Abstract
Introduction:
The placenta is one of the least understood, yet arguably one of the most important organs for human health and development. While there have been numerous research efforts dedicated to understanding the placenta’s critical role, these studies and the data they produced remain separated and largely disparate. In order to facilitate placental research, the Eunice Kennedy Shriver National Institute of Child and Human Development (NICHD) released in October 2018 the Placental Atlas Tool (PAT) (https://pat.nichd.nih.gov/), an internet-based platform offering users a centralized placental database of molecular datasets, analytic tools, and images.
Methods:
PAT is a cloud-based system developed by the business requirements defined by NICHD leadership and extramural placental researchers. PAT employs a metadata-driven web interface to provide curated placental datasets and images, enriched with structured, descriptive metadata to enhance data discoverability. PAT also incorporates open source molecular data analytical tools to provide a flexible analytics workflow for placental researchers.
Results:
PAT launched with 426 analyzable molecular placental datasets consisting of over 12,500 samples from 10 distinct species, all systematically annotated and processed for enhanced research utility. 828 placental images, consisting of 7 imaging modalities across 47 species, and nearly 300 annotated linked publications supplement the datasets to facilitate knowledge integration and hypothesis generation across disparate molecular studies.
Discussion:
PAT will maximize the NICHD’s investment in placental research by reinforcing open scientific inquiry, facilitating reuse of datasets, promoting novel research and testing of new hypotheses and analytic methods, and facilitating education of new researchers.
Introduction
The placenta is one of the least understood human organs, yet is considered to be one of the most important organs for human health and disease [1–3]. The placenta is implicated in many pregnancy disorders and even subtle placental abnormalities can impact the health of both mother and her fetus later in life [1,3]. The Eunice Kennedy Shriver National Institute of Child and Human Development (NICHD) launched the Human Placenta Project [2,4,5] as a collaborative research effort dedicated to addressing the critical need for a better, real-time assessment of human placental structure and function in health and disease during the course of pregnancy [6]. While this and other efforts are fueling much-needed attention and funding for placental research [2,4,7], there is no substantial, internet-based resource for the robust existing molecular placental research data and analytic tools [8], as there are for other research fields [9–12]. This effectively slows the progress of placental research by placing the responsibility on researchers to spend precious time and resources scanning through disparate data resources to perform rigorous scientific investigations.
The Placental Atlas Tool (PAT) (https://pat.nichd.nih.gov/) has been developed to create, maintain, and update a shared research analytics platform to serve as an important resource for the placental research community. This effort relies heavily on input from a panel of non-government, placental researchers, called the External Scientific Consultant Panel (ESCP), to align development of PAT with the needs and expectations of the placental research community. The ESCP is made up of scientists and clinicians who are experts in placental biology and creative thinkers in adjacent fields, such as biotechnology, imaging, and data science. The ESCP members are listed in the Acknowledgements section. Together the panel provides subject matter expertise throughout the PAT development process, evaluates progress, and assists with refinement of the system.
PAT provides technologies that allow researchers to visualize molecular characteristics of placental function and development, from gross anatomy to the molecular biology. PAT leverages existing resources to identify knowledge gaps, develop new hypotheses to address those gaps, and identify potential therapeutic targets and biomarkers. PAT has been created as a cloud-based resource for the scientific community leveraging applicable components, both technical and non-technical, of another NICHD internet-based resource called the Data and Specimen Hub (DASH) [13,14], as well as developing new functionality to enable molecular data investigation.
PAT addresses a need for scalable and agile systems integration, supporting NICHD’s organizational goals and their user community, and maximizing research investment through data discoverability and reuse. This work closely aligns with goals of the new National Institutes of Health (NIH) Strategic Plan for Data Science [15]. This paper details the system functionality that users will encounter when accessing the PAT system and sheds light on the software, terminology, and content development processes to better inform the placental research community of users.
