Abstract
Background
Clostridium (Clostridioides) difficile infection (CDI) is a health care–associated infection that can lead to serious complications. Potential complications include intensive care unit (ICU) admission, development of toxic megacolon, need for colectomy, and death. However, identifying the patients most likely to develop complicated CDI is challenging. To this end, we explored the utility of a machine learning (ML) approach for patient risk stratification for complications using electronic health record (EHR) data.
Methods
We considered adult patients diagnosed with CDI between October 2010 and January 2013 at the University of Michigan hospitals. Cases were labeled complicated if the infection resulted in ICU admission, colectomy, or 30-day mortality. Leveraging EHR data, we trained a model to predict subsequent complications on each of the 3 days after diagnosis. We compared our EHR-based model to one based on a small set of manually curated features. We evaluated model performance using a held-out data set in terms of the area under the receiver operating characteristic curve (AUROC).
Results
Of 1118 cases of CDI, 8% became complicated. On the day of diagnosis, the model achieved an AUROC of 0.69 (95% confidence interval [CI], 0.55–0.83). Using data extracted 2 days after CDI diagnosis, performance increased (AUROC, 0.90; 95% CI, 0.83–0.95), outperforming a model based on a curated set of features (AUROC, 0.84; 95% CI, 0.75–0.91).
Conclusions
Using EHR data, we can accurately stratify CDI cases according to their risk of developing complications. Such an approach could be used to guide future clinical studies investigating interventions that could prevent or mitigate complicated CDI.
Keywords: Clostridium (Clostridioides) difficile infection, complications, electronic health records, machine learning, patient risk stratification
Clostridium (Clostridioides) difficile infection (CDI) is a prevalent condition [1–3] that often arises in health care settings [4]. As a pathogen, C. difficile is genetically diverse [1], with some strains more associated with the development of complicated disease, including polymerase chain reaction (PCR) ribotypes 027, 078/126, 056, and 018 [5, 6]. Clinically, the outcome of CDI varies across patients, ranging from complete recovery to mortality [7]. Currently, treatment decisions (eg, antimicrobial selection, admission to intensive care, and use of adjuvant therapies) are not informed by data-driven models of patient risk for complications, constituting a critical need [8].
Current clinical guidelines recommend treatment with vancomycin (125 mg orally 4 times per day) or fidaxomicin (200 mg twice daily for 10 days) on initial diagnosis. The recently revised guidelines no longer recommend treatment with metronizadole, but data exist demonstrating that compared with vancomycin this antibiotic may be adequate for mild/moderate disease at a lower cost and potentially lower risk of selecting for antibiotic resistance in clinically important bacteria such as Enterococcus species [9–11]. Given the complexity in treating patients with CDI, researchers have sought risk stratification models in support of individualized treatments. To quantify patient risk for developing complicated CDI, previous models have used expert-curated sets of patient characteristics [12–15]. In light of the fact that many of the factors driving patient risk for complications are not well understood, we sought a more comprehensive approach.
We used the structured contents of the EHR and a machine learning approach to develop an intelligible predictive model for complicated CDI. Such a model has the capacity to make a prediction on the day of diagnosis and each day thereafter. Though, as the patient’s disease progresses, we expect the task to become easier as it shifts from prediction to recognition. We compared the discriminative performance of this model with a previously published, expert-curated model for the same outcome [13]. In parallel, we investigated patient characteristics associated with increased risk for complications. Insights from these analyses may guide the development of interpretable machine learning applications for supporting CDI treatment decisions in real time.
METHODS
Study Population
We considered a cohort of 1144 cases of CDI, as described in Rao et al. [13]. This study population included adult inpatients diagnosed with CDI between October 2010 and January 2013 at the University of Michigan hospitals (UM). We linked each case of CDI from this cohort with structured EHR data available in the Research Data Warehouse at UM. We excluded a small fraction of CDI cases for whom data were not readily available. This study was approved by the Institutional Review Board at the University of Michigan.
