Abstract
OBJECTIVES:
Women have a 20% risk of developing a urinary tract infection (UTI) following urogynecologic surgery. This study assessed the association of post-operative UTI with bacteria in pre-operative samples of catheterized urine.
METHODS:
Immediately before surgery, vaginal swabs, perineal swabs and catheterized urine samples were collected and the V4 region of the 16S rRNA gene was sequenced. The cohort was dichotomized in two ways: 1) standard day-of-surgery urine culture result (positive/negative) and 2) occurrence of post-operative UTI (positive/negative). Characteristics of the bladder, vaginal and perineal microbiomes were assessed to identify factors associated with post-operative UTI.
RESULTS:
87% of the 104 POP/UI surgical patients were white; the mean age was 57 years. The most common genus was Lactobacillus with a mean relative abundance of 39.91% in catheterized urine, 53.88% in vaginal swabs, and 30.28% in perineal swabs. Two distinct clusters, based on dispersion of catheterized urine (i.e., bladder) microbiomes, had highly significant (p<2.2e-16) differences in age, microbes and post-operative UTI risk. Post-operative UTI was most associated with the bladder microbiome; microbes in adjacent pelvic floor niches also contributed to UTI risk. UTI risk was associated with depletion of Lactobacillus iners and enrichment of a diverse mixture of uropathogens.
CONCLUSIONS:
Post-operative UTI risk appears associated with pre-operative bladder microbiome composition, where an abundance of L. iners appears to protect against post-operative UTI.
Keywords: Urinary tract infection, Urobiome, Surgical infection, Post-operative infection
Brief Summary:
In adult women undergoing urogynecologic surgery post-operative UTI risk appears associated with pre-operative bladder microbiome composition.
INTRODUCTION:
Hospital-acquired urinary tract infections (UTI) are an important clinical quality metric. Many adult women with pelvic organ prolapse and/or stress urinary incontinence undergo urogynecologic surgery. Approximately 7-24% of these surgical patients will develop a post-operative UTI despite prophylactic antibiotics and other preventive measures 1–3. There is a transient increase in UTI risk in the early post-operative period following retropubic tension-free vaginal tape, regardless of concomitant prolapse repair 1. Post-operative UTI is commonly attributed to the introduction of a uropathogen into the lower urinary tract during instrumentation (catheterization and/or cystoscopy). The recent discovery and confirmation of live bacteria in the bladders of adult women (bladder microbiota) 4–10, however, provides a basis for alternate etiologies for post-operative UTI, a potentially preventable adverse event.
Multiple peri-operative events have potential to influence the bladder microbiota, including urinary tract instrumentation and systemic antibiotics used to prevent surgical infection. Resilience and recovery of the post-operative bladder microbiota have not been described; however, the resilience/recovery process is likely to vary across individuals based on the unique characteristics of their bladder microbiota and their overall physiology. Pre-surgical disruption (dysbiosis) of the microbiota is likely to be associated with an increased risk of post-operative dysbiosis, potentially increasing post-operative UTI risk. We have previously described the use of a simple standard urine culture as a tool for identifying a subgroup of women at increased risk for post-operative UTI 11. Compared to patients with negative standard urine cultures on the day of surgery, patients with a positive (>1000 CFU/ml) culture were almost six times more likely to develop a post-operative UTI.
Here, we expand our analysis of this cohort of women, using 16S rRNA gene sequencing to characterize day-of-surgery (DOS) bladder, vaginal and perineal microbiomes and to correlate these findings with UTI risk following urogynecological surgery.
METHODS
Study design
Recruitment and initial characterization of the primary study cohort was previously described11. Briefly, after institutional review board approval, we approached women undergoing a varetiy of common urogynecologic procedures (slings and prolapse repair, both transvginal and laparoscopically) to treat pelvic organ prolapse (POP) and/or stress urinary incontinence (UI) at Loyola University Medical Center. The primary objective of this analysis was to determine correlations of pelvic floor bacteria with UTI risk. Therefore, we restricted this analytic cohort to the 104 participants with sequence-positive samples from urine, vagina and perineum.
Clinical data, such as age, BMI, medical co-morbidities, type of POP/UI surgery, DOS status, and hormone status was extracted from the electronic medical record. Hormone status was determined clinically and categorized as pre-menopausal, post-menopausal on hormones, or post-menopausal not on hormones.
Following induction of anesthesia and prior to systemic antibiotic administration or any surgery, we collected vaginal and perineal samples using cotton swabs and a standardized collection protocol; we collected the urine sample via a urinary catheter placed consistent with surgical protocols. A portion of the DOS urine sample, obtained pre-operatively, was sent for standard clinical urine culture, and a portion was stored at −80C in 10% AssayAssure (Sierra Molecular, Incline Village, NV) prior to DNA isolation for sequencing. Each swab was suspended in 1 ml of PBS and the suspension was stored at −80°C in 10% AssayAssure prior to DNA isolation for sequencing.