Methods
PAT Technical Implementation
The PAT system employs a microservice architecture methodology to be scalable, extensible, and data-driven to adapt to changing scientific and business needs of the placental research community. The technical components utilize an Amazon Web Services (AWS) cloud environment, ElasticSearch for data cataloging, metadata indexing and rapid querying, and a JavaScript-based User Interface using open source software for analytics and visualizations. An Apache Jena Triple database is used as the semantic framework for terminologies, and a Postgres database is used to store metadata describing scientific content as well as information related to workspaces and user management.
Terminology-based Placental Content Integration
PAT was designed to be metadata-driven and built around standardized terminologies of concepts relevant to placental research. The process of content extraction, annotation and validation is described in Figure 1. All terminologies utilized in PAT were either extracted from publicly available resources, developed by a PAT ESCP member, or developed by the PAT content team. The Placental Pathology Lesion Classification terminology was provided by placental pathologist Dr. Raymond Redline as a continuation of the Amsterdam Placental Lesion Consensus Group [16,17]. Custom PAT terminologies were developed by the PAT content team based on existing ontological resources, supplemented with additional placental concepts, and vetted by members of the ESCP. Following annotation of data entities, modifications to existing terminologies were informed by rounds of manual annotation validation. Additional terminologies were extracted from external ontological sources including the NCI Thesaurus and BioPortal [18]. These external source ontologies were pared down to focus on terms most relevant to placental research. See Table 1 for a full listing of the placental terminologies developed and implemented as of November 2018.
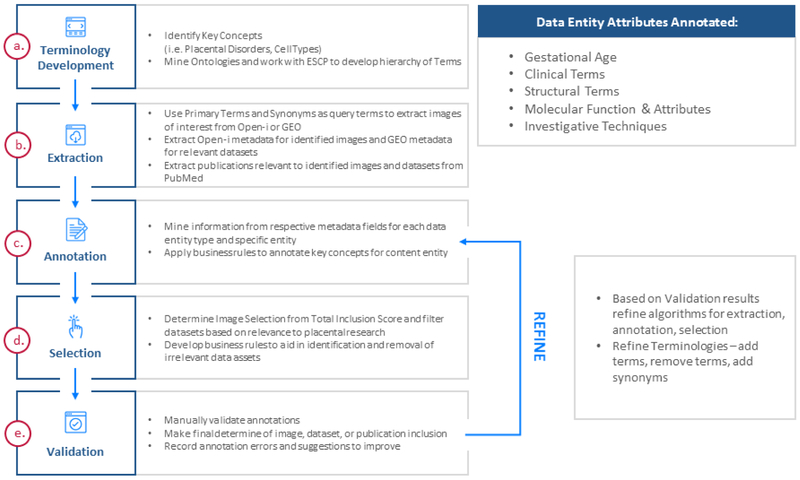
Figure 1.
PAT Content Extraction, Annotation, and Validation Process. The PAT content ingest process starts with terminology development and involved manual validation of all steps in the process. Findings during the manual validation process are incorporated into revisions of the automated content identification, extraction, and annotation process for future iterations.
Table 1.
