Abstract
The subcellular mechanism by which nonsteroidal anti-inflammatory drugs (NSAIDs) induce apoptosis in gastric cancer and normal mucosal cells is elusive because of the diverse cyclooxygenase-independent effects of these drugs. Using human gastric carcinoma cells (AGSs) and a rat gastric injury model, here we report that the NSAID indomethacin activates the protein kinase Cζ (PKCζ)–p38 MAPK (p38)–dynamin-related protein 1 (DRP1) pathway and thereby disrupts the physiological balance of mitochondrial dynamics by promoting mitochondrial hyper-fission and dysfunction leading to apoptosis. Notably, DRP1 knockdown or SB203580-induced p38 inhibition reduced indomethacin-induced damage to AGSs. Indomethacin impaired mitochondrial dynamics by promoting fissogenic activation and mitochondrial recruitment of DRP1 and down-regulating fusogenic optic atrophy 1 (OPA1) and mitofusins in rat gastric mucosa. Consistent with OPA1 maintaining cristae architecture, its down-regulation resulted in EM-detectable cristae deformity. Deregulated mitochondrial dynamics resulting in defective mitochondria were evident from enhanced Parkin expression and mitochondrial proteome ubiquitination. Indomethacin ultimately induced mitochondrial metabolic and bioenergetic crises in the rat stomach, indicated by compromised fatty acid oxidation, reduced complex I– associated electron transport chain activity, and ATP depletion. Interestingly, Mdivi-1, a fission-preventing mito-protective drug, reversed indomethacin-induced DRP1 phosphorylation on Ser-616, mitochondrial proteome ubiquitination, and mitochondrial metabolic crisis. Mdivi-1 also prevented indomethacin-induced mitochondrial macromolecular damage, caspase activation, mucosal inflammation, and gastric mucosal injury. Our results identify mitochondrial hyper-fission as a critical and common subcellular event triggered by indomethacin that promotes apoptosis in both gastric cancer and normal mucosal cells, thereby contributing to mucosal injury.
Keywords: inflammation, apoptosis, oxidative stress, mitochondrial membrane potential, mitochondrial respiratory chain complex, dynamin-related protein 1 (DRP1), kinase signaling
Introduction
Nonsteroidal anti-inflammatory drugs (NSAIDs)3 are the most effective medicines for treating pain and inflammation (1, 2). In addition to their anti-nociceptive action, NSAIDs are also gaining significant importance because of their anti-neoplastic effects against a wide spectrum of cancers. In fact, prolonged NSAID users are at lower risk of developing cancers (3, 4), and these noncanonical anti-cancer drugs are now included in a combination–chemotherapy regimen as they potentiate chemotherapy and radiotherapy (5). Although prostaglandin depletion due to cyclooxygenase (COX) inhibition is primarily responsible for both anti-inflammatory as well as cytotoxic anti-cancer action of NSAIDs (6), COX-independent targets, including cGMP phosphodiesterase, peroxisome proliferator-activated receptors, retinoid X receptor, IKKβ, AMP kinase, and other targets of these drugs as well as their metabolites (7), help to trigger cell death by apoptosis while blocking proliferation. Hence, NSAIDs are gaining immense importance and have been under exploration in various diseases, including cancer (7–10). Despite their multidimensional health benefits, the toxic actions of NSAIDs are observed against various normal cells of the body that compromise metabolic homeostasis and tissue integrity (6, 11, 12). Of the several organs affected by long-term NSAID usage (13–16), the gastrointestinal system and predominantly the stomach are the most prominent targets that suffer severe gastric mucosal injury, bleeding and abdominal pain (10, 17). All these effects strongly discourage their rampant usage and warrant thorough mechanistic studies to reveal precise subcellular effects of NSAIDs, which in turn can lead to better and rational therapeutic exploitation of these drugs, bypassing their side effects.
Mitochondrial structural and functional integrity underlies the maintenance of bioenergetic homeostasis and cellular health. Constant dynamic equilibrium (18) between extended filamentous network and small punctate particles (19) plays a vital role in the mitigation of myriad metabolic duties, including ATP production, intracellular [Ca2+] regulation, endoplasmic reticulum-assisted [Ca2+] buffering, and cell death. At any instance, the predominance of a particular structural form determines the status of cellular health. Moreover, changes in the cellular microenvironment and pathogenic conditions in turn pose a significant effect on mitochondrial structure, including fission–fusion dynamics and turnover (18). Although filamentous mitochondria are frequently associated with enhanced cellular respiration, mitochondrial ATP production, distribution of metabolites, and mitochondrial gene products all over the organelle (20), fragmented mitochondria either characterize cell division or elimination of damaged mitochondria during stress and diseases (21–25). Damaged and fragmented mitochondria are selectively culled from the cell by a process of specialized organellar autophagy called mitophagy (26) to prevent their incorporation back into the healthy mitochondrial pool during acute and persistent damage. However, excess mitochondrial fragmentation coupled with mitophagy and proteasomal degradation result in severe bioenergetic deficit and consequent tissue damage (27).
Alterations in mitochondrial dynamics and the predominance of abnormal phenotypes involving either excessively fragmented mitochondria or hyper-fused elongated tubules are exclusively disease-specific (21–25, 28–31). At the same time, inhibition of mitochondrial fission may prove detrimental (32–34) due to its importance in clearance of damaged organelles or segregation of mitochondria during cell division (35). Interestingly, all these pathologies have oxidative stress as a common etiologic factor wherein mitochondrial oxidative stress (MOS) and consequent apoptosis act as major perpetrators governing the severity of diseases associated with mitochondrial functional derangement (36). Although much explored in various preclinical models, the putative role of aberrant mitochondrial dynamics in gastropathy and the impact of NSAIDs therein are still lacking.
In this study, we have studied the effect of NSAIDs on mitochondrial dynamics using indomethacin as the representative NSAID because of its efficacy in expanding the life span of cancer patients with metastasis (37). We report that indomethacin activates the PKCζ–p38–DRP1 pathway to trigger mitochondrial hyper-fission in both gastric cancer cells and normal mucosal cells. The activation of mitochondrial hyper-fission is a common subcellular event involved in antineoplastic and gastro-damaging activities of this NSAID.
Results
Indomethacin (an NSAID) interferes with human AGS proliferation and induces mitochondrial hyper-fission
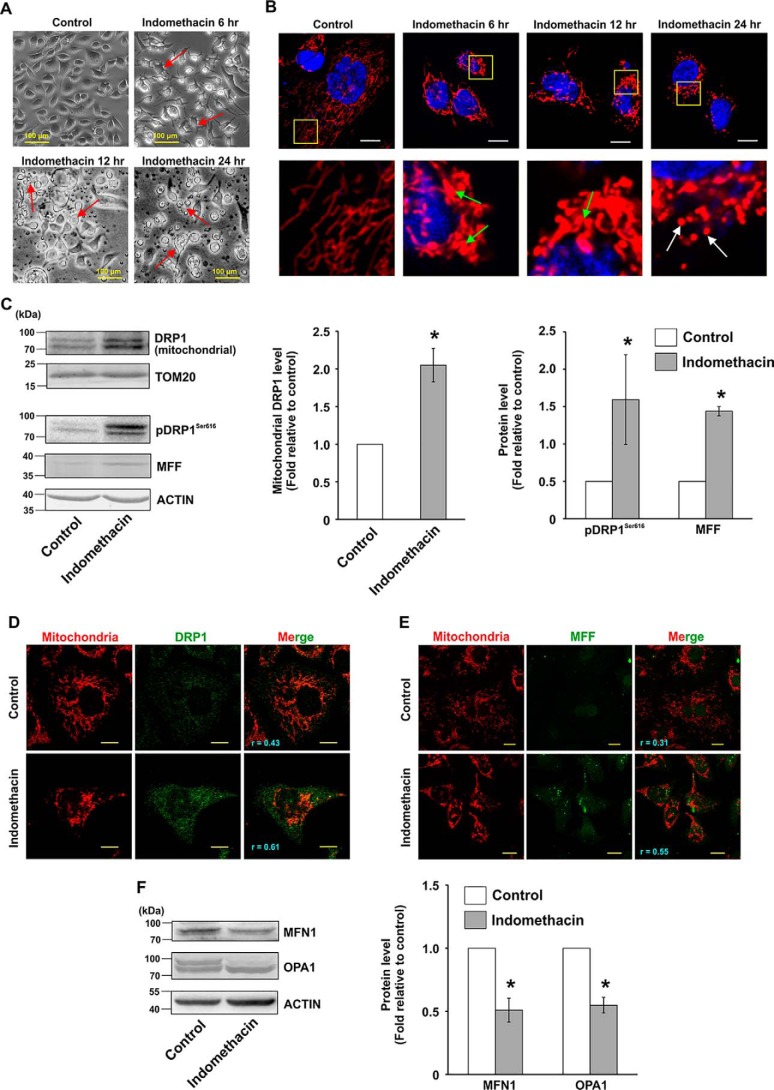
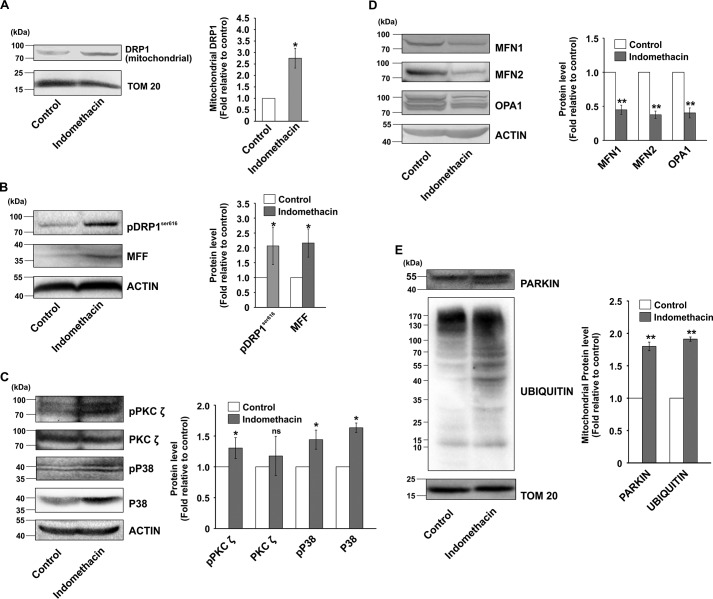
Initially, the optimum concentration required for anti-proliferative activity of indomethacin was determined by following [3H]thymidine uptake and cellular dehydrogenase activity in AGS cells. Data indicated that indomethacin concentration-dependently reduced AGS cell proliferation (Table 1). Indomethacin at a concentration of 0.5 mm significantly reduced the DNA content (≈81%) and inhibited cell viability (≈ 50%) as evident from [3H]thymidine incorporation and cellular dehydrogenase assays, respectively. At further higher doses (0.8 and 1.5 mm), the cytotoxic effects were extremely high. Because we wanted to check the subtle subcellular alterations induced by indomethacin on gastric cancer cells, we deliberately used the IC50 concentration (0.5 mm) for all the subsequent in vitro studies. Next, we were keen to check any plausible cytoarchitectural alterations that indomethacin might cause to the AGS cells. Direct visualization of control and indomethacin-treated cells by phase-contrast microscopy revealed a remarkable deterioration of cellular architecture starting early at 6 h of treatment (Fig. 1A). At 12 and 24 h, indomethacin induced apoptotic changes in the cells as evident from cytoplasmic shrinkage, membrane blebbings, and pinching off of cellular structures (indicated by red arrows) (Fig. 1A). An early detrimental change in the cytoarchitecture further warranted the examination of mitochondrial structure. Confocal microscopy revealed that indomethacin time-dependently rendered the transformation of uniformly distributed filamentous mitochondria into clumped, peri-nuclearly clustered and fragmented puncta. Insets containing magnified portions of the micrographs clearly indicated abundance of tubular mitochondrial structures in control cells, compared with indomethacin-treated cells enriched in fragmented mitochondria (Fig. 1B). Furthermore, to explore the status of the molecular regulators of fission, subcellular localization of dynamin-related protein 1 (DRP1, pro-fission master-switch) was checked along with other fission mediators by immunoblotting (Fig. 1C). Densitometric analyses of the immunoblots indicated that the DRP1 level was found to be significantly high in the mitochondrial fractions implying mitochondrial translocation (Fig. 1C). Moreover, the level of pDRP1Ser-616, a marker for fissogenic activation of DRP1, was found to be significantly increased in the indomethacin-treated cells compared with control. In addition, mitochondrial fission factor (MFF), which is one of the predominant DRP1 adapters on the mitochondrial surface, was also up-regulated upon indomethacin treatment (Fig. 1C). The data were fairly supported by confocal super-resolution (STED) immunocytographs, which pin-pointed focal localization of DRP1 (green) on mitochondria (red) along with the extent of mitochondrial translocation under the influence of indomethacin (r = 0.61 in indomethacin treated cells compared with control, where r = 0.43) (Fig. 1D). Immunocytochemistry of MFF similarly revealed up-regulation with high mitochondrial colocalization (r = 0.55) upon indomethacin treatment (Fig. 1E). In accordance with the expressional pattern of the fission mediators, mitochondrial fusion mediators, including the outer mitochondrial membrane fusion mediator mitofusin 1 (MFN1) and inner mitochondrial membrane (IMM) fusion regulator optic atrophy 1 (OPA1), exhibited depleted expression in the presence of indomethacin. The band patterns in the immunoblot data of OPA1 were indicative of relative depletion of higher molecular weight isoforms of OPA1 (Fig. 1F). Next, it was imperative to check the effect of altered structural dynamics on mitochondrial integrity. Data indicated that indomethacin induced moderate mitochondrial depolarization even after 6 h of treatment (Fig. 2A). Depolarization was however severely elevated at the 24 h post treatment with indomethacin indicating compromised mitochondrial integrity. Next, to check the effect of indomethacin on mitochondrial metabolic status, an oxygen consumption ratio was measured. Data indicated that mitochondrial metabolic parameters were severely compromised upon indomethacin treatment compared with control (Table 2). Basal respiration was considerably reduced along with severely compromised ATP production in the presence of indomethacin. The effect was further reflected in decreased maximal respiration and loss of cellular reserve capacity (Table 2). Collective effect of all these metabolic crises led to gradual mitochondrial depolarization and induction of apoptosis, which initiated at 6 h after indomethacin treatment and significantly increased by 24 h (Fig. 2, A and B). We were next eager to check the status of mitochondrial fission at this early time point (6 h), when mitochondria were metabolically compromised, yet apoptosis had not set in significantly. Therefore, we followed the status of DRP1, pDRP1Ser-616 and OPA1 as major markers of fission and fusion, respectively. Data indicated that DRP1 activation already started 6 h after indomethacin treatment, and it further increased until 24 h (Fig. 2C) as evident from immunoblot data performed using total cell lysates. Notably, down-regulation of OPA1 was also initiated after 6 h of indomethacin treatment, and the level remained low at 24 h as well.
