Abstract
Antibiotic resistance is a public health issue with links to environmental sources of antibiotic resistance genes (ARGs). ARGs from nonviable sources may pose a hazard given the potential for transformation whereas ARGs in viable sources may proliferate during host growth or conjugation. In this study, ARGs in the effluent from three municipal wastewater treatment plants (WWTPs) and the receiving surface waters were investigated using a viability-based qPCR technique (vPCR) with propidium monoazide (PMA). ARGs sul1, tet(G), and blaTEM, fecal indicator marker BacHum, and 16S rRNA gene copies/mL were found to be significantly lower in viable-cells than in total concentrations for WWTP effluent. Viable-cell and total gene copy concentrations were similar in downstream samples except for tet(G). Differences with respect to season in the prevalence of nonviable ARGs in surface water or WWTP effluent were not observed. The results of this study indicate that qPCR may overestimate viable-cell ARGs and fecal indicator genes in WWTP effluent but not necessarily in the surface water >1.8km downstream.
Keywords: Antibiotic resistance genes, wastewater, propidium monoazide, chlorination, vPCR
1. Introduction
Antibiotic resistance is a major modern public health issue. In the United States at least two million illnesses and 23,000 deaths are caused by antibiotic-resistant bacteria annually (Centers for Disease Control and Prevention, 2013). Given that antibiotic resistant infections in humans have been linked to environmental sources of antibiotic resistance (Casey et al., 2013), fully addressing the risk posed by ARGs requires understanding environmental hotspots and determining the relative importance of ARGs from viable and nonviable sources for different mechanisms of proliferation.
Wastewater treatment plants (WWTPs) have been described as hotspots for the spread of antibiotic resistant bacteria and ARGs in the urban environment (Rizzo et al., 2013) and correlations have been observed with respect to land use and ARG concentrations (Pruden et al., 2012). The oxidizing disinfectants applied in secondary treatment at WWTPs, such as chlorine, can inactivate antibiotic resistant bacteria but do not necessarily destroy ARGs (Fahrenfeld et al., 2013; Yuan et al., 2015). The release of extracellular ARGs and nonviable cells containing ARGs to the environment from activities such as wastewater treatment presents a potential risk because of the possibility for these genes to be incorporated into competent host genomes by transformation (Chang et al., 2017; Mao et al., 2014). Understanding the relative proportion of ARGs that are present in viable versus nonviable cells or as extracellular DNA (eDNA) can provide insight into their availability for different mechanisms of ARG proliferation (i.e., via growth and/or conjugation versus transformation) in a given environmental compartment.
eDNA, including ARG-carrying plasmids, persists in the environment after the host bacteria dies, and is therefore potentially available for transformation (Mao et al., 2014). Likewise, the total DNA of fecal indicators has been shown to persist long after the decay of culturable E. coli and E. faecalis, which occurs within days in surface water (Gutierrez-Cacciabue et al., 2016). In contrast to ARGs, 16S rRNA genes from fecal indicators are not generally considered a hazard if the host cell is not viable. The relative risk posed by ARGs in viable cells compared to extracellular ARGs are not well characterized because the rates of conjugation and growth versus transformation are poorly understood. Understanding the ratios of these ARG pools is one step towards better characterizing the hazard posed by ARGs in different compartments. The viability qPCR (vPCR) method used in this study reduces the qPCR signal from DNA originating from nonviable sources in environmental samples (Bae and Wuertz, 2009; Eramo et al., 2017b; Nocker et al., 2007b). This is achieved using propidium monoazide (PMA), a dye that irreversibly intercalates extracellular DNA or DNA in cells with compromised membranes, thereby preventing amplification by qPCR. Recent applications of this method for targeting ARGs include (1) determining disinfection efficiency of combined sewer overflow effluent with peracetic acid (PAA) (Eramo et al., 2017b) and (2) measuring microbial decay rates in aerobic and anaerobic sludge (Mantilla-Calderon, 2017).
The objective of this study was to investigate the relative proportion of ARGs in cells with intact membranes to total ARGs observed in WWTP effluent and receiving waters towards understanding their availability for proliferation via different mechanisms. The partitioning of ARGs in WWTP effluent and receiving waters was evaluated by differentiating viable-cell ARGs by vPCR from total gene copies by qPCR, the latter of which has been shown to serve as a conservative proxy for ARG transformation (Chang et al., 2017). Other bacterial genes (BacHum, a human fecal marker, and 16S rRNA genes) were evaluated because these target genes are not considered functional genes and therefore may have different fates with respect to transformation compared to ARGs. The sampling design also allowed for determination of whether the vPCR results varied by season or WWTP. Overall, the results presented have implications for the understanding the relative pool of ARGs available for horizontal gene transfer (HGT) via transformation in different aqueous environments.
2. Materials and Methods
2.1. Field sampling and water quality analysis
Samples were collected on three days in the summer (7/30 or 8/5, 8/24, and 8/31/2015) and three days in the winter (1/25 or 3/2, 3/7, and 3/14/2016) from the treated effluent of three WWTPs (WWTP-A, B, and C in the US mid-Atlantic region) and from surface water upstream and downstream of the WWTPs’ discharge locations. WWTP flow rates, upstream, and downstream sampling distances are summarized in Tables S1-S2. Surface water and treatment plant effluent samples were collected on the same day for as many sampling events as possible. Variation from the paired sampling design was due to precipitation that occurred before sampling could be completed on 7/30/2015 for WWTP-A downstream samples, a plant issue on 8/24/2015 preventing effluent sampling of WWTP-B, and inaccessibility to field sites due to snow and ice on 1/25/2016. Composite effluent samples (500-mL each) were collected at 8 am, 10 am, and 12 pm. All samples were collected during baseflow conditions in sterile 500-mL Nalgene bottles, transported to the lab on ice and stored at 4 °C prior to processing. WWTP-A and WWTP-B serve municipalities with major hospitals and/or medical facilities within the sewershed, while WWTP-C does not have major hospitals and/or medical facilities within the sewershed. Surface water sampling locations were chosen based on availability of public access. Further details including sampling times for the surface water samples are presented in Table S2.
pH and conductivity of the downstream water samples were measured in the field with an Orion Star A329 multimeter (Thermo Scientific, Waltham, MA). Downstream and composite effluent samples were analyzed for total suspended solids (TSS) using Environmental Sciences Section Method 340.2 (Wisconsin State Lab of Hygiene, 1993). Field blanks consisting of autoclaved deionized water were carried in the field on each sampling date for QA/QC.