Model Outcome
As in Rao et al., we considered a binary prediction task, in which CDI cases that became complicated were labeled 1, and 0 otherwise [13]. Building on previous work, we defined CDI as complicated if it led to any of 3 adverse outcomes within 30 days: admission to intensive care, colectomy, or mortality [12, 13, 16]. All cases were independently labeled through chart review by 2 clinicians, and adjudicated by a third if there was disagreement [13]. In this way, cases were labeled complicated only if the complication(s) were judged to have been caused by the CDI and not by other factors. Using data available before the time of prediction, we sought to learn a model that could accurately sort cases from low to high risk for this composite outcome. To facilitate comparison with the model produced by Rao et al., we evaluated predictions made 2 days after diagnosis. However, recognizing that earlier predictions are more useful, we considered 2 additional prediction times: the day of CDI diagnosis and the day after diagnosis. We hypothesized that prediction will be more difficult near the beginning of a patient’s infection course, as fewer clinical indicators for mild or severe disease will have manifested. We identified the day of CDI diagnosis using the time-stamped laboratory test result for presence of toxigenic C. difficile.
Data Preprocessing
We extracted EHR data describing each patient admission in our study population from the Research Data Warehouse at UM using the patient’s medical record number and date of CDI diagnosis. Specifically, we extracted patient demographics (eg, age), patient history within the past 90 days (eg, diagnosis of diabetes within the past 90 days), admission details (eg, scheduled, urgent, or emergency admission), and daily hospitalization details (eg, prescribed inpatient medications) (Supplementary Table 1). To focus more on patient state than clinician suspicion, we removed variables that clearly encoded clinical suspicion of complicated CDI or could act as a proxy for any elements of our composite outcome, including those related to administration of intravenous metronidazole, sodium chloride bolus, vancomycin enema, vancomycin-resistant Enterococcus culture (VRE), and methicillin-resistant Staphylococcus aureus culture (MRSA), as well as assignment to medical critical care units/wards. We removed VRE and MRSA cultures, as they are strong proxies for any-cause admission to the intensive care unit.
For every case, we considered data collected on the day of the prediction, in addition to the 2 days leading up to the prediction, with the goal of capturing recent trends. We represented all the data from this time period as a vector of binary features. These data included time-invariant features such as county of residence and time-varying features such as systolic blood pressure. In the case of categorical data like hospital unit/ward, we mapped this information to binary features indicating whether the patient had been in that hospital location during the 3-day period. For continuous vital signs (eg, respiratory rate), we used expert-defined ranges to map the variable to binary features indicating the presence of any low, normal, or high measurements during the 3-day period. We discretized other continuous variables into quintiles, mapping quintiles to a binary feature (eg, having an age in the first quintile was mapped to a binary feature). Some variables exhibited a significant amount of skew (eg, many patients with 0 previous encounters in the past 90 days). In such cases, we grouped any nonunique quintiles together. EHR data are typically not missing at random. Thus, we included additional features encoding missingness for each variable and did not perform data imputation or case-wise deletion.
In addition to these EHR data, we also considered the manually curated set of variables used in Rao et al. [13]. These data included 1 demographic characteristic (age) and information about the current hospitalization (eg, high-creatinine flag and metastatic cancer comorbidity). The curated feature set did not include information about previous hospitalizations, prior CDI exposure, or patient location within the hospital. However, it did include information about cancer diagnoses more than 90 days before the current admission, unlike the EHR model. For vital signs and laboratory results, each feature corresponded to either the minimum or maximum measurement taken within 48 hours of diagnosis. We processed these features like in the EHR model and concatenated them into a binary feature vector to be used as input to the predictive model. This yielded 2 different data representations (curated vs EHR) that could be used in making predictions.
Model Training and Evaluation
We trained models using the curated data, the EHR data, and a combination of both. To train and evaluate the predictive models, we separated cases into train and held-out test partitions. We split the cases temporally, training the model on the earliest 80% of the data and evaluating on the most recent 20%. Compared with separating the cases randomly into train and held-out partitions, a temporal split better emulates how a predictive model may perform during prospective deployment.