Table 1.
Effect of different concentrations of indomethacin on human gastric cancer cell proliferation and viability
AGS cells were treated with indomethacin at the indicated doses for 24 h. Cells were analyzed for DNA content and cellular dehydrogenase activity as described under “Experimental procedures.” All experiments were done in triplicate. Data are presented as means ± S.D. **, p < 0.01; ***, p < 0.001 versus indomethacin 0 mm.
| Sample | [3H]Thymidine incorporation (counts/min) | Cellular dehydrogenase activity (MTT reduction, OD570 nm) |
|---|---|---|
| Indomethacin 0 mm | 87,859.0 ± 6505.9 | 0.93 ± 0.03 |
| Indomethacin 0.25 mm | 28,501.7 ± 4132.7*** | 0.65 ± 0.02** |
| Indomethacin 0.50 mm | 16,130.3 ± 2344.8*** | 0.48 ± 0.02*** |
| Indomethacin 0.80 mm | 7319.7 ± 428.5*** | 0.26 ± 0.01*** |
| Indomethacin 1.5 mm | 3515.0 ± 472.7*** | 0.18 ± 0.01*** |
Figure 1.
Indomethacin enhances mitochondrial fragmentation in human gastric cancer cells. A, phase-contrast micrographs of AGS cells treated with indomethacin (0.5 mm) for 6, 12, and 24 h; size bar indicates 100 μm. Red arrows indicate blebbings and pinching off of cellular structures. B, high-resolution confocal micrographs to demonstrate time-dependent effect of indomethacin on AGS cells; white scale bars correspond to 10 μm. 80–100 cells were randomly screened, and a single cell was randomly selected for demonstration of mitochondrial fission at the single-cell level. Enlarged images of the region of interest (ROI) were prepared by digital zooming of the selected region for clear visualization of mitochondrial filaments. Green arrows indicate clumped and peri-nuclearly clustered mitochondria, and white arrows indicate fragmented and punctate mitochondrial particles. C, immunoblot analysis of DRP1 in the mitochondrial extract and phosphorylation level of DRP1 at Ser-616 along with expression of MFF in the whole-cell lysates from control and indomethacin-treated AGS cells. Actin and TOM20 were used as the loading control for cell lysates and mitochondrial fractions, respectively. Numerical values at the side of the blots indicate the positions of corresponding molecular mass markers. Bar graphs adjacent to the blots represent the densitometric analyses of the immunoblot data after normalization with respective loading controls. D, STED microscopy to precisely follow mitochondrial translocation of DRP1; scale bars correspond to 10 μm. 80–100 cells were randomly screened, and a single cell was randomly selected for precise demonstration of mitochondrial localization of DRP1 at the single-cell level. Mitochondria were immunostained by anti-TOM20 antibody. A representative image of one of several experiments performed has been presented. Corresponding values in the figure represent Pearson's correlation coefficient (r) of the red and green signals corresponding to TOM20 and DRP1, respectively. E, confocal microscopy to follow expression and localization of MFF. Corresponding values in the figure represent Pearson's correlation coefficient (r) of the red and green signals corresponding to TOM20 and MFF, respectively. F, immunoblot analysis of MFN1 and OPA1. Actin was used as the loading control. Numerical values at the side of the blots indicate the positions of corresponding molecular mass markers. Bar graphs adjacent to the blots represent the densitometric analyses of the immunoblot data after normalization with the respective loading controls. All experiments were done in triplicate. A detail of each method is described under “Experimental procedures.” *, p < 0.05 versus control calculated by unpaired Student's t test.
Figure 2.
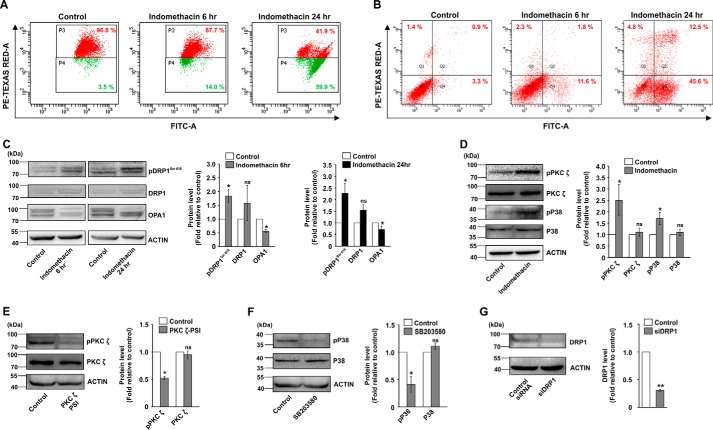
Indomethacin induces mitochondrial depolarization, fragmentation, and gastric cancer cell apoptosis through PKCζ–p38–MAPK pathway. A, flow cytometric analysis to follow mitochondrial transmembrane potential (ΔΨm) in control and indomethacin-treated AGS cells at 6 and 24 h of incubation; 10,000 events were screened per experimental set. Experiments were performed in triplicate, and a representative image has been presented. Red signal in the scatterplot indicates JC-1 aggregates fluorescing at 590 nm, and the green signal indicates JC-1 monomers (corresponding to depolarized mitochondria) fluorescing at 530 nm; % values in the P3 and P4 quadrants correspond to number of cells with polarized and depolarized mitochondria, respectively. B, FACS analysis to show apoptosis in control and indomethacin-treated cells at 6 and 24 h. Values in the Q4 and Q2 quadrants represent percentage of cells showing apoptotic and late apoptotic–necrotic cell death, respectively; dot plot representations of FACS analysis are representative of one of the three independent experiments repeated under similar experimental conditions. C, immunoblot analysis of pDRP1Ser-616, DRP1, and OPA1. Actin was used as the loading control. D, immunoblot analysis for phospho-PKCζ, PKCζ, phospho-p38, and p38 in the total cell lysates of control and indomethacin-treated AGS cells. Actin was used as the loading control. E, immunoblot analysis for phospho-PKCζ and PKCζ in control and PKCζ–PSI (PKCζ pseudo substrate inhibitor)-treated cells to show the basal effect of PKCζ–PSI on PKCζ activation. Actin was used as the loading control. F, immunoblot analysis for phospho-p38 and p38 in control and SB203580 (p38 inhibitor)-treated cells to show the basal effect of SB203580 on p38 activation. Actin was used as the loading control. G, immunoblot analysis to document the efficiency of DRP1 silencing in the siDRP1-treated AGS cells compared with control siRNA treatment for 72 h. Actin was used as the loading control. Numbers at the side of the blots indicate the positions of corresponding molecular mass markers. Bar graphs adjacent to the blots represent the densitometric analyses of the immunoblot data after normalization with actin. A detail of each method is described under “Experimental procedures.” *, p < 0.05; **, p < 0.001 versus control calculated by unpaired Student's t test; ns, nonsignificant.
Table 2.
Indomethacin-induced alteration in viability and respiration of AGS cells as well as effect of p38 MAPK inhibition or DRP1 silencing in pathology
AGS cells were pretreated with p38 inhibitor (SB203580) or siDRP1 followed by treatment with indomethacin at the IC50 dose (0.5 mm). Cells were subsequently analyzed for viability by measuring MTT reduction and metabolic integrity by measuring oxygen consumption ratio (OCR) in extracellular flux analyzer as described under “Experimental procedures.” All experiments were done in triplicate. Data are presented as means ± S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus control; ##, p < 0.01; ###, p < 0.001 versus indomethacin; NS, not significant.
| Cell viability (MTT reduction, OD570 nm) | Basal respiration (pmol O2/min) | ATP production (pmol O2/min) | Maximal respiration (pmol O2/min) | Reserve capacity (pmol O2/min) | |
|---|---|---|---|---|---|
| Control | 0.96 ± 0.03 | 144.28 ± 5.9 | 107.17 ± 6.4 | 265.49 ± 3.4 | 121.21 ± 8.5 |
| Indomethacin | 0.45 ± 0.04*** | 28.04 ± 3.4*** | 16.48 ± 3.6*** | 47.61 ± 4.8*** | 19.56 ± 4.4*** |
| SB203580 + indomethacin | 0.64 ± 0.05### | 93.34 ± 3.9### | 50.26 ± 3.7## | 132.25 ± 3.6### | 38.87 ± 7.3NS |
| SB203580 | 0.88 ± 0.04NS | 120.29 ± 2.6* | 90.99 ± 3.6* | 236.83 ± 15.7NS | 116.54 ± 14.4NS |
| siDRP1 + indomethacin | 0.62 ± 0.06## | 74.21 ± 6.9### | 48.96 ± 4.5## | 108.91 ± 3.2## | 34.70 ± 3.7NS |
| siDRP1 | 0.87 ± 0.05NS | 99.61 ± 4.6** | 79.28 ± 5.9** | 211.26 ± 9.7* | 111.66 ± 5.1NS |
Activation of PKCζ–p38 MAPK–DRP1 pathway by indomethacin to stimulate excess mitochondrial fission
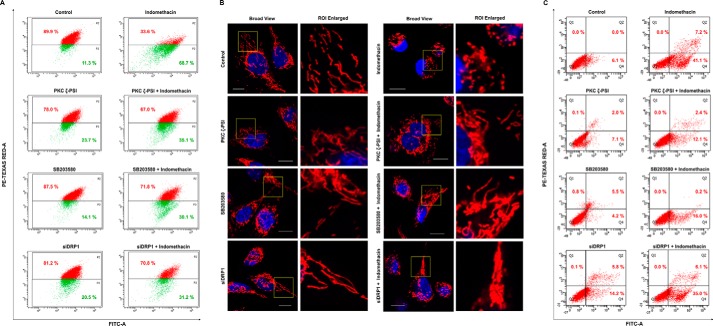
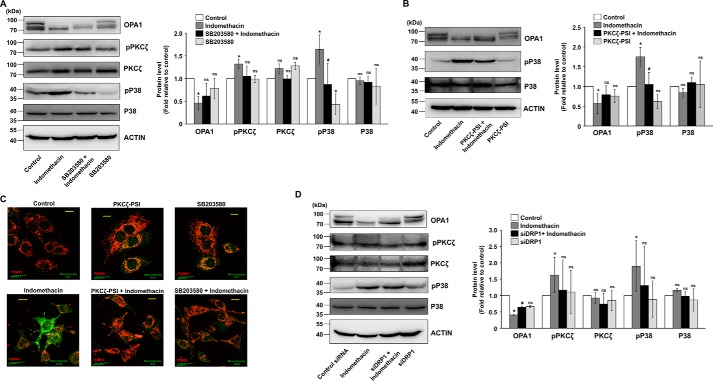
We next focused on identifying the plausible upstream mediators that regulate DRP1 activation by indomethacin. Data indicate that indomethacin activated p38 MAPK as evident from phosphorylation of p38 (Fig. 2D). The event was concurrent with DRP1Ser-616 phosphorylation. Further exploration revealed that p38 MAPK activation was positively associated with phosphorylation of the atypical protein kinase C (PKC) isoform, PKCζ (Fig. 2D). To check the effect of MAPK inhibition on indomethacin-induced pathology, we pretreated cells with a p38-specific inhibitor, SB203580, or a PKCζ pseudo-substrate inhibitor peptide (PKCζ–PSI) prior to indomethacin treatment. The activation by phosphorylation and the expression levels of p38 and PKCζ were monitored in the presence and absence of the respective inhibitors (Fig. 2, E and F) used at an optimum dose. Furthermore, to check the role of DRP1 in indomethacin-induced pathology, DRP1 was knocked down, and the expression level of DRP1 was monitored (Fig. 2G). Interestingly, pretreatment with the p38-specific inhibitor, SB203580, rescued cells from indomethacin-mediated deterioration in mitochondrial structural and functional integrity to a considerable extent as evident from the reduction of ΔΨm collapse measured by flow cytometry (Fig. 3A) and reduction in mitochondrial fragmentation as observed by live-cell imaging (Fig. 3B). A similar effect was observed by blocking PKCζ activation by PKCζ–PSI or DRP1 knockdown (DRP1 KD) prior to indomethacin administration (Fig. 3, A and B). Cell death by indomethacin was also reduced by SB203580 and PKCζ–PSI as evident from flow cytometric analysis of apoptosis (Fig. 3C). Furthermore, the protective effect of SB203580 and siDRP1 pretreatment on the indomethacin-induced metabolic crisis was further reflected from the preservation of dehydrogenase activity, mitochondrial respiration, oxygen consumption, and ATP production (Table 2). In addition, immunoblot analysis indicated that indomethacin-induced OPA1 depletion and increased p38 phosphorylation were both rectified by SB203580 pretreatment or PKCζ–PSI pretreatment (Fig. 4, A and B). Reduction in indomethacin-induced fissogenic DRP1 activation was also clearly evident from the depletion of the pDRP1Ser-616 level upon pretreatment with SB203580 or PKCζ–PSI as revealed by immunocytochemical analysis (Fig. 4C). However, being downstream in the signaling pathway, siDRP1 offered negligible protection against indomethacin-induced PKCζ and p38 activation by phosphorylation (Fig. 4D), although the reduced OPA1 level was moderately rectified. Overall, in the present model, an association of PKCζ–p38 signaling with mitochondrial fission was evident from the fact that a considerable rescue effect was observed upon PKCζ–PSI treatment, which moderately inhibited indomethacin-induced p38 and DRP1 activation (Fig. 4, B and C).
