Abstract
As limits on O2 availability during submergence impose severe constraints on aerobic respiration, the oxygen binding globin proteins of marine mammals are expected to have evolved under strong evolutionary pressures during their land-to-sea transition. Here, we address this question for the order Sirenia by retrieving, annotating, and performing detailed selection analyses on the globin repertoire of the extinct Steller’s sea cow (Hydrodamalis gigas), dugong (Dugong dugon), and Florida manatee (Trichechus manatus latirostris) in relation to their closest living terrestrial relatives (elephants and hyraxes). These analyses indicate most loci experienced elevated nucleotide substitution rates during their transition to a fully aquatic lifestyle. While most of these genes evolved under neutrality or strong purifying selection, the rate of nonsynonymous/synonymous replacements increased in two genes (Hbz-T1 and Hba-T1) that encode the α-type chains of hemoglobin (Hb) during each stage of life. Notably, the relaxed evolution of Hba-T1 is temporally coupled with the emergence of a chimeric pseudogene (Hba-T2/Hbq-ps) that contributed to the tandemly linked Hba-T1 of stem sirenians via interparalog gene conversion. Functional tests on recombinant Hb proteins from extant and ancestral sirenians further revealed that the molecular remodeling of Hba-T1 coincided with increased Hb–O2 affinity in early sirenians. Available evidence suggests that this trait evolved to maximize O2 extraction from finite lung stores and suppress tissue O2 offloading, thereby facilitating the low metabolic intensities of extant sirenians. In contrast, the derived reduction in Hb–O2 affinity in (sub)Arctic Steller’s sea cows is consistent with fueling increased thermogenesis by these once colossal marine herbivores.
Keywords: ancient DNA, aquatic adaptation, gene conversion, hemoglobin, oxygen affinity, molecular evolution, myoglobin, neuroglobin, cytoglobin, pseudogene
Introduction
The impact of pseudogenization events on gene expression and protein function are a critical component of phenotypic evolution. While a loss of function is the most obvious outcome of a gene inactivation event, the vestiges of such loci may also influence nearby paralogs via gene conversion. Because of an accelerated mutation rate and lack of selection acting on pseudogenes, drastic phenotypic changes may arise in the event a functional gene segment is converted by an inactivated paralog. While such incidents are often deleterious (Chen et al. 2007), they can also enhance genetic diversity (Sharon et al. 1999). However, linking genetic diversity derived from a pseudogene to environmental adaptation is challenging. The hemoglobin (Hb) family of genes is ideally suited to address questions regarding the evolutionary significance of pseudogenes, as it has a well-established evolutionary history of gene duplications, deletions, inactivations, and conversions (Lam and Jeffreys 2006; Storz et al. 2007; Hoffmann et al. 2008; Opazo et al. 2008; Gaudry et al. 2014; Natarajan et al. 2015). Moreover, numerous studies have revealed how Hb proteins—the primary oxygen carrier in vertebrate red blood cells—have been adaptively modified to optimize O2 uptake and release in diverse terrestrial environments that vary widely in O2 availability (Weber 2007; Storz and Moriyama 2008; Campbell, Storz, et al. 2010).
In contrast, specializations in Hb function with respect to fully aquatic lifestyles have received relatively little research attention. This discrepancy is surprising as O2 storage and management are the foundations of successful underwater foraging by aquatic mammals. As finite O2 availability during submergence imposes severe constraints on aerobic respiration, diving mammals have evolved physiological traits that maximize internal O2 storage for periods of submergence apnea when atmospheric gas exchange has ceased (Ponganis 2011). The muscle protein myoglobin (Mb) is of paramount importance in this regard, with diving species having convergently evolved Mb proteins with an elevated net surface charge and increased protein stability, allowing for markedly increased Mb concentrations, and hence aerobic dive times (Mirceta et al. 2013; Samuel et al. 2015). Although some work has focused on the molecular evolution of morphological adaptations in cetaceans (McGowen et al. 2014), little attention has been directed to the molecular foundations underlying the evolution of O2 uptake and exchange strategies that are essential for diving success.
Mb and Hb were long thought to be the only globins present in mammalian genomes, but several additional anciently diverged globin lineages have been discovered in the past 15 years. These include neuroglobin (Ngb) and cytoglobin (Cygb), which are primarily expressed in neurons and the cytoplasm of all cells, respectively (Burmester and Hankeln 2014). The primary sequence of these proteins is highly conserved among living vertebrates, suggesting that each serves important physiological functions (Burmester et al. 2004). While their exact roles remain elusive, evidence suggests that these proteins may be associated with O2 storage, reactive oxygen species detoxification, nitric oxide production/scavenging, cell signaling, and the regulation of apoptosis (Burmester and Hankeln 2014), making them attractive subjects to study the molecular foundations of mammalian diving.
Aquatic breath-hold foraging has independently evolved multiple times within Mammalia, but living members of only two groups, cetaceans and sirenians, have completely forgone terrestrial life. The morphological and physiological adaptations that accompanied this drastic lifestyle change allow for maximal dive times ranging from 5 to 138 min (Ponganis 2011). While the cetacean clade encompasses ∼90 species (McGowen et al. 2014), the strictly herbivorous sirenian clade is small by comparison in that it consists of only four species divided amongst two living families. The family Trichechidae contains three extant species, the Amazonian manatee (Trichechus inunguis), the West Indian manatee (Trichechus manatus), and the African manatee (Trichechus senegalensis), whereas the dugong (Dugong dugon) became the sole living member of the family Dugongidae following the extinction of Steller’s sea cow (Hydrodamalis gigas) in 1768 (Stejneger 1887). Extant species inhabit shallow tropical and subtropical waters along coastlines throughout the Atlantic, western Indian, and southwest Pacific Oceans (Marsh et al. 2011). In the case of manatees, this distribution may reflect their nutritional and physiological restrictions, as food availability and poor thermogenic abilities confine this genus to shallow tropical waters (Scholander and Irving 1941; Blessing 1972). While less is known regarding dugongs, their tropical/subtropical distribution suggests a similarly suppressed metabolic capacity (Marsh et al. 2011). Conversely, the extinct Steller’s sea cows last inhabited the frigid waters surrounding the Commander Islands in the Bering Sea, with recent distributions extending along the Aleutian archipelago and even north of the Arctic Circle to St. Lawrence Island (Crerar et al. 2014). This ecological shift was presumably initiated by a period of global cooling in the middle Miocene (∼15 Ma) that decimated sea grass populations along the coast of California in favor of cryophilic kelps, which were adopted as a primary food source by the ancestors of Steller’s sea cows (Domning 1976). Morphological adaptations accompanying this dietary transition included the loss of teeth and the development of cornified pads on the upper palate and lower jaw that aided kelp consumption. These changes coincided with marked reductions in the forelimb elements (including a complete loss of fingers; Domning 1976), development of blubber and a thick bark-like skin (Steller 1751), and massive increases in size (up to 10 m in length and over 11,000 kg in mass; Domning 1976). Nothing, however, is known regarding physiological or molecular adaptations underlying this thermal transition, despite its likely significance for oxygen unloading to cold peripheral tissues (Weber and Campbell 2011).
To address these shortcomings, we used hybridization capture coupled with Illumina sequencing to retrieve and annotate the complete globin gene repertoire of the extinct Steller’s sea cow (H. gigas) and the extant dugong (D. dugon). These loci were combined with those mined from the genome of the Florida manatee (T. manatus latirostris) and the molecular evolution of this collection of genes was compared with that of their closest terrestrial relatives (elephants and hyraxes). We employed pairwise sequence similarity, synteny analysis, phylogenomics, and selection analyses to identify the emergence of a chimeric Hba-T2/Hbq pseudogene and provide evidence that it influenced the evolution of the expressed α-type hemoglobin gene (Hba-T1) via gene conversion during the secondary aquatic transition of Sirenia. Recombinant expression and functional tests of extant and ancestral sirenian adult-type Hb proteins reveal that the advent of the chimeric pseudogene and accelerated molecular evolution of the upstream Hba-T1 locus were temporally linked with an increased Hb–O2 affinity in this lineage.
Results and Discussion
Genomic Organization of Paenungulate Hb Gene Clusters
The α- and β-type Hb genes of mammals are arranged into clusters on (human) chromosomes 16 and 11, respectively. The 5′ to 3′ order of these genes determines at which life stage they are expressed, whereby genes at the 5′ end (Hbz, Hbe, and Hbg) are expressed prenatally and genes at the 3′ end (Hba, Hbd, and Hbb) are expressed in adults (Hardison 1999, 2012; Storz 2016). Hb proteins are tetramers composed of two α-type and two β-type polypeptide chains. Consequently, in early human development, the α-type product of the Hbz (ζ) gene first dimerizes with the β-type product of Hbe (ε) prior to forming the ζ2ε2 (Gower I) Hb isoform. As development continues, expression of Hbz is replaced by Hba (α), which sequentially dimerizes with the products of the β-type Hbe and Hbg (γ) chains prenatally, and then the products of Hbd (δ) and Hbb (β) after birth (Wood 1976). Paenungulates (sirenians, elephants, hyraxes) exhibit a slight modification to this paradigm, as both the Hbb and Hbd genes became inactivated following the emergence and ascendancy of an “antiLepore” Hbb/Hbd (β/δ) chimeric fusion mutant early in their evolution (Opazo et al. 2009), that is, they only express an α2β/δ2 (HbA/A2) isoform in adulthood (Campbell, Roberts, et al. 2010).