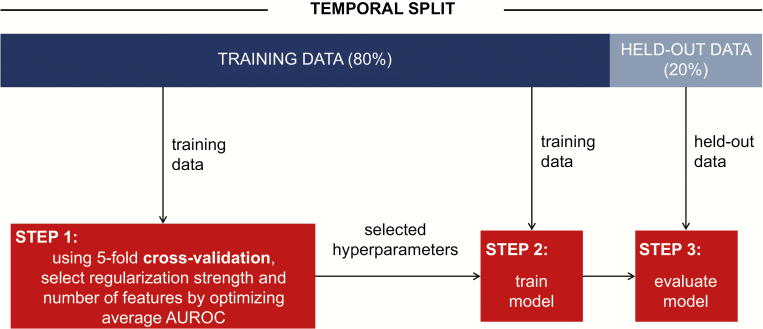
Given the high dimensionality of our data set and the potential to overfit, we used L2 regularization and k-best feature selection (explained below). We selected the regularization hyperparameter C from 10–5, 10–4, …, 105 using 5-fold cross-validation on the training set with 5 temporally defined folds [17]. We used the same process to fit k, a hyperparameter selecting the number of features to keep, after ranking features based on how related they were with the outcome (calculated with Pearson’s chi-square statistic). We optimized k over a range that spanned 25–100 with step size 25, 100–1500 with step size 100, and 1500–4000 with step size 500. The final model was trained using the C and k that led to the best average discriminative performance across training set folds. Applied to the held-out test data, we measured the discriminative performance of the model in terms of the area under the receiver operating characteristic curve (AUROC). We also compared the models’ sensitivity, specificity, and average precision. We computed empirical 95% confidence intervals using 10 000 bootstrapped samples of the held-out set. We repeated the process above for each feature representation (curated, EHR, and a combination) and each possible prediction time. This resulted in 5 different models. Figure 1 illustrates our model training and evaluation pipeline. All analyses were performed in Python (Python 3.6), and our code is available at https://gitlab.eecs.umich.edu/mld3/complicated_cdi_prediction.
Figure 1.
Predictive model training and evaluation flowchart. We split the data 80%/20% temporally, performed 5-fold cross-validation on the training data to select hyperparameters (step 1), and then, using these hyperparameters and all of the training data, learned a model (step 2). The model was evaluated on a held-out set of data (not used in selecting the model). Abbreviation: AUROC, area under the receiver operating characteristic curve.
RESULTS
Patient Characteristics
Of the cases in the original cohort from Rao et al. [13], 26 (<2.5%) could not be matched with data in the Research Data Warehouse. After excluding these cases, the final patient cohort involved 1118 cases of CDI, 89 (8%) of which were complicated CDI. The median length of stay was 9 days, and median time of CDI diagnosis was on the third day of hospitalization (Table 1).
Table 1.
Selected Patient Characteristics of Our Study Cohort
| Characteristic | Median (IQR) or No. (%) |
|---|---|
| No. of CDI cases | 1118 |
| No. of patients | 966 |
| Age, y | 59 (46–69) |
| Female gender | 611 (54.7) |
| LOS, d | 9 (5–17) |
| Day of CDI diagnosis | 3 (2–7) |
| Charlson-Deyo score | 2 (0–3) |
| Inflammatory bowel disease diagnosed in the past 90 da | 20 (1.8) |
| Solid organ transplant | 179 (16.0) |
| Concurrent non-CDI antimicrobial use | 745 (66.6) |
| Fluoroquinolone use from admission to diagnosis | 378 (33.8) |
| Proton pump inhibitor use | 775 (69.3) |
| Prior CDI within the past year | 262 (23.4) |
| Prior CDI within the past 90 d | 182 (16.3) |
| Failed initial CDI therapy within the past 14 d | 22 (2.0) |
| BMI, kg/m2; missing No. (%) | 26.3 (22.6–31.6); 131 (11.7) |
aICD-9-CM codes 555 or 556.
Abbreviations: BMI, body mass index; CDI, Clostridium difficile infection; IQR, interquartile range; LOS, length of stay.
Model Training
Models were trained on a training set of 894 cases and tested on a held-out set of 224 cases. As in Rao et al., the curated model used 23 features [13]. We extracted 4271 features from the EHR. After feature selection, the EHR models for making a prediction at diagnosis, 1 day after, and 2 days after, retained 3000, 800, and 900 features, respectively. Regarding regularization strength, the EHR models used C values of 10–3, 10–4, and 10–3, respectively.