Figure 3.
Blocking PKCζ or p38 or silencing DRP1 reduces indomethacin-induced mitochondrial membrane depolarization, fragmentation, and apoptosis. A, flow cytometric analysis to follow mitochondrial transmembrane potential (ΔΨm) in AGS cells treated with indomethacin in presence and/or absence of PKCζ inhibitor, p38 inhibitor, or siDRP1; 10,000 events were screened per experimental set. % values in the P2 and P3 quadrants correspond to number of cells with polarized and depolarized mitochondria, respectively. Experiments were performed in triplicate, and a representative image has been presented. B, high-resolution confocal micrographs to demonstrate the effect of PKCζ blocking or p38 blocking or DRP1 silencing against indomethacin-induced mitochondrial fragmentation in AGS cells; white scale bars correspond to 10 μm; 80–100 cells were randomly screened, and a single cell was randomly selected for demonstration of mitochondrial fission at the single-cell level; enlarged images of the ROI were prepared by digital zooming of the selected region for clear visualization of mitochondrial filaments. Experiments were performed in triplicate, and a representative image has been presented. C, flow cytometric analysis to follow apoptosis in AGS cells treated with indomethacin in presence and/or absence of PKCζ inhibitor, p38 inhibitor, or siDRP1; 10,000 events were screened per experimental set. Values in the Q4 and Q2 quadrants represent percentage of cells showing apoptotic and late apoptotic–necrotic cell death, respectively; dot plot representations of FACS analysis are representative of one of the three independent experiments repeated under similar experimental conditions. A representative flow cytometry scatterplot of gated cell population was presented. All experiments were done in triplicate. A detail of each method is described under “Experimental procedures.”
Figure 4.
Blocking p38 activation or PKCζ activation affects indomethacin-induced PKCζ–p38 MAPK signaling and prevents fissogenic DRP1 activation. A, immunoblot analysis of OPA1, phospho-PKCζ, PKCζ, phospho-p38 and p38 in the whole-cell lysates from control and SB203580-treated AGS cells. Actin was used as the loading control for cell lysates. Bar graphs adjacent to the blots represent the densitometric analyses of the immunoblot data after normalization with actin. B, immunoblot analysis of OPA1, phospho-p38, and p38 in the whole-cell lysates from control and PKCζ–PSI-pretreated, indomethacin-treated AGS cells. Actin was used as the loading control for cell lysates. Numerical values at the side of the blots indicate the positions of corresponding molecular mass markers. Bar graphs adjacent to the blots represent the densitometric analyses of the immunoblot data after normalization with actin. C, confocal microscopy to follow the level of Ser-616 phosphorylation of DRP1 in control, indomethacin-treated, PKCζ–PSI-treated, PKCζ–PSI-pretreated indomethacin-treated, SB203580-treated, and SB203580-pretreated indomethacin-treated AGS cells. Scale bars correspond to 10 μm, 80–100 cells were randomly screened, and a single cell was randomly selected for precise demonstration of mitochondrial localization of pDRPSer-616 (green) at the single-cell level. Mitochondria (red) were immunostained by anti-TOM20 antibody. A representative image of one of several experiments performed has been presented. Mean intensity values corresponding to p pDRP1Ser-616 are indicated in each image. D, immunoblot analysis of OPA1, phospho-PKCζ, PKCζ, phospho-p38, and p38 in the whole-cell lysates from control and siDRP1-treated AGS cells. Actin was used as the loading control for cell lysates. Numerical values at the side of the blots indicate the positions of corresponding molecular mass markers. Bar graphs adjacent to the blots represent the densitometric analyses of the immunoblot data after normalization with actin. All experiments were done in triplicate. *, p < 0.05 versus control, and #, p < 0.05 versus indomethacin calculated by ANOVA followed by Bonferroni's post hoc test. ns, not significant. A detail of each method is described under “Experimental procedures.”
Indomethacin negatively modulates mitochondrial structural dynamics and functional integrity in rat gastric mucosa
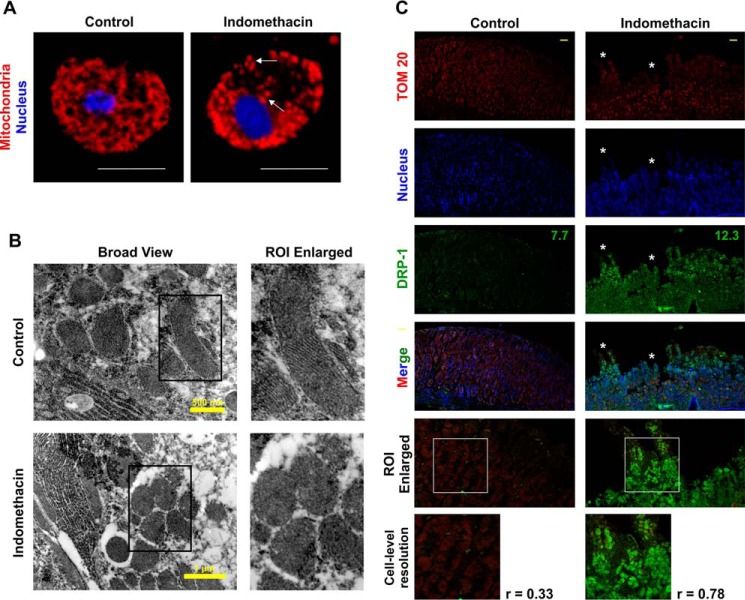
After checking the effect of indomethacin on gastric carcinoma cells in vitro, we were keen to see whether it also targets mitochondrial dynamics in gastric mucosa in vivo. Therefore, gastric mucosal cells isolated from control and indomethacin-treated rats were stained with Mitotracker Red and observed under a high-resolution confocal microscope. Data indicated that although the control cells displayed uniformly distributed mitochondria throughout the cells, cells from indomethacin-treated rat stomachs displayed clumped and punctate mitochondrial particles (Fig. 5A). We next zoomed deeper into the gastric mucosal ultra-structure, in situ, to visualize the finer structural alterations induced by indomethacin. Direct visualization by TEM revealed that architecture of mitochondrial cristae was significantly compromised in indomethacin-exposed injured tissues as evident from an overall loss of stacked structures (Fig. 5B). Because DRP1 is the ultimate mito-fragmenting mediator, its status during gastric mucosal injury was immediately analyzed along with other fission/fusion mediators. In situ analysis by confocal fluorescent immunohistochemistry further indicated that DRP1 expression was moderately higher in the injured gastric mucosa (Fig. 5C, white asterisks) obtained from indomethacin-treated rats. Notably, the colocalization of signals corresponding to DRP1 and TOM20 (mitochondria) implied that indomethacin treatment resulted in elevated mitochondrial translocation of DRP1 compared with control (Fig. 5C). Immunohistochemical data were also supported by immunoblot analysis of DRP1 (Fig. 6, A and B) and its predominant adapter MFF (Fig. 6B). Interestingly in corroboration with the observations in AGS cells, immunoblot analysis in rat gastric mucosa also revealed DRP1 activation by indomethacin, as evident from enhanced DRP1Ser-616 phosphorylation (Fig. 6B). This DRP1 activation was found to be associated with phosphorylation of both PKCζ and p38 MAPK (Fig. 6C). Moreover, depleted expression of mitochondrial fusion mediators, namely MFN1, MFN2, and OPA1, was also documented upon indomethacin treatment relative to control (Fig. 6D).
Figure 5.
Indomethacin induces mitochondrial clumping and fission in vivo in rat gastric mucosa. A, high-resolution confocal micrographs of gastric mucosal primary cells isolated from control and indomethacin-treated rats after 4 h; white scale bars correspond to 10 μm; white arrows indicate clumped, punctated, and peri-nuclearly clustered mitochondria. 80–100 cells were screened, and experiments have been replicated three times; a representative image has been presented. Size bar corresponds to 10 μm. B, ultrastructural analysis of gastric mucosa by TEM to visualize mitochondrial cristae upon indomethacin treatment. Magnified ROI have been presented as “ROI enlarged” adjacent to the “broad view” to clearly demonstrate loss of stacked structures (mitochondrial cristae) after indomethacin treatment. A representative image of one of the several micrographs captured during independent experiments performed has been presented. C, immunohistochemical analysis of mitochondrial translocation of DRP1 upon indomethacin treatment; white asterisks shows the region of mucosal surface erosion/injury. Numerical values presented in green correspond to respective mean intensity values of DRP1. Size bars represent 50 μm. Experiments have been repeated three times, and a representative image for each experimental set is provided. Corresponding values adjacent to the micrographs represent Pearson's correlation coefficient (r) of the red and green signals corresponding to TOM20 and DRP1, respectively. All experiments were done in triplicate. A detail of each method is described under “Experimental procedures.”
Figure 6.
Indomethacin-induced mitochondrial fission in vivo is associated with activation of PKCζ–p38–DRP1 pathway, depletion of fusogenic mediators, and elevation of mitochondrial ubiquitination. A, immunoblot analysis of DRP1 in the mitochondrial extracts isolated from control and indomethacin-treated rat gastric mucosa. TOM20 was used as the loading control. B, immunoblot analysis of pDRP1Ser-616 and MFF in the total tissue lysates from control and indomethacin-treated rat gastric mucosa. Actin was used as the loading control. C, immunoblot analysis of phospho-PKCζ, PKCζ, phospho-p38, and p38 in control and indomethacin-treated rat gastric mucosal extracts. Actin was used as the loading control. D, immunoblot analysis of fusogenic mediators MFN1, MFN2, and OPA1 in control and indomethacin-treated rat gastric mucosal extracts. Actin was used as the loading control. E, immunoblot analysis of Parkin and protein ubiquitination in the mitochondrial extracts from control and indomethacin-treated rat gastric mucosa. TOM20 was used as the loading control. Numerical values at the side of the blots indicate the positions of corresponding molecular mass markers. Bar graphs adjacent to the blots represent the densitometric analyses of the immunoblot data after normalization with the respective loading controls. A representative image of one of the several experiments performed has been presented. *, p < 0.05; **, p < 0.001 versus control calculated by unpaired Student's t test; ns, nonsignificant. A detail of each method is described under “Experimental procedures.”
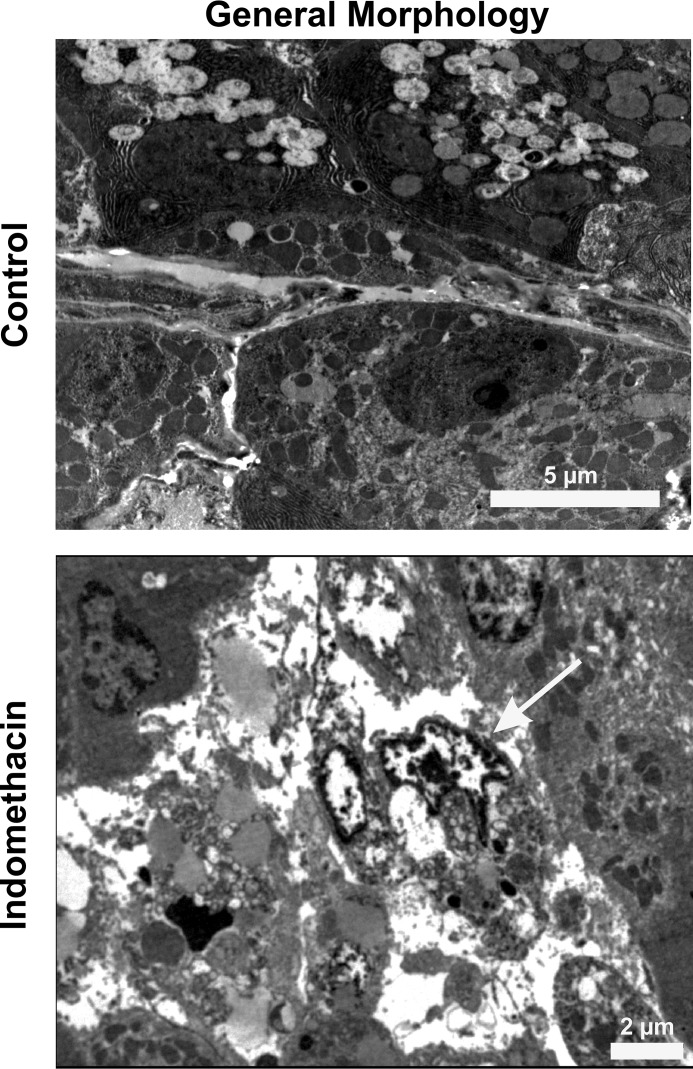
Furthermore, Western blot analysis also revealed increased levels of Parkin and elevated ubiquitination of the mitochondrial proteome in the indomethacin-treated gastric mucosal mitochondrial fractions (Fig. 6E) indicating onset of mitophagy to dispose damaged mitochondria. To study the consequence of these events on mitochondrial physiology and metabolism in vivo, a series of metabolic assays were performed (Table 3). Data indicated that indomethacin-induced elevated mitochondrial fission and clearance was concurrent with retarded ETC complex-I activity and complex-I–driven mitochondrial respiration, which was followed by analyzing the respiratory control ratio (RCR) (measured as a ratio of state 3 and state 4 respirations). Data clearly indicated mitochondrial respiratory defect (Table 3). Because fatty acid oxidation and mitochondrial dehydrogenase activity majorly contributes to the supply of reducing equivalents for the ETC, we next checked these two parameters and found that indomethacin treatment resulted in a significant reduction of dehydrogenase activity, fatty acid oxidation (Table 3). These detrimental changes to the mitochondrial metabolism were further concurrent with indomethacin-induced loss of mitochondrial integrity, as evident from the collapse of ΔΨm and associated bio-energetic deficit, as evident from ATP depletion (Table 3). Furthermore, ultrastructural analysis of the tissue at this time point by TEM revealed that indomethacin caused significant damage to the gastric mucosal tissue architecture with prominent nuclear chromatin condensation and loss of structural integrity of plasma membrane (Fig. 7). Induction of mitochondrial oxidative stress and mitochondrial disintegration upon indomethacin treatment, as evident from the elevated level of macromolecular oxidation end product and reduced cardiolipin content (Table 4), significantly potentiated the damage. These detrimental reactions initiated by indomethacin finally led to the activation of caspase 9 and caspase 3 (Table 4) for triggering the intrinsic pathway of apoptosis.