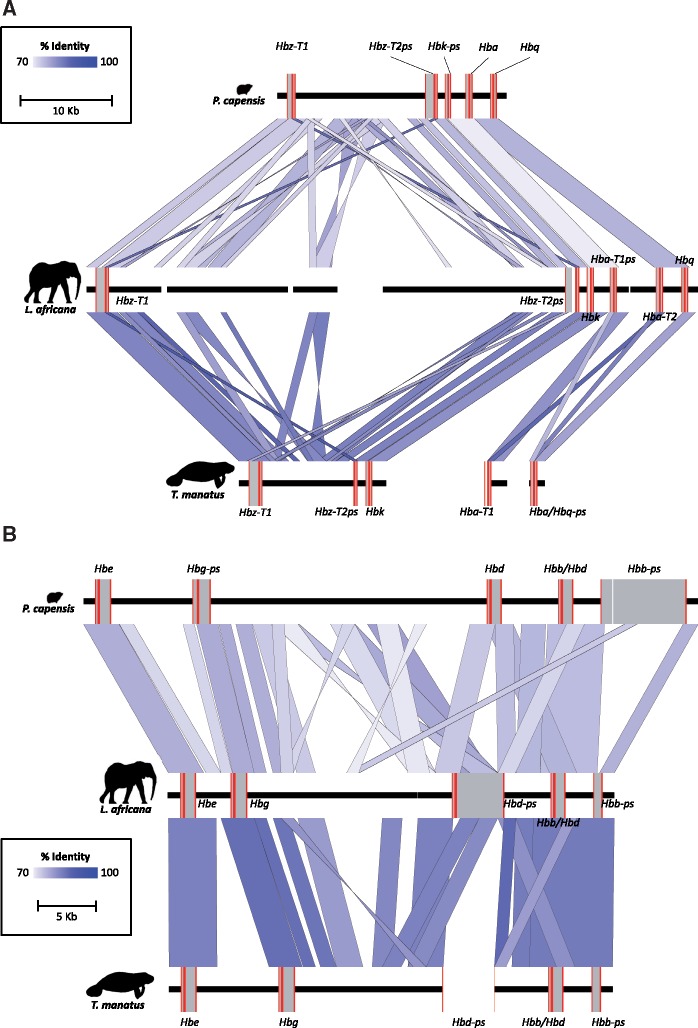
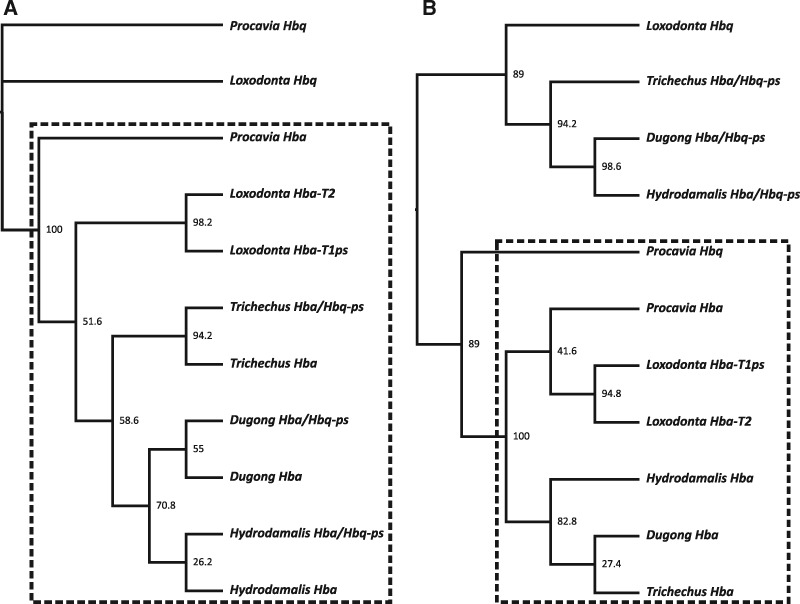
Orthology between sirenian, elephant, and hyrax Hb genes was identified by synteny analysis and pairwise sequence similarity (fig. 1) based on the draft genomes of the Florida manatee (Foote et al. 2015), African elephant (Loxodonta africana), and Cape rock hyrax (Procavia capensis). The arrangement of manatee α- and β-type Hb genes within their respective clusters resembles those of elephants (fig. 1), but with one important exception: sirenians do not possess Hba-T2 or Hbq genes, and instead have a pseudogenized chimeric fusion hybrid (Hba-T2/Hbq-ps) of the two genes (fig. 1A). Accordingly, the adult expressed α-type gene in sirenians (Hba-T1) is orthologous to the Hba-T1ps locus in elephants, rather than to Hba-T2, which is expressed in adult elephant blood (Campbell, Roberts, et al. 2010). The mammalian Hbq gene is derived from a tandem duplication of an ancestral Hba gene and is typically expressed in fetal erythroid cells, but does not appear to be translated into a functional globin protein (Hardison 2012). Maximum likelihood phylogenetic trees using representative paenungulate Hba, Hbq, and Hba-T2/Hbq-ps genes (fig. 2A and B) corroborate presence of a Hba-T2/Hbq-ps hybrid in that they illustrate that the 5′ regions (exon 1 and intron 1) of sirenian Hba-T2/Hbq-ps and Hba-T1 genes form a highly supported monophyletic clade (bootstrap value = 100) that is sister to Hbq, whereas the 3′ regions (intron 2 and exon 3) of all paenungulate Hba genes form a strongly supported monophyletic clade to the exclusion of sirenian Hba-T2/Hbq-ps and other paenungulate Hbq genes (i.e., the 5′ and 3′ ends of sirenian Hba-T2/Hbq-ps genes are more similar to Hba and Hbq genes, respectively).
Fig. 1.
(A) Organization and synteny analysis of rock hyrax (Procavia capensis), African elephant (Loxodonta africana), and Florida manatee (Trichechus manatus) α-globin gene clusters. Red and grey boxes represent exonic and intronic sequences, respectively. The “ps” suffix denotes pseudogenes, whereas “-T1” and “T2” suffices indicate the 5′–3′ linkage order of tandemly duplicated genes. Thick black lines represent intergenic sequences. Gaps in the contiguous sequence denote missing data. (B) Organization and synteny analysis of rock hyrax (P. capensis), African elephant (L. africana), and Florida manatee (T. manatus) β-globin gene clusters. Red and grey boxes represent exonic and intronic sequences, respectively. The “ps” suffix denotes pseudogenes. Thick black lines represent intergenic sequences. Gaps in the contiguous sequence denote missing data.
Fig. 2.
Maximum likelihood phylogenies depicting relationships between (A) the first exon and intron (bp 1–353) and (B) the second intron and third exon (bp 593–848) of paenungulate Hba- and Hbq-type genes. Values adjacent to each node represent maximum likelihood bootstrap support. The “ps” suffix denotes pseudogenes, whereas “-T1” and “-T2” suffices indicate the 5′–3′ linkage order of tandemly duplicated genes. Boxes highlight noteworthy gene groupings (see text for details).
Genomic deletions and duplications are a common occurrence in the evolutionary history of mammalian α- and β-type Hb clusters (Lam and Jefferys 2006; Hoffmann et al. 2008; Opazo et al. 2008). These changes in gene copy number often arise via unequal crossing over between mispaired paralogous genes on homologous chromosomes during meiosis or on sister chromatids in mitotic germ cells (Ohta 1980), and result in a gene duplication on one chromosome and a deletion on the other. However, if the crossover point interrupts the coding region of the mispaired genes, the resulting duplicate (and the surviving gene on the opposite chromosome) will be a hybrid of the two parent genes (Opazo et al. 2009). This mechanism is the underlying cause of hemoglobin Lepore syndrome in humans, where a Hbd/Hbb hybrid of the Hbb and Hbd genes supplants the parental Hbd and Hbb genes on one chromosome, and the corresponding “antiLepore” duplication (a Hbb/Hbd mutant) is inserted between them on the other (Forget 2001). The presence of functional antiLepore chimeric Hbb/Hbd genes in the genomes of paenungulates, eulipotyphlans, carnivorans, and cetartiodactyls (which arose independently in the ancestors of each lineage) further attests that the resulting fusion genes may become fixed in the genome and even supplant expression of the parental loci (Opazo et al. 2009; Gaudry et al. 2014). The hybrid Hba-T2/Hbq-ps locus found in the sirenian α-globin cluster is fundamentally similar to the (less commonly fixed) Hb Lepore deletion mutant, as the parental Hba-T2 and Hbq genes are no longer present. Examination of the Hba-T2/Hbq-ps nucleotide sequence reveals that the breakpoint of the unequal crossover event in the sirenian Hba-T2/Hbq-ps locus (between codons 76 and 87) is similar to the breakpoint of both the Hb Lepore Baltimore variant (between codons 66 and 81; Metzenberg et al. 1991) and the paenungulate antiLepore Hbb/Hbd locus (between codons 78 and 87; Opazo et al. 2009). Unlike these previously described chimeric recombinants, the sirenian Hba-T2/Hbq-ps locus does not produce a functional product, as a 34 bp deletion in the second exon removes codons 77–88 and introduces a frameshift mutation. It is likely that this 34 bp deletion is a result of the initial unequal crossover event that created the Hba-T2/Hbq-ps chimera (as it occurs at the chimeric breakpoint). Importantly, this recombination event appears to have contributed to the subsequent evolution of the intact upstream Hba-T1 gene—which encodes both pre and postnatal sirenian Hbs—via gene conversion.