2.2. Cell collection, viability cross-linking and biomolecular analysis
Bacteria from 210 mL aliquots of effluent composite and downstream samples were harvested by sequential centrifugation at 4,000×g for 15 min and removing the supernatant except for the last 10 mL, as demonstrated by others (Banihashemi et al., 2012; Contreras et al., 2011; Luo et al., 2010; Nocker et al., 2007a). Aliquots of centrifuge-concentrated cells were either treated with 50 µM propidium monoazide (PMA) or preserved for DNA extraction. The former allowed for quantification of the “viable” cell fraction of DNA in the samples, defined as DNA in cells with intact membranes, and the latter for quantification of total DNA in the samples. A PMA concentration of 50 µM was selected because it was sufficient to suppress qPCR signals from heat-inactivated cells without impacting viability. Additional preliminary testing showed that 20 µM PMA was not sufficient (data not shown). This PMA concentration has been used by others for cells from wastewater, estuarine benthic mud, and marine sediment (Nocker et al., 2007a). PMA-treated samples were incubated in the dark for 5 min and photoactivated for 15 min using a PMA-Lite™ LED Photolysis Device (Biotium, Fremont, CA) to facilitate cross linking of the dye with DNA. PMA methods were adapted from Nocker et al. (2007a and Nocker et al. (2010. A schematic of the methodology is provided (Figure S1). Upstream and downstream samples (~200–700 mL) were also filter concentrated (0.22 µm nitrocellulose) and preserved for DNA extraction. Samples were stored at −20 °C until DNA extraction.
DNA was extracted from centrifuge-concentrated cells (500 µL) with and without PMA treatment and filter concentrated cells using a FastDNA® SPIN Kit for Soil (MP Biomedicals, Solon, OH). Concentrations of select ARGs [sul1 (Pei et al., 2006), tet(G) (Aminov et al., 2002), blaTEM (Narciso-da-Rocha et al., 2014)], fecal indicator marker BacHum (Kildare et al., 2007) and 16S rRNA gene copies for total bacterial population (Muyzer et al., 1993)] were quantified by qPCR. A standard SybrGreen (5 µL SsoFast EvaGreen, BioRad, Hercules, CA) chemistry with 0.4 µM forward and reverse primers, and 1 µL diluted DNA extract (1:50 dilution to reduce inhibition) in a 10 µL total reaction volume was used for all targets except BacHum. Hydrolysis probe chemistry (5 μL SsoFastProbes SuperMix, BioRad, Hercules, CA) for BacHum consisted of 0.07 μM probe, 0.22 μM forward and reverse primers, and 1 μL diluted DNA extract (1:50). Thermocycler (BioRad CFX96 Touch, Hercules, CA) conditions are summarized in Table S3. Samples and standards were analyzed in triplicate (technical replicates). A seven-point calibration curve and negative control were included in each run. Average amplification efficiency across all assays was 89±15 (mean ± SD) and average R2 was 0.99±0.01. Standards were generated from 10-fold serial dilutions of cloned and sequenced genes originating from environmental samples. A melt curve and/or gel electrophoresis were used to verify the specificity of qPCR products. Based on the lowest standard on the curve and factoring in dilution implementing during sample processing the limits of quantification were 4.1–4.9 log gene copies/mL for the genes tested.
2.3. Matrix spike study
To explore whether environmental matrix effects were interfering with the PMA performance in the downstream samples, centrifuge-concentrated river water was spiked with heat-inactivated E. coli cells (100ºC, 10 min). E. coli were plated on LB agar prior to the heat inactivation to confirm viability and after heat treatment to confirm inactivation. Samples were split and treated with PMA (0, 50, or 100 µM) or preserved for DNA extraction followed by vPCR or qPCR for 16S rRNA gene copies, as described above.
2.4. Statistical Analysis
All statistical analyses were performed in R (http://www.r-project.org). Except for blaTEM and 16S rRNA, which were log transformed, qPCR data were Box-Cox transformed prior to analysis given that ≥20% samples had ARGs below detection. A paired Wilcoxon rank sum test was used to determine differences (α< 0.05) in transformed absolute ARG (copies/mL) or 16S rRNA gene normalized ARG concentrations between the viable-cell and total gene copies in samples and between effluent and downstream viable-cell to total ratios for all WWTPs. In order to pair viable-cell to total ratios, the second WWTP-B downstream sample was excluded from this analysis because the corresponding effluent sample was not collected. A Kruskal Wallis test was used to determine differences between effluent and downstream concentrations and seasonal differences in viable or total gene copies for a given WWTP. A post-hoc pairwise t-test with a Bonferroni correction for multiple comparison was then applied. All sample results were included in the analyses including zeros and outliers. The same approach was used to compare pH, TSS, and conductivity data. A one-way test for equal means was performed on data from the matrix spike experiment, followed by a post-hoc pairwise t-test with a Bonferroni correction for multiple comparison.