Table 3.
Indomethacin-induced alteration in mitochondrial functional integrity in rat gastric mucosa and effect of Mdivi-1 in the pathology
Rats were pretreated with Mdivi-1 at 2 mg/kg body weight followed by oral gavaging with indomethacin for 4 h. Control rats were treated with vehicle. Mitochondria were isolated for analyzing β-oxidation of fatty acids, mitochondrial dehydrogenase activity, ETC complex-I, RCR, ATP content, and ΔΨm as described under “Experimental procedures.” All experiments were done in triplicate (with n = 6–8 in each group). Data are presented as means ± S.D. **, p < 0.01; ***, p < 0.001 versus control; #, p < 0.05; ##, p < 0.01 versus indomethacin; NS, not significant.
| Fatty acid oxidation | Mitochondrial dehydrogenase activity (MTT reduction, OD570 nm) | ETC complex I activity | Respiratory control ratio (state 3/state 4) | ATP content | ΔΨm (fluorescence 590/530 nm) | |
|---|---|---|---|---|---|---|
| pmol CO2/g/h | nmol/min/mg | relative luciferase units | ||||
| Control | 1675.2 ± 126.9 | 0.87 ± 0.02 | 55.15 ± 4.0 | 5.29 ± 0.93 | 6355.00 ± 241.89 | 7.10 ± 0.35 |
| Indomethacin | 698.7 ± 137.3** | 0.48 ± 0.05** | 14.06 ± 1.4*** | 1.78 ± 0.14** | 2835.33 ± 465.54*** | 3.10 ± 0.1** |
| Mdivi-1 + indomethacin | 1270.9 ± 121.3## | 0.76 ± 0.04## | 38.28 ± 1.1## | 3.060 ± 0.43# | 4582.67 ± 285.55# | 6.02 ± 0.52# |
| Mdivi-1 | 1483.3 ± 107.4NS | 0.86 ± 0.03NS | 51.12 ± 5.9NS | 5.18 ± 0.80NS | 5727.32 ± 533.59NS | 6.72 ± 0.37NS |
Figure 7.
Indomethacin damages the gastric mucosal ultrastructure. TEM analysis of the gastric mucosal sections from control and indomethacin-treated rats to visualize cytoarchitectural damage. The arrow indicates nuclear chromatin condensation. A representative image of one of the several micrographs captured during independent experiments performed has been presented. All experiments were replicated. A detail of each method is described under “Experimental procedures.”
Table 4.
Indomethacin-induced mitochondrial oxidative stress and apoptosis in rat gastric mucosa and rescue effect of Mdivi-1
Rats were pretreated with Mdivi-1 at 2 mg/kg body weight followed by oral gavaging with indomethacin for 4 h. Control rats were treated with vehicle. Mitochondria were isolated for analyzing MOS, cardiolipin content, and caspase activation as described under “Experimental procedures.” All experiments were done in triplicate (with n = 6–8 in each group). Data are presented as means ± S.D. **, p < 0.01; ***, p < 0.001 versus control; #, p < 0.05; ##, p < 0.01 versus indomethacin.
| Macromolecular oxidation end product | Cardiolipin content | Caspase 9 activity (absorbance, OD405 nm) | Caspase 3 activity (fluorescence, OD460 nm) | |
|---|---|---|---|---|
| nmol/mg wet tissue | fluorescence/mg wet tissue | |||
| Control | 0.68 ± 0.04 | 7288.12 ± 363.2 | 0.198 ± 0.01 | 205.7 ± 12.41 |
| Indomethacin | 1.69 ± 0.05*** | 2800.29 ± 385.3*** | 0.427 ± 0.04** | 524.88 ± 30.02*** |
| Mdivi-1 + indomethacin | 1.06 ± 0.05## | 5189.47 ± 623.3## | 0.264 ± 0.03# | 292.41 ± 33.35## |
Mitochondrial division inhibitor-1 (Mdivi-1) prevents indomethacin-induced mitochondrial fission and metabolic crisis
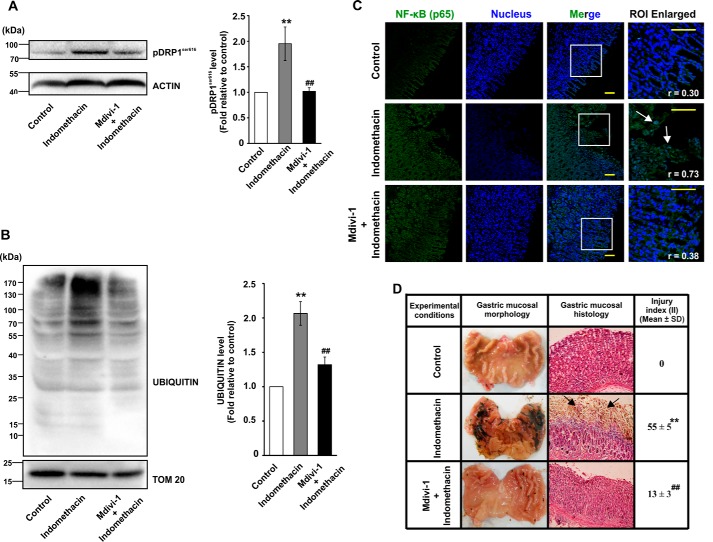
So far, the data clearly indicated that enhanced mitochondrial fission was associated with indomethacin-induced gastric injury. Therefore, we next asked the following question. What would be the effect of blocking enhanced mitochondrial fission? With this aim, we checked whether Mdivi-1 (a small molecule with several positive effects on mitochondrial metabolism (38) and fission-reducing action (39, 40)) could offer mitochondrial protection in vivo. Data indicated that pretreatment with Mdivi-1 at 2 mg/kg body weight resulted in a significant rectification of indomethacin-induced decrease in [14C]palmitate oxidation, mitochondrial dehydrogenase activity, ETC complex-I activity, RCR, and ΔΨm collapse (Table 3). The protective effects of Mdivi-1 were further positively associated with rectification of indomethacin-induced DRP1Ser-616 phosphorylation (Fig. 8A) as well as increased mitochondrial proteome ubiquitination (Fig. 8B). Notably, Mdivi-1 did not significantly affect basal mitochondrial metabolic parameters (Table 3). Thus, the data suggested that Mdivi-1 salvaged gastric mucosal mitochondria from NSAID-induced acute metabolic dysfunction.
Figure 8.
Mdivi-1 prevents indomethacin-induced mitochondrial damage, inflammation, and gastric injury. Immunoblot analyses for DRPSer-616 phosphorylation (A) and mitochondrial proteome ubiquitination (B) in control, indomethacin-treated and Mdivi-1 + indomethacin-treated rat gastric mucosa. Actin and TOM20 were used as the loading control for total cell lysates and mitochondrial fractions respectively. Numerical values at the side of the blots indicate the positions of corresponding molecular mass markers. Bar graphs adjacent to the blots represent the densitometric analyses of the immunoblot data after normalization with the respective loading controls. C, immunohistochemical analysis of nuclear translocation of NF-κB upon indomethacin treatment or Mdvi-1 + indomethacin treatment; white arrows indicate mucosal epithelial cells with higher degree of nuclear NF-κB. NF-κB (green) was immunostained by anti-NF-κB (p65) primary and Alexa Fluor 488-tagged anti-rabbit secondary antibodies. Nucleus (blue) was stained by DAPI. Scale bars correspond to 50 μm. Corresponding values in the inset represent Pearson's correlation coefficient (r) of the blue and green signals corresponding to nucleus and NF-κB, respectively. Random tissue sections were screened, and a representative image of one of the several experiments has been presented. D, gastric mucosal morphology and histology by hematoxylin–eosin staining. Black arrows indicate injured mucosal surface. For all animal experiments n = 6–8, and experiments were performed three times. **, p < 0.01 versus control, and ##, p < 0.01 versus indomethacin as calculated by ANOVA followed by Bonferroni's post hoc test. A detail of each method is described under “Experimental procedures.”
Mdivi-1 also attenuates indomethacin-induced mitochondrial oxidative stress (MOS), apoptosis, and mucosal inflammation to prevent gastropathy
Because indomethacin-induced excess fission and MOS were positively associated, we next checked the effect of Mdvi-1 on mitochondrial macromolecular integrity. Data indicated that Mdivi-1 significantly reduced indomethacin-induced mitochondrial oxidative damage as evident from reduced macromolecular oxidation as well as preservation of cardiolipin content in gastric mucosa of Mdivi-1–treated rats (Table 4). Notably, Mdivi-1 also prevented apoptotic tissue damage as evident from rectification of indomethacin-induced caspase activation (Table 4). Alteration in redox homeostasis activates NF-κB signaling that is found to be associated with proinflammatory tissue damage (41). Therefore, we subsequently checked the impact of Mdivi-1 on gastric inflammation. Data indicated that Mdivi-1 significantly inhibited indomethacin-induced nuclear translocation of NF-κB (Fig. 8C). Subsequently, gene expression analysis of some common proinflammatory biomarkers indicated that Mdivi-1 significantly corrected indomethacin-induced up-regulation of cytokines, IL1β, IL1α, IL17A, chemokines, CXCL2 and CXCL3, tissue-remodeling matrix metalloproteases, including MMP3 and MMP13 and FtMt, a classical marker of MOS (Table 5). Finally, morphological examination followed by the scoring of injury index and histological evaluation strongly indicated that Mdivi-1 offered significant gastroprotection by reducing indomethacin-induced epithelial denudation due to mucosal cell shedding (Fig. 8D).
Table 5.
Gene expression profiling by real-time RT-PCR represented in fold change in gene expression (relative to control, after normalization by Gapdh)
Data are presented as means ± S.D. All experiments were done in triplicate. ***, p < 0.001 versus control; ##, p < 0.01; ###, p < 0.001 versus indomethacin.
| IL1α | Il1β | Il17A | Cxcl2 | Cxcl3 | Mmp3 | Mmp13 | FtMt | |
|---|---|---|---|---|---|---|---|---|
| Indomethacin | 3.45 ± 0.12*** | 11.36 ± 0.68*** | 9.19 ± 0.31*** | 7.15 ± 0.39*** | 7.55 ± 0.17*** | 8.51 ± 0.25*** | 6.66 ± 0.16*** | 5.83 ± 0.47*** |
| Mdivi-1/ indomethacin | 1.82 ± 0.18## | 5.34 ± 0.61### | 2.76 ± 0.62### | 5.77 ± 0.62## | 3.55 ± 0.39### | 5.57 ± 0.56## | 3.05 ± 0.31### | 1.96 ± 0.08### |
Discussion
In this study, we report that indomethacin, a typical NSAID, shifts mitochondrial dynamics toward enhanced fission to induce bio-energetic crisis and subsequently triggers apoptosis in human gastric cancer cells in vitro and rat gastric mucosal cells in vivo. We further provide evidence that blocking excess mitochondrial fragmentation prevents NSAID-induced cellular damage.
Indomethacin, used here as a representative NSAID, is an indoleacetic acid derivative with a pKa of 4.5 (42) that elicits a nonselective inhibitory effect on COX and helps in extending the life span of cancer patients with diagnosed metastases. Moreover, the anti-nociceptive action of NSAIDs makes their usage mandatory despite the negative effects of PG depletion on various systems (43–48). About 54.4 million adults are affected with arthritis in the United States (49), and NSAIDs are rampantly used for therapeutic intervention (50, 51). Apart from arthritis, NSAIDs are extensively used to manage musculoskeletal pain, sprains, fractures, abdominal pains associated with menstrual cramps, and bone pains in certain forms of cancer (52–54). The emerging role of NSAIDs as potential candidates of combinatorial chemotherapy has drawn sufficient attention leading to repurposing the use of these drugs (55–58). Apart from aspirin, various other NSAIDs, including indomethacin, are documented with potential anti-cancer effects. In addition, diclofenac potentiates the anti-neoplastic efficacy of chemotherapy and radiotherapy upon combinatorial usage against various cancers (10, 57). Despite their multiple beneficial attributes, NSAIDs are infamous for their PG-dependent and -independent effects that account for the toxic actions on various cells and organs (6, 11, 12). The gastrointestinal system is most severely harmed, and stomach injury by chronic NSAID consumption is characterized by mucosal perforation, bleeding, tissue wasting, and abdominal pain (10, 17, 59).