Gene conversion events, whereby a gene copies itself onto a nearby paralogous gene, are quite common among Hb encoding loci (Nagylaki 1984; Papadakis and Patrinos 1999). This type of concerted evolution homogenizes sequence variation among tandemly duplicated genes, resulting in identical sequences in both gene fragments involved. We used GENECONV analyses (Sawyer 1989) to identify significant (P < 0.001) gene conversion tracts between the Hba-T1 and Hba-T2/Hbq-ps genes of the dugong, manatee, Steller’s sea cow, and the reconstructed dugongid and sirenian ancestors (table 1). If the inactive Hba-T2/Hbq-ps gene (whose evolution is unconstrained) converted the functional Hba-T1 gene in sirenians, dramatic functional changes to the expressed protein may result (see, e.g., Natarajan et al. 2015). To test this premise, we employed the Cluster History Analysis Package (Song et al. 2011) to determine the polarity of the conversion event between Hba-T1 and Hba-T2/Hbq-ps genes within the dugong, manatee, Steller’s sea cow, and the reconstructed dugongid and sirenian ancestors (table 2). These analyses identified conversion tracts congruent with those found by the GENECONV analysis (tables 1). The most recent conversion events between Hba-T1 and Hba-T2/Hbq-ps genes within the α-like Hb clusters of dugong, manatee, and Steller’s sea cow reveal that the adult expressed Hba-T1 gene is the donor and Hba-T2/Hbq-ps is the acceptor (table 2). Conversely, these analyses indicate that Hba-T2/Hbq-ps was the donor gene for the conversion events in the reconstructed dugongid and sirenian ancestors (table 2). These results are consistent with expectations, as the accumulation of mutations in the downstream pseudogene make it progressively less likely for converted segments to become fixed upon being transferred to the functional Hba-T1 gene (Chen et al. 2007).
Table 1.
Identification of Significant (P < 0.001) Gene Conversion Events from an Alignment of All Hba-like and Hbq-like Genes from the Dugong, Manatee, Steller’s Sea Cow, Reconstructed Sirenian Common Ancestor (Anc. Sirenian) and Reconstructed Dugongid Common Ancestor (Anc. Dugongid), Using GENECONV.
| Species | Converted Gene Pair | Tract Start | Tract End |
|---|---|---|---|
| T. manatus | Hba-T1 and Hba-T2/Hbq-ps | 1 | 462 |
| D. dugon | Hba-T1 and Hba-T2/Hbq-ps | 1 | 487 |
| H. gigas | Hba-T1 and Hba-T2/Hbq-ps | 1 | 405 |
| Anc. Sirenian | Hba-T1 and Hba-T2/Hbq-ps | 15 | 462 |
| Anc. Dugongid | Hba-T1 and Hba-T2/Hbq-ps | 5 | 462 |
Table 2.
Gene Conversion Tracts Identified by the Cluster History Analysis Package (CHAP2) among the Genomic Regions Containing Hba-like and Hbq-like Sequences from the Dugong, Manatee, Steller’s Sea Cow, Reconstructed Sirenian Common Ancestor (Anc. Sirenian), and Reconstructed Dugongid Ancestor (Anc. Dugongid).
| Species | Hba-T1Tract Start | Hba-T1Tract End | Conversion Polarity | Hba-T2/Hbq-psTract Start | Hba-T2/Hbq-psTract End |
|---|---|---|---|---|---|
| T. manatus | 12 | 414 | → | 12 | 414 |
| D. dugon | 12 | 472 | → | 12 | 438 |
| H. gigas | 14 | 348 | → | 14 | 351 |
| Anc. Dugongid | 12 | 519 | ← | 15 | 442 |
| Anc. Sirenian | 79 | 449 | ← | 79 | 403 |
The earliest known fossil sirenians (prorastomids, e.g., Prorastomus and Pezosiren) are from the middle Eocene (∼47 Ma) and were fully capable of terrestrial locomotion (Domning 2001). However, they exhibited a number of aquatic specializations that suggest they spent appreciable time foraging underwater, such as retracted nares to aid respiration while breaching water, a jaw modified for bottom feeding, and a slender body with prominent tail that allowed locomotion via dorsoventral spinal undulation combined with hindlimb thrusts (Domning 2001; Springer et al. 2015). These features are more pronounced in the next phase of sirenian evolution (protosirenids, e.g., Protosiren), which show further streamlining of the body, reduced limbs, and an enlarged tail that implies a mostly aquatic lifestyle, but likely capable of dragging itself short distances on land (Domning and Gingerich 1994). The eventual loss of hindlimb locomotion in primitive dugongs suggests that this aquatic transition was completed by the Bartonian (∼41 Ma; Domning 2000). Thus, the emergence of Hba-T2/Hbq-ps and subsequent conversion events between this locus and Hba-T1 in early sirenians may have played a key role in modifying their Hb–O2 affinity to accommodate an exclusive underwater foraging strategy.
Molecular Evolution of Paenungulate Globin Genes
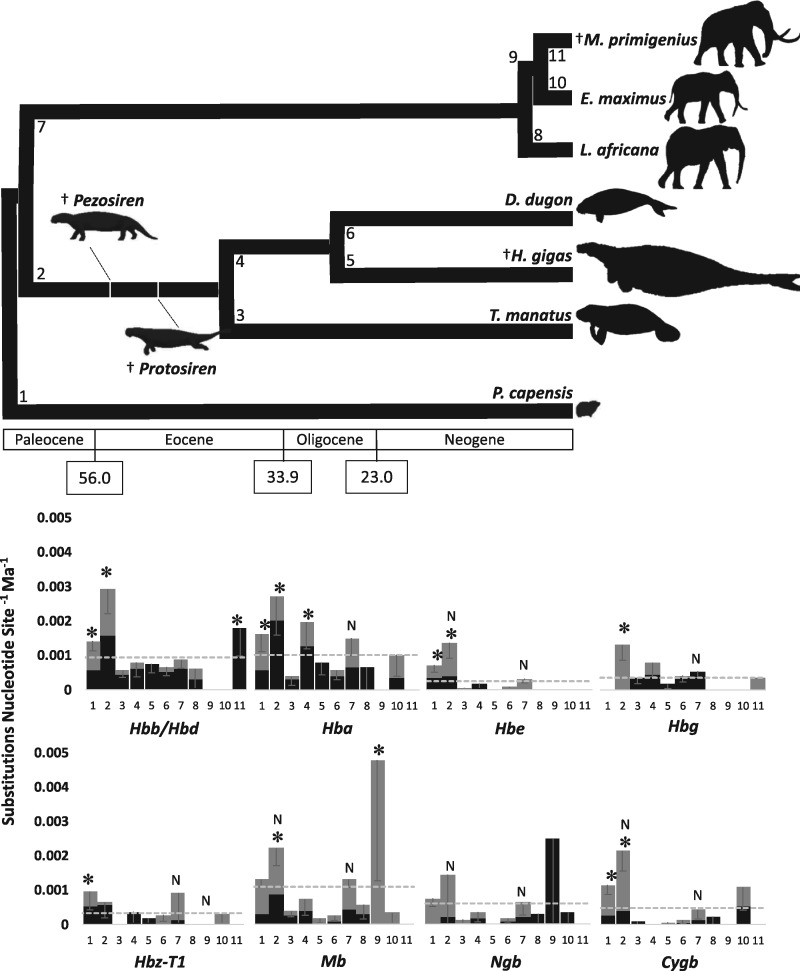
As a first step in testing this hypothesis, we sequenced the coding regions for five α-type Hb genes (Hbz-T1, Hbz-T2ps, Hbk, Hba-T1, and Hba-T2/Hbq-ps), four β-type Hb genes (Hbe, Hbg, Hbb/Hbd, and Hbb-ps), and three other globin genes (Mb, Ngb, and Cygb) encoding noncirculating monomeric proteins from two dugongs and three Steller’s sea cow specimens, together with partial coverage of Hbd-ps from both of these species (GenBank accessions: MK547529–MK547535, MK550610–MK550614, MK559326–MK559329, MK559338–MK559344, MK562072–MK562085). Complete coverage for all functional loci was obtained from both dugong individuals and a single Steller’s sea cow, though each Steller’s sea cow specific nucleotide replacement was corroborated by at least one additional specimen (see, e.g., supplementary table S1, Supplementary Material online for Hba-T1 and Hbb/Hbd). These data were combined with publicly available paenungulate globin sequences (supplementary tables S2–S10, Supplementary Material online) and the rate of molecular evolution (nucleotide substitutions site−1 Ma−1) for each nonpseudogenized gene was calculated. The average rate of nucleotide substitution across all branches in the paenungulate phylogeny (fig. 3) was appreciably higher for Mb and the adult-expressed Hb genes (Hba, Hbb/Hbd) than for Ngb, Cygb, and the prenatal Hb loci (Hbz-T1, Hbe, and Hbg; fig. 3). Evolutionary rate values for paenungulate Hba, Hbb/Hbd, and Mb (0.00101, 0.00093, and 0.00110 substitutions nucleotide site−1 Ma−1, respectively) are in close agreement to those previously estimated for adult Hb genes (0.001 substitutions nucleotide site−1 Ma−1; Efstratiadis et al. 1980). As the in utero environment likely does not vary appreciably between mammals, the evolution of prenatal Hb isoforms may be constrained by strong stabilizing selection (Koop and Goodman 1988). Thus, the genes encoding these polypeptide chains generally evolve at a lower rate with respect to adult Hbs, as is reflected in our rate estimates for Hbe, Hbg, and Hbz-T1 (0.00025, 0.00035, and 0.00033, respectively; fig. 3). However, the rate of molecular evolution was not constant across all branches at each of the eight globin loci tested, as tests of a constant molecular clock were rejected for all loci (P ≤ 0.01). For example, five of eight globin loci evolved significantly faster than average on the hyrax branch, which we attribute to the much shorter generation time and lifespans relative to sirenians and elephants. Similarly, six of eight globin genes (all but Hbz-T1 and Ngb) evolved significantly faster than average along the stem sirenian branch; that is, during their secondary aquatic transition.