Model Performance
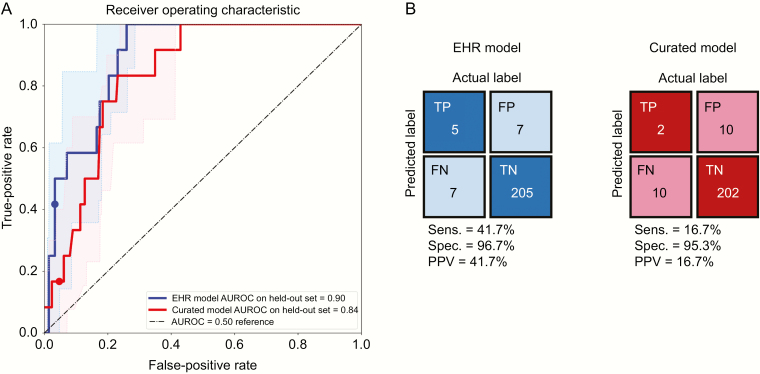
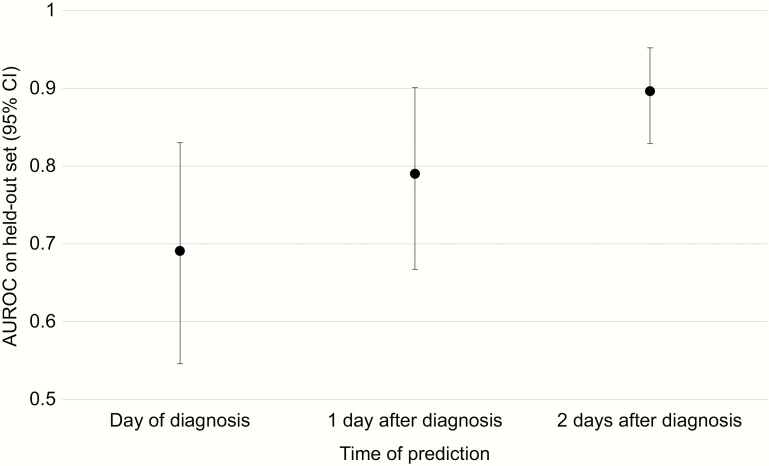
Two days after CDI diagnosis, the EHR model resulted in better discriminative performance compared with the curated model (0.90; 95% CI, 0.83–0.95; vs 0.84; 95% CI, 0.75–0.92). The EHR model also had greater average precision (AUROC, 0.30; 95% CI, 0.15–0.60; vs AUROC, 0.25; 95% CI, 0.09–0.47) (Supplementary Figure 1). At a threshold based on the 95th percentile of risk for each model, the EHR model had greater specificity (96.7% vs 95.3%) and sensitivity (41.7% vs 16.7%) (Figure 2). Combining the curated features with the EHR features did not improve discriminative performance 2 days after diagnosis (Table 2). Model performance decreased when tasked with making predictions earlier: AUROC, 0.79; 95% CI, 0.67–0.90 1 day after diagnosis and AUROC, 0.69; 95% CI, 0.55–0.83 at diagnosis (Figure 3). Removing several variables clearly encoding clinical suspicion of complicated CDI did not substantially change model performance at any prediction time point (Supplementary Figure 2). Examining the learned coefficients, on the day of CDI diagnosis, high respiratory rate and obtaining an adult blood culture were strongly associated with development of complicated CDI (Table 3). Two days later, the factors most associated with risk included high and low respiratory rate, low systolic blood pressure, and low blood CO2. At both time points, normal respiratory rate and young age were associated with protection.
Figure 2.
Discriminative performance (A) and confusion matrices (B) of predictive models for a complicated course of CDI 2 days after diagnosis using either electronic health record (EHR)–based or curated features. Shaded regions in (A) represent empirical 95% confidence intervals. The EHR model dominates the curated model for nearly all values of specificity (1-FPR). At a threshold based on the 95th percentile of risk for each model (marked with dots), the EHR-based model yields better sensitivity, specificity, and positive predictive value. Abbreviations: CDI, Clostridium difficile infection; FN, false negative; FP, false positive; FPR, false-positive rate; PPV, positive predictive value; TN, true negative; TP, true positive.
Table 2.
Comparison of EHR, Curated, and Merged Models for Complicated CDI 2 Days After Diagnosis
| Model | No. of Features | AUROC on Held-Out Set (95% CI) |
|---|---|---|
| Curated | 23 | 0.84 (0.75–0.92) |
| EHR | 900 | 0.90 (0.83–0.95) |
| EHR + curated | 923 | 0.88 (0.81–0.95) |
Abbreviations: AUROC, area under the receiver operating characteristic curve; CDI, Clostridium difficile infection; CI, confidence interval; EHR, electronic health record.
Figure 3.
Discriminative performance of electronic health record–based complicated CDI model across time. As more data become available and are integrated into the model, the model achieves better discriminative performance. Error bars represent empirical 95% confidence intervals. Abbreviations: AUROC, area under the receiver operating characteristic curve; CDI, Clostridium difficile infection; CI, confidence interval.