Although there is some evidence of different COX-independent cytotoxic actions of NSAIDs (7, 9, 60), it is yet unknown whether these drugs affect mitochondrial structural dynamics in the stomach to create the subcellular havoc. Diverse COX-independent molecular targets of NSAIDs and their metabolites have been found to mediate the effect of these drugs (7). This study is an attempt to find a common COX-independent cytotoxic pathway that may be responsible for inducing apoptosis in cancer as well as normal gastric mucosal cells.
A sublethal dose of indomethacin was selected based on the dose-response study to take into account the subtle effects on mitochondria, which would have otherwise been missed at a higher acute dose. Mitochondrial structural destabilization in gastric cancer cells initiated as early clumping and perinuclear clustering that progressed toward hyper-fission and ultimately fragmentation. OPA-1 is associated with cristae remodeling, mitochondrial fusion, and mitochondrial retention of cytochrome c against pro-apoptotic release (18, 61, 62). In this context, mitochondrial dynamics regulators reacted to indomethacin treatment in a way indicative of enhanced fission. DRP1 oligomerization on mitochondria is mandatory for the fragmentation of mitochondrial tubules (63, 64). Various protein kinases and phosphatases control the phosphorylation of DRP1 on the serine residues 616 and 637, which in turn affects the function of the protein in opposing ways (65–71). Disbalanced phosphorylation ratio at Ser-616 and Ser-637 is implicated in several pathologies (72). Therefore, we checked the status of DRP1 in the NSAID-treated AGS cells. Up-regulation of pDRP1Ser-616 is evident in oxidative stress-associated pathologies (73, 74). Because excess mitochondrial fission is reportedly detrimental to functional integrity and cellular health (75), the metabolic data in this study were in close corroboration with reports of oxidative stress-induced mitochondrial damage during multiple pathologies (74, 76–79). An obvious question is whether the effects pertaining to NSAID-induced metabolic failure are an attribute of cell death or is it indeed the faulty mitochondrial metabolism that increases the predisposition to cell death. To this end, OCR analysis revealed that drastic deterioration of mitochondrial metabolic parameters was evident much earlier even when severe cell death has not set in, implying the contributing role of aberrant fission that initiates at the early hours of indomethacin treatment. In fact, alteration in mitochondrial dynamics via enhanced DRP1 activation along with loss of higher isoforms of OPA1 were found to start much earlier than cell death due to apoptosis. Next, it was mandatory to deduce the mechanistic basis of NSAID-induced DRP1 activation. Indomethacin is known to activate MAPK signaling in various experimental systems (80). Of the three MAPKs (ERK, JNK, and p38), the latter two kinases were more attributed to the cytotoxic actions of the NSAIDs (81, 82). Interestingly, in this study a probable linear relation between NSAID treatment and mitochondrial fission was observed from the sequential phosphorylation-dependent activation of PKCζ followed by p38 and DRP1 along with notable rescue of ΔΨm collapse, mitochondrial hyper-fission, DRP1 activation, and cell death upon pretreatment with the specific p38 inhibitor SB203580. It has been also established that indomethacin increases the permeability of gastric epithelial cells at the tight junctions by p38 MAPK activation (80) and that p38 MAPK activation is positively associated with PKCζ activation (83) under the influence of ROS (84). Thus it may be presumed that NSAIDs trigger PKCζ phosphorylation, which plausibly activates p38 that finally acts as the probable DRP1-activating kinase. In fact, in the aforementioned context, the PKCζ inhibitor PKCζ–PSI offered significant protection against NSAID-induced mitochondrial depolarization, fragmentation, and apoptosis comparable with the p38 inhibitor. Moreover, blockage of p38 activation and DRP1 phosphorylation by PKCζ inhibition, in this study, further indicated a probable regulatory action of PKCζ on p38–DRP1 signaling in response to indomethacin-induced oxidative stress in the AGS cells. It is worth mentioning that various PKC isoforms, including PKCζ, are associated with mitochondrial ROS production; however, the signaling pathways triggered by ROS-induced activated PKCs are essentially isoform- and cell type-specific in addition to the differences based on intracellular sites of ROS generation (85). We also confirmed that mitochondrial fission was indeed a cause and not simply a consequence of NSAID-induced cancer cell death because DRP1-knockdown cells treated with indomethacin exhibited better metabolic integrity along with reduced fission.
Apart from cancer, the toxic effects of NSAIDs are observed as a side effect in various organs in which stomach is the most prominent target (10). However, it is still unknown whether mitochondrial structural dynamics are affected by NSAIDs during the induction of gastric injury and, if it happens at all, whether aberrant mitochondrial dynamics can be targeted for therapeutic management of NSAID gastropathy. In this study, we show that as in the gastric cancer cells, indomethacin similarly affects mitochondrial dynamics to elicit the gastrodamaging effects. Fissogenic modification of DRP1 and its concerted association with PKCζ–p38 MAPK activation by NSAID may be justified because oxidative stress-induced p38 MAPK and JNK pathway activation is associated with stress responses, including inflammation and apoptosis (86, 87), whereas the ERK–MAPK pathway is associated with cell survival, proliferation, and mitotic division (88). Ultrastructural micrographs of gastric mucosa after NSAID-induced damage to mitochondrial cristae logically explained cardiolipin depletion due to its location in the IMM. Clearance of metabolically unfit mitochondria commences with mitophagic activation and ubiquitination-induced degradation (89, 90). However, if the intensity of the cytodamaging signal and the extent of damage surpass the limit of mitochondrial quality control machinery, the cells are sensitized to undergo apoptosis (89, 91). ROS operates in a positive feedback cycle by causing perpetual mitochondrial and tissue damage. In fact, accumulation of damaged and fragmented mitochondria potentiates the intrinsic pathway of apoptosis (92). In this context, it is worth mentioning that pharmacological blocking of altered mitochondrial dynamics by a well-established potent inhibitor of mitochondrial fission (93), Mdivi-1, offered significant protection against NSAID-induced mitopathology and gastropathy. Mdivi-1 has been extensively validated in several preclinical disease models of oxidative stress–associated mitochondrial pathologies (31, 94–96). It has also been shown that the dose of Mdivi-1 is very crucial because a higher dose may induce cytotoxicity instead of offering cytoprotection (97). Moreover, whereas Mdivi-1 offers protection against various oxidative stress-associated diseases (both in vitro and in vivo), it is toxic to cancer cells (98, 99). Hence, we used Mdivi-1 only in the in vivo study, whereas DRP1 was knocked down in vitro by using siDRP1. The dose used in this study is in accord with the diverse reports of the protective effect of Mdivi-1 in various diseases (39). Moreover, prevention of indomethacin-induced DRP1 activation and mitochondrial proteome ubiquitination confirmed the anti-fission action of Mdivi-1 in this study, which is also in accordance with the reports of oxidative stress-associated brain tissue injury (72, 100). Fragmented and depolarized mitochondria have been reported to aggravate ROS generation, which further damages the organellar macromolecules in cardiovascular injury where mitochondrial fragmentation is positively correlated with elevated mitochondrial ROS generation (101). Thiobarbituric acid-reactive mitochondrial macromolecule oxidation end products are highly detrimental irreversible chemical changes induced by O2˙̄ (102–105). Cardiolipin peroxidation (106) is associated with cytochrome c release (107) through the permeability transition pores formed during oxidative stress (108). In this study, we observed that despite not being a classical antioxidant, Mdivi-1 successfully prevented mitochondrial macromolecular oxidation and cardiolipin depletion, which is most likely via stabilizing aberrant structural dynamics and preventing the onset of self-propagating oxidative damage. Furthermore, free heme released upon ROS-induced damage to mitochondrial heme-containing proteins, in concert with gastric acid–dependent erosion of microvasculature and consequent hemolysis, promotes localized pro-inflammatory cell recruitment (109–114). Activated phagocytes secrete more cytokines and chemokines to aggravate inflammation (115). NF-κB is a master regulator of inflammation (116) that controls the expression of several pro-inflammatory cytokines (117). Moreover cytokines like IL-1β and IL-17A regulate NF-κB activation (118–120). It is often observed that cytokine-induced activation of the tissue-remodeling matrix metalloproteases like MMP3 and MMP13 occurs via NF-κB in various chronic inflammatory diseases (121, 122). Moreover, IL-1β–induced CXCL2 and CXCL3 expression during vascular remodeling, leukocyte infiltration, and inflammation is also regulated by NF-κB (123). FtMt, a classical marker of MOS, is up-regulated upon intra-mitochondrial free iron accumulation during ROS-induced damage of mitochondrial iron–sulfur cluster-containing proteins (124, 125). A redox-active milieu with active inflammation thereby sets in to perpetuate tissue injury. In this study, besides inhibiting caspase activation, Mdivi-1 was also effective in preventing gastric mucosal inflammation and inflammation-associated detrimental tissue remodeling. It may be speculated that ROS and mitochondrial fission proceeds in a self-perpetuating positive feedback loop in which a defect in the mitochondrial ETC and subsequent ROS generation may trigger the initial activation of fissogenic signaling for segregation of damaged mitochondria. However, excess fission becomes detrimental due to generation of more ROS in the subsequent stages. In addition, inflammation-associated ROS further amplifies the detrimental chain reactions (126). In this regard, Mdivi-1 is presumed to block excess fission thereby preventing the vicious feed-forward loop of ROS-fission chain reaction. It is worth mentioning that no significant change was found in the basal state of mitochondrial metabolic parameters when treated only with Mdivi-1. However, a slight increase in the ΔΨm in “Mdivi-1” sample might be due to the reduction of fission. Hence, it may be considered that Mdivi-1 is potentially safe for use in murine experimental gastric injury models. Another notable fact is that a mitochondrially targeted antioxidant, mitoTEMPO, has been found to prevent pathological fission of mitochondria (127, 128). Interestingly, mitoTEMPO also offers gastroprotection from NSAIDs (129). Hence, the contribution of ROS in initiating fissogenic activation of DRP1 is unequivocal. However, from this study it may be concluded that aberrant mitochondrial fission is apparently one of the most significant events that underlie NSAID-induced gastric cell death, predominantly due to the detrimental activation of DRP1 that participates both in triggering fission while contributing to apoptosis.
Therefore, this study for the first time demonstrates that NSAIDs induce mitochondrial hyper-fission to induce apoptosis in both gastric cancer as well as normal gastric mucosal cells, whereas targeting aberrant mitochondrial dynamics and contextual modulation of hyper-fission can certainly help in increasing the therapeutic utility of NSAIDs.
Experimental procedures
Materials
Indomethacin, thiobarbituric acid, 5,5′-dithiobis(nitrobenzoic acid), reduced glutathione (GSH), DMSO, paraformaldehyde, and mitoTEMPO were obtained from Sigma. Fetal bovine serum was purchased from Gibco, Invitrogen. MitoTracker Red CMX-ROS, nonyl acridine orange (NAO), and Hoechst 33342 were purchased from Invitrogen. 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazol-carbocyanine iodide (JC-1) was procured from Molecular Probes (Eugene, OR). The mitochondria isolation kit was purchased from the Biochain Institute (Hayward, CA). Alexa Fluor 647- and Alexa Fluor 488–tagged antibodies were purchased from Invitrogen. MFN-1, OPA-1, DRP1, MFF, TOM20, and NF-κB (p65) antibodies were procured from Abcam. Actin antibody was obtained from Biovision. pDRPSer-616 and pPKCζ antibodies were obtained from Cell Signaling Technology. MFN2, TOM20, p38, phospho-p38, and ubiquitin antibodies as well as siRNAs, SB203580, and PKCζ pseudo substrate inhibitor peptide (PKCζ–PSI) were obtained from Santa Cruz Biotechnology. Mdivi-1 and sodium fluoride were purchased from Merck. 14C-Labeled palmitate was procured from BRIT (Mumbai, India). F-12 Ham Kaighn's modification medium was procured from HiMedia Laboratories. All other reagents were of analytical grade purity.
Human gastric epithelial cell culture
AGS cells (ATCC-CRL-1739) were procured from American Type Culture Collection (Manassas, VA). These cells were cultured in nutrient mixture F-12 Ham, Kaighn's modification with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin, at 37 °C in a 5% CO2 incubator. The cells were split by 1× trypsin/EDTA solution once every 3 days. All the experiments were done with the cells within 5–8 passages. The cells were plated at a density of 1 × 106 in a 60-mm culture plate for most of the experiments and allowed to grow overnight, after which experiments were performed in fresh media. For p38 MAPK inhibition, cells were treated with SB203580 (10 μm) initially for a period of 1.5 h following which the cells were treated with 0.5 mm indomethacin. For PKCζ inhibition, cells were pretreated with PKCζ–PSI (20 μm) initially for a period of 1.5 h following which the cells were treated with 0.5 mm indomethacin. For immunofluorescence studies, cells were grown on polylysine-coated glass coverslips or glass-bottom culture dishes.