Fig. 3.
Time-calibrated phylogeny modified from Springer et al. (2015) and Rohland et al. (2007) (top panel). Column plots representing the rate of nucleotide substitutions (substitutions per nucleotide site per million years ± SE) among the eight functional globin genes of paenungulate mammals (bottom panel). Black and grey regions of each column represent the contribution of nonsynonymous and synonymous substitutions to the overall substitution rate, respectively. Column numbers correspond to specific branches in the above time-calibrated phylogeny. Asterisks denote sirenian branches that have a rate of nucleotide substitution significantly higher than the locus average (denoted by grey dotted lines). Columns marked “N” denote negative selection.
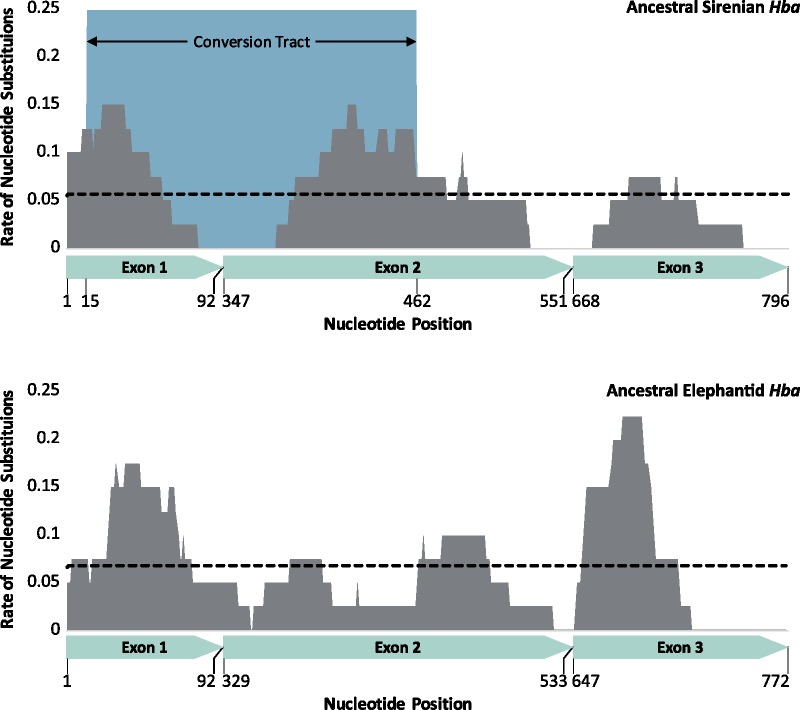
The evolutionary transition of mammals to the aquatic environment generally entails a suite of morphological and physiological specializations to improve O2 storage and management thereby allowing them to maximize underwater foraging (Ponganis 2011). However, the limited information available on sirenian diving physiology indicates that this lineage is not typical of mammalian divers. Indeed, cetaceans and pinnipeds greatly enhance O2 stores by increasing the concentration of Mb in muscle cells and Hb in red blood cells (Ponganis 2011), whereas these levels in manatees are more typical of terrestrial mammals (Blessing 1972). Pinnipeds and large cetaceans generally also have Hb–O2 affinities equal to or lower than terrestrial mammals (Snyder 1983), which is presumed to facilitate O2 transfer to their large Mb stores. While not well characterized, the O2 affinity of manatee and dugong blood appears to be elevated (McCabe et al. 1978; Farmer et al. 1979). An increased Hb–O2 affinity would likely permit more complete depletion of lung O2 stores during submergence (Snyder 1983; Meir and Ponganis 2009), and may be expected to have evolved during the prorastomid/protosirenian stages of sirenian evolution. This contention is consistent with the pronounced increases in evolutionary rate for the genes encoding the adult expressed Hb isoform (0.00291 and 0.00271 substitutions nucleotide site−1 Ma−1, respectively, for Hbb/Hbd and Hba-T1) on the stem sirenian branch. Of note, the abrupt rate increase in Hba-T1 appears to be mechanistically linked with the emergence of the Hba-T2/Hbq-ps chimeric gene that contributed to its rapid evolution via interparalog gene conversion (see above). Accordingly, the converted region of the reconstructed Hba-T1 gene of the ancestral sirenian contains a disproportionate number of nucleotide substitutions relative to the nonconverted 3′ region, as the conversion tract covers 48.4% of the ancestral sirenian Hba-T1 coding sequence, yet it contains 62.5% of nucleotide substitutions (fig. 4). Conversely, the same 5′ region of the ancestral elephant Hba-T2 gene contains only 48.2% (i.e., a roughly proportional number) of nucleotide substitutions (fig. 4). The increased nucleotide substitution rate in the converted region of the ancestral sirenian Hba-T1 gene has resulted in 12 nonsynonymous nucleotide substitutions, nine of which appear to be derived from the Hba-T2/Hbq-ps gene. Unfortunately, the epistatic nature of Hb function prevents drawing parallels between the individual and combined effects of these mutations on Hb–O2 affinity relative to homologous mutations that have been characterized in other Hbs (Storz 2018). Any functional changes resulting from the accumulation of nonsynonymous substitutions in the stem sirenian Hba-T1 branch may further have been expected to drive evolutionary change in other genes, as Hba-T1 pairs with Hbe, Hbg, and Hbb/Hbd to form tetrameric Hb isoforms (Hb Gower II, HbF, and HbA, respectively) from early embryonic through to adult life.
Fig. 4.
Rates of nucleotide substitution (substitutions per nucleotide site) for ancestral sirenian and ancestral elephant Hba genes. Gene wide substitution rate (total substitutions/423 nt) is represented by black dotted lines. Vertical grey bars represent the substitution rate within a 40 bp sliding window. The blue shaded area denotes the location of the gene conversion event (identified by GENECONV; table 1) that took place between Hba-T1 and Hba-T2/Hbq-ps genes in the sirenian ancestor and the green arrows denote exon boundaries.
Selection Analysis of Sirenian Globin Genes
To examine the possibility that Hba-T2/Hbq-ps may have influenced the evolution of Hba-T1 (and indirectly, the β-type globins it pairs with throughout development) on the stem sirenian branch we performed selection analyses on all globin loci at each branch of sirenian and elephant lineages. We found no instances of positive selection (ω > 1), as all globin genes of the stem sirenian exhibited ω values equal to one (neutral evolution) or less than one (purifying selection; fig. 3; supplementary table S11, Supplementary Material online). The ω values of globin genes in the stem elephant branch are generally similar to those on the stem sirenian branch, with the exception of Hba-T1 and Hbz-T1, which are elevated in the latter (fig. 3; supplementary table S11, Supplementary Material online). The close interactions between amino acid residues along and between globin chains of Hb make it vulnerable to epistatic and pleiotropic effects, which more often than not render substitutions—even those that seem beneficial in isolation—deleterious (Tufts et al. 2015). This is especially prominent for the prenatal globin genes (Hbe and Hbg), which are strongly conserved at the protein level (fig. 3). Conversely, the Hba-T1 and Hbz-T1 genes of the stem sirenian exhibited increased ω values with respect to the paralogous globins within this branch as well as those of the stem elephant (supplementary table S11, Supplementary Material online). These findings mirror those of deep diving cetaceans (i.e., those with maximum submergence times >40 min), where ω values for the Hbb gene had increased (but remained ≤ 1) relative to species with shorter maximum submergence times (<20 min; Tian et al. 2016). To test for intensified positive selection or relaxed purifying selection on the ancestral sirenian branch, relative to the ancestral elephant branch, we performed RELAX analysis on all globin loci (Wertheim et al. 2015). These tests are consistent with our codeml analyses (table 3), as they suggest selection pressures on both Hba-T1 and Hbz-T1 genes of stem sirenians were relaxed following their divergence from proboscideans (P = 0.024 and 0.006 for Hba-T1 and Hbz-T1, respectively). As Hba-T1 and Hbz-T1 encode the only functional α-type globin chains in sirenians, relaxed selection on these genes may have impacted both the O2 affinity of stem sirenian adult blood and O2 transfer to the embryo/fetus. However, the functional consequences of this evolutionary shift were probably greater for the Hba-T1 gene, as it exhibited 20 nonsynonymous nucleotide substitutions (with respect to the paenungulate ancestor) compared with only five in the Hbz-T1 gene. As noted earlier, this difference is likely attributed in part to the emergence of a chimeric Hba-T2/Hbq-ps pseudogene in stem sirenians, which our evidence suggests converted the expressed Hba-T1 gene repeatedly during the early stages of sirenian evolution. To identify potentially significant amino acid replacements imparted by the relaxed purifying selection on sirenian Hba-T1 and Hbz-T1 genes, we employed branch-site analyses similar to those which identified significant amino acid residues in cetacean Hba and Hbb genes (Nery et al. 2013). However, our tests identified no positively selected sites among sirenian globin genes at any point in their evolutionary history (supplementary table S12, Supplementary Material online).