Table 3.
Predictive Features for Complicated CDI at Diagnosis and 2 Days After
| Day of Diagnosis | 2 Days After Diagnosis | ||
|---|---|---|---|
| Rank | Associated With Higher Estimated Risk | Rank | Associated With Higher Estimated Risk |
| 1 | Obtaining adult blood culture | 1 | High respiratory rate (>20 breaths/min) |
| 2 | High respiratory rate (>20 breaths/min) | 2 | Low respiratory rate (<12 breaths/min) |
| 3 | Low blood CO2 (<23 mmol/L; Q 1/5) | 3 | Low systolic BP (<90 mmHg) |
| Rank | Associated With Lower Estimated Risk | Rank | Associated With Lower Estimated Risk |
| 1 | Normal respiratory rate (12–20 breaths/min) | 1 | Missing PTT measurement |
| 2 | Low RDW (<14.6%; Q 1/5) | 2 | Normal respiratory rate (12–20 breaths/min) |
| 3 | Age (<41 years old; Q 1/5) | 3 | Missing phosphorous level measurement |
Please see Supplementary Table 2 for the model coefficients associated with these features.
Abbreviations: BP, blood pressure; CDI, Clostridium difficile infection; PTT, partial thromboplastin time; RDW, red cell distribution width.
DISCUSSION
Automated patient risk stratification techniques that leverage EHR data have the potential to provide critical support for clinical decision-making. In contrast to risk estimates that depend on manual chart review, risk estimates derived from the EHR can be automatically generated and updated throughout a patient’s hospitalization. In the context of CDI, predictive models have been developed for identifying patients at high risk for infection, recurrence, and complications [18–20]. We have previously shown how a model that leverages the structured contents of the EHR can outperform a curated model in predicting CDI, and we have demonstrated how these techniques can generalize across hospitals with different patient populations and different EHR systems [21, 22]. In the context of recurrent CDI, Escobar et al. compared machine learning and curated risk stratification models, concluding that neither approach could reliably predict first recurrence [18]. In this work, we focus on estimating patient risk for a complicated course of CDI.
The task of predicting complicated CDI is difficult, and current treatment guidelines do not incorporate data-driven estimates of risk. To this end, we sought to develop a systematic way to improve our ability to identify complicated cases by considering all structured data available in the EHR. This work represents a first step in that direction. We demonstrate that, using a machine learning approach, it is possible to accurately risk-stratify patients for complications early on in the course of disease.
The development and validation of EHR-based risk stratification models for predicting complicated CDI could eventually help clinicians tailor treatments to individuals. On the day of CDI diagnosis, a patient’s estimated risk for complications could serve as an adjunct, easily obtainable resource for clinical decision support. Treatment decisions such as whether to use high-dose vancomycin or perform a loop ileostomy with antegrade vancomycin infusions [23] often do not occur until complicated CDI has already set in. In severe cases, early aggressive therapy can positively impact the course. However, invasive treatments such as enemas (fecal microbiota transplantation or vancomycin) and surgery are optimally used in only select patients, and such decisions lack the rigorous guidelines associated with initial treatment.
In addition to potentially guiding more drastic and invasive treatment, accurate risk prediction for complicated CDI could guide initial antibiotic therapy. The new US guidelines on treating CDI promote vancomycin or fidaxomicin over metronidazole [24]. In the inpatient setting, this shifts nearly all cases to vancomycin therapy due to the expense of fidaxomicin. This, in turn, could increase VRE selection pressure and disrupt the microbiome, leading to subsequent infections [25]. As even more narrow-spectrum CDI treatments in the pipeline become available [26], a model that could enable targeted vancomycin or fidaxomicin use in a cost-effective way could help alleviate these potential adverse consequences.
Though identifying effective interventions that prevent or mitigate complicated CDI would require extensive additional studies, tools like the one developed here play an important role. The statistical power of cohort studies and randomized controlled trials (RCTs) is often limited when the primary outcome is infrequent. Risk stratification tools can help target patients with a higher risk of adverse outcomes and increase the feasibility of such important trials.