: PAT Terminologies
| Group | Category | Terminology | Source |
|---|---|---|---|
| Clinical | Pathology | Placental Lesion Classification | Dr. Raymond Redline |
| Disorder | Disease Ontology | http://disease-ontology.org/ | |
| NCI Thesaurus Placental Disorders | https://ncit.nci.nih.gov/ | ||
| NCI Thesaurus Pregnancy Disorders | https://ncit.nci.nih.gov/ | ||
| SNOMED Placental Disorders | http://browser.ihtsdotools.org/ | ||
| SNOMED Pregnancy Complications & Disorders | http://browser.ihtsdotools.org/ | ||
| SNOMED Trophoblastic Neoplasm | http://browser.ihtsdotools.org/ | ||
| Finding | Human Phenotype Ontology | https://hpo.jax.org/app/ | |
| NCI Thesaurus Placental Findings | https://ncit.nci.nih.gov/ | ||
| NCI Thesaurus Pregnancy Findings | https://ncit.nci.nih.gov/ | ||
| SNOMED Placental Findings | http://browser.ihtsdotools.org/ | ||
| Structural | Anatomy | NCI Thesaurus Placental Anatomy | https://ncit.nci.nih.gov/ |
| SNOMED Placental Anatomy | http://browser.ihtsdotools.org/ | ||
| Cell Type | PAT Cell Type Ontology | PAT Content Team and ESCP | |
| Molecular Function & Attributes | Genetic Variance | PAT Genetic Variance | PAT Content Team and ESCP |
| Biological Process | NCI Thesaurus Biological Processes | https://ncit.nci.nih.gov/ | |
| Methods, Procedures, & Techniques | Investigative Technique | PAT Investigative Technique | PAT Content Team and ESCP |
After terminologies were developed and loaded into the semantic database, the terminology concepts were used to query publicly available databases for relevant placental content (Figure 1b). PAT datasets were derived from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) [19,20] through a query for all datasets containing placenta-related terminology terms such as “placenta”, “choriocarcinoma”, and “trophoblast”. For inclusion in PAT, datasets were required to be from placental species, have at least two samples for each of at least two experimental conditions, and have identifiable transcribed genes associated with each microarray probe. Thresholds for methodological rigor or data quality are not considered in selection of datasets for PAT. Publications associated with extracted molecular datasets were derived from the PubMed repository and linked in PAT to provide context for those datasets. Images were extracted from National Library of Medicine (NLM) Open-i [21] using terminology concepts as query terms. Additional images were provided by Dr. Raymond Redline from the recent Placental and Gestational Pathology textbook [22].
Programmatic Annotation of Placental Content
Datasets, images, and publications were annotated using the terminologies described above. Terminologies were grouped into broad categories, such as Disorders or Anatomy, and annotations of data entities for each category followed a distinct set of business rules (Figure 1c). For example, when annotating datasets with Investigative Technique concepts only the Experiment Type, Overall Design, and select Sample metadata fields were considered. Annotation began at the most granular concepts of each terminology and proceeded in reverse topological order. Parental lineage nodes were excluded for found concepts to represent ideas by the most granular terminological concepts. If “microRNA Microarray” and “Microarray” were both annotated for a single dataset, only the “microRNA Microarray” annotation was kept as it is a descendant of “Microarray”. Equivalent concepts from other terminologies were automatically added to the set of annotations.
Following text annotation, a selection step filtered out irrelevant images and datasets based on an inclusion score calculated by the automated annotation tool (Figure 1d). Annotated concept sets were then augmented via a custom set of cross-category rules which considered the resource metadata other than text, such as image modality or dataset experiment type. Across all manual validation efforts, automated annotation errors were recorded, and annotation business rule and terminology refinements were implemented to improve subsequent rounds of data ingest and annotation (Figure 1e).
Genetic Analysis of Placental Datasets
Molecular datasets were run through a backend preprocessing pipeline, followed by ingestion of experimental data and metadata that is served up to cloud-based analytical tools and user interface. Placental gene expression datasets were preprocessed using a combination of open-source R software packages, Snakemake, and Python. For example, Figure 2 illustrates the process for RNA sequencing (RNA-Seq) datasets. Preprocessed data were then loaded into the PAT database to be easily discovered and analyzed by users with embedded web-based analytical tools.
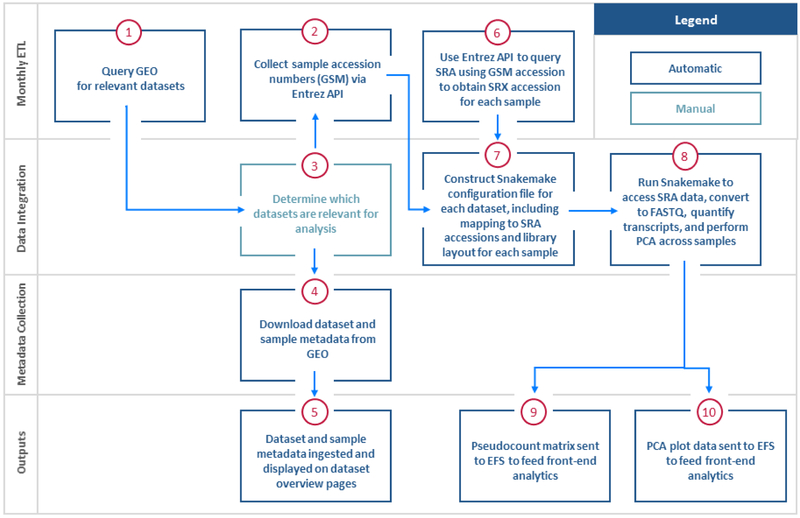
Figure 2.