Table 3.
Tests of Relaxed Purifying Selection on Ancestral Sirenian Globin Genes, Relative to the Ancestral Elephant, Using RELAX Analyses.
| Gene | P-value |
|---|---|
| Hbb/Hbd | 0.1260 |
| Hba | 0.0224* |
| Hbe | 0.0757 |
| Hbg | 0.8895 |
| Hbz-T1 | 0.0090* |
| Mb | 0.8845 |
| Ngb | 0.9120 |
| Cygb | 0.5143 |
Note.—P-values are the result of pairwise tests between ancestral sirenian and ancestral elephant globin genes, values marked with an asterisk represent a significant relaxation of purifying selection in ancestral sirenian globins (P < 0.05).
Oxygen Affinity of Sirenian Hbs
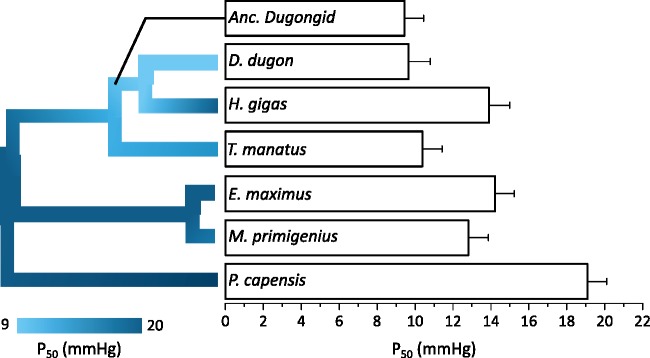
To confirm and extend previous work suggesting that sirenians possess a high O2-affinity Hb phenotype (McCabe et al. 1978; Farmer et al. 1979), we produced recombinant adult-type sirenian Hbs, measured their O2 binding properties at physiologically relevant effector concentrations and temperature (see methods), and compared them with those of native rock hyrax Hb and previously published values for the Asian elephant and woolly mammoth measured under identical test conditions (Campbell, Roberts, et al. 2010; supplementary table S13, Supplementary Material online). The P50 values (i.e., the PO2 at which half of Hb molecules are saturated with O2) for dugong and manatee Hb in the presence of physiologically relevant effector molecules at pH 7.2 and 37 °C (9.7 and 10.4 mmHg, respectively) were distinctly lower (corresponding to higher Hb–O2 affinities) than those of the Asian elephant, woolly mammoth, and hyrax (14.2, 12.8, and 19.1 mmHg, respectively), while the P50 of Steller’s sea cow Hb (13.9 mmHg) was comparable to those of elephantids (fig. 5). The observed difference in P50 is consistent with either a high Hb–O2 affinity phenotype having evolved independently in the dugong and manatee lineages or a high Hb–O2 affinity phenotype evolving in a common sirenian ancestor that was subsequently lowered in the Steller’s sea cow branch. We thus produced a recombinant Hb protein matching the last common ancestor of Steller’s sea cow and the dugong (branch 4 in fig. 3) and measured its Hb–O2 affinity (we were unable to unambiguously determine Hb sequences for the stem sirenian; differential inactivation of Hba-T1 [in proboscideans] and Hba-T2 [in sirenians] further precluded determination and hence production of the ancestral paenungulate Hb). These tests revealed that the ancestral dugongid Hb exhibited a P50 of 9.5 mmHg, supporting the hypothesis that a high Hb–O2 affinity phenotype had already evolved prior to the divergence of Trichechidae and Dugongidae. This evolutionary scenario is supported by a phenotypic reconstruction of Hb–O2 affinity using the phytools package in R (fig. 5 and supplementary fig. S1, Supplementary Material online), which further reconstructs a marked increase in Hb–O2 in stem sirenians following their divergence from proboscideans and hyraxes. Notably, whole blood O2 affinity of mammals scales with body mass, presumably to match O2 offloading with tissue requirements (Schmidt-Neilsen and Larimer 1958; Lahiri 1975). Thus, the P50 of the proboscidean/sirenian ancestor—which were presumably no larger than the earliest 6–17 kg paenungulates, Eritherium azzourzorum and Phosphatherium escuilliei (Larramendi 2015)—was likely even higher than that reconstructed by the phytools package (i.e., similar to that of the rock hyrax). As the lungs of manatees constitute a larger proportion of body oxygen stores (∼33%) than most other diving mammals, it has accordingly been suggested that their high blood–O2 affinity (P50 = 16.5 mmHg; White et al. 1976) maximizes the amount of O2 extracted from lung stores while submerged (Gallivan et al. 1986). However, manatee blood–O2 affinity is well above that of both closely related elephants (P50 = 23.2–25.2 mmHg; Dhindsa et al. 1972) and other diving mammals (P50 = 26–31 mmHg; Snyder 1983), though comparable to sloths and pangolins (P50 = ∼19–21 mmHg; Lahiri 1975; Weber et al. 1986) that, like manatees (Scholander and Irving 1941), have low mass-specific rates of metabolism. While an elevated Hb–O2 affinity may have initially evolved to maximize O2 extraction from finite lung stores during prolonged submergence in stem sirenians, the relatively low blood and muscle O2 stores of extant sirenians (Gallivan et al. 1986) suggests its primarily role may be to help foster the low metabolic intensities characteristic of these species (Scholander and Irving 1941, Blessing 1972).
Fig. 5.
Oxygen tensions at half saturation (P50) for representative paenungulate hemoglobins (0.0625 mM Hb4) in the presence of both 0.1 M KCl and 0.125 mM 2, 3-diphosphoglycerate at 37 °C, pH 7.2. Values are shown as P50 ± SE. The phylogenetic tree is colored according to P50 value. The P50 at branches without in vitro measurements were estimated using the phytools package in R.
A second notable finding of this analysis is the sharp reduction in Hb–O2 affinity in the ancestors of Steller’s sea cows. Fortunately, the naturalist Georg Wilhelm Steller recorded detailed observations of their foraging behavior in 1741 (Steller 1751), that allows us to interpret this shift within an ecological context. Steller noted that “these animals are fond of shallow sandy places along the seashore,” do not fully submerge their bodies while feeding, and simply “raise their noses above the water, as they do every four or five minutes, [to] blow out the air.” While it is conceivable that the reduction in O2 affinity of Steller’s sea cow Hb may have been the by-product of selection on another aspect of Hb function or simply arisen via genetic drift, this phenotype is consistent with expectations in that it would have promoted O2 offloading necessary to fuel higher thermoregulatory costs associated with its frigid (sub)Arctic marine environment. High thermoregulatory costs are in line with Steller’s observations as “These animals are very voracious and eat incessantly, and because they are so greedy they keep their heads always under water, without regard to life and safety” (Steller 1751). Primary sequence comparisons further suggest that this secondary decrease in Hb–O2 affinity largely arose from a single rare charge-altering residue replacement (β/δ82Lys→Asn; supplementary table S1, Supplementary Material online) within the central cavity of the Steller’s sea cow Hb tetramer. In human HbA, the strongly cationic β82Lys residue is known to be a key effector binding site that is able to form five concurrent salt bridges with anionic charges on DPG and also constitutes a Cl– binding site in the T-state protein (Richard et al. 1993). In contrast, the human Hb Providence mutant (β82Lys→Asn) deletes this key binding site for both Hb–O2 affinity modulators, and predictably displays a reduced Cl– effect, a strongly impaired DPG effect, and a reduction in Hb–O2 affinity (Bonaventura et al. 1976) similar in magnitude to that found for Steller’s sea cow Hb relative to dugongs. The presence of this substitution within a (sub)Arctic aquatic mammal is surprising however, as reductions in effector binding are also expected to increase the thermal sensitivity of the protein and hinder O2 delivery to poorly insulated appendages (Weber and Campbell 2011). Interestingly, the protein encoded by the Hbb/Hbd gene (β/δ) of the dugong also has evolved a rare amino acid replacement (β/δ101Glu→Gln; supplementary table S12, Supplementary Material online) adjacent to the highly conserved sliding interface of the Hb tetramer, that is only known to occur in two lemur species and the woolly mammoth(Campbell, Roberts, et al. 2010). The mammoth replacement has been studied extensively (Campbell, Roberts, et al. 2010; Yuan et al. 2011; Noguchi et al. 2012; Yuan et al. 2013) and has been speculated to confer a selective advantage for this extinct Arctic species by lowering the effect of temperature on its O2 binding properties. It is therefore surprising to find this mutation in the dugong, as there is presumably little selective advantage for a reduced thermal sensitivity Hb phenotype in a subtropical species. In light of the unusual substitutions in the Hbb/Hbd genes of dugong and Steller’s sea cow, detailed analyses of red blood cell effector and temperature effects on sirenian Hbs are warranted in order to further assess the physiological ramifications of these residue exchanges.