Based on thousands of variables, the EHR-based model provided better risk estimates than one that relied on a curated set alone. In this study, we sought to test the feasibility of a machine learning approach to generate early and accurate predictions of a patient’s risk for complicated CDI; reassuringly, the model features with highest weight align with those a clinician would identify. Even if such models do not identify new risk factors, they consider a much larger set of patient characteristics than any 1 clinician can consider simultaneously. As the amount of data collected in hospitals continues to increase, it will be important to equip clinicians, who are already faced with many competing priorities, with the tools necessary to identify patterns and distill the data into actionable knowledge. For example, operating in the background, a model for predicting complications could alert the care team if a patient’s risk increases unexpectedly. In parallel, if many patients in the same hospital unit are identified as being at heightened risk, this could inform hospital unit/ward cleaning practices or strategic transferring of patients to private rooms.
These results should be interpreted in the context of several limitations. First, given that these are merely associations, additional investigation is required to establish the direction of the true underlying relationships (eg, through RCTs). Moreover, despite the removal of many variables that could serve as proxies for our composite outcome, some model features may not be true risk factors but rather markers for the beginning of complicated CDI itself. This may be elucidated by tracking the evolution of a patient’s risk over multiple days of their hospitalization; automated EHR models enable these kinds of future analyses. Second, our analysis is retrospective. As a following step, prospective studies are necessary for understanding how such models perform in real time. Third, the results are based on a small sample size from a single institution. Though this has implications regarding the generalizability of the model, it does not diminish the generalizability of the approach. As patient populations, clinical protocols, and in turn risk factors can vary across institutions, we encourage researchers to focus on tailoring models to the populations in which the model will ultimately be deployed [22]. On a related note, the definition of a complicated course of CDI is not universal. We used the CDC surveillance definition [16], the same definition as in Rao et al., which includes a stipulation that the specific adverse outcomes be attributable to CDI (requiring clinical review). Future work should investigate the appropriateness of definitions of complicated CDI derived solely from the EHR. Fourth, we cannot exclude the possibility that patients may experience the outcome at another hospital, and thus we may have potentially underestimated the extent of complications from CDI. In addition, although CDI testing was recommended only for symptomatic patients during our study period and this was further validated by chart review, some positive CDI tests might reflect asymptomatic carriers. We hypothesize that our model learns, in part, to differentiate asymptomatic carriers from those who will experience complicated disease. Finally, beyond the structured content, the EHR contains data from pathogen genomic sequencing, free-text clinical notes, and/or wearable technology that could provide a more complete picture of a patient’s clinical state. For example, the Xpert C. difficile diagnostic test has a gene target that can report whether the strain is NAP1 or non-NAP1. In the future, integrating both biological and administrative data like these has potential for improving models of disease progression.
In summary, we demonstrated how a machine learning approach could be used to learn an EHR-based predictive model for accurately estimating a patient’s risk of developing complicated CDI. Our approach leverages thousands of variables that can be readily extracted from the EHR. This approach has many potential applications, including guiding future clinical studies. Prospective evaluation and deployment of models such as these offer important opportunities to aid clinicians in real time and tailor patient therapy.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (grant No. U01AI124255).
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Centers for Disease Control and Prevention. 2015 Annual Report for the Emerging Infections Program for Clostridium difficile Infection. Atlanta: Centers for Disease Control and Prevention; 2017. [Google Scholar]
- 2. Lessa FC, Mu Y, Bamberg WM, et al. . Burden of Clostridium difficile infection in the United States. N Engl J Med 2015; 372:825–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ma GK, Brensinger CM, Wu Q, Lewis JD. Increasing incidence of multiply recurrent Clostridium difficile infection in the United States: a cohort study. Ann Intern Med 2017; 167:152–8. [DOI] [PubMed] [Google Scholar]