PAT Genomic Dataset Preprocessing: RNA-Seq. RNA-Seq pre-processing begins with the identification of datasets that are of relevance to placental research through a structured query in GEO. The data and metadata for relevant datasets are extracted from GEO. High-throughput data from SRA and output through PAT. This output includes the ability for users to download read/transcript quantification files, PCA widget data, and gene level abundance matrix data for analytics.
The private analytics backend microservice serves up R functions as API calls to the public analytics frontend microservice, allowing for seamless integration of R computations and JavaScript visualizations. This type of architecture is highly extensible, allowing for easy implementation of additional open source packages, tools, and quantitative analysis algorithms. A full list of external software libraries and tools utilized for PAT analytics can be found under Resources on the PAT website.
Results
PAT was released in October 2018 as a resource freely accessible to all researchers. PAT is based on user-centric design principles and places emphasis on a flexible, data-driven experience that enables discovery from multiple user perspectives. All users are welcomed by the PAT homepage when visiting the site (Figure 3). From the homepage, users may navigate to the resource that best fits their needs. Placental researchers can view the Terminology Browser to explore standardized placental terminologies and their associated data, view images related to specific placental diseases through the Image Explorer or analyze placental gene expression datasets through the Dataset Explorer. In addition, there are educational and informational resource pages available to aid placental researchers.
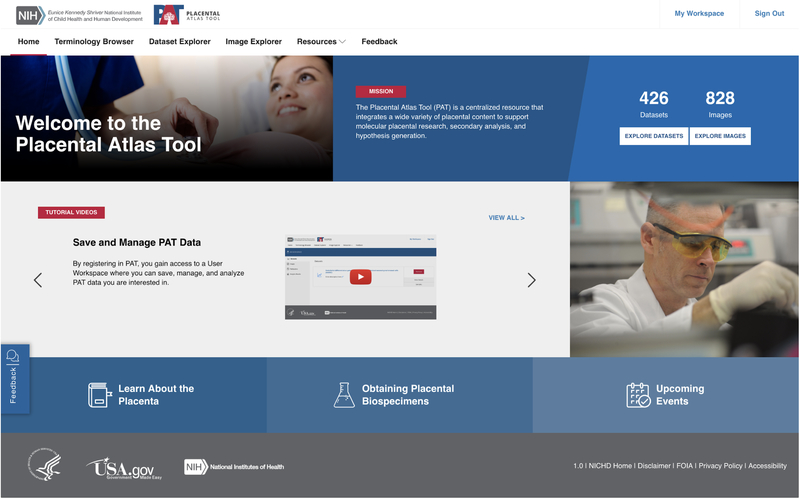
Figure 3.
PAT Homepage. Users will be brought to the PAT Homepage to start their journey. The panels of the homepage display important information such as the tutorials, placental related resources, and information on the PAT tool. The navigation bar at the top left allows users to explore different aspects of PAT functionality, including the Terminology Browser, Image Explorer, Dataset Explorer, and additional PAT resources.
Terminology Browser
The terminologies within the Terminology Browser form the basis of the metadata-driven explorers and content utilized throughout the site. These concepts are used to query and extract relevant datasets and images, to annotate extracted data entities, and can be used by researchers to create informative linkages between distinct data entities. The terminologies within cover an array of broad terminological categories including clinical, structural, molecular, and methodological terms. Each terminology contains a list of concepts. When a user selects a given concept, they are presented with information on that concept including a definition, term source, and synonyms. The preferred terms and synonyms are used for annotation of all data entities in the PAT system, thereby semantically linking them to a given concept. Datasets and images that have been tagged with a given concept can be accessed through the concept page in the terminology browser. See Table 1 for a complete breakdown by terminology grouping.