3. Results
3.1. Total and viable-cell quantification of ARGs, BacHum fecal indicator marker, and 16S rRNA in WWTP effluent and downstream samples
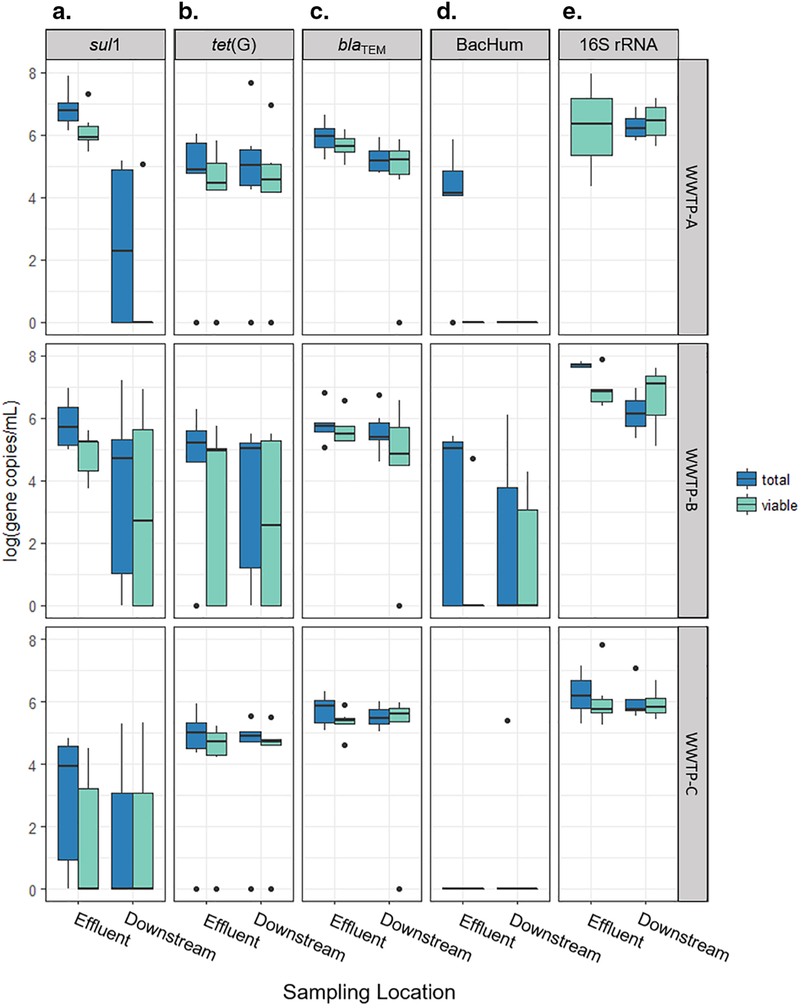
A viability-based approach was used to quantify viable-cell gene copies of ARGs sul1, tet(G), and blaTEM, human fecal marker BacHum, and 16S rRNA genes in WWTP effluent and surface water to better understand the potential availability of these genes for growth/conjugation versus transformation. Total concentrations of the target genes were quantified by qPCR and concentrations of viable-cell copies were quantified by first pretreating the samples with PMA, a membrane impermeable dye. There were significantly lower concentrations of all genes tested in the viable-cell fraction compared to the total measurements in the paired WWTP effluent samples (p<0.02 for each gene across plants and seasons), as would be expected for WWTP effluent disinfected with chlorine (Fig. 1). ARGs sul1 and tet(G) were detected in both the viable-cell and total fractions of 76% of effluent samples (n=17), where the average ratio of viable-cell to total concentrations (vPCR/qPCR) was 0.43 ± 0.52 for sul1 and 0.46 ± 0.29 for tet(G) (Fig. 2). In the other 24% of samples, ARGs were either not detected in the viable-cell fractions but present in the total ARG measurement or were not detected in either measurement in treated effluent (Fig. 1a, b). The ARG blaTEM was detected in both the viable-cell and total measurement in 100% of effluent samples at a ratio of viable-cell to total ARGs of 0.69 ± 0.46 (Fig. 1c).
Figure 1.
Concentrations of ARGs (a.) sul1, (b.) tet(G), and (c.) blaTEM, (d.) fecal indicator marker BacHum, and (e.) 16S rRNA present overall (total) and in viable cells only (viable) measured in effluent and downstream samples from wastewater treatment plant A, B, and C during summer and winter seasons. Boxes represent upper and lower quartiles, whiskers extend to high and low data points excluding outliers, and dots indicate outliers.
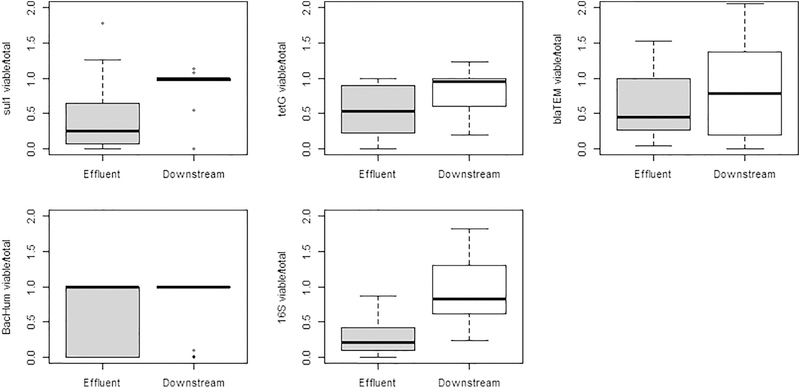
Figure 2.
Ratios of viable-cell gene concentrations measured by vPCR to total gene concentrations measured by qPCR in effluent and downstream samples.
In addition to the ARG vPCR and qPCR, two other genes were quantified with both methods: BacHum human fecal marker and 16S rRNA genes. These genes would presumably be less likely to be transformed and maintained in the host cell’s genome compared to ARGs which serve as functional genes. In contrast to the ARGs, the BacHum marker was detected in the viable-cell fraction of only one effluent sample, where a ratio of 0.51 viable to total gene copies per mL (vPCR/qPCR) was observed. BacHum was below detection in 53% of effluent samples and observed in the total measurement but not viable cells in the remaining 41% of samples (Fig. 1d). The average ratio of 0.24 viable-cell to total 16S rRNA gene copies in effluent samples suggests, as expected, that most bacteria are inactivated with a membrane sufficiently damaged to allow PMA dye entry prior to leaving the WWTPs sampled in this study (Fig. 1e).
Towards understanding the fate of environmental ARGs, vPCR and qPCR were next applied in the receiving surface water. In contrast to the WWTP effluent, viable-cell concentrations of sul1, blaTEM, BacHum, and 16S rRNA gene copies via vPCR were not different from the total measurement of these genes via qPCR in the downstream samples (all p>0.18). sul1, blaTEM, BacHum, and 16S rRNA targets were detected in both the viable-cell and total gene copy concentrations in 33%, 83%, 12%, and 100% of downstream samples (n=18), respectively. sul1 was below detection in both the total and viable-cell fraction for 50% of samples and observed only in total cells in the remaining three (17%) samples. In the 3/18 samples where viable-cell blaTEM was not observed, the gene was detected in the total measurement via qPCR.
tet(G) was the only gene tested where viable-cell gene copies were significantly lower than total gene copies in the downstream samples (p<4.7×10−3). In 76% of downstream samples where tet(G) was detected in both the viable-cell fraction and the total measurement via qPCR, the ratio of viable-cell to total gene copies was 0.71 ± 0.35. In the remaining samples, tet(G) was below detection in both the viable-cell fractions and the total measurement or only detected in the total measurement.