Similar to our prior report on postoperative UTI risk11 patients were queried on UTI symptoms during the 6 week post-operative period and catheterized urine samples were obtained as clinically indicated. A positive urine culture was defined as at least 1,000 bacterial colony-forming units per milliliter of catheterized urine identified on standard microbial culture techniques.
DNA isolation from Urine
DNA isolation, polymerase chain reaction (PCR) amplification, and 16S rRNA gene sequencing of urine cultures has been described previously 10. To avoid contamination, isolation of DNA was performed in a laminar flow hood. Genomic DNA was extracted from 1 ml of urine, using previously validated protocols developed for the Human Microbiome Project 4, 10, 12. To isolate genomic DNA from urine samples, this protocol includes the addition of the peptidoglycan degrading enzymes, mutanolysin and lysozyme, that ensure robust lysis of Gram-positive and Gram-negative species 12. Briefly, 1 ml of urine was centrifuged at 13,500 rpm for 10 min, and the resulting pellet was resuspended in 200 μl of filter-sterilized buffer consisting of 20 mM Tris-CI (pH 8), 2 mM EDTA, 1.2% Triton X-100, and 20 μg/ml lysozyme and supplemented with 30 μl of filter-sterilized mutanolysin (5,000 U/ml; Sigma-Aldrich, St. Louis, MO). The mixture was incubated for 1 h at 37°C, and the lysates were processed through the DNeasy blood and tissue kit (Qiagen, Valencia, CA), according to the manufacturer’s protocol. The DNA was eluted into 50 μl of buffer AE, pH 8.0, and stored at −20° C.
The hyper-variable region 4 (V4) of the bacterial 16S rRNA gene was amplified via a two-step PCR protocol, as described previously 4, 10. Briefly, in the first amplification, the V4 region was amplified using illumina MiSeq modified universal primers 515F and 806R. Extraction negative controls (no urine or swab suspension) and PCR-negative controls (no template) were included to assess the contribution of extraneous DNA from reagents. Ten-microliter aliquots of each reaction mixture were run on a 1% agarose gel. Samples containing a band of approximately 360 bp were considered PCR-positive and subjected to further library preparation. Samples with no visible amplified product were considered PCR-negative and not processed further. The PCR-positive reaction mixtures were diluted 1:50 and amplified for an additional 10 cycles, using primers encoding the required adapter sequences for illumina MiSeq sequencing and an 8-nucleotide sample index. The PCR reaction was purified and size selected using Agencourt AMPure XP-PCR magnetic beads (Beckman Coulter, Pasadena, CA). Each sample was quantified using the Qubit fluorometeric system (Thermo-Fisher, Waltham, MA). The samples were pooled, quantified to a standard volume, and placed in the 2 X 250 bp sequencing reagent cartridge, according to the manufacturer’s instructions (illumina, San Diego, CA).
Because urine samples typically contain small amounts of bacteria (i.e., low biomass), amplification and sequencing was performed in duplicate (technical replicates) and samples were classified as either sequence-positive or sequence-negative. A sequence-positive sample was one in which DNA was amplified from both replicas and, if present, the dominant taxon (representing >50% of sequences from the sample) was the same in both replicas. For each sequence-positive urine sample, both replicas were used for further analysis. Because they usually contain high biomass, vaginal and perineal swabs were sequenced a second time only if the first attempt was negative. For this analysis, sequence-negative urines and swabs were not analyzed further.
Data analysis
Sample barcodes and sequencing primers were removed using the Illumina proprietary MiSeq post-sequencing software. The mothur program (v1.37.4) was used to process the raw sequences by following the recommended MiSeq standard operating procedure 13. Briefly, mothur produced 16S contigs by combining the paired end reads based on overlapping nucleotides in the sequence reads; contigs of incorrect length for the V4 region (<290 bp, >300 bp) and/or contigs containing ambiguous bases were removed. Chimeric sequences were removed using UCHIME within the mothur package 14. Subsampling at a depth of 5000 sequences was performed to correct for different sequencing depth of each sample. The sequences were clustered into species-level operational taxonomic units (OTUs) with identity cutoff at 97% 15. The OTUs were classified using RDP classifier (v2.11) at the genus level 15 and BLCA 16 at the species level. All the statistical analyses were performed with open-source R packages. For each sample, the alpha diversity indices, Chao1, ACE, Shannon, and Simpson, were calculated using the vegan 17 package in R. Principal coordinate analysis (PCoA) (ape package in R 18) was applied to cluster microbiome samples based on Bray-Curtis dissimilarities (ecodist package in R 19). K-means clustering was applied to confirm PCoA clustering results. Dispersion of the PCoA clusters was compared using the betadisper command in R vegan package 20, 21. The PERMANOVA test, using the vegan 17 package in R, was performed to determine whether there are statistically significantly separation between different sample groups (e.g., urine vs. vagina). Student’s t-test and chi-squared test were applied to compare the demographic and clinical variables (e.g., age and premenopausal status) among different urogynecologic surgical groups (i.e., SUI and POP). Using the glmmADMB package 22 in R, generalized linear regression with a negative binomial model was applied to identify the association between bacterial abundance and post-operative UTI status, DOS result, and PCoA clusters. Fisher’s exact test was applied to examine the association between DOS/UTI status and PCoA clusters. Multiple testing correction was performed with the standard Benjamini-Hochberg procedure.