Conclusions
Our analyses revealed that the genes encoding the O2-binding globin proteins of sirenians generally displayed high nucleotide substitution rates during the early aquatic transition of this lineage. While most of these loci evolved under neutrality or strong purifying selection, the Hba-T1 and Hbz-T1 genes—which encode Hb isoforms at all sirenian life stages—exhibited elevated rates of nonsynonymous/synonymous amino acid replacements on the stem sirenian branch. Notably, the rate change in Hba-T1 is temporally linked to the emergence of a chimeric Hba-T2/Hbq-ps pseudogene in sirenians, which contributed to the upstream Hba-T1 locus via gene conversion. Intriguingly, this molecular remodeling of the ancestral sirenian Hba-T1 locus coincided temporally with a strong increase in Hb–O2 affinity, which likely aided the sirenian’s secondary aquatic transition by maximizing O2 extraction from finite lung stores during prolonged dives while promoting a relatively low rate of metabolism. In contrast, the cold-adapted Steller’s sea cow lineage evolved amino acid substitutions that would have markedly lowered its blood O2 affinity. This derived phenotype provides just the second example of a physiological specialization in an extinct species in that it is expected to have fostered the elevated metabolic/thermal needs of this once colossal (sub)Arctic species.
Materials and Methods
Genome Mining
The complete α- and β-globin clusters, Mb, Ngb, and Cygb were mined from the draft genomes of the Florida manatee, African elephant, woolly mammoth, and Cape rock hyrax. Additionally, all available globin gene sequences for these species and the Asian elephant (Elephas maximus) were mined from the National Center for Biotechnology Information’s (NCBI) Sequence Read Archive and Nucleotide Collection (supplementary table S2, Supplementary Material online).
Globin Sequencing
We obtained complete coding sequences of Mb, Ngb, Cygb, and the full complement of α- and β-globin genes from two dugongs and three ∼1,000 year old Steller’s sea cow specimens using microarray hybridization enrichment coupled with Illumina sequencing. Details outlining the construction of DNA libraries, target enrichment, Illumina sequencing, and sequence assembly are described elsewhere (Springer et al. 2015). Briefly, barcoded DNA libraries suitable for Illumina sequencing were prepared from dugong and Steller’s sea cow extracts, and subsequently enriched for globin sequences using Agilent SureSelect Capture arrays primarily designed based on African elephant sequence information. The resulting enriched library was sequenced on an Illumina GAIIx genome analyzer using a 55-bp single-end sequencing protocol. Raw Illumina reads were then trimmed of adapter sequences and those <20 bp were removed from the data set. These reads were mapped to reference sequences (based on previously published dugong, manatee or African elephant globin genes; supplementary table S2, Supplementary Material online) using Geneious R6.1 software (Biomatters Ltd, Auckland, New Zealand). The coding sequences of the various globin genes were deduced from these assemblies and used in subsequent analyses. DNA damage artifacts (C→T and G→A) in the Steller’s sea cow assemblies were identified and corrected following Springer et al. (2015).
Identification of Gene Conversion
Gene conversion events between sirenian Hba-T1 and Hba-T2/Hbq-ps genes were initially identified by phylogenetic analysis, similar to previous works on chimeric globin genes (Opazo et al. 2009; Gaudry et al. 2014). Paenungulate Hba and Hbq genes were aligned using MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/; last accessed March 11, 2019. 848 bp total length). In order to test for independent evolutionary origins of the 5′ and 3′ ends of the Hba-T2/Hbq-ps chimeric gene, the alignment was divided into two sections: 1) start codon to end of the first intron (bp 1–353) and 2) beginning of the second intron to the stop codon (bp 593–848). Separate maximum likelihood trees were constructed from each of these partial alignments using PhyML under the TN93 + G model of nucleotide substitution (determined by FindModel as the best fit for both alignments). Trees were constructed using ten replicate heuristic searches with random number seeds and refined using both nearest neighbor interchange and subtree pruning and regrafting algorithms. The best tree produced by the two competing algorithms was subjected to branch support analysis by performing 500 bootstrap replicates.
To test for the presence of a gene conversion event in the stem sirenian, paenungulate Hba, and Hba-T2/Hbq-ps genes were aligned separately using MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/) and the ancestral sirenian genes were reconstructed using baseml, as implemented in PAML 4.7 (see Substitution Rates for parameters). Reconstructed sirenian Hba-T1 and Hba-T2/Hbq-ps genes were aligned together with modern paenungulate Hba, Hbq, and Hba-T2/Hbq-ps genes and pairwise tests for gene conversion events were performed using GENECONV (Sawyer 1989). Analyses were run with default parameters, except that mismatch penalties (gscale) were set to 3. To test the directionality of sirenian gene conversion events, genomic regions containing Hba- and Hbq-type genes for L. africana, P. capensis, T. manatus, D. dugon, H. gigas, the reconstructed dugongid ancestor, and the reconstructed sirenian ancestor were input into the Cluster History Analysis Package (CHAP2; Song et al. 2011). Sequence orthology and gene conversion events were detected using the phylogenetic tree in figure 3 (with E. maximus and Mammuthus primigenius pruned out) and the T. manatus genome sequence as a reference.
Substitution Rates
The protein coding regions of each globin gene were aligned separately using MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/). Using gene specific models of nucleotide substitution (as determined by FindModel, http://www.hiv.lanl.gov/) and a user generated timetree adapted from previous studies (Rohland et al. 2007; Springer et al. 2015), the number of nucleotide substitutions per nucleotide site was calculated for each alignment using baseml, as implemented in the software package PAML 4.7 (Yang 2007). Substitution rates were calculated both with and without the assumption of a constant molecular clock. Likelihood ratio tests (LRT) were then used to determine if a constant molecular clock best fit the data set (Yang 2007). The above baseml analyses for Hbb/Hbd, Hba and Hba-T1/Hbq-ps were run with RateAncestor = 1 to reconstruct the sequences of the ancestral sirenian, ancestral dugongid, and ancestral elephant braches. The relative contribution of nonsynonymous and synonymous substitutions to the overall substitution rate was calculated from the above data using codeml (see Selection Analyses), as implemented in the software package PAML 4.7 (Yang 2007).
Selection Analyses
The modes of natural selection acting on sirenian globin genes were determined using codeml, as implemented in the PAML 4.7 software package (Yang 2007). The same nucleotide alignments and phylogenetic tree as above (see Substitution Rates) were tested under the M2 model using the F3x4 codon frequency model (Yang 2007). Lineages were independently designated as foreground branches and their ω value was estimated for each globin gene. These tests were then repeated with the ω parameter fixed to 1.0, rather than being estimated. LRT were then used to determine if the estimated ω value for each branch was significantly different from 1.0. Benjamini–Hochberg multiple testing correction was applied to the LRTs where ω values were considered significantly different from 1.0 if P-values were lower than the Benjamini–Hochberg threshold (Benjamini and Hochberg 1995). To detect instances of intensified positive selection or relaxed purifying selection in the globin genes of the stem sirenian, with respect to the stem elephantid, RELAX (Wertheim et al. 2015) analysis was used, as implemented on the Adaptive Evolution Server (Weaver et al. 2018). For each globin gene, the stem sirenian branch was designated the “test branch” and the stem elephant branch was used as a reference.
Branch-site analyses were performed using the MA model to test for individual amino acids evolving under positive selection (Yang 2007). These tests were then repeated with the ω parameter fixed to 1.0, rather than being estimated. LRT were then used to determine if each branch had significant sites under positive selection (P < 0.05). All tests were run with the parameter “method = 0” which calculates branch length simultaneously, rather than individually.
Construction of Recombinant Hb Expression Vectors
Sirenian Hba-T1 and Hbb/Hbd genes were optimized for expression in Escherichia coli and synthesized by GenScript (Piscataway, NJ). These constructs were digested with restriction enzymes and tandemly ligated into a custom Hb expression vector (Natarajan et al. 2011) using a New England BioLabs Quick Ligation Kit as recommended by the manufacturer. Chemically competent JM109 (DE3) E. coli (Promega) were prepared using a Z-Competent E. coli Transformation Kit and Buffer Set (Zymo Research). Hb expression vectors were cotransformed into JM109 (DE3) chemically competent E. coli alongside a plasmid expressing methionine aminopeptidase (Natarajan et al. 2011), plated on LB agar containing ampicillin (50 µg/ml) and kanamycin (50 µg/ml), and incubated for 16 h at 37 °C. A single colony from each transformation was cultured in 50 ml of 2xYT broth for 16 h at 37 °C while shaking at 200 rpm. Postincubation, 5 ml of the culture was pelleted by centrifugation and plasmid DNA was isolated using a GeneJET Plasmid Miniprep Kit (Thermo Scientific). The plasmid sequence was verified using BigDye 3.1 sequencing chemistry and an ABI3130 Genetic Analyzer. The remainder of the culture was supplemented with glycerol to a final concentration of 10%, divided into 25 ml aliquots and stored at –80 °C until needed for expression.
Expression and Purification of Recombinant Hb
Twenty five milliliter of starter culture (above) was added to 1,250 ml of TB media containing ampicillin (100 µg/µl) and kanamycin (50 µg/µl) and distributed evenly amongst five 1 l Erlenmeyer flasks. Cultures were grown at 37 °C while shaking at 200 rpm until the absorbance at 600 nm reached 0.6–0.8. Hb expression was induced by supplementing the media with 0.2 mM isopropyl β-d-1-thiogalactopyranoside, 50 µg/ml of hemin and 20 g/l of glucose and the culture was incubated at 28 °C for 16 h while shaking at 200 rpm. Once expression had completed, dissolved O2 was removed by adding sodium dithionite (1 mg/ml) to the culture, which was promptly saturated with CO for 15 min. Bacterial cells were then pelleted by centrifugation and the Hb was purified by ion exchange chromatography according to Natarajan et al. (2011).