3.2. Comparison of WWTP effluent and downstream gene concentrations
Concentrations of viable-cell and total gene copies were also compared between effluent and downstream sampling locations to further understand the fate of ARGs and fecal markers released from WWTPs. Effluent concentrations of total sul1 were higher than the receiving water where the total sul1 gene copies were below detection limits in many (9/17) downstream samples (p=0.02; Fig. 1a). When above detection limits, total sul1 gene copies downstream were on average 98 ± 6% less than the total sul1 concentration detected in the WWTP effluent. Both the 16S rRNA normalized concentrations of the viable-cell and total sul1 gene copies (viable-cell sul1/viable-cell 16S rRNA genes and total sul1/total 16S rRNA genes), were higher in WWTP effluent samples compared to the downstream samples (all p<1.1 ×10−3; Fig. S2a). The concentrations of the other genes measured in the viable-cell fractions via vPCR and total measurement via qPCR were similar in effluent and downstream samples, both on a volume concentration (gene copies/mL) and a 16S rRNA gene copy normalized basis (all p>0.06). For most sampling events, if gene targets were detected in the viable fraction of cells downstream, they were also detected in viable cell fraction in the corresponding effluent sample. However, on the first winter sampling event, sul1 was detected in the viable-cell fraction downstream but not in the viable-cell fraction of effluent at WWTP-C, and tet(G) at WWTP-A. (Effluent and downstream samples were not paired for this event given sampling access issues.) Detection in viable-cells downstream but not in effluent also occurred on the second day of winter sampling for sul1 and tet(G) at WWTP-C and WWTP-B, respectively. BacHum exhibited this phenomenon on the last summer day of sampling at WWTP-B.
Comparing the ratio of viable-cell to total gene copies observed in WWTP effluent and the downstream surface water, the ratio of viable-cell to total gene concentrations (vPCR/qPCR) was greater in surface water samples compared to effluent for sul1, tet(G), and 16S (all p<0.05; Fig. 2). The ratio of viable-cell to total concentrations was <0.5 for these genes after chlorination, but this impact of wastewater disinfection on viable ARG-carrying microbes was no longer observed in downstream surface water because there was generally no difference between the viable-cell and total concentrations. Similar viable-cell to total concentration ratios were observed for both blaTEM and BacHum in effluent and downstream samples (p>0.08). In contrast to the other genes, blaTEM was the most frequently detected ARG in viable cells and had the highest ratio of viable-cell to total concentrations leaving the WWTPs, while BacHum was detected infrequently in both effluent and downstream samples. These results indicate some gene-to-gene differences in ARG fate.
3.3. Matrix spike experiment
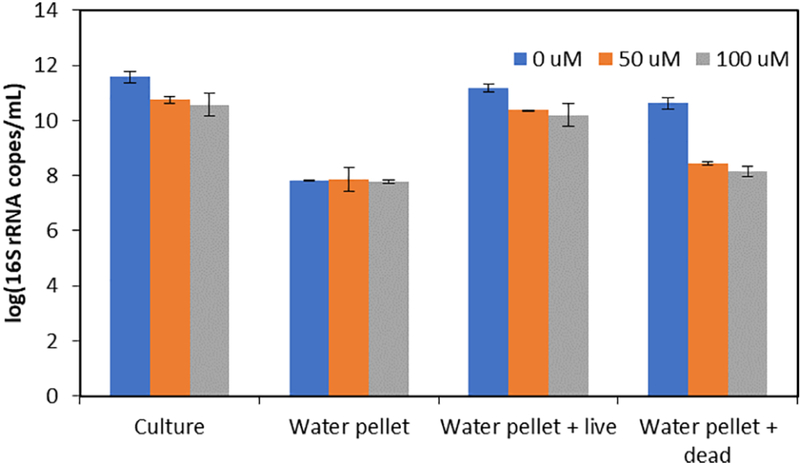
Given that differences between vPCR and qPCR results were observed less frequently in downstream samples, a separate experiment was performed to determine if matrix effects were interfering with the PMA performance in the downstream samples. PMA was found to successfully suppress 16S rRNA qPCR signals in centrifuge-concentrated river water cells spiked with inactivated (heat-treated) cell cultures, which represented free DNA and cells with compromised membranes (Fig. 3). These samples exhibited the same 16S rRNA gene copy concentrations as concentrated water only controls (p=0.30). Concentrated water samples spiked with the same culture but not heat-treated served as a second control. As would be expected, samples spiked with viable cultures were greater than river water (p=5.9×10−3) because PMA does not interfere with qPCR for cells with intact membranes. Plates with the live spike (not heat-treated) were too numerous to count but plates with heat-inactivated cultures had no growth, confirming that heat treatment inactivated the E. coli culture.
Figure 3.
Concentrations of 16S rRNA in centrifuge-concentrated E. coli culture at exponential phase, river samples only (water pellet), centrifuge-concentrated river samples with a live cell culture spike (water pellet + live), and centrifuge-concentrated river samples with an inactivated (heat-treated) cell culture spike (water pellet + dead). Samples were analyzed by qPCR (0 µM PMA) and vPCR (50 or 100 µM PMA), which represent total and viable-cell concentrations, respectively.
3.4. WWTP and seasonal impacts
No differences in total or viable-cell concentrations between the two seasons sampled were observed in effluent and downstream samples for the genes tested (all p>0.06). Downstream surface water pH, conductivity and TSS and effluent TSS were also consistent between seasons (p>0.45; Table 2). Thus, seasonality did not impact concentrations of genes or water quality measurements observed in this study (Fig. S3).