RESULTS
Patient characterization
The 104 surgical patients in this analytic cohort includes 94 women who were described in our earlier report 11 and an additional 10 women from the same IRB-approved study of post-operative infection in urogynecologic patients. Other than an expected higher proportion of positive urine cultures in these women with sequence-positive urine samples (18.3% versus 9.5 %, p=0.02), we did not detect significant clinical differences variables between this analytic cohort and the originally described larger cohort (n=284).
Most (87%) women in this analysis were white; the mean age was 57 years (Table 1). The group had a typical distribution of indications for urogynecologic surgical: 31 (30%) women had surgery for SUI only, 42 (40%) had POP only and 31 (30%) had combined SUI and POP. Women undergoing POP or SUI/POP surgeries were older than those undergoing surgery for SUI (p<0.05). Consistent with clinical practice, women who had isolated SUI surgery were more likely to be premenopausal and those having isolated POP surgery were more likely to be postmenopausal (p<0.05). Of the 17 patients reporting hormone therapy use, 16 reported oral hormone treatment, 1 reported transdermal patch use, and 2 reported vaginal estrogen cream use.
Table 1.
Baseline Participant Demographic and Clinical Characteristics
| POP (N =42) | SUI (N = 31) | POP/SUI (N=31) | P value | |
|---|---|---|---|---|
| Age (years) | 62 (37-85) | 50 (25-75) | 57 (38 −50) | <0.05 |
| Race | Caucasian = 36 (86%) African American =5 (12%) Other= 1 (2%) |
Caucasian = 28 (90%) African American = 1 (3%) Latina =1 (3%) Other= 1 (3%) |
Caucasian = 26 (84%) African American = 3 (10%) Latina =1 (3%) Other= 1 (3%) |
0.8 |
| BMI (kg/m2) | 28 (20-44) | 29 (20-50) | 28 (20-46) | 0.5 |
| Smoking | 0 | 1 (3%) | 0 | 0.3 |
| Hypertension | 18 (43%) | 6 (20%) | 11 (36%) | 0.1 |
| Coronary Artery Disease | 4 (10%) | 1 (3%) | 1 (3%) | 0.4 |
| Hormone Status | Pre-menopausal = 9 (21%) Postmenopausal on Hormones = 7 (17%) Postmenopausal no Hormones = 26 (62%) |
Pre-menopausal = 19 (61%) Postmenopausal on Hormones = 6 (19%) Postmenopausal no Hormones =6 (19%) |
Pre-menopausal = 12 (39%) Postmenopausal on Hormones = 4 (13%) Postmenopausal no Hormones = 15 (48%) |
<0.05 |
Microbiome characterization
The bacterial sequences detected were classified into 22 phyla, 45 classes, 102 orders, 199 families, and 646 genera. The most common genus was Lactobacillus with a mean abundance of 39.91% in catheterized urine, 53.88% in vaginal swabs, and 30.28% in perineal swabs (Table 2). Gardnerella was the only other genus to be amongst the 5 most common genera in all three niches with a mean abundance of 10.2% in the urine, 11.5% in the vagina and 5.5% in the perineum. Anaerococcus was common in the vagina (3.9%) and the perineum (9.2%), but was not amongst the 5 most common genera in urine, whereas Corynebacterium was common in the perineum (9.1%) and urine (6.0%), but not the vagina.
Table 2.
Five Most Abundant Genera Detected in the Three Pelvic Floor Niches
| Sample | Genera Relative Abundance (mean) | ||||
|---|---|---|---|---|---|
| Urine | Lactobacillus (36.9%) | Gardnerella (10.2%) | Escherichia_Shlgella (6.9%) | Corynebacterium (6.0%) | Staphylococcus (4.3%) |
| Vagina | Lactobacillus (53.9%) | Gardnerella (11.5%) | Streptococcus (4.6%) | Anaerococcus (3.9%) | Prevotella (3.4%) |
| Perineum | Lactobacillus (30.3%) | Anaerococcus (9.2%) | Corynebacterium (9.1%) | Peptoniphilus (5.6%) | Gardnerella (5.5%) |
The bladder microbiomes of the participants grouped into two distinct clusters based on their bacterial compositions at the genus level: a More Dispersed Cluster (MDC) and a Less Dispersed Cluster (LDC) (Figure 1). The difference in dispersion was highly significant (p<2.2×10−16). Women in the LDC were younger (average age 51 years) and their urinary microbiome was more likely to contain Lactobacillus iners (Table 3). The women in the MDC were older (average age 59 years) and their urinary microbiomes were enriched in a diverse set of pathogens, including members of the family Enterobacteriaceae, the genera Pseudomonas and Staphylococcus, and the species L. delbrueckii, Actinotignum (formerly Actinobaculum) schaalii, Anaerococcus obesiensis, Corynebacterium tuberculostearicum, Streptococcus anginosus, Aerococcus christensenii and Anaerococcus murdochii (Table 3).