Future functionality will include global search across all terminologies in the PAT system and explicit linkages between concepts of different terminologies to help streamline the automated annotation process and increase the knowledge integration functionality of metadata annotations.
Image Explorer
The PAT Image Explorer contains 828 placental images from 47 different placental species. Seven broad categories of image types, including microscopy and ultrasound images, are annotated with concepts from the PAT Terminologies. Enriched metadata allows users to search and sort through images of everything from images of a trophoblast cell line expressing markers of interest to histological cases of placenta previa. These images are currently derived from either the Placental and Gestational Pathology textbook [22] or NLM Open-i [21], with plans to expand to additional images sources.
Planned future functionality for the image explorer includes secondary analysis of whole-mount immunofluorescence (IF) images by segmenting and quantitating images, displaying them in the browser, and adding an overlay for selection and analysis. Additional plans include development of an interactive 3D placenta for visualization of gene expression in the healthy and diseased placenta.
Dataset Explorer
The Dataset Explorer presently allows users to search through 426 analyzable molecular datasets through either direct query or faceted browsing. These datasets contain data from more than 12,500 individual samples produced on nearly 180 different molecular profiling platforms. The dataset overview page contains basic information on each dataset, including dataset title and overall design, linked publications details, dataset sample data, and a PCA overview widget to aid sample selection for analytics. New datasets will be added monthly as they become available.
Currently, PAT only supports analysis of microarray and RNA-Seq datasets, but functionality will continue to expand to support additional dataset types in the future, including chromatin immunoprecipitation sequencing (ChIP-Seq) and single cell RNA-Seq. PAT will also integrate datasets from other sources, such as summary level transcript abundances from the database of Genotypes and Phenotypes (dbGaP).
Analytics Workflow
PAT makes bioinformatic analysis of large molecular datasets accessible to all users from the placental research community, regardless of bioinformatic experience. After dynamically selecting two groups of samples, users can run the dataset analytics, which provides bioinformatic outputs including differential gene expression, interactive Clustergrammer [23] gene expression heatmaps, enriched gene sets, as well as visualizations of enriched Reactome pathways [24]. Users can download results, tables and images for each analytics process for reuse in applications such as scientific talks, publications, or grant applications. While the PAT analytics service has been designed to facilitate analysis of a wide variety of datasets and datatypes the quality of datasets is not directly assessed as a prerequisite for inclusion. For this reason, users are encouraged to treat these services as supportive for hypothesis generation for designing more tailored experiments and analyses to better test these hypotheses for their own research purposes.
In the future, we plan to support the ability to perform multiple pair-wise comparisons within a single dataset and meta-analysis of samples across multiple datasets. Coupled with the Workspace functionality discussed below, this will further enhance the investigative and integrative nature of the PAT platform.
Workspace
Users can save entities of interest from the Image and Dataset Explorers by adding them to their personal Workspace. Once saved to the Workspace users can assign personalized titles, descriptions, and annotations to each of their saved items for subsequent reference. In the future, the workbench will provide storage and annotation of analytic results as well as storage and reuse of previous search parameters. This will allow users to more easily monitor changes to their area of interest throughout periodic content expansions. We expect that future enhancements will enable researchers to form shared workspaces that facilitate collaboration and interactive discovery.
Informational Resources
While the focus of PAT is on allowing users to access a wealth of bioinformatic and secondary research utilization resources, PAT has also incorporated additional resources to aid broader aspects of the placental research community. These include information on upcoming events, such as conferences and webinars, information on obtaining placental biospecimens for reuse in their own research, and placental knowledge resources to help scientists, professionals, and students new to the placental research space.