[3H]Thymidine incorporation to follow cell proliferation
DNA synthesis during cell proliferation was monitored by labeling the cells using [3H]thymidine. Briefly, AGS cells (1 × 104 cells/ml) were seeded in triplicate for each set of treatments in 96-well plates and allowed to grow overnight. The next day, the medium was changed, and the cells were treated with indomethacin at the indicated concentrations for 24 h. Next, an aqueous solution of [3H]thymidine (1 mCi) (PerkinElmer Life Sciences) diluted with media (final concentration 5 μCi/well) was added to each well. After 24 h of incubation, cells were harvested and transferred to scintillation liquid. The level of incorporated [3H]thymidine was assessed using the PerkinElmer Life Sciences (Tricarb 2810TR) liquid scintillation counter. Radioactivity of the experimental samples was expressed as disintegrations/min, which was proportional to the amount of [3H]thymidine incorporated into the DNA during synthesis.
Isolation of mitochondria
Mitochondria were isolated from both control and experimental sets of rat stomachs using a commercially available kit from Biochain. Briefly, gastric mucosal scrapings were isolated from the rats, after which they were weighed, and an equal amount of tissue (200 mg) from various experimental sets was finely minced in mitochondria isolation buffer. The contents were finally homogenized in a Dounce homogenizer. To get the mitochondrial fraction, the nuclei and the unbroken cells were first eliminated from the homogenate by centrifuging at 600 × g for 10 min, and the supernatant was further centrifuged at 12,000 × g for 15 min. After this, the entire process was repeated, and the mitochondrial pellet was washed three times by centrifugation. For future use, the pellet that was formed as a result of the above step was resuspended in mitochondrial storage buffer. Mitochondria from cells were isolated using a commercially available kit from Qiagen following the manufacturer's instructions. Percoll density gradient centrifugation was done to further purify the mitochondrial pellet for mitochondrial respiration assay (130).
Cellular dehydrogenase assay
Cellular dehydrogenase activity was evaluated by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction. The principle of the assay was that cellular dehydrogenases are capable of reducing MTT into purple-colored formazan dye which upon dissolution in DMSO can be estimated spectrophotometrically at 570 nm. AGS cells were plated in 48-well tissue culture plates and allowed to grow to 60–70% confluency after which they were treated with indomethacin at the indicated concentrations for 24 h. After treatment, the cells were incubated with 1% MTT in PBS and incubated for 3.5 h under a 37 °C, 5% CO2 condition. The MTT solution was subsequently aspirated out, and the formazan precipitates were solubilized in DMSO and spectrophotometrically measured at 570 nm. For measuring mitochondrial dehydrogenase activity of rat gastric mucosa, isolated mitochondria from control and treated gastric tissues were measured by the Lowry method, and an equal amount of mitochondrial protein was incubated with MTT (dissolved in PBS) for 3.5 h at 37 °C/5% CO2. After incubation was over, samples were centrifuged, and the formazan precipitate was dissolved in equal amount of DMSO and measured at 570 nm.
Phase-contrast and confocal microscopy for live cells, immunocytochemical and immunohistochemical analysis
Sample preparation for live-cell imaging was done as mentioned earlier with minor modifications (131). Live-cell phase–contrast microscopy was done by treating AGS cells with indomethacin in a 35-mm culture plate followed by washing and viewing under an inverted phase-contrast microscope (Leica Microsystems). Representative images of control and indomethacin-treated cells have been provided. For live-cell imaging to document mitochondrial fragmentation, the cells were washed with prewarmed PBS and loaded with MitoTracker Red CMX ROS (100 nm) for 20 min at 37 °C. Nucleus was counterstained by Hoechst 33342. The cells were next washed with PBS and viewed under the microscope on a thermo-regulated stage in a 5% CO2 environment at the indicated time points. Samples were viewed under ×63 oil immersion lens, and digital zooming was performed. Image cropping and global adjustments to brightness and contrast were done in Adobe Photoshop CS6. The data presented are a typical representation of individually repeated experiments with similar results. Approximately 100 cells were screened per experimental set. The image acquisition times were kept as low as possible (not exceeding 30 s), and laser intensities were kept at <2% to avoid any possible laser-induced toxicity. Immunofluorescent staining of DRP1, MFF, or p-Drp1Ser-616 was performed as mentioned previously (132). Briefly, AGS cells were plated on polylysine-coated coverslips. After treatment, the cells were washed with prewarmed PBS and subsequently fixed in 2% buffered paraformaldehyde for 10 min and next blocked in a buffer containing 2% BSA in PBS/Tween 20 for 1 h. Immunostaining was done using antibodies against TOM20 (sc-136211, 1:250) developed in mice and DRP1 (ab54038, 1:100) or MFF (ab81127, 1:250) or p-Drp1Ser-616 (4494S, 1:400) developed in rabbits. Goat anti-rabbit Alexa Fluor 488–conjugated secondary antibody (A-11034, 1:500; Invitrogen) was used for staining DRP1 or MFF, whereas goat anti-mouse Alexa Fluor 647–conjugated secondary antibody (A-21245, 1:1000; Invitrogen) was used for staining TOM20 to visualize mitochondria. For STED super-resolution microscopy, the samples were imaged by excitation with 592-nm depletion laser under ×100 oil immersion objective lens. Precise location of DRP1 on mitochondrial filaments was visualized as discrete dotted fluorescent signals. Colocalization of the red (mitochondria) with the green (DRP1) signals were quantified using the LAS-X software taking into account the Pearson's correlation coefficient, and images provided were a representative of three independent experiments performed. About 100 cells were scanned, and representative images of different fields were provided. For immunohistochemical analysis of DRP1 and NF-κB p65 translocation in gastric mucosal tissues, formalin-fixed and paraffin-embedded tissue sections were deparaffinized and finally heated in sodium citrate buffer for antigen retrieval. 10% goat serum and 1% BSA, prepared in TBS, was used as a blocking solution to block the tissue sections for 2 h. For DRP1 immunohistochemistry, mitochondria were stained by TOM20 primary antibody (sc-136211, 1:250) and DRP1 by anti-DRP1 primary antibody (ab54038, 1:100). For nuclear translocation of NF-κB, the fixed and permeabilized tissues were stained with antibody against NF-κB p65 (ab7970, 1:400), and the nuclei were stained with DAPI. All experiments were performed three times, and confocal micrographs presented were representatives of randomly chosen cells or a portion of the gastric mucosal sections. Digital zooming of the obtained micrographs was performed as and when required. Images were cropped and processed globally for enhancement of brightness and contrast using Adobe Photoshop CS6. Images were assembled in Corel DRAW X7 software to prepare the figure.
Immunoblot analysis
Gastric cancer cells were extracted by trypsinization after treatment with indomethacin, and total protein or mitochondrial extract was prepared. Cellular mitochondria were isolated using mitochondria isolation kit (Qiagen) following the manufacturer's instructions. Tissue mitochondria were isolated, as mentioned previously (133), using a commercially available kit from Biochain. Isolated mitochondrial fractions were lysed in the lysis buffer supplemented with the protease inhibitor mixture that was provided along with the kit. Protein quantification was done by the Lowry method, and equal amounts of protein were loaded in each well of the 10% SDS-polyacrylamide gels. The electrophoresis was performed, and subsequently immunoblotting was done at a constant voltage of 100 V. Pre-stained molecular weight markers were used as a reference. Proteins were transferred in nitrocellulose membrane in a transfer buffer (190 mm glycine, 20 mm Tris base buffer, pH 8.3, containing 20% methanol). The membranes were blocked in 5% BSA or nonfat milk in TBS solution. The membranes were incubated in respective primary antibody solutions overnight followed by washing in TBS containing 0.1% Tween 20. Next, the blots were incubated in secondary antibody solution for 2 h at room temperature. The membranes were again washed, and blots were developed by staining with either 3,3′-diaminobenzidine (DAB) solution containing H2O2 or developed in Bio-Rad ChemiDoc MP imaging system. The documentation of DAB-based developed bands was done by scanning at high resolution (300 ppi) in a scanner. For total protein, cells were lysed in mammalian cell lysis buffer and used for Western immunoblotting. For phosphoprotein immunoblotting, cell lysates were also supplemented with phosphatase inhibitor mixture (Sigma) immediately after cell harvesting. TOM20 and actin were used as the internal controls for mitochondrial extract and total cell lysates, respectively. Densitometric analyses of the developed bands were done by ImageJ software (134), and a representative immunoblot image of one of the several independently performed experiments has been provided in the image with calculation of fold change relative to control after normalization with actin or mitochondria as required for total lysates or mitochondria, respectively. Furthermore, densitometric analyses of the immunoblot data are provided as bar graphs adjacent to the respective blots. The antibodies from Abcam were as follows: Drp1, ab54038, 1:1000; Mfn1, ab104274, 1:1000; Mfn2, ab56889, 1:1000; OPA1, ab42364, 1:1000; and MFF, ab81127, 1:1000. The antibodies from Cell Signaling Technology were as follows: p-Drp1Ser-616, 4494S, 1:1000; phospho-p38, 9211S, 1:1000; PKCζ, 9368S, 1:1000; and pPKCζ, 9378S, 1:1000. The antibodies from Santa Cruz Biotechnology were as follows: ubiquitin, sc-8017, 1:500; p38, sc7972, 1:1000; TOM20, sc-11415, 1:1000; actin, 3598R, 1:2000; Parkin, P6248, 1:1000; anti-rabbit, IgG, A0545, 1:40,000; and anti-mouse IgG, A9044, 1:25,000.
OCR by extracellular flux analyzer
Oxygen consumption ratio was measured as mentioned previously (135, 136) with minor modifications. AGS cells were counted in a TC-10 cell counter (Bio-Rad), plated at a density of 30,000/well in an XF24 plate (Seahorse Biosciences, North Billerica, MA), and allowed to grow for 24 h in 5% CO2 at 37 °C. Next day, the medium was changed, and cells were treated with 0.5 mm indomethacin for 6 h in presence/absence of SB203580. In siDRP1-treated cells, transfection was done 48 h before indomethacin treatment. After the treatment, the cells were washed and incubated for 1 h in 500 μl of sodium bicarbonate free XF Assay media (143 mm NaCl, 5.4 mm KCl, 0.91 mm Na2HPO4, 20 mm glucose, 2 mg/ml BSA, 1 mm sodium pyruvate, 0.8 mm MgSO4, and 1 mm glutamine) at pH 7.4 for 1 h. Subsequently, oligomycin (2 mm), carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP) (1 mm), and rotenone + antimycin A (1 mm each) were injected over a span of 100 min (via loading in the injection ports A, B, and C, respectively) to measure the different stages of mitochondrial respiration at 37 °C. The analysis was performed with XFe 2.0.0 software (Seahorse Biosciences). Oxygen consumption ratio for each sample was normalized with the respective protein concentrations. The formulae for calculations were as follows.
Basal respiration = (measurement prior to oligomycin addition) − nonmitochondrial respiration.
Proton leak = (first measurement after oligomycin injection through measurement before FCCP) − nonmitochondrial respiration.
ATP production = basal respiration − proton leak.
Maximal respiration = (first measurement after oligomycin addition through measurement prior to rotenone + antimycin A injection) − nonmitochondrial respiration.
Spare/reserve respiratory capacity = maximal respiration − basal respiration.
Analysis of mitochondrial transmembrane potential (ΔΨm)
After treatment, the cells were rinsed in prewarmed media (37 °C) and incubated in JC-1 (5 μg/ml) diluted in cell culture media for 15 min at 37 °C, 5% CO2 in darkness. After incubation, the cells were dissociated and diluted in PBS to a density of 1 × 106 cells/ml and analyzed by fluorescence-activated cell sorter (FACS) LSR Fortessa (BD Biosciences) using FACS DIVA software under standard parameters. Experiments were repeated three times, and representative images of the FACS scatterplots corresponding to each set were presented in the image. For analysis of ΔΨm in vivo, mitochondria were isolated from control and treated stomachs. Mitochondrial protein was estimated, and equal amounts of mitochondria from different experimental sets were incubated in darkness for 15 min in 500 μl of JC-1 assay buffer containing 300 nm JC-1. For measuring fluorescence, an F-7000 fluorescence spectrophotometer (Hitachi High-Technologies Corp.) was used with single excitation–dual emission format. ΔΨm was expressed as fluorescence ratio of 590:530 nm.
FITC-annexin V staining for cell death determination
Annexin V staining was done to measure the apoptosis in AGS cell after DCF treatment. Briefly, in complete medium 1 × 106 cells were plated in 6 cm culture plates and grown overnight. The next day, cells were treated with different concentrations of indomethacin for 24 h by replacing the existing media with fresh media. After the treatment, the media were removed, and the cells were harvested by trypsinization. Cells were then counted, and 1 × 106 cells were taken in 2-ml microcentrifuge tubes. Cells were then stained with FITC–annexin V in annexin V binding buffer as instructed in the manufacturer's protocol (Abcam, Cambridge, MA). The cells were next counter-stained with propidium iodide and finally analyzed in FACS as mentioned above. Experiments were repeated three times, and representative images of the FACS scatterplots corresponding to each set were presented in the image.
Transient transfection for DRP1 silencing
Reverse transfection protocol was followed. The lyophilized siRNA duplex was resuspended to make a 10 μm solution in the resuspension buffer (10 μm Tris-HCl, 20 mm NaCl, 1 mm EDTA, pH 8.0). Cells were plated and allowed to grow until 50–60% confluency, and then they were transfected. For each transfection, Lipofectamine RNAiMAX was diluted in Opti-MEM media. siRNA was also diluted individually to 80 pmol in Opti-MEM media. Both the incubations were of 5 min each and mixed by gentle tapping. The diluted siRNA was added dropwise to the Lipofectamine solution and gently mixed by tapping following which the mixture was incubated at room temperature for 30 min. In the meantime, the cells were also acclimatized in Opti-MEM for 30 min. Finally, the siRNA/Lipofectamine mixture was added to the cells and incubated for 16–18 h, after which the transfection medium was replaced by complete growth media. After 48 h from the initiation of transfection process, the cells were treated with indomethacin for 24 h and finally analyzed for dehydrogenase activity.