Figure 1. PCoA using Bray-Curtis dissimilarities at OTU level.
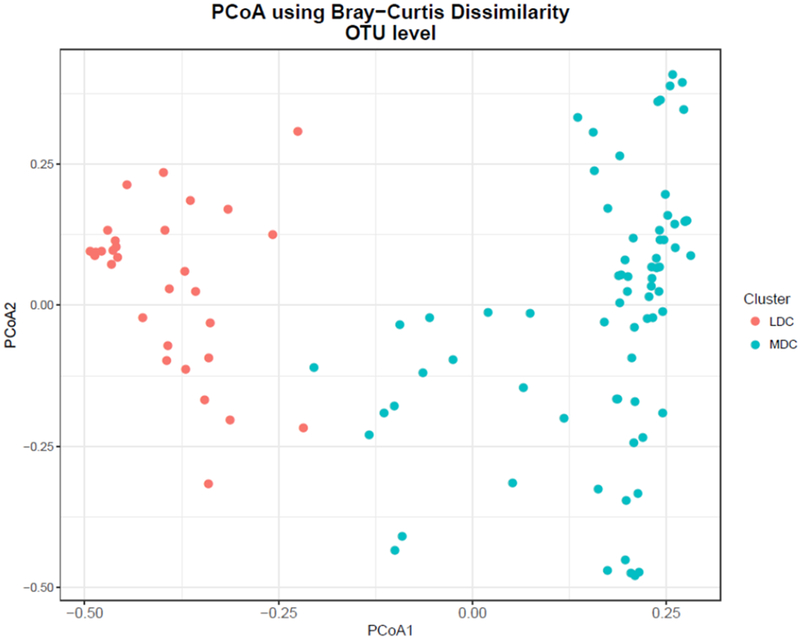
Based on the Bray-Curtis dissimilarity distance of microbiome composition, urine samples sorted into two clusters (Less Dispersed Cluter [LDC] and More Dispersed Cluster [MDC]), p < 0.05 (see main text for details).
Table 3.
OTU Level Comparison of the LDC and MDC
| OTU | Taxonomic classification a | P-value b | Relative Abundance c in LDC (%) | Relative Abundance c in MDC (%) |
|---|---|---|---|---|
| Otu000001 | Lactobacillus iners (species) | 2.19E-11 | 67.10(17.27) | 4.89 (8.30) |
| Otu000012 | Pseudomonas (genus) | 5.26E-09 | 0.02(0.05) | 5.06 (19.39) |
| Otu000008 | Staphylococcus (genus) | 5.13E-08 | 0.38(0.63) | 5.96(21.99) |
| Otu000032 | Lactobacillus delbrueckii (species) | 1.31E-04 | 0.001(0.01) | 1.47(11.69) |
| Otu000018 | Actinotignum schaalii (species) | 3.26E-04 | 0.22(0.49) | 2.84(11.86) |
| Otu000021 | Enterobacteriaceae (family) | 3.89E-04 | 0.09(0.16) | 1.44(11.67) |
| Otu000014 | Anaerococcus obesiensis (species) | 8.03E-03 | 0.25(0.49) | 1.16(3.00) |
| Otu000006 | Enterobacteriaceae (family) | 1.14E-02 | 1.52(3.01) | 9.27(25.73) |
| Otu000016 | Corynebacterium tuberculostearicum (species) | 1.33E-02 | 0.28(0.47) | 1.01(2.55) |
| Otu000013 | Streptococcus anginosus (species) | 1.49E-02 | 0.38(1.20) | 2.60(12.78) |
| Otu000031 | Aerococcus christensenii (species) | 1.56E-02 | 0.16(0.39) | 1.03(4.35) |
| Otu000005 | Anaerococcus murdochii (species) | 2.45E-02 | 0.45(0.81) | 1.36(2.71) |
Taxonomic classifications are based on BLCA annotation with its default threshold of confidence score being 80. If species-level classification cannot be confidently assigned OTUs (i.e., below the threshold), higher-level taxonomic classifications are provided instead. The lowest taxonomic levels being assigned are indicated in the parentheses.
p values adjusted by the Benjamini-Hochberg multiple test correction.
mean (standard deviation)
Table 4 compares the LDC and MDC in terms of DOS culture results and post-operative UTI status. Almost all of the women in the LDC were DOS-negative and none experienced a post-operative UTI. Thus, most of the DOS-positive and/or UTI-positive women aligned with the MDC group.
Table 4.