Discussion
PAT is poised to maximize the investment NICHD and the broader research community has made in placental research by reinforcing open scientific inquiry, expediting formation of new insights by combining data sources, promoting the testing of new or alternative hypotheses and analytic methods, and facilitating education of new researchers. PAT accomplishes this by coupling cutting-edge data-driven, flexible development principles with rigorous content curation practices, ultimately creating a fluid, accessible, and robust investigational resource. The placental terminologies form the backbone of the system and create connections between previously disparate entities, exposing links between concepts or between datasets and images from separate studies. The Image Explorer allows users to explore placenta-related images and compare anatomical and molecular properties in a streamlined manner not previously possible. The Dataset Explorer brings the vast genomic resources produced by the placental research community into one accessible platform equipped with built-in analytics and visualization, thereby enriching secondary utility of datasets and facilitating discovery and hypothesis generation. All aspects of discovery and hypothesis generation are further amplified through the dedicated Workspace, which allows users to track datasets and images of interest and annotate their findings for future inquiries.
At present, the NICHD has envisioned a 5-year timeline for PAT development. Now, at the start of the third year of development the focus is shifting to ongoing system enhancements, operations and maintenance, and integration into the placental research community. In addition to new content added through monthly updates, existing content will be continually reannotated and updated as the terminologies and tools evolve over time. New focus is being placed on developing site administration and moderation features to allow for functionality such as user terminology suggestions, networking features, and Q&A sessions. See Table 2 for an overview of planned system enhancements. These planned enhancements are subject to change pending feedback from the user community and the availability of funds. Future PAT functionality is also liable to benefit from advances in machine intelligence (MI) and machine learning (ML). At present, ML is used to score and rank images for inclusion and could be further utilized to score relevance of publication annotations or detect placenta-related anomalies in clinical images. We will keep a close eye on the rapidly evolving MI space, and welcome feedback and ideas for future implementation.
Table 2.
In Development and Planned Future Functionality
| Group | Functionality | Status |
|---|---|---|
| Terminology Browser | Additional Terminologies | In Development |
| Global Search Across Terminologies | In Development | |
| Image Explorer | Secondary image analysis | Planned |
| 3D placenta | Planned | |
| Dataset Explorer | New Datasets | Updated Monthly |
| ChIP-Seq Datasets | In Development | |
| Single Cell RNA-Seq Datasets | Planned | |
| Dataset Analytics | ChIP-Seq Analysis | In Development |
| Single Cell RNA-Seq Analysis | Planned | |
| Dataset Meta-analysis | Planned | |
| Workspace | Collaborative Workspaces | Planned |
| General | Site Administration and Moderation Enhancements | Planned |
The NICHD hopes that PAT will facilitate the discovery of molecular targets that would lead to effective therapies to improve pregnancy outcomes and, ultimately, human health. As PAT has been built from the ground up as a resource for the placental research community, the PAT team encourages the community to not only utilize its resources, but also to become engaged in the evolution of the platform. Periodic user testing has been critical in guiding the development of the system, and the PAT team will continue soliciting and incorporating user feedback through the site Feedback feature and user surveys to improve functionality, scope, and user-friendliness. Frequent functionality feedback and terminology and content suggestions will be an integral part in shaping what PAT offers the placental research community over time.
Highlights:
The Placental Atlas Tool (PAT) is a free resource for the research community.
PAT is a tool for studying molecular features of placental development and function.
PAT bridges diverse datasets and images into a single research resource.
PAT incorporates open source molecular analytical tools for analytics workflows.
Investigators from all backgrounds can use PAT to generate and test new hypotheses.