Comparisons were made among the three WWTPs to determine if there were plant-to-plant differences. While all WWTP have similar treatment trains consisting of primary clarification, activated sludge treatment and secondary clarification followed by chlorination, WWTP-A and B have larger influent flow rates (>100 MGD) compared to WWTP-C (<100 MGD; Table S1). In addition, WWTP-A and WWTP-B serve municipalities with combined collection systems while WWTP-C receives water from a separate sanitary collection system. Stormwater did not impact the sampling because all samples were collected during baseflow conditions. There were no differences observed in the effluent concentrations of the viable-cell via vPCR or the total measurement via qPCR of tet(G) and blaTEM between the three plants sampled (both p>0.59). In contrast, WWTP-A and WWTP-B had higher concentrations of viable-cell and total sul1 compared to WWTP-C (all p<0.02). Additionally, WWTP-A had higher BacHum marker and 16S rRNA gene copy total concentrations than WWTP-C (p<4.0×10−3). Higher concentrations of TSS were also observed at WWTP-A compared to WWTP-C (p=0.02), but no other differences between plants were observed (p>0.27; Table 2). The select inter-plant effluent differences observed were not preserved downstream: no differences were observed between the three surface water bodies sampled for any of the genes tested (all p>0.27).
4. Discussion
4.1. Total and viable-cell gene concentrations in WWTP effluent and downstream samples
A viability-based qPCR method was applied demonstrating that the ARGs and other target genes were significantly lower in the viable-cell fraction of effluent samples compared to total gene concentrations. This was expected given that all three WWTPs sampled in this study disinfected by chlorine, an oxidant whose disinfectant mechanisms includes the disruption of cell membranes by reactive oxygen (McFadden et al., 2017). Cultivation-based methods have also been applied to demonstrate that some viable antibiotic resistant bacteria are released from wastewater treatment to surface water (Huang et al., 2012; Li et al., 2015). While the vPCR method used in this study reduces signal from extracellular DNA and cells with compromised membranes, this method may be expected to overestimate the viable fraction of cells given that cells may be inactivated without having damaged membranes, which may explain the observations of vPCR/qPCR values above zero. The same viability-based method applied here was recently used to demonstrate the disinfection efficiency of PAA on ARG-carrying cells in simulated combined sewer overflow (Eramo et al., 2017b). With PAA disinfection, the fractions of viable-cell sul1 and tet(G) concentrations compared to total sul1 and tet(G) concentrations reached as low as 5.3×10−3 ± 3.5×10−3, or less than 1% (Eramo et al., 2017b), while in this study, the percentage was slightly less than 50%. Differences in disinfectant, concentration, contact time, and the presence of organic matter can affect the disinfection efficiency and may explain the differences in these observations.
The lack of observed differences between vPCR and qPCR results for sul1 and blaTEM in the water column downstream of WWTPs is consistent with other reports of ARGs in river water samples where the majority of DNA was intracellular DNA (iDNA) rather than eDNA (Mao et al., 2014). Those researchers accounted for the loss of eDNA in the water column through observations that eDNA was much higher in river sediment samples, suggesting deposition was a loss mechanism for extracellular ARGs in the water column (LaPara et al., 2015; Mao et al., 2014). Sediment samples were not collected in the present study to confirm this result, but it is worth noting that vPCR may not perform well at differentiating nonviable sources in sediment due to matrix interferences (Kim et al., 2014). In addition to deposition, the lack of differences for viable-cell and total gene copies in the water column may also be explained by degradation (LaPara et al., 2015), dilution (LaPara et al., 2015), cellular repair (Huang et al., 2011), and/or transformation of extracellular gene copies or those associated with nonviable cells in WWTP effluent (Mao et al., 2014). In the Mississippi River, it was reported that the flow rate relative to WWTP effluent flow resulted in a large dilution effect on ARGs, minimizing the measurable impact on receiving water (LaPara et al., 2015). Likewise, in this study, no differences were observed between total ARG concentrations in surface water samples collected upstream and downstream of the three WWTPs during both the summer and winter seasons (p=1.0; Fig. S4). ARGs in these samples were measured by filtering of water samples through 0.22 µM nitrocellulose filters followed DNA extraction from filters, rather than collection of cells by centrifugation applied for the total and viable-cell concentrations. The results of upstream and downstream sampling support the observation that WWTPs are point sources of nonviable and viable-cell ARG but the concentrations of the monitored ARGs present in these WWTP effluents do not significantly impact concentrations >1.8km downstream and indicate that dilution and/or other loss/decay mechanisms could be responsible for this phenomenon. The lack of gages in the vicinity of sampling locations made it difficult to estimate a dilution rate for the WWTP effluent in this study. Another potentially confounding factor, tide direction, did not result in differences in ARG concentrations (Table S2). It is not known whether these observations would be consistent for the full range of ARGs present in wastewater or whether the hosts of the ARGs from WWTPs present a greater human health risk than surface water ARG hosts do. Therefore, further study of WWTPs with methods that address these issues are warranted.
BacHum human fecal marker and 16S rRNA genes were also measured in effluent and downstream samples to provide information on the fate of non-functional genes to compare to the ARG data. Their fate was expected to be different than that of ARGs given than ARGs can provide a selective advantage and therefore may be more likely to be transformed. BacHum was observed in only one viable-cell effluent sample. This observation is consistent with other studies where viable fecal indicator organisms were not detected in secondary effluent (Li et al., 2014) or were detected at very low concentrations by vPCR (Varma et al., 2009). Viability qPCR methods have been used to target pathogens and fecal indicators with positive correlations observed between vPCR and cultivation methods (Li et al., 2014; Varma et al., 2009). Thus, the utility of this fecal marker gene as an indicator gene for ARGs is not clear, consistent with our previous observations during combined sewer overflows (Eramo et al., 2017a). The BacHum marker was also observed infrequently in the water column downstream from WWTPs. Fecal indicator genes not detected in viable cells may have settled, degraded, or, most likely, been too dilute for detection. The presence of BacHum suggests human fecal pollution, which is most likely to originate from WWTPs during dry weather sampling of urban environments without septic tanks or from cross amplification with select other non-human fecal sources (Kildare et al., 2007). As expected, the low ratio of viable-cell to total BacHum marker downstream does not provide evidence of growth or HGT.