The sequence of DRP1 siRNA was as follows: sense, 5′-AACGCAGAGCAGCGGAAAGAGtt-3′, and antisense, 5′-CUUUCCGCUGCUCUGCGUUtt-3′.
Animals and NSAID-induced gastric mucosal injury
All the in vivo studies were performed following the protocol approved by the Animal Ethics Committee of CSIR–Indian Institute of Chemical Biology, Kolkata, India, and registered with the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA), India (Permit no. 147/1999/CPCSEA). Maximum care was used while handling the animals to minimize their suffering. Sprague-Dawley rats weighing 180–220 g were used. For proper maintenance, the animals were kept in the institutional animal house at a temperature of 24 ± 2 °C with a 12-h light and dark cycle and were provided standard rat chow. To avoid unnecessary food-induced acid secretion and its adverse aggravating effects on gastric injury, animals were fasted for 24 h (with ad libitum access to water) prior to the experiments. All the animals were divided into following experimental groups (with n = 6–8 in each group); “control,” “indomethacin-treated,” and “Mdivi-1 + indomethacin treated.” The vehicle controls were treated separately each time. Indomethacin was administered orally to the rats at a dose of 48 mg/kg body weight, and Mdivi-1 (2 mg/kg body weight) was administered 1 h prior to NSAID treatment by intravenous injection. The stock solution of Mdivi-1 (dissolved in DMSO) was diluted in sterile saline during injection so as to keep the final concentration of DMSO at 0.1%. At this concentration, DMSO did not impose any effect on gastric injury caused by the NSAIDs (data not shown). A separate set of rats was treated only with Mdivi-1, and mitochondrial metabolic parameters were measured to follow the effect of Mdivi-1 on basal mitochondrial metabolism. After 4 h of NSAID administration, the rats were sacrificed upon proper euthanasia. The stomachs of the rats were dissected out after washing and gathered. An individual ignorant of the treatment was called for scoring the ulceration that was indicated as injury index (II). This is calculated as a percentage of injured area in the gut of the rats formed by bleeding lesions and clots. Mean II was calculated as the sum of the total scores in each group of rats divided by the number of individuals in that group.
TEM
Sample preparation for TEM analysis was done as mentioned previously (133). Control and indomethacin-treated gastric mucosal tissues were cut into small pieces (1 mm3) and fixed with 4% paraformaldehyde and 2% glutaraldehyde in 0.1 m sodium phosphate buffer, pH 7.4, for 4 h at room temperature. The tissues were then washed in 0.1 m sodium phosphate buffer and subsequently incubated in 2% osmium tetraoxide for 2 h. The post-fixed tissues were then dehydrated in ascending grades of ethanol and embedded in Epon 812 by polymerization at 60 °C for 24 h. Ultracut ultra-microtome (Leica Microsystems GmbH, Wetzlar, Germany) was used for tissue sectioning (50–70 nm). The sections were then positioned onto 200 mesh copper grids and double-stained with uranyl acetate and lead citrate followed by visualization under FEI Tecnai-12 twin transmission electron microscope at 80 kV (FEI Co., Hillsboro, OR).
Measurement of MOS
Mitochondrial oxidative stress was evaluated by measuring thiobarbituric acid–reactive macromolecule oxidation products, including malondialdehyde, which is one of the major aldehyde-adducts formed during oxidative modification of cellular as well as organellar macromolecules like proteins, lipids, and nucleic acids. Therefore, in vivo MOS was measured by analyzing the extent of accumulated oxidized mitochondrial macromolecules in the rat gastric mucosa. For this, equal amounts of mitochondrial fraction (100 μg) were taken from both the control and treated groups and then homogenized in ice-cold 0.9% saline. Next, 0.5 ml of the homogenate was taken and mixed with 1 ml of a thiobarbituric acid/TCA mixture (0.375% w/v, 15% w/v, respectively) in 0.25 n HCl. Then, the mixture was boiled for 15 min. The solutions were then cooled to room temperature and spun at high speed for 10 min. The clear supernatant was collected and measured at 535 nm with tetraethoxypropane considered as standard.
Measurement of fatty acid oxidation
Oxidation of 14C-labeled palmitate to 14CO2 was measured via the assay that was described previously (73) with minor change. In brief, an equal amount of gastric mucosal tissue was taken to perform the assay. Assay mixture contained 1 μCi of 1-14C, which was preincubated in 5% BSA. 14CO2 was trapped on Whatman chromatography paper that was soaked in 3 m NaOH. The papers were allowed to dry overnight, and finally the 14C levels were measured in a scintillation counter (PerkinElmer Life Sciences). The assay was performed in triplicate and normalized by tissue weight.
Mitochondrial respiration
Estimation of oxygen consumption rate was performed to evaluate the mitochondrial respiration. Clark-type oxygen electrode was used in a liquid-phase oxygen measurement system (Oxytherm System, Hansatech Instruments Ltd., UK). RCR was derived from the ratio of state 3 and state 4 respiration that was measured in terms of nanomoles of O2 consumed. Briefly, malate (5 mm) and glutamate (5 mm) were added to the respiratory medium (5 mm KH2PO4, 250 mm sucrose, 0.1 mm EDTA, 5 mm MgCl2, and 0.1% BSA in 20 mm HEPES, pH 7.2). After adding 1 mm ADP, state 3 respiration was measured, and state 4 respiration was recorded in the presence of 15 μm oligomycin (acts as an ATP synthase blocker). Mitochondrial respiration was normalized by protein content of the respective mitochondrial fractions in control and different experimental sets.
Mitochondrial ETC complex I assay
Mitochondrial complex I activity was measured spectrophotometrically as described (137, 138). Equal amounts of isolated mitochondria (15 μg), isolated from control as well as treated rat stomachs, were exposed to two rounds of freeze–thaw cycles in hypotonic buffer. Complex I activity was assayed based on change in OD340 nm caused by oxidation of NADH along with ubiquinone as the substrate. In a separate reaction, rotenone was used for control as well as each treatment sets comprising all the reagents. Decrease in absorbance was recorded in a Shimadzu UV-visible spectrometer for a period of 2 min (Shimadzu Corp., Japan). Complex 1 activity was estimated by measuring rotenone-sensitive NADH–ubiquinone oxidoreductase activity.
Assays of caspase-9 and caspase-3 activities
Caspase-9 and caspase-3 activities in gastric tissue homogenate were measured by commercially available kit from Calbiochem (Merck, Germany) following the protocol of the manufacturer. For both the assays, 30 mg of gastric mucosa was homogenized in the lysis buffer provided with the respective kit. The homogenate was spun at 16,000 × g for 15 min. The supernatant was collected, and the protein quantity was measured by Lowry's method. Finally, the samples containing equal amounts of proteins from both the control and the treated groups were used to perform the assay. The respective substrates provided in the kit were added in the reaction mixture and incubated in darkness at 37 °C for 2 h. The assay was kinetically monitored; and the end point reading was considered to depict the respective caspase activities in the tissue samples from control as well as the treated rats.
Measurement of cardiolipin content
NAO was used to measure the content of mitochondrial cardiolipin as mentioned previously (133) with minor modifications. In brief, mitochondria were isolated from the gastric tissue of control and treated rats, and equal amounts of mitochondria (25 μg) were used to perform the assay. After the incubation with NAO, the fluorescence of each sample was measured in a spectrofluorimeter microplate reader (BioTek Instruments, Inc., BioTek India, Mumbai) (excitation 495 nm; emission 519 nm, gain 50%). Mean fluorescence was calculated per mg of mitochondrial protein and presented as a fold decrease relative to control for both control and treated gastric mitochondrial fractions.
Measurement of tissue ATP content
ATP determination kit (Invitrogen) was used to measure the ATP. Briefly, 50 mg of gastric mucosal tissue from both the control and NSAID-treated rat was minced and lysed in 5% sulfosalicylic acid. Next, the lysate was centrifuged. The supernatant was collected and used for the measurement of ATP in a Varioskan Flash luminometer (Thermo Fisher Scientific Inc.). The ATP content was represented as relative luciferase unit after the blank was subtracted from the samples.
RNA isolation and qPCR
Total RNA were isolated from control and treated tissues using TRIzol reagent (Invitrogen) following the manufacturer's protocol. DNA contamination was eliminated by applying rDNase (DNA-free kit, Invitrogen) to 10 μg of RNA. cDNA was then synthesized from the total RNA (2 μg) by using oligo(dT)18 primer with the help of RevertAid First Strand cDNA synthesis kit (Thermo Fisher Scientific Inc.). Next, the synthesized cDNA was qPCR-amplified by using gene-specific primers (Integrated DNA Technologies, Inc., San Diego) listed in Table 1 to study the gene expression profile using Power SYBR Green (Applied Biosystems), in an ABI 7500 real-time PCR system (Applied Biosystems). The reaction conditions were maintained as follows: 1 cycle of 95 °C for 10 min and then 40 cycles of 15 s at 95 °C and 60 °C for 1 min. Gapdh was chosen as an internal control. 2−ΔΔCT method (134) was opted for the analysis of data that were normalized to the Gapdh levels and expressed as fold change compared with control. The primer sequences are provided in Table 6.
Table 6.
Primer sequences used in PCR
FP is forward primer, and RP is reverse primer.
| Genes | Primer sequence (5′→3′) | Product size |
|---|---|---|
| bp | ||
| Ftmt | ||
| FP | CGTGTACCTGTCCATGGCTTAC | 84 |
| RP | AGGGACTGGCGAAGGAAATAC | |
| IL1α | ||
| FP | GTCAGCAACATCAAACAAAGGGAAG | 80 |
| RP | CCAGGTCATCTTCAGTAAAGGGC | |
| Mmp3 | ||
| FP | CTGTCTTTGAAGCATTTGGGTTTC | 82 |
| RP | GTCACTTTCCCTGCATTTGGATC | |
| Mmp13 | ||
| FP | TTTGATGGGCCTTCTGGTCTTC | 97 |
| RP | TGCTTGTCCAGGTTTCATCATCATC | |
| Cxcl2 | ||
| FP | GGGTTGTTGTGGCCAGTGAG | 84 |
| RP | CCGTCAAGCTCTGGATGTTC | |
| Cxcl3 | ||
| FP | GGACTGTTGTGGCCCGTG | 86 |
| RP | CACCGTCAAGCTCTGGATGTTC | |
| Il17a | ||
| FP | GGAGCCTGAGAAGTGCCC | 90 |
| RP | GCATGGCGGACAATAGAGGAAAC | |
| Il1β | ||
| FP | CCCCAACTGGTACATCAGCAC | 72 |
| RP | TCCCGACCATTGCTGTTTCC | |
| Gapdh | ||
| FP | AGTATGACTCTACCCACGGC | 165 |
| RP | TGAAGACGCCAGTAGACTCC |
Histological study of gastric mucosal tissue sections and Soret spectroscopy to detect mucosal bleeding
Gastric mucosal tissues were collected each from control as well as drug-treated rats and were fixed in 10% buffered formalin, pH 7.4, for 12 h followed by passage through graded ethanol and finally embedded in paraffin wax. Next, semi-thin sections (5 μm) were made and collected on poly-l-lysine–coated glass slides. The tissue sections were ultimately double-stained using eosin–hematoxylin and visualized under microscope (Leica DM-2500, Germany). Soret spectroscopy was used to check gastric mucosal bleeding by following hemoglobin release. Briefly, the stomachs from control and treated rats were washed in PBS, pH 7.4. The clarified PBS (mucosal washing) was analyzed for hemoglobin content by wavelength scanning (360–600 nm) in UV-visible spectrophotometer (Shimadzu, UV-1700 PharmaSpec).
Statistical analysis
All animal experiments were done in triplicate with at least six to eight animals in each group (n = 6–8). All in vitro experiments were done in triplicate and repeated. Data obtained from all experiments were expressed as mean ± S.D. Unpaired t test was used to calculate the levels of significance while comparing “control” and “indomethacin” groups while using one-way ANOVA followed by Bonferroni post hoc test used for comparing multiple goups. p value less than 0.05 (p < 0.05) was considered as statistically significant. Statistical analysis of the data were performed with the help of GraphPad Prism 6 and Microsoft Office Excel 2007 software.
Author contributions
S. M. and U. B. conceptualization; S. M., R. D., S. B., and U. B. data curation; S. M., R. D., S. B., and U. B. formal analysis; S. M., R. D., S. D., P. M., S. S., S. J. S., S. N., D. S., and U. B. investigation; S. M., R. D., S. D., P. M., A. A. S., C. B., S. P., and K. M. visualization; S. M., R. D., S. D., P. M., S. S., S. J. S., S. N., D. S., and K. M. methodology; S. M. and R. D. writing-original draft; S. M., R. D., S. B., A. A. S., and U. B. writing-review and editing; C. B. and U. B. validation; U. B. resources; U. B. supervision; U. B. funding acquisition; U. B. project administration.
Acknowledgment
We thank Debalina Chakrabarty for assisting in FACS experiments.
This work was supported by a Council of Scientific and Industrial Research, New Delhi, India, fellowship (to S. M. and R. D.), Research Grants BEnD, BSC 0206, and DST (J. C. Bose Fellowship) Grant SB/S2/JCB-54/2014. The authors declare that they have no conflicts of interest with the contents of this article.