Acknowledgements
This project owes its success to the dedication and commitment of the ESCP Committee: Julie Baker, Ph.D. (Stanford University), Brian Cox, Ph.D. (University of Toronto), Susan Fisher, Ph.D. (University of California, San Francisco), Raymond Redline, M.D. (Case Comprehensive Cancer Center), Antonis Rokas, Ph.D. (Vanderbilt University), Yoel Sadovsky, M.D. (University of Pittsburgh), Geetu Tuteja, Ph.D. (Iowa State University). In addition, the authors would also like to thank the members of the Booz Allen PAT Development Team and PAT Content Team for their instrumental role in the development and implementation of PAT. This project is funded under a contract mechanism (contract number GS-35F-0306J) by the NICHD to Booz Allen Hamilton, Inc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of Interest: None
References
- [1].Erlebacher A, Fisher SJ, Baby’s First Organ, Sci. Am 317 (2017) 46–53. doi: 10.1038/scientificamerican1017-46. [DOI] [PubMed] [Google Scholar]
- [2].Guttmacher AE, Maddox YT, Spong CY, The Human Placenta Project: placental structure, development, and function in real time, Placenta. 35 (2014) 303–304. doi: 10.1016/j.placenta.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cross JC, Adaptability and potential for treatment of placental functions to improve embryonic development and postnatal health, Reprod. Fertil. Dev 28 (2016) 75–82. doi: 10.1071/RD15342. [DOI] [PubMed] [Google Scholar]
- [4].Guttmacher AE, Spong CY, The human placenta project: it’s time for real time, Am. J. Obstet. Gynecol 213 (2015) S3–5. doi: 10.1016/j.ajog.2015.08.037. [DOI] [PubMed] [Google Scholar]
- [5].Human Placenta Project, Http://Www.Nichd.Nih.Gov/. (n.d.). http://www.nichd.nih.gov/research/supported/HPP/default.
- [6].Maltepe E, Fisher SJ, Placenta: The Forgotten Organ, Annu. Rev. Cell Dev. Biol 31 (2015) 523–552. doi: 10.1146/annurev-cellbio-100814-125620. [DOI] [PubMed] [Google Scholar]
- [7].Sadovsky Y, Clifton VL, Burton GJ, Invigorating placental research through the “Human Placenta Project,” Placenta. 35 (2014) 527. doi: 10.1016/j.placenta.2014.06.367. [DOI] [PubMed] [Google Scholar]
- [8].Knott JG, Paul S, Transcriptional regulators of the trophoblast lineage in mammals with hemochorial placentation, REPRODUCTION. 148 (2014) R121–R136. doi: 10.1530/REP-14-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Heng TSP, Painter MW, Consortium TIGP, Elpek K, Lukacs-Kornek V, Mauermann N, Turley SJ, Koller D, Kim FS, Wagers AJ, Asinovski N, Davis S, Fassett M, Feuerer M, Gray DHD, Haxhinasto S, Hill JA, Hyatt G, Laplace C, Leatherbee K, Mathis D, Benoist C, Jianu R, Laidlaw DH, Best JA, Knell J, Goldrath AW, Jarjoura J, Sun JC, Zhu Y, Lanier LL, Ergun A, Li Z, Collins JJ, Shinton SA, Hardy RR, Friedline R, Sylvia K, Kang J, The Immunological Genome Project: networks of gene expression in immune cells, Nat. Immunol (2008). https://www.nature.com/articles/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- [10].Pickett B, Greer D, Zhang Y, Stewart L, Zhou L, Sun G, Gu Z, Kumar S, Zaremba S, Larsen C, Jen W, Klem E, Scheuermann R, Pickett BE, Greer DS, Zhang Y, Stewart L, Zhou L, Sun G, Gu Z, Kumar S, Zaremba S, Larsen CN, Jen W, Klem EB, Scheuermann RH, Virus Pathogen Database and Analysis Resource (ViPR): A Comprehensive Bioinformatics Database and Analysis Resource for the Coronavirus Research Community, Viruses. 4 (2012) 3209–3226. doi: 10.3390/v4113209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Pickett BE, Sadat EL, Zhang Y, Noronha JM, Squires RB, Hunt V, Liu M, Kumar S, Zaremba S, Gu Z, Zhou L, Larson CN, Dietrich J, Klem EB, Scheuermann RH, ViPR: an open bioinformatics database and analysis resource for virology research, Nucleic Acids Res. 40 (2012) D593–D598. doi: 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, Conrad N, Dietrich EM, Disz T, Gabbard JL, Gerdes S, Henry CS, Kenyon RW, Machi D, Mao C, Nordberg EK, Olsen GJ, Murphy-Olson DE, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Vonstein V, Warren A, Xia F, Yoo H, Stevens RL, Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center, Nucleic Acids Res. 45 (2017) D535–D542. doi: 10.1093/nar/gkw1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hazra R, Tenney S, Shlionskaya A, Samavedam R, Baxter K, Ilekis J, Weck J, Willinger M, Grave G, Tsilou K, Songco D, DASH, the data and specimen hub of the National Institute of Child Health and Human Development, Sci. Data 5 (2018). doi: 10.1038/sdata.2018.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].NICHD DASH - Eunice Kennedy Shriver National Institute of Child Health and Human Development Data and Specimen Hub, (n.d.). https://dash.nichd.nih.gov/.