To explore whether environmental matrix effects were interfering with the PMA performance in the downstream samples, concentrated river water was spiked with heat-inactivated cells and analyzed by vPCR and qPCR. PMA was found to successfully suppress 16S rRNA qPCR signals in centrifuge-concentrated river water cells spiked with inactivated (heat-treated) cell cultures, which represented free DNA and cells with compromised membranes. This result is attributed to PMA suppression of DNA amplification from the heat-treated culture. During PMA treatment, the light exposure inactivates any excess PMA that has not entered cells, so that it cannot affect DNA when it is released during DNA extraction (Nocker et al., 2010). Also, upon light treatment, the free DNA that binds to PMA is rendered insoluble and lost during subsequent DNA extraction (Nocker et al., 2006). This result indicates that matrix interference does not explain the different results observed in WWTP effluent compared to surface water. The lower concentrations of viable-cell tet(G) concentrations compared to total concentrations in this study also supports that differences in viability are detectable in surface water samples.
4.2. Comparison of WWTP effluent and receiving water
While the volume-based concentrations of viable-cell sul1 gene copies (gene copies/mL) in the WWTP effluent and downstream samples were similar, sul1 gene carrying cells comprised a smaller fraction of the viable-cell microbial community downstream. The sul1 viable-cell concentration in WWTP effluent was similar to both the viable-cell and the total concentration of this gene in downstream samples. This suggests that the nonviable cells carrying sul1 and extracellular sul1 genes were, on net, not remaining suspended in the water column. Community analysis was not performed in this study therefore any shifts in bacterial community composition of ARG hosts between WWTP effluent and receiving water that could help explain the fate of the investigated genes are not known.
4.3. vPCR versus eDNA extraction methods
Other methods have been used to investigate nonviable sources of ARGs within aqueous samples by qPCR. An extraction procedure (Corinaldesi et al., 2005) has also been used to differentiate ARGs in extracellular DNA (eDNA) and intracellular DNA (iDNA) in the sludge of livestock waste management operations (Zhang et al., 2013) and surface water (Mao et al., 2014). This method allows for simultaneous separation of the fractions. However, Liu et al. (2018 noted that eDNA concentrations were too low harvest in treated water and utilized an adsorption-elution method. This method does not allow for simultaneous comparison of the two DNA fractions but allows for observation over time or treatments. The benefits of the vPCR method used in this study are that it allows for paired analysis of viable-cell and total gene concentrations and that it is simple to apply directly to collected cells in ~15min. eDNA/iDNA extractions procedures require multiple washing and pelleting steps, while PMA pretreatment only requires collecting bacterial cells and particles, here accomplished by centrifuging to generate a pellet. However, the vPCR method may not perform well in sediment (Kim et al., 2014) which represents a major limitation compared to the extraction methods which were developed for sediment. Even in aqueous samples it is recommended that the PMA dye used for vPCR should be titrated to determine the correct concentration: if too much is added it can be toxic to cells resulting in artificially low vPCR results, and if too little is added one would expect results artificially similar to the qPCR. In this study we tested 20–100 µM PMA and found 20 µM was insufficient and that results were similar for 50 or 100 µM. In this study select samples, mostly in surface water, had vPCR to qPCR ratios (copies/mL : copies/mL) of greater than two up to ten (Fig. 2), which was not expected. These anomalies may be due to the variability in the environmental split samples for vPCR and qPCR. As stated above, the eDNA extraction and vPCR methods have not been previously applied to paired samples, to the authors knowledge, but could provide further insight into the fate of eDNA and DNA from nonviable cells.
To the authors’ knowledge, the vPCR and extraction methods have not been compared on paired samples for ARGs, but have the potential to provide slightly different information: extraction is dependent on physically separating iDNA and eDNA, vPCR targets eDNA and the fraction of iDNA in cells with compromised membranes. While there is no consensus on which methods to apply to establish the risk posed by environmental ARGs and antibiotic resistance (Vikesland et al., 2017), the published literature applying either extraction or vPCR provides a basis for defining which environments have or do not have significant concentrations or fractions of extracellular/nonviable cell ARGs [i.e., livestock waste management structures (Zhang et al., 2013), surface water sediments (Mao et al., 2014), PAA-treated combined sewer overflow (Eramo et al., 2017b)]. But, given that these studies applied the different methods across different environments, questions remain about the relative abundance of extracellular and nonviable sources of ARGs in environmental hotspots, including surface waters receiving treated wastewater effluent.
4.4. Implications for understanding the mechanisms driving ARG fate
A critical question with respect to nonviable sources of ARGs measured in this study is determining the potential pool of DNA available for HGT via transformation in different environments. A risk assessment requires accounting for the relative importance of different mechanisms of HGT (Vikesland et al., 2017). Once these environments are identified, kinetic investigations can be performed to determine the rates of HGT. At the downstream sampling locations in this study located more than 1.8 kilometers away from the WWTPs, qPCR and vPCR results were similar for sul1, blaTEM, BacHum marker, and 16S rRNA genes indicating nonviable DNA was not present in high abundance, unlike in the WWTP effluent, and thus no longer available in high proportion for transformation into viable cells in the water column. For tet(G), the nonviable concentrations were a significant proportion of the ARGs in the water column. These gene-to-gene differences may be driven by differences in the fate of the host cells, although, sul1, tet(G), and blaTEM all encode for resistance to antibiotics used to control Gram negative bacteria. Microbial community analysis was not performed in this study therefore the shifts in bacterial community composition of ARG hosts between WWTP effluent and receiving water that could also explain the fate of the genes investigated are not known. Testing for a broader range of ARGs may also provide further insight if this approach were repeated with high throughput qPCR arrays targeting a broader range of ARGs.
5. Conclusions
Overall results from this study confirm that qPCR is a conservative proxy (i.e., overestimate) for analyzing viable-cell ARGs in chlorinated WWTP effluent. In contrast, in downstream water, qPCR represented a reasonably accurate measure of sul1 and blaTEM concentrations 1.8–3.9 km from WWTP outfalls but overestimated the viable-cell concentrations of tet(G). How close to the WWTP outfalls this observation would be consistent is not clear but likely is a function of dilution factor and the background prevalence of these genes in the receiving waterbody. Therefore, it would be advisable to sample the water column closer to the WWTP outfalls in future research to look for evidence of significant fractions of eDNA and nonviable cells with sul1 and blaTEM in the water column that would be available for transformation (the lack of public access points prevented that in this study) or to focus on bed sediments, which were found by others to have significant amounts of eDNA (Mao et al., 2014). The lack of seasonal differences in these observations indicates treatment and fate/transport process maybe consistent across seasons. Analyses of a broader suite of ARGs are needed to determine how consistent this finding is across resistance types. Quantifying the risk posed by ARGs in the environment will require understanding the relative hazard posed by viable versus nonviable-cell ARG. In particular, there is a need to compare the rates of horizontal gene transfer from these ARG reservoirs to pathogenic organisms and determine if these rates are concentration dependent (if so, vPCR could have utility towards understanding the risk posed by environmental ARG).