Day-Of-Surgery (DOS) Result and Urinary Tract Infection (UTI) Status in the LDC and MDC.
| LDC | MDC | |
| DOSneg/UTIneg | 30 | 53 |
| DOSpos/UTIneg | 1 | 12 |
| DOSneg/UTIpos | 0 | 2 |
| DOSpos/UTIpos | 0 | 6 |
Association of microbes with risk of post-operative UTI
Table 5 depicts a PERMANOVA analysis of differences in bacteria composition using Bray-Curtis Dissimilarity between the three sampled sites. It reveals that bladder microbiomes differed significantly by post-operative UTI status with or without consideration of the DOS result; in contrast, the vaginal and perineal microbiomes did not differ. These findings were consistent at both the genus and OTU levels.
Table 5.
PERMANOVA TEST on the bacteria composition differences among four cohorts (Neg/Neg, Pos/Neg, Neg/Pos, Pos/Pos) and UTI positive and negative at OTU and genus levels, at three body sites.
| OTU | genus | |||
|---|---|---|---|---|
| Four cohorts P-value |
UTI+/− P value |
Four cohorts P-value |
UTI+/− P value |
|
| urine | 0.001* | 0.011* | 0.001* | 0.045* |
| vagina | 0.325 | 0.969 | 0.928 | 0.882 |
| perineum | 0.192 | 0.768 | 0.486 | 0.705 |
P values below the 0.05 threshold.
Table 6 displays the taxa that were associated with post-operative UTI. In urine, L iners and an OTU from another Lactobacillus species were abundant and enriched in women who did not experience a post-operative UTI. A less abundant OTU of the genus Peptoniphilus was also enriched in the urine of UTI-negative women. No taxa detected in the vagina were associated with UTI status; however, a few low abundance taxa detected in the perineum were associated with post-operative UTI, including the species Bacteroides coagulans and Bacteroides fragilis and an OTU in the class β-proteobacteria.
Table 6.
Taxa associated with post-operative UTI status
| Urine | Mean Relative Abundance (UTI status) | ||||
|---|---|---|---|---|---|
| OTU | Taxonomic classification a | Neg | Pos | P_Value b | Adjusted P_Value c |
| OTUOOOOOl | Lactobacillus iners (species) | 25.09 ± 31.47 | 3.56±9.30 | 0.01* | 0.25 |
| OTU000002 | Lactobacillus (genus) | 9.95±20.75 | 0.95±1.88 | 0.01* | 0.25 |
| OTU000011 | Peptonlphllus (genus) | 1.17±1.79 | 0.12±0.21 | 0.05* | 0.67 |
| Perineum | |||||
| OTU000098 | Bacteroides coagulans (species) | 0.09±0.19 | 0.45±1.28 | 0.01* | 0.36 |
| OTU000108 | Bacteroides fragilis (species) | 0.10±0.35 | 0.39±0.55 | 0.01* | 0.36 |
| OTU000054 | β-Proteobacteria (class) | 0.07±0.24 | 0.43±1.16 | 0.03* | 0.81 |
Taxonomic classifications are based on BLCA annotation with its default threshold of confidence score being 80. If species-level classification cannot be confidently assigned OTUs (i.e., below the threshold), higher-level taxonomic classifications are provided instead. The lowest taxonomic levels being assigned are indicated in the parentheses.
p values below the 0.05 threshold.
p values adjusted by the Benjamini-Hochberg multiple test correction.
In urine (Supplemental Table 1), the abundant species L. iners was most enriched in women with a negative DOS result and a negative post-operative UTI status and most depleted in women with a positive DOS result and a positive post-operative UTI status. Several other abundant taxa were associated with a negative DOS result. In contrast, a few taxa were associated with a positive DOS result; OTUs in the family Enterobacteriaceae and the genus Pseudomonas were abundant and enriched in women that were DOS positive whether or not they developed a post-operative UTI. A few other taxa were associated with a positive DOS result, including members of the species Staphylococcus aureus, Klebsiella pneumoniae, Aerococcus christenesenii and Prevotella denticola (Supplemental Table 1).
Although post-operative UTI was most associated with the urine microbiome, the microbiomes of the vagina and perineum also might be potential sources of pathogenic contribution. In the vagina (Supplemental Table 2), a few taxa were associated with a positive DOS result, but not a positive UTI status. The species L. gasseri and an OTU of the family Enterobacteriaceae were both abundant and enriched in DOS-positive women; the less abundant genus Veillonella and the species Bacteroides coagulans also were enriched in DOS-positive women. The genus Ureaplasma was enriched in DOS-positive women relative to DOS-negative women, but most enriched in DOS-positive women with a positive post-operative UTI status. In the perineum (Supplemental Table 3), two taxa were enriched in women with a negative DOS result: an OTU in the genus Lactobacillus and the species Corynebacterium tuberculostearicum. Others were enriched in women with a positive DOS result, including OTUs in the family Enterobacteriaceae and the genera Bifidobacterium and Peptoniphilus. Finally, a few taxa were enriched in women with a positive DOS result and a positive post-operative UTI status relative to women that were negative for both: the species Megasphaera massiliensis, Bacteroides coagulans, and Bacteroides fragilis and an OTU in the genus Enterococcus.
