following remission: proof of concept. Am J Psychiatry 2016; 173:1140–1146 [DOI] [PubMed] [Google Scholar]
- 88.Zeisler H, Llurba E, Chantraine F, et al. : Predictive value of the sFlt-1:PlGF ratio in women with suspected preeclampsia. N Engl J Med 2016; 374:13–22 [DOI] [PubMed] [Google Scholar]
- 89.Goodsaid FM, Mendrick DL: Translational medicine and the value of biomarker qualification. Sci Transl Med 2010; 2:47ps44. [DOI] [PubMed] [Google Scholar]
- 90.Khleif SN, Doroshow JH, Hait WN: AACR-FDA-NCI Cancer Biomarkers Collaborative consensus report: advancing the use of biomarkers in cancer drug development. Clin Cancer Res 2010; 16:3299–3318 [DOI] [PubMed] [Google Scholar]
- 91.Hlatky MA, Greenland P, Arnett DK, et al. : Criteria for evaluation of novel markers of cardiovascular risk: a scientific statement from the American Heart Association. Circulation 2009; 119:2408–2416 [DOI] [PMC free article] [PubMed] [Google Scholar]