- 4. Lessa FC, Gould CV, McDonald LC. Current status of Clostridium difficile infection epidemiology. Clin Infect Dis 2012; 55(Suppl 2):S65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Depestel DD, Aronoff DM. Epidemiology of Clostridium difficile infection. J Pharm Pract 2013; 26:464–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barbut F, Rupnik M. Editorial commentary: 027, 078, and others: going beyond the numbers (and away from the hypervirulence). Clin Infect Dis 2012; 55:1669–72. [DOI] [PubMed] [Google Scholar]
- 7. Evans CT, Safdar N. Current trends in the epidemiology and outcomes of Clostridium difficile infection. Clin Infect Dis 2015; 60(Suppl 2):S66–71. [DOI] [PubMed] [Google Scholar]
- 8. Bagdasarian N, Rao K, Malani PN. Diagnosis and treatment of Clostridium difficile in adults: a systematic review. JAMA 2015; 313:398–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stevens VW, Nelson RE, Schwab-Daugherty EM, et al. . Comparative effectiveness of vancomycin and metronidazole for the prevention of recurrence and death in patients with Clostridium difficile infection. JAMA Intern Med 2017; 177:546–53. [DOI] [PubMed] [Google Scholar]
- 10. Deshpande A, Hurless K, Cadnum JL, et al. . Effect of fidaxomicin versus vancomycin on susceptibility to intestinal colonization with vancomycin-resistant enterococci and Klebsiella pneumoniae in mice. Antimicrob Agents Chemother 2016; 60:3988–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gerding DN. Is there a relationship between vancomycin-resistant enterococcal infection and Clostridium difficile infection? Clin Infect Dis 1997; 25(Suppl 2):S206–10. [DOI] [PubMed] [Google Scholar]
- 12. Na X, Martin AJ, Sethi S, et al. . A multi-center prospective derivation and validation of a clinical prediction tool for severe Clostridium difficile infection. PLoS One 2015; 10:e0123405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rao K, Micic D, Natarajan M, et al. . Clostridium difficile ribotype 027: relationship to age, detectability of toxins A or B in stool with rapid testing, severe infection, and mortality. Clin Infect Dis 2015; 61:233–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Henrich TJ, Krakower D, Bitton A, Yokoe DS. Clinical risk factors for severe Clostridium difficile-associated disease. Emerg Infect Dis 2009; 15:415–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cobo J, Merino E, Martínez C, et al. ; Nosocomial Infection Study Group. Prediction of recurrent Clostridium difficile infection at the bedside: the GEIH-CDI score. Int J Antimicrob Agents 2018; 51:393–8. [DOI] [PubMed] [Google Scholar]
- 16. McDonald LC, Coignard B, Dubberke E, et al. Ad Hoc Clostridium difficile Surveillance Working Group. Recommendations for surveillance of Clostridium difficile-associated disease. Infect Control Hosp Epidemiol 2007; 28:140–5. [DOI] [PubMed] [Google Scholar]
- 17. Roberts DR, Bahn V, Ciuti S, et al. . Cross validation strategies for data with temporal, spatial, hierarchical, or phylogenetic structure. Ecography 2017; 40:913–29. [Google Scholar]
- 18. Abou Chakra CN, Pepin J, Sirard S, Valiquette L. Risk factors for recurrence, complications and mortality in Clostridium difficile infection: a systematic review. PLoS One 2014; 9:e98400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Escobar GJ, Baker JM, Kipnis P, et al. . Prediction of recurrent Clostridium difficile infection using comprehensive electronic medical records in an integrated healthcare delivery system. Infect Control Hosp Epidemiol 2017; 38:1196–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hebert C, Du H, Peterson LR, Robicsek A. Electronic health record-based detection of risk factors for Clostridium difficile infection relapse. Infect Control Hosp Epidemiol 2013; 34:407–14. [DOI] [PubMed] [Google Scholar]
- 21. Wiens J, Campbell WN, Franklin ES, et al. . Learning data-driven patient risk stratification models for Clostridium difficile. Open Forum Infect Dis 2014; 15;1(2)ofu045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Oh J, Makar M, Fusco C, et al. . A generalizable, data-driven approach to predict daily risk of Clostridium difficile infection at two large academic health centers. Infect Control Hosp Epidemiol 2018; 39:425–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Neal MD, Alverdy JC, Hall DE, et al. . Diverting loop ileostomy and colonic lavage: an alternative to total abdominal colectomy for the treatment of severe, complicated Clostridium difficile associated disease. Ann Surg 2011; 254:423–7; discussion 7–9. [DOI] [PubMed] [Google Scholar]
- 24. McDonald LC, Gerding DN, Johnson S, et al. . Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis 2018; 66:987–94. [DOI] [PubMed] [Google Scholar]
- 25. Baggs J, Jernigan JA, Halpin AL, et al. . Risk of subsequent sepsis within 90 days after a hospital stay by type of antibiotic exposure. Clin Infect Dis 2018; 66:1004–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dieterle MG, Rao K, Young VB. Novel therapies and preventative strategies for primary and recurrent Clostridium difficile infections. Ann N Y Acad Sci 2019; 1435:110–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.