Common OTUs in women with and without post-operative UTI
As expected, some taxa were detected both in women who experienced a post-operative UTI and in women who did not. For the family Enterobactericeae and the genera Pseudomonas and Staphylococcus, one OTU was dominant regardless of UTI status (Figure 2). However, this was not true of the genus Lactobacillus; multiple OTUs were often detected in the same woman and the composition of those OTUs differed by UTI status.
Figure 2. Distribution of OTUs from selected genera in our cohorts of study.
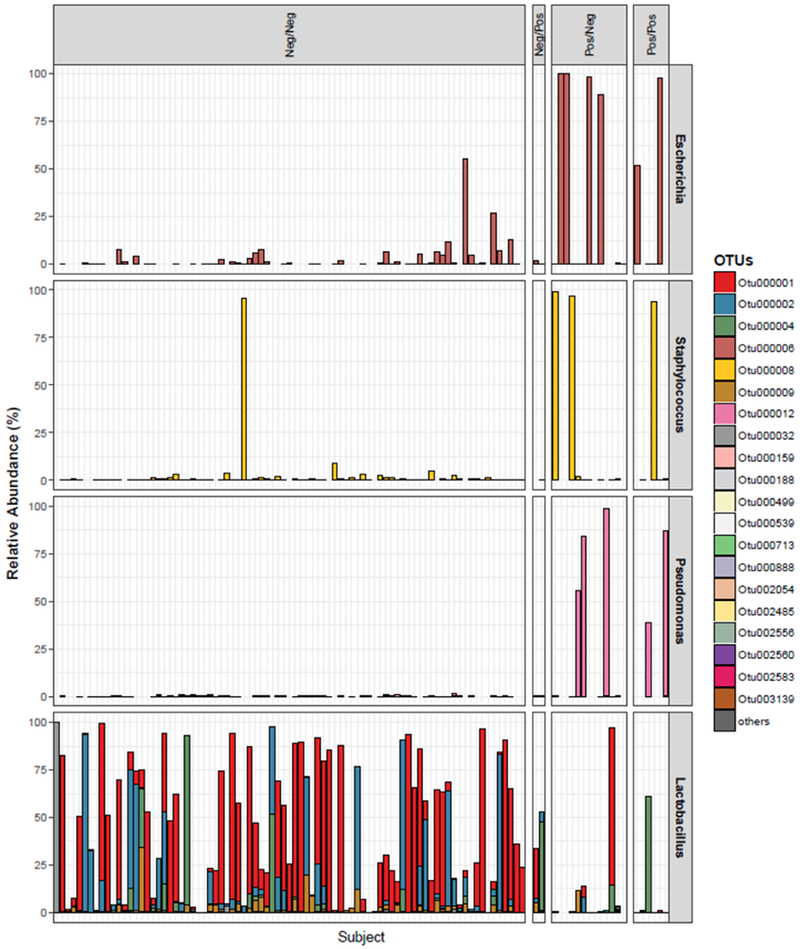
The relative abundance of all the OTUs within four genera, Escherichia, Staphylococcus, Pseudomonas, and Lactobacillus, were displayed in stacked bar-plots in our study cohorts (each color represents a distinct OTU, each column represents an unique urine sample). For Escherichia, Staphylococcus and Pseudomonas, a single OTU was dominant in urine regardless of DOS result or UTI status. For Lactobacillus, multiple OTUs were often detected in the same urine.
DISCUSSION:
Our findings support the hypothesis that risk of post-operative UTI is most associated with the composition of the day-of-surgery microbiome of catheterized urine, suggesting a potential etiologic role for the bladder microbiome in post-operative UTI, a condition that may be more preventable than previously recognized. Risk of post-operative UTI was most associated with depletion of certain Lactobacillus species, especially L. iners, and enrichment of certain uropathogens, especially members of the family Enterobacteriaceae (which includes Escherichia coli and Klebsiella pneumoniae) and the genus Pseudomonas (which includes P. aeruginosa).
Consistent with clinical observations, older patient age and absence of estrogen were clearly associated with an increased risk for post-operative UTI in urogynecologic surgical patients. This analysis provides important insight into the associated microbial mechanisms that may allow effective risk-stratification through pre-operative phenotyping. For most clinicians, absence of UTI-associated symptoms (variably defined) and/or negative urine testing (variably performed) has provided confidence that the patient does not have a current UTI. Yet, clearly, there is an opportunity to reduce the occurrence of post-operative UTI given our finding that most post-operative UTIs are related to depletion of Lactobacillus and enrichment of uropathogens in the bladder prior to surgery. The findings from this analysis extend the impact of our prior analysis 11, which demonstrated that a pre-surgical DOS standard urine culture effectively identifies a group of urogynecologic patients at increased short-term risk for UTI.