Purification of Native Rock Hyrax Hb
A whole blood sample (∼1 ml) was obtained from a (male) rock hyrax (P. capensis abyssinica) housed at the Assiniboine Park Zoo (Winnipeg, Canada) during routine veterinary testing, and stored at –80 °C. Thawed blood aliquots (∼200 μl) were added to a 5× volume of ice cold water and incubated on ice for 30 min to lyse the red blood cells. Samples were centrifuged at 20,000 × g for 10 min to remove cell debris. Buffer was added to the supernatants to a final concentration of 0.01 M HEPES/0.2 M NaCl (pH 7.4) and passed through a PD-10 desalting column (GE Healthcare) equilibrated with 20 mM HEPES/0.5 mM EDTA (pH 6.8). Samples were then loaded onto a HiTrap SP HP cation exchange column (GE Healthcare) and eluted with a linear pH gradient (pH 6.8–8.4). Eluted Hb factions were concentrated using Amicon Ultra-4 Centrifugal Filter Units (Millipore).
Functional Analyses of Hbs
O2-equillibrium curves for Hb solutions (62.5 μM Hb, 0.1 M HEPES buffer, 0.1 M KCl, 0.125 mM 2, 3-diphosphoglycerate) were measured at 37 °C using the thin film modified diffusion chamber technique described by Weber (1992). Each Hb solution was sequentially equilibrated with three to five different gas mixtures that cause Hb–O2 saturations between 30% and 70%. Hill plots (log[fractional saturation/[1 – fractional saturation]] vs. logPO2) constructed from these measurements were used to determine the PO2 and co-operativity coefficient at half saturation (P50 and n50, respectively) from the zero-intercept and slope of these plots, respectively. P50 values were measured at three different pH levels (6.9, 7.4, and 7.9). A linear regression was fit to plots of logP50 versus pH wherein the slope of this plot represents the Bohr effect and the resulting equation was used to correct P50 values to pH 7.2 (± SE of the regression estimate). These P50 values were then used to reconstruct those of all ancestral paenungulates using the phytools package in R (Revell 2012).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank M. Gaudry and E. Ellebæk Petersen for technical assistance, G. Glover for the hyrax blood sample, and J. Storz for constructive comments. This study was supported by National Sciences and Engineering Research Council (NSERC) of Canada Discovery (RGPIN/238838-2011) and Accelerator Supplement Grants (RGPIN/412336-2011) to K.L.C., an NSERC Postgraduate Scholarship to A.V.S., an NSF grant (EF0629860) to M.S.S., a National Science Foundation DEB award (1132229) to M.H., an Aarhus University, Faculty of Science and Technology Grant to R.E.W., and the Danish Council for Independent Research Grant (DFF-4181-00094) to A.F.
References
- Benjamini Y, Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 57:289–300. [Google Scholar]
- Blessing MH. 1972. Studies on the concentration of myoglobin in the sea-cow and porpoise. Comp Biochem Physiol A. 413:475–480. [DOI] [PubMed] [Google Scholar]
- Bonaventura J, Bonaventura C, Sullivan B, Ferruzzi G, McCurdy PR, Fox J, Moo-Penn WF.. 1976. Hemoglobin providence. Functional consequences of two alterations of the 2, 3-diphosphoglycerate binding site at position beta 82. J Biol Chem. 25123:7563–7571. [PubMed] [Google Scholar]
- Burmester T, Haberkamp M, Mitz S, Roesner A, Schmidt M, Ebner B, Gerlach F, Fuchs C, Hankeln T.. 2004. Neuroglobin and cytoglobin: genes, proteins and evolution. IUBMB Life 56(11–12):703–707. [DOI] [PubMed] [Google Scholar]
- Burmester T, Hankeln T.. 2014. Function and evolution of vertebrate globins. Acta Physiol. 2113:501–514. [DOI] [PubMed] [Google Scholar]
- Campbell KL, Roberts JEE, Watson LN, Stetefeld J, Sloan AM, Signore AV, Howatt JW, Tame JRH, Rohland N, Shen T-J, et al. 2010. Substitutions in woolly mammoth hemoglobin confer biochemical properties adaptive for cold tolerance. Nat Genet. 426:536–540. [DOI] [PubMed] [Google Scholar]
- Campbell KL, Storz JF, Signore AV, Moriyama H, Catania KC, Payson AP, Bonaventura J, Stetefeld J, Weber RE.. 2010. Molecular basis of a novel adaptation to hypoxic–hypercapnia in a strictly fossorial mole. BMC Evol Biol. 101:214–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-M, Cooper DN, Chuzhanova N, Férec C, Patrinos GP.. 2007. Gene conversion: mechanisms, evolution and human disease. Nat Rev Genet. 810:762–775. [DOI] [PubMed] [Google Scholar]
- Crerar LD, Crerar AP, Domning DP, Parsons E.. 2014. Rewriting the history of an extinction—was a population of Steller's sea cows (Hydrodamalis gigas) at St Lawrence Island also driven to extinction? Biol Lett. 1011:20140878–20140878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhindsa DS, Sedgwick CJ, Metcalfe J.. 1972. Comparative studies of the respiratory functions of mammalian blood. VIII. Asian elephant (Elephas maximus) and African elephant (Loxodonta africana africana). Resp Physiol. 143:332–342. [DOI] [PubMed] [Google Scholar]
- Domning DP, Gingerich PD.. 1994. Protosiren smithae, new species (Mammalia, Sirenia), from the late middle Eocene of Wadi Hitan, Egypt. Vol. 29 Ann Arbor, Michigan: Contrib. Mus. Paleontol. Univ. Mich; p. 69–87. [Google Scholar]
- Domning DP. 1976. An ecological model for late tertiary sirenian evolution in the North Pacific Ocean. Syst Biol. 25:352–362. [Google Scholar]
- Domning DP. 2000. The readaptation of Eocene sirenians to life in water. Hist Biol. 14(1–2):115–119. [Google Scholar]
- Domning DP. 2001. The earliest known fully quadrupedal sirenian. Nature 4136856:625–627. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A, Posakony JW, Maniatis T, Lawn RM, O'Connell C, Spritz RA, DeRiel JK, Forget BG, Weissman SM, Slightom JL, et al. 1980. The structure and evolution of the human β-globin gene family. Cell 213:653–668. [DOI] [PubMed] [Google Scholar]
- Farmer M, Weber RE, Bonaventura J, Best RC, Domning D.. 1979. Functional properties of hemoglobin and whole blood in an aquatic mammal, the Amazonian manatee (Trichechus inunguis). Comp Biochem Physiol A. 621:231–238. [Google Scholar]
- Foote AD, Liu Y, Thomas GW, Vinař T, Alföldi J, Deng J, Dugan S, van Elk CE, Hunter ME, Joshi V, et al. 2015. Convergent evolution of the genomes of marine mammals. Nat Genet. 473:272–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forget BG. 2001. Molecular mechanisms of β thalassemia In: Steinberg M, Forget B, Higgs D, Nagel R, editors. Disorders of hemoglobin: genetics pathophysiology, and clinical management. Cambridge: Cambridge University Press; p. 252–276. [Google Scholar]
- Gallivan GJ, Kanwisher JW, Best RC.. 1986. Heart rates and gas exchange in the Amazonian manatee (Trichechus inunguis) in relation to diving. J Comp Physiol B. 1563:415–423. [DOI] [PubMed] [Google Scholar]
- Gaudry MJ, Storz JF, Butts GT, Campbell KL, Hoffmann FG.. 2014. Repeated evolution of chimeric fusion genes in the β-globin gene family of laurasiatherian mammals. Genome Biol Evol. 65:1219–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison R. 1999. The evolution of hemoglobin studies of a very ancient protein suggest that changes in gene regulation are an important part of the evolutionary story. Am Sci. 872:126. [Google Scholar]
- Hardison RC. 2012. Evolution of hemoglobin and its genes. Cold Spring Harb Perspect Med. 212:a011627.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann FG, Opazo JC, Storz JF.. 2008. Rapid rates of lineage-specific gene duplication and deletion in the alpha-globin gene family. Mol Biol Evol. 253:591–602. [DOI] [PubMed] [Google Scholar]
- Koop BF, Goodman M.. 1988. Evolutionary and developmental aspects of two hemoglobin beta-chain genes (epsilon M and beta M) of opossum. Proc Natl Acad Sci U S A. 8511:3893–3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam KW, Jeffreys AJ.. 2006. Processes of copy-number change in human DNA: the dynamics of α-globin gene deletion. Proc Nat Acad Sci U S A. 10324:8921–8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri S. 1975. Blood oxygen affinity and alveolar ventilation in relation in body weight in mammals. Am J Physiol. 2292:529–536. [DOI] [PubMed] [Google Scholar]
- Larramendi A. 2015. Proboscideans: shoulder height, body mass and shape Acta Palaeontol. Pol. 61:537–574. [Google Scholar]
- Marsh H, O'Shea TJ, Reynolds JE III. 2011. Ecology and conservation of the Sirenia: dugongs and manatees. Cambridge: Cambridge University Press; p. 1–521. [Google Scholar]
- McCabe M, Hamilton R, Marsh H.. 1978. Some studies on the oxygen affinity of haemoglobin from the dugong. Comp Biochem Physiol A. 611:19–22. [Google Scholar]