- [15].NIH Strategic Plan for Data Science | Data Science at NIH, (n.d.). https://datascience.nih.gov/strategicplan (accessed November 29, 2018). [Google Scholar]
- [16].Redline RW, Classification of placental lesions, Am. J. Obstet. Gynecol 213 (2015) S21–S28. doi: 10.1016/j.ajog.2015.05.056. [DOI] [PubMed] [Google Scholar]
- [17].Khong TY, Mooney EE, Ariel I, Balmus NCM, Boyd TK, Brundler M-A, Derricott H, Evans MJ, Faye-Petersen OM, Gillan JE, Heazell AEP, Heller DS, Jacques SM, Keating S, Kelehan P, Maes A, McKay EM, Morgan TK, Nikkels PGJ, Parks WT, Redline RW, Scheimberg I, Schoots MH, Sebire NJ, Timmer A, Turowski G, van der Voorn JP, van Lijnschoten I, Gordijn SJ, Sampling and Definitions of Placental Lesions: Amsterdam Placental Workshop Group Consensus Statement, Arch. Pathol. Lab. Med 140 (2016) 698–713. doi: 10.5858/arpa.2015-0225-CC. [DOI] [PubMed] [Google Scholar]
- [18].Whetzel PL, Noy NF, Shah NH, Alexander PR, Nyulas C, Tudorache T, Musen MA, BioPortal: enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications, Nucleic Acids Res. 39 (2011) W541–545. doi: 10.1093/nar/gkr469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Edgar R, Domrachev M, Lash AE, Gene Expression Omnibus: NCBI gene expression and hybridization array data repository, Nucleic Acids Res. 30 (2002) 207–210. http://www.ncbi.nlm.nih.gov/pubmed/11752295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A, NCBI GEO: archive for functional genomics data sets—update, Nucleic Acids Res. 41 (2013) D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Demner-Fushman D, Antani S, Simpson M, Thoma GR, Design and Development of a Multimodal Biomedical Information Retrieval System, J. Comput. Sci. Eng 6 (2012) 168–177. doi: 10.5626/JCSE.2012.6.2.168. [DOI] [Google Scholar]
- [22].Redline RW, Boyd TK, Roberts DJ, Placental and Gestational Pathology Hardback with Online Resource, Cambridge University Press, New York, NY, 2018. https://www.amazon.com/Placental-Gestational-Pathology-Diagnostic-Pediatric/dp/1316632539/ref=sr_1_1?ie=UTF8&qid=1530803978&sr=8-1&keywords=redline+placenta. [Google Scholar]
- [23].Fernandez NF, Gundersen GW, Rahman A, Grimes ML, Rikova K, Hornbeck P, Ma’ayan A, Clustergrammer, a web-based heatmap visualization and analysis tool for high-dimensional biological data, Sci. Data 4 (2017) 170151 10.1038/sdata.2017.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B, Milacic M, Roca CD, Rothfels K, Sevilla C, Shamovsky V, Shorser S, Varusai T, Viteri G, Weiser J, Wu G, Stein L, Hermjakob H, D’Eustachio P, The Reactome Pathway Knowledgebase, Nucleic Acids Res. 46 (2018) D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]