What specifically is different about older urogynecologic surgical patients that increases their post-operative UTI risk? Our findings demonstrate that there is a relative depletion of certain Lactobacillus species in their bladder microbiomes. Although the Lactobacillus OTUs that are present in the bladder are similar in women with and without UTI, there is a significant difference in the frequency and abundance, especially of L. iners. Clinicians have prescribed estrogen (often topical vaginal estrogen) with the intent of increasing or restoring vaginal Lactobacillus; these findings suggest that vaginal estrogen may also be a potential therapy aimed at improving the bladder microbiome in order to reduce post-operative UTI risk. The depletion of L. iners also was associated with a more diverse microbial community with a higher risk of uropathogenic taxa. Further research will be necessary to determine whether pre-surgical vaginal estrogen in hypoestrogenic, older women is associated with a reduced risk of post-operative UTI. If so, one would hypothesize that the risk-reduction mechanism may be associated with a healthier peri-operative bladder microbiome (more L. iners, fewer uropathogenic taxa and the appropriate microbial diversity). Given the possibility of uropathogen reservoirs in the vagina and perineum, such future studies should also include analyses of effects on these adjacent pelvic niches.
As our knowledge of the bladder microbiome continues to expand, the simple dichotomous definition of “UTI” may need to be reconsidered 23. We were intrigued to detect potential uropathogens in women who did not develop UTI. This may be related to a variety of biologic scenarios: (1) her immune system could resist infection, (2) the standard surgical prophylaxis effectively eradicated the organism, and/or (3) the bacterium detected by standard culture was a non-pathogenic strain of a known uropathogenic species. For example, we found that the OTUs of Pseudomonas, a genus that includes a known uropathogen, were identical in women with and without UTI. This was also true for Staphylococcus. This finding should cause us to reconsider “pathogenicity” since, at the genomic level these bacteria are identical (at least by the sequence of the 16S rRNA gene). For E. coli, other investigators have recently shown that there is no genomic difference between strains associated with UTI or no UTI; the investigators report that the differences appear related to transcription rather than core genetics 24.
As with all studies, there are limitations to this study, including the use of clinical characterization of hormonal status and a sample size that limited certain analysis for less common microbes. In addition, this cohort was established prior to the development of enhanced culture protocols that more effectively identify urinary bacteria compared to the standard urine culture protocol, which has a high false negative rate 4, 25, 26. There are known limitations to 16S rRNA gene sequencing, including taxonomic classification resolution for V4 regions and detection threshold for low biomass samples; however, sequencing did generally detect the bacteria identified by standard culture, as well as numerous other bacteria that standard culture cannot detect.
Until additional information is available for UTI prevention efforts, clinicians should be cautious in modifying their current clinical practice. Although clinicians could initiate routine or selective pre-operative (or day-of-surgery) urine culture testing to screen for women at increased UTI risk, standard urine culture results typically do not become available to the clinician until 24-48 hour following initial plating. The window for optimal intervention may close prior to the availability of urine culture results. Culture-independent testing of urine is still a research tool that is not used for clinical care in the pre-operative setting. There is currently no evidence for the clinical utility of rapid culture-independent diagnostics (sequencing or PCR of known or suspected uropathogens) that could expedite identification of individuals at increased risk for post-operative UTI.
These insights into the microbial mechanism of post-operative UTI may help other investigators test hypothesis that can advance our understanding of post-operative UTI and further prevention efforts.
Supplementary Material
ACKNOWLEDGMENTS:
We kindly thank Mary Tulke RN for her assistance with participant recruitment and sample collection; we thank Noriko Shibata MS for her assistance with sample analysis. We also thank Dr. Michael Zilliox and Gina Kuffel of the Loyola Genomics Facility for performing the DNA sequencing.
FUNDING: This study was supported by a grant from the Falk Foundation (LU#202567) and by NIH grants R21 DK097435 and P20 DK108268.
Footnotes
CONFLICT OF INTEREST STATEMENT:
Dr. Wolfe discloses research support from Astellas and Kimberly Clark; Dr. Mueller discloses research support from Astellas and Boston Scientic. The remaining authors (Thomas-White, Gao, Lin, Fok, Ghanayem, Dong and Brubaker) report no disclosures.