Supplementary Material
Table 1.
Average pH, conductivity and TSS (+/− standard deviation) collected from upstream, influent, effluent, and downstream samples from three wastewater treatment plants during the summer and winter seasons (n=2* or 3).
| Plant A | Plant B | Plant C | |||||
|---|---|---|---|---|---|---|---|
| Summer | Winter | Summer | Winter | Summer | Winter | ||
| pH | Upstream | 7.55 ± 0.11 | 7.63 ± 0.06 | 7.40 ± 0.40 | 7.61 ± 0.22 | 7.45 ± 0.05 | 7.7 ± 0.3 |
| Downstream | 7.84 ± 0.07 | 7.77 ± 0.34 | 7.65 ± 0.18 | 7.63 ± 0.34 | 7.49 ± 0.27 | 7.38 ± 0.54 | |
| Conductivity (mS/cm) | Upstream | 249 ± 397 | 6.63 ± 9.49 | 278 ± 454 | 11.3 ± 10.6 | 3.36 ± 4.23 | 0.54 ± 0.49 |
| Downstream | 341 ± 529 | 28.4 ± 13.4 | 367 ± 580 | 25.7 ± 9.36 | 2.50 ± 3.55 | 1.43 ± 599 | |
| TSS (mg/L) | Upstream | 74.9 ± 32.9 | 58.7 ± 35 | 75.8 ± 28.5 | 90.7 ± 63.9 | 5.00 ± 1.67 | 21.7 ± 26.4 |
| Influent | 95.5 ± 33.1 | 211 ± 93 | 252 ± 19 | 262 ± 76 | 276 ± 58 | 244 ± 32 | |
| Effluent | 20.6 ± 4.1 | 20.0 ± 12.2 | 8.33 ± 7.07 | 14.3 ± 3.5 | 3.33 ± 5.77 | 10.1 ± 10.5 | |
| Downstream | 118 ± 108 | 179 ± 192 | 499 ± 488 | 223 ± 300 | 6.67 ± 7.64 | 56.3 ± 77.3 | |
Highlights.
Viability qPCR demonstrated in wastewater effluent, downstream surface water
Effluent viable-cell copies < total for antibiotic resistance genes, fecal markers
Differences were generally not preserved downstream
Results improve understanding of prevalence of nonviable sources of ARG
Acknowledgements
Laboratory assistance was provided by Hannah Delos Reyes, Michelle Yam, and Sophia Blanc. Thanks to our utility partners for providing access to treatment plant samples. Funding for this project was largely provided by a grant from the New Jersey Water Resources Research Institute, with additional support provided by the National Science Foundation (#1510461), a Mark B. Bain Fellowship from the Hudson River Foundation, the Aresty Program at Rutgers, and NIH Bridges to the Doctorate Program (R25GM058389).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aminov RI, Chee-Sanford JC, Garrigues N, Teferedegne B, Krapac IJ, White BA, et al. Development, Validation, and Application of PCR Primers for Detection of Tetracycline Efflux Genes of Gram-Negative Bacteria. Appl Environ Microbiol 2002; 68: 1786–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S, Wuertz S. Discrimination of viable and dead fecal Bacteroidales bacteria by quantitative PCR with propidium monoazide. Appl Environ Microbiol 2009; 75: 2940–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banihashemi A, Van Dyke MI, Huck PM. Long-amplicon propidium monoazide-PCR enumeration assay to detect viable Campylobacter and Salmonella. J Appl Microbiol 2012; 113: 863–73. [DOI] [PubMed] [Google Scholar]
- Casey JA, Curriero FC, Cosgrove SE, Nachman KE, Schwartz BS. High-density livestock operations, crop field application of manure, and risk of community-associated methicillin-resistant Staphylococcus aureus infection in Pennsylvania. JAMA Intern Med 2013; 173: 1980–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2013 In: Department of Health and Human Services, editor, 2013. [Google Scholar]
- Chang PH, Juhrend B, Olson TM, Marrs CF, Wigginton KR. Degradation of extracellular antibiotic resistance genes with UV254 treatment. Environ Sci Technol 2017; 51: 6185–6192. [DOI] [PubMed] [Google Scholar]
- Contreras PJ, Urrutia H, Sossa K, Nocker A. Effect of PCR amplicon length on suppressing signals from membrane-compromised cells by propidium monoazide treatment. J Microbiol Methods 2011; 87: 89–95. [DOI] [PubMed] [Google Scholar]
- Corinaldesi C, Danovaro R, Dell’Anno A. Simultaneous recovery of extracellular and intracellular DNA suitable for molecular studies from marine sediments. Appl Environ Microbiol 2005; 71: 46–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eramo A, Delos Reyes H, Fahrenfeld NL. Partitioning of Antibiotic Resistance Genes and Fecal Indicators Varies Intra and Inter-Storm during Combined Sewer Overflows. Front Microbiol 2017a; 8: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eramo A, Medina WM, Fahrenfeld NL. Peracetic acid disinfection kinetics for combined sewer overflows: indicator organisms, antibiotic resistance genes, and microbial community. Environ Sci Water Res Technol 2017b; 3: 1061–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenfeld NL, Ma Y, O’Brien M, Pruden A. Reclaimed water as a reservoir of antibiotic resistance genes: distribution system and irrigation implications. Frontiers in Microbiology 2013; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Cacciabue D, Cid AG, Rajal VB. How long can culturable bacteria and total DNA persist in environmental waters? The role of sunlight and solid particles. Sci Total Environ 2016; 539: 494–502. [DOI] [PubMed] [Google Scholar]
- Huang JJ, Hu HY, Lu SQ, Li Y, Tang F, Lu Y, et al. Monitoring and evaluation of antibiotic-resistant bacteria at a municipal wastewater treatment plant in China. Environ Int 2012; 42: 31–6. [DOI] [PubMed] [Google Scholar]
- Huang JJ, Hu HY, Tang F, Li Y, Lu SQ, Lu Y. Inactivation and reactivation of antibiotic-resistant bacteria by chlorination in secondary effluents of a municipal wastewater treatment plant. Water Res 2011; 45: 2775–81. [DOI] [PubMed] [Google Scholar]