- McGowen MR, Gatesy J, Wildman DE.. 2014. Molecular evolution tracks macroevolutionary transitions in Cetacea. Trend Ecol Evol. 296:336–346. [DOI] [PubMed] [Google Scholar]
- Meir JU, Ponganis PJ.. 2009. High-affinity hemoglobin and blood oxygen saturation in diving emperor penguins. J Exp Biol. 212(Pt 20):3330–3338. [DOI] [PubMed] [Google Scholar]
- Metzenberg AB, Wurzer G, Huisman TH, Smithies O.. 1991. Homology requirements for unequal crossing over in humans. Genetics 1281:143–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirceta S, Signore AV, Burns JM, Cossins AR, Campbell KL, Berenbrink M.. 2013. Evolution of mammalian diving capacity traced by myoglobin net surface charge. Science 3406138:1234192.. [DOI] [PubMed] [Google Scholar]
- Nagylaki T. 1984. The evolution of multigene families under intrachromosomal gene conversion. Genetics 1063:529–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan C, Hoffmann FG, Lanier HC, Wolf CJ, Cheviron ZA, Spangler ML, Weber RE, Fago A, Storz JF.. 2015. Intraspecific polymorphism, interspecific divergence, and the origins of function-altering mutations in deer mouse hemoglobin. Mol Biol Evol. 324:978–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan C, Jiang X, Fago A, Weber RE, Moriyama H, Storz JF.. 2011. Expression and purification of recombinant hemoglobin in Escherichia coli. PLoS One 65:e20176.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nery MF, Arroyo JI, Opazo JC.. 2013. Accelerated evolutionary rate of the myoglobin gene in long-diving whales. J Mol Evol. 766:380–387. [DOI] [PubMed] [Google Scholar]
- Noguchi H, Campbell KL, Ho C, Unzai S, Park S-Y, Tame J.. 2012. Structures of haemoglobin from woolly mammoth in liganded and unliganded states. Acta Crystallogr D Biol Crystallogr. 68(Pt 11):1441–1449. [DOI] [PubMed] [Google Scholar]
- Ohta T. 1980. Evolution and variation of multigene families. Genetics 106:529–548. [Google Scholar]
- Opazo JC, Hoffmann FG, Storz JF.. 2008. Differential loss of embryonic globin genes during the radiation of placental mammals. Proc Natl Acad Sci U S A. 10535:12950–12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo JC, Sloan AM, Campbell KL, Storz JF.. 2009. Origin and ascendancy of a chimeric fusion gene: the beta/delta-globin gene of paenungulate mammals. Mol Biol Evol. 267:1469–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadakis MN, Patrinos GP.. 1999. Contribution of gene conversion in the evolution of the human β-like globin gene family. Hum Genet. 1042:117–125. [DOI] [PubMed] [Google Scholar]
- Ponganis PJ. 2011. Diving mammals. Compr Physiol. 11:447–465. [DOI] [PubMed] [Google Scholar]
- Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Met Ecol Evol. 32:217–223. [Google Scholar]
- Richard V, Dodson GG, Mauguen Y.. 1993. Human deoxyhaemoglobin-2, 3-diphosphoglycerate complex low-salt structure at 2·5 Å resolution. J Mol Biol. 2332:270–274. [DOI] [PubMed] [Google Scholar]
- Rohland N, Malaspinas AS, Pollack JL, Slatkin M, Matheus P, Hofreiter M.. 2007. Proboscidean mitogenomics: chronology and mode of elephant evolution using mastodon as outgroup. PLoS Biol. 58:e207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel PP, Smith LP, Phillips GN Jr, Olson JS.. 2015. Apoglobin stability is the major factor governing both cell-free and in vivo expression of holomyoglobin. J Biol Chem. 29039:23479–23495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer S. 1989. Statistical tests for detecting gene conversion. Mol Biol Evol. 65:526–538. [DOI] [PubMed] [Google Scholar]
- Schmidt-Neilsen K, Larimer JL.. 1958. Oxygen dissociation curves of mammalian blood in relation to body size. Am J Physiol. 1952:424–428. [DOI] [PubMed] [Google Scholar]
- Scholander PF, Irving L.. 1941. Experimental investigations on the respiration and diving of the Florida manatee. J Cell Comp Physiol. 172:169–191. [Google Scholar]
- Sharon D, Glusman G, Pilpel Y, Khen M, Gruetzner F, Haaf T, Lancet D.. 1999. Primate evolution of an olfactory receptor cluster: diversification by gene conversion and recent emergence of pseudogenes. Genomics 611:24–36. [DOI] [PubMed] [Google Scholar]
- Snyder GK. 1983. Respiratory adaptations in diving mammals. Respir Physiol. 543:269–294. [DOI] [PubMed] [Google Scholar]
- Song G, Hsu C-H, Riemer C, Zhang Y, Kim HL, Hoffmann F, Zhang L, Hardison RC, Green ED, Miller W.. 2011. Conversion events in gene clusters. BMC Evol Biol. 11:226.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer MS, Signore AV, Paijmans JLA, Velez-Juarbe J, Domning DP, Bauer CE, He K, Crerar L, Campos PF, Murphy WJ, et al. 2015. Interordinal gene capture, the phylogenetic position of Steller’s sea cow based on molecular and morphological data, and the macroevolutionary history of Sirenia. Mol Phylogenet Evol. 91:178–193. [DOI] [PubMed] [Google Scholar]
- Stejneger L. 1887. How the great northern sea-cow (Rytina) became exterminated. Am Nat. 2112:1047–1054. [Google Scholar]
- Steller GW. 1751. De bestiis marinis [The beasts of the sea]. Novi Commentarii Acad Sci Imp Petropoli. 2:289–398. [Google Scholar]
- Storz JF, Baze M, Waite JL, Hoffmann FG, Opazo JC, Hayes JP.. 2007. Complex signatures of selection and gene conversion in the duplicated globin genes of house mice. Genetics 1771:481–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Moriyama H.. 2008. Mechanisms of hemoglobin adaptation to high altitude hypoxia. H Alt Med Biol. 92:148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF. 2016. Causes of molecular convergence and parallelism in protein evolution. Nat Rev Genet. 174:239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF. 2018. Compensatory mutations and epistasis for protein function. Curr Opin Struct Biol. 50:18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian R, Wang Z, Niu X, Zhou K, Xu S, Yang G.. 2016. Evolutionary genetics of hypoxia tolerance in cetaceans during diving. Gen Biol Evol. 8:827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufts DM, Natarajan C, Revsbech IG, Projecto-Garcia J, Hoffmann FG, Weber RE, Fago A, Moriyama H, Storz JF.. 2015. Epistasis constrains mutational pathways of hemoglobin adaptation in high-altitude pikas. Mol Biol Evol. 322:287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver S, Shank SD, Spielman SJ, Li M, Muse SV, Kosakovsky Pond SL.. 2018. Datamonkey 2.0: a modern web application for characterizing selective and other evolutionary processes. Mol Biol Evol. 353:773–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber RE, Campbell KL.. 2011. Temperature dependence of haemoglobin-oxygen affinity in heterothermic vertebrates: mechanisms and biological significance. Acta Physiol. 2023:549–562. [DOI] [PubMed] [Google Scholar]
- Weber RE, Heath ME, White FN.. 1986. Oxygen binding functions of blood and hemoglobin from the Chinese pangolin, Manis pentadactyla: possible implications of burrowing and low body temperature. Resp Physiol. 641:103–112. [DOI] [PubMed] [Google Scholar]
- Weber RE. 1992. Use of ionic and zwitterionic (Tris/BisTris and HEPES) buffers in studies on hemoglobin function. J Appl Physiol. 724:1611–1615. [DOI] [PubMed] [Google Scholar]
- Weber RE. 2007. High-altitude adaptations in vertebrate hemoglobins. Resp Physiol Neurobiol. 158(2–3):132–142. [DOI] [PubMed] [Google Scholar]
- Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, Scheffler K.. 2015. RELAX: detecting relaxed selection in a phylogenetic framework. Mol Biol Evol. 323:820–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JR, Harkness DR, Isaacks RE, Duffield DA.. 1976. Some studies on blood of the Florida manatee, Trichechus manatus latirostris. Comp Biochem Physiol A. 55(4A):413–417. [DOI] [PubMed] [Google Scholar]
- Wood WG. 1976. Haemoglobin synthesis during human fetal development. Br Med Bull. 323:282–287. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic Analysis by Maximum Likelihood. Mol Biol Evol. 248:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Byrd C, Shen T-J, Simplaceanu V, Tam TCS, Ho C.. 2013. Role of β/δ101Gln in regulating the effect of temperature and allosteric effectors on oxygen affinity in woolly mammoth hemoglobin. Biochemistry 5249:8888–8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Shen T-J, Gupta P, Ho NT, Simplaceanu V, Tam TCS, Hofreiter M, Cooper A, Campbell KL, Ho C.. 2011. A biochemical–biophysical study of hemoglobins from woolly mammoth, Asian elephant, and humans. Biochemistry 50:7350–7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.