References
- [1].Karram MM, Segal JL, Vassallo BJ, Kleeman SD: Complications and untoward effects of the tension-free vaginal tape procedure. Obstet Gynecol 2003, 101:929–32. [DOI] [PubMed] [Google Scholar]
- [2].Nygaard I, Brubaker L, Chai TC, Markland AD, Menefee SA, Sirls L, Sutkin G, Zimmern P, Arisco A, Huang L, Tennstedt S, Stoddard A: Risk factors for urinary tract infection following incontinence surgery. Int Urogynecol J 2011, 22:1255–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gehrich AP, Patzwald JR, Kern ME, Squires CC, Lustik MB: The incidence of early and recurrent urinary tract infections after midurethral sling operations. Mil Med 2014, 179:1301–6. [DOI] [PubMed] [Google Scholar]
- [4].Hilt EE, McKinley K, Pearce MM, Rosenfeld AB, Zilliox MJ, Mueller ER, Brubaker L, Gai X, Wolfe AJ, Schreckenberger PC: Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol 2014, 52:871–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Fouts DE, Pieper R, Szpakowski S, Pohl H, Knoblach S, Suh MJ, Huang ST, Ljungberg I, Sprague BM, Lucas SK, Torralba M, Nelson KE, Groah SL: Integrated next-generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury. J Transl Med 2012, 10:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wolfe AJ, Toh E, Shibata N, Rong R, Kenton K, Fitzgerald M, Mueller ER, Schreckenberger P, Dong Q, Nelson DE, Brubaker L: Evidence of uncultivated bacteria in the adult female bladder. J Clin Microbiol 2012, 50:1376–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Karstens L, Asquith M, Davin S, Stauffer P, Fair D, Gregory WT, Rosenbaum JT, McWeeney SK, Nardos R: Does the Urinary Microbiome Play a Role in Urgency Urinary Incontinence and Its Severity? Front Cell Infect Microbiol 2016, 6:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Thomas-White KJ, Hilt EE, Fok C, Pearce MM, Mueller ER, Kliethermes S, Jacobs K, Zilliox MJ, Brincat C, Price TK, Kuffel G, Schreckenberger P, Gai X, Brubaker L, Wolfe AJ: Incontinence medication response relates to the female urinary microbiota. Int Urogynecol J 2016, 27:723–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Brubaker L, Nager CW, Richter HE, Visco A, Nygaard I, Barber MD, Schaffer J, Meikle S, Wallace D, Shibata N, Wolfe AJ: Urinary bacteria in adult women with urgency urinary incontinence. Int Urogynecol J 2014, 25:1179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Pearce MM, Hilt EE, Rosenfeld AB, Zilliox MJ, Thomas-White K, Fok C, Kliethermes S, Schreckenberger PC, Brubaker L, Gai X, Wolfe AJ: The female urinary microbiome: a comparison of women with and without urgency urinary incontinence. MBio 2014, 5:e01283–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Fok CS, McKinley K, Mueller ER, Kenton K, Schreckenberger P, Wolfe A, Brubaker L: Day of surgery urine cultures identify urogynecologic patients at increased risk for postoperative urinary tract infection. J Urol 2013, 189:1721–4. [DOI] [PubMed] [Google Scholar]
- [12].Yuan S, Cohen DB, Ravel J, Abdo Z, Forney LJ: Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS One 2012, 7:e33865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF: Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 2009, 75:7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R: UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27:2194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wang Q, Garrity GM, Tiedje JM, Cole JR: Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007, 73:5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gao X, Lin H, Revanna K, Dong Q: A Bayesian taxonomic classification method for 16S rRNA gene sequences with improved species-level accuracy. BMC Bioinformatics 2017, 18:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jari Oksanen FGB, Kindt Roeland, Legendre Pierre, Minchin Peter R., O’Hara GLS RB, Solymos Peter, Stevens M. Henry H. and Wagner Helene vegan: Community Ecology Package. R package version 2.2-1. 2015, http://CRAN.R-project.org/package=vegan.
- [18].Paradis E, Claude J, Strimmer K: APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 2004, 20:289–90. [DOI] [PubMed] [Google Scholar]
- [19].Goslee SC, Urban DL: The ecodist package for dissimilarity-based analysis of ecological data. J Stat Softw 2007, 22:1–19. [Google Scholar]
- [20].Anderson MJ, Walsh DCI: Permanova, anosim, and the mantel test in the face of heterogeneous dispersions: what null hypothesis are you testing? . Ecol Monogr 2013, 83:557–74. [Google Scholar]
- [21].Anderson MJ: Distance-based tests for homogeneity of multivariate dispersions. . Biometrics 2006, 62:245–53. [DOI] [PubMed] [Google Scholar]
- [22].Bolker B, Skaug H, Magnusson A, Nielsen A: Getting started with the glmmADMB 2012. [Google Scholar]
- [23].Price TK, Hilt EE, Dune TJ, Mueller ER, Wolfe AJ, Brubaker L: Urine trouble: should we think differently about UTI? Int Urogynecol J 2017, 29:205–10. [DOI] [PubMed] [Google Scholar]
- [24].Schreiber HLt, Conover MS, Chou WC, Hibbing ME, Manson AL, Dodson KW, Hannan TJ, Roberts PL, Stapleton AE, Hooton TM, Livny J, Earl AM, Hultgren SJ: Bacterial virulence phenotypes of Escherichia coli and host susceptibility determine risk for urinary tract infections. Sci Transl Med 2017, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Coorevits L, Heytens S, Boelens J, Claeys G: The resident microflora of voided midstream urine of healthy controls: standard versus expanded urine culture protocols. Eur J Clin Microbiol Infect Dis 2016. [DOI] [PubMed] [Google Scholar]
- [26].Khasriya R, Sathiananthamoorthy S, Ismail S, Kelsey M, Wilson M, Rohn JL, Malone-Lee J: Spectrum of bacterial colonization associated with urothelial cells from patients with chronic lower urinary tract symptoms. J Clin Microbiol 2013, 51:2054–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