- Kildare BJ, Leutenegger CM, McSwain BS, Bambic DG, Rajal VB, Wuertz S. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: a Bayesian approach. Water Res 2007; 41: 3701–15. [DOI] [PubMed] [Google Scholar]
- Kim M, Gutierrez-Cacciabue D, Schriewer A, Rajal VB, Wuertz S. Evaluation of detachment methods for the enumeration of Bacteroides fragilis in sediments via propidium monoazide quantitative PCR, in comparison with Enterococcus faecalis and Escherichia coli. J Appl Microbiol 2014; 117: 1513–22. [DOI] [PubMed] [Google Scholar]
- LaPara TM, Madson M, Borchardt S, Lang KS, Johnson TJ. Multiple Discharges of Treated Municipal Wastewater Have a Small Effect on the Quantities of Numerous Antibiotic Resistance Determinants in the Upper Mississippi River. Environ Sci Technol 2015; 49: 11509–15. [DOI] [PubMed] [Google Scholar]
- Li D, Tong T, Zeng S, Lin Y, Wu S, He M. Quantification of viable bacteria in wastewater treatment plants by using propidium monoazide combined with quantitative PCR (PMA-qPCR). J Environ Sci 2014; 26: 299–306. [DOI] [PubMed] [Google Scholar]
- Li J, Cheng W, Xu L, Jiao Y, Ali Baig S, Chen H. Occurrence and removal of antibiotics and the corresponding resistance genes in wastewater treatment plants: effluents’ influence to downstream water environment. Enviorn Sci Pollut Res 2015: 10. [DOI] [PubMed] [Google Scholar]
- Liu S-S, Qu H-M, Yang D, Hu H, Liu W-L, Qiu Z-G, et al. Chlorine disinfection increases both intracellular and extracellular antibiotic resistance genes in a full-scale wastewater treatment plant. Water Res 2018; 136: 131–136. [DOI] [PubMed] [Google Scholar]
- Luo JF, Lin WT, Guo Y. Method to detect only viable cells in microbial ecology. Appl Microbiol Biotechnol 2010; 86: 377–84. [DOI] [PubMed] [Google Scholar]
- Mantilla-Calderon D, Hong P, . Fate and Persistence of a Pathogenic NDM-1-Positive Escherichia coli Strain in Anaerobic and Aerobic Sludge Microcosms. Appl and Environ Microbiol 2017; 83: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao D, Luo Y, Mathieu J, Wang Q, Feng L, Mu Q, et al. Persistence of extracellular DNA in river sediment facilitates antibiotic resistance gene propagation. Environ Sci Technol 2014; 48: 71–8. [DOI] [PubMed] [Google Scholar]
- McFadden M, Loconsole J, Schockling A, Nerenberg R, Pavissich J. Comparing peracetic acid and hypochlorite for disinfection of combined sewer overflows: Effects of suspended-solids and pH. Sci Total Environ 2017; 599: 533–539. [DOI] [PubMed] [Google Scholar]
- Muyzer G, De Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis or polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 1993; 59: 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narciso-da-Rocha C, Varela AR, Schwartz T, Nunes OC, Manaia CM. blaTEM and vanA as indicator genes of antibiotic resistance contamination in a hospital-urban wastewater treatment plant system. J Glob Antimicrob Resist 2014; 2: 309–315. [DOI] [PubMed] [Google Scholar]
- Nocker A, Cheung CY, Camper AK. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J Microbiol Methods 2006; 67: 310–20. [DOI] [PubMed] [Google Scholar]
- Nocker A, Richter-Heitmann T, Montijn R, Schuren F, Kort R. Discrimination between live and dead cellsin bacterial communities from environmental water samples analyzed by 454 pyrosequencing. Int Microbiol 2010; 13: 59–65. [DOI] [PubMed] [Google Scholar]
- Nocker A, Sossa-Fernandez P, Burr MD, Camper AK. Use of propidium monoazide for live/dead distinction in microbial ecology. Appl Environ Microbiol 2007a; 73: 5111–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nocker A, Sossa KE, Camper AK. Molecular monitoring of disinfection efficacy using propidium monoazide in combination with quantitative PCR. J Microbiol Methods 2007b; 70: 252–60. [DOI] [PubMed] [Google Scholar]
- Pei R, Kim SC, Carlson KH, Pruden A. Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res 2006; 40: 2427–35. [DOI] [PubMed] [Google Scholar]
- Pruden A, Arabi M, Storteboom H. Correlation between upstream human activities and riverine antibiotic resistance genes. Environ Sci Tech 2012; 46: 11541–11549. [DOI] [PubMed] [Google Scholar]
- Rizzo L, Manaia C, Merlin C, Schwartz T, Dagot C, Ploy MC, et al. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci Total Environ 2013; 447: 345–60. [DOI] [PubMed] [Google Scholar]
- Varma M, Field R, Stinson M, Rukovets B, Wymer L, Haugland R. Quantitative real-time PCR analysis of total and propidium monoazide-resistant fecal indicator bacteria in wastewater. Water Res 2009; 43: 4790–801. [DOI] [PubMed] [Google Scholar]
- Vikesland PJ, Pruden A, Alvarez PJJ, Aga D, Burgmann H, Li XD, et al. Toward a Comprehensive Strategy to Mitigate Dissemination of Environmental Sources of Antibiotic Resistance. Environ Sci Technol 2017; 51: 13061–13069. [DOI] [PubMed] [Google Scholar]
- Wisconsin State Lab of Hygiene. ESS Method 340.2: Total Suspended Solids, Mass Balance (Dried at 103–105 C) Volatile Suspended Solids (Ignited at 550_C), Madison, WI, 1993. [Google Scholar]
- Yuan QB, Guo MT, Yang J. Fate of antibiotic resistant bacteria and genes during wastewater chlorination: implication for antibiotic resistance control. PLoS One 2015; 10: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Snow DD, Parker D, Zhou Z, Li X. Intracellular and extracellular antimicrobial resistance genes in the sludge of livestock waste management structures. Environ Sci Technol 2013; 47: 10206–13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.