- NSAID
- nonsteroidal anti-inflammatory drug
- MFF
- mitochondrial fission factor
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- STED
- stimulated emission depletion microscopy
- ETC
- electron transport chain
- ROS
- reactive oxygen species
- OCR
- oxygen consumption ratio
- ANOVA
- analysis of variance
- TEM
- transmission EM
- COX
- cyclooxygenase
- IMM
- inner mitochondrial membrane
- MOS
- mitochondrial oxidative stress
- RCR
- respiratory control ratio
- qPCR
- quantitative PCR
- NAO
- nonyl acridine orange
- AGS
- human gastric epithelial cell
- PSI
- pseudo-substrate inhibitor
- FCCP
- carbonyl cyanide p-trifluoromethoxyphenylhydrazone
- II
- injury index
- ROI
- region of interest
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular signal-regulated kinase
- JNK
- c-Jun N-terminal kinase
- PG
- prostaglandin
- FtMt
- mitochondrial ferritin
- DAPI
- 4′,6-diamidino-2-phenylindole
- DAB
- 3,3′-diaminobenzidine.
References
- 1. Ong C. K., Lirk P., Tan C. H., and Seymour R. A. (2007) An evidence-based update on nonsteroidal anti-inflammatory drugs. Clin. Med. Res. 5, 19–34 10.3121/cmr.2007.698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. da Costa B. R., Reichenbach S., Keller N., Nartey L., Wandel S., Jüni P., and Trelle S. (2017) Effectiveness of non-steroidal anti-inflammatory drugs for the treatment of pain in knee and hip osteoarthritis: a network meta-analysis. Lancet 390, e21–e33 10.1016/S0140-6736(17)31744-0 [DOI] [PubMed] [Google Scholar]
- 3. Zhao X., Xu Z., and Li H. (2017) NSAIDs use and reduced metastasis in cancer patients: results from a meta-analysis. Sci. Rep. 7, 1875 10.1038/s41598-017-01644-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wakabayashi K. (2000) NSAIDs as cancer preventive agents. Asian Pac. J. Cancer Prev. 1, 97–113 [PubMed] [Google Scholar]
- 5. de Groot D. J., de Vries E. G., Groen H. J., and de Jong S. (2007) Non-steroidal anti-inflammatory drugs to potentiate chemotherapy effects: from lab to clinic. Crit. Rev. Oncol. Hematol. 61, 52–69 10.1016/j.critrevonc.2006.07.001 [DOI] [PubMed] [Google Scholar]
- 6. Hunter T. S., Robison C., and Gerbino P. P. (2015) Emerging evidence in NSAID pharmacology: important considerations for product selection. Am. J. Manag. Care 21, S139–S147 [PubMed] [Google Scholar]
- 7. Gurpinar E., Grizzle W. E., and Piazza G. A. (2013) COX-independent mechanisms of cancer chemoprevention by anti-inflammatory drugs. Front. Oncol. 3, 181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Soh J. W., and Weinstein I. B. (2003) Role of COX-independent targets of NSAIDs and related compounds in cancer prevention and treatment. Prog. Exp. Tumor Res. 37, 261–285 10.1159/000071377 [DOI] [PubMed] [Google Scholar]
- 9. Gurpinar E., Grizzle W. E., and Piazza G. A. (2014) NSAIDs inhibit tumorigenesis, but how? Clin. Cancer Res. 20, 1104–1113 10.1158/1078-0432.CCR-13-1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Matsui H., Shimokawa O., Kaneko T., Nagano Y., Rai K., and Hyodo I. (2011) The pathophysiology of non-steroidal anti-inflammatory drug (NSAID)-induced mucosal injuries in stomach and small intestine. J. Clin. Biochem. Nutr. 48, 107–111 10.3164/jcbn.10-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marcum Z. A., and Hanlon J. T. (2010) Recognizing the risks of chronic nonsteroidal anti-inflammatory drug use in older adults. Ann. Longterm. Care 18, 24–27 [PMC free article] [PubMed] [Google Scholar]
- 12. Smolinske S. C., Hall A. H., Vandenberg S. A., Spoerke D. G., and McBride P. V. (1990) Toxic effects of nonsteroidal anti-inflammatory drugs in overdose. An overview of recent evidence on clinical effects and dose-response relationships. Drug Saf. 5, 252–274 10.2165/00002018-199005040-00003 [DOI] [PubMed] [Google Scholar]
- 13. Tarnawski A. S., and Jones M. K. (2003) Inhibition of angiogenesis by NSAIDs: molecular mechanisms and clinical implications. J. Mol. Med. 81, 627–636 10.1007/s00109-003-0479-y [DOI] [PubMed] [Google Scholar]
- 14. Monnier Y., Zaric J., and Rüegg C. (2005) Inhibition of angiogenesis by non-steroidal anti-inflammatory drugs: from the bench to the bedside and back. Curr. Drug Targets Inflamm. Allergy 4, 31–38 10.2174/1568010053622975 [DOI] [PubMed] [Google Scholar]
- 15. Schafer A. I. (1995) Effects of nonsteroidal antiinflammatory drugs on platelet function and systemic hemostasis. J. Clin. Pharmacol. 35, 209–219 10.1002/j.1552-4604.1995.tb04050.x [DOI] [PubMed] [Google Scholar]
- 16. Coxib and traditional NSAID Trialists' (CNT) Collaboration, Bhala N., Emberson J., Merhi A., Abramson S., Arber N., Baron J. A., Bombardier C., Cannon C., Farkouh M. E., FitzGerald G. A., Goss P., Halls H., Hawk E., Hawkey C., et al. (2013) Vascular and upper gastrointestinal effects of non-steroidal anti-inflammatory drugs: meta-analyses of individual participant data from randomised trials. Lancet 382, 769–779 10.1016/S0140-6736(13)60900-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cryer B. (2001) Nonsteroidal anti-inflammatory drug gastrointestinal toxicity. Curr. Opin. Gastroenterol. 17, 503–512 10.1097/00001574-200111000-00005 [DOI] [PubMed] [Google Scholar]
- 18. van der Bliek A. M., Shen Q., and Kawajiri S. (2013) Mechanisms of mitochondrial fission and fusion. Cold Spring Harb. Perspect. Biol. 5, a011072 10.1101/cshperspect.a011072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Skulachev V. P. (2001) Mitochondrial filaments and clusters as intracellular power-transmitting cables. Trends Biochem. Sci. 26, 23–29 10.1016/S0968-0004(00)01735-7 [DOI] [PubMed] [Google Scholar]
- 20. Westermann B. (2012) Bioenergetic role of mitochondrial fusion and fission. Biochim. Biophys. Acta 1817, 1833–1838 10.1016/j.bbabio.2012.02.033 [DOI] [PubMed] [Google Scholar]
- 21. Archer S. L. (2013) Mitochondrial dynamics–mitochondrial fission and fusion in human diseases. N. Engl. J. Med. 369, 2236–2251 10.1056/NEJMra1215233 [DOI] [PubMed] [Google Scholar]
- 22. Zhan M., Brooks C., Liu F., Sun L., and Dong Z. (2013) Mitochondrial dynamics: regulatory mechanisms and emerging role in renal pathophysiology. Kidney Int. 83, 568–581 10.1038/ki.2012.441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sanchis-Gomar F., Lippi G., and Lucia A. (2016) 'Mitotherapy' for heart failure. Trends Mol. Med. 22, 267–269 10.1016/j.molmed.2016.02.007 [DOI] [PubMed] [Google Scholar]
- 24. Marín-García J., and Akhmedov A. T. (2016) Mitochondrial dynamics and cell death in heart failure. Heart Fail. Rev. 21, 123–136 10.1007/s10741-016-9530-2 [DOI] [PubMed] [Google Scholar]
- 25. Mohanty K., Dada R., and Dada T. (2016) Neurodegenerative eye disorders: role of mitochondrial dynamics and genomics. Asia Pac. J. Ophthalmol. 5, 293–299 10.1097/APO.0000000000000203 [DOI] [PubMed] [Google Scholar]
- 26. Mao K., and Klionsky D. J. (2013) Participation of mitochondrial fission during mitophagy. Cell Cycle 12, 3131–3132 10.4161/cc.26352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Renault T. T., and Chipuk J. E. (2014) Death upon a kiss: mitochondrial outer membrane composition and organelle communication govern sensitivity to BAK/BAX-dependent apoptosis. Chem. Biol. 21, 114–123 10.1016/j.chembiol.2013.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang Q., Zhan L., Cao H., Li J., Lyu Y., Guo X., Zhang J., Ji L., Ren T., An J., Liu B., Nie Y., and Xing J. (2016) Increased mitochondrial fission promotes autophagy and hepatocellular carcinoma cell survival through the ROS-modulated coordinated regulation of the NFKB and TP53 pathways. Autophagy 12, 999–1014 10.1080/15548627.2016.1166318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee S., and Kim J. S. (2014) Mitophagy: therapeutic potentials for liver disease and beyond. Toxicol. Res. 30, 243–250 10.5487/TR.2014.30.4.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu T., Wang L., Lee H., O'Brien D. K., Bronk S. F., Gores G. J., and Yoon Y. (2014) Decreasing mitochondrial fission prevents cholestatic liver injury. J. Biol. Chem. 289, 34074–34088 10.1074/jbc.M114.588616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brooks C., Wei Q., Cho S. G., and Dong Z. (2009) Regulation of mitochondrial dynamics in acute kidney injury in cell culture and rodent models. J. Clin. Invest. 119, 1275–1285 10.1172/JCI37829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ferreira-da-Silva A., Valacca C., Rios E., Pópulo H., Soares P., Sobrinho-Simões M., Scorrano L., Máximo V., and Campello S. (2015) Mitochondrial dynamics protein Drp1 is overexpressed in oncocytic thyroid tumors and regulates cancer cell migration. PLoS ONE 10, e0122308 10.1371/journal.pone.0122308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xie Q., Wu Q., Horbinski C. M., Flavahan W. A., Yang K., Zhou W., Dombrowski S. M., Huang Z., Fang X., Shi Y., Ferguson A. N., Kashatus D. F., Bao S., and Rich J. N. (2015) Mitochondrial control by DRP1 in brain tumor initiating cells. Nat. Neurosci. 18, 501–510 10.1038/nn.3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tao H., Zhang Y., Zeng X., Shulman G. I., and Jin S. (2014) Niclosamide ethanolamine-induced mild mitochondrial uncoupling improves diabetic symptoms in mice. Nat. Med. 20, 1263–1269 10.1038/nm.3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mishra P., and Chan D. C. (2014) Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell Biol. 15, 634–646 10.1038/nrm3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fernández-Checa J. C., Fernández A., Morales A., Marí M., García-Ruiz C., and Colell A. (2010) Oxidative stress and altered mitochondrial function in neurodegenerative diseases: lessons from mouse models. CNS Neurol. Disord. Drug Targets 9, 439–454 10.2174/187152710791556113 [DOI] [PubMed] [Google Scholar]
- 37. Lundholm K., Gelin J., Hyltander A., Lönnroth C., Sandström R., Svaninger G., Körner U., Gülich M., Kärrefors I., and Norli B. (1994) Anti-inflammatory treatment may prolong survival in undernourished patients with metastatic solid tumors. Cancer Res. 54, 5602–5606 [PubMed] [Google Scholar]
- 38. Bordt E. A., Clerc P., Roelofs B. A., Saladino A. J., Tretter L., Adam-Vizi V., Cherok E., Khalil A., Yadava N., Ge S. X., Francis T. C., Kennedy N. W., Picton L. K., Kumar T., Uppuluri S., Miller A. M., et al. (2017) The putative Drp1 inhibitor mdivi-1 is a reversible mitochondrial complex I inhibitor that modulates reactive oxygen species. Dev. Cell 40, 583–594.e6 10.1016/j.devcel.2017.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rosdah A. A., K Holien J., Delbridge L. M., Dusting G. J., and Lim S. Y. (2016) Mitochondrial fission–a drug target for cytoprotection or cytodestruction? Pharmacol. Res. Perspect. 4, e00235 10.1002/prp2.235 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Smith G., and Gallo G. (2017) To mdivi-1 or not to mdivi-1: is that the question? Dev. Neurobiol. 77 10.1002/dneu.22519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li X., Fang P., Mai J., Choi E. T., Wang H., and Yang X. F. (2013) Targeting mitochondrial reactive oxygen species as novel therapy for inflammatory diseases and cancers. J. Hematol. Oncol. 6, 19 10.1186/1756-8722-6-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Barbato F., La Rotonda M. I., and Quaglia F. (1997) Interactions of nonsteroidal antiinflammatory drugs with phospholipids: comparison between octanol/buffer partition coefficients and chromatographic indexes on immobilized artificial membranes. J. Pharm. Sci. 86, 225–229 10.1021/js960233h [DOI] [PubMed] [Google Scholar]
- 43. Gargaun S., and Gargaun L. (2014) A novel method aimed at counteracting the side effects caused by prostaglandin E2 deficiency during non-steroidal anti-inflammatory drug therapy. Antiinflamm. Antiallergy Agents Med. Chem. 13, 121–127 10.2174/1871523012666131229132519 [DOI] [PubMed] [Google Scholar]
- 44. Burian M., and Geisslinger G. (2005) COX-dependent mechanisms involved in the antinociceptive action of NSAIDs at central and peripheral sites. Pharmacol. Ther. 107, 139–154 10.1016/j.pharmthera.2005.02.004 [DOI] [PubMed] [Google Scholar]
- 45. Hörl W. H. (2010) Nonsteroidal anti-inflammatory drugs and the kidney. Pharmaceuticals 3, 2291–2321 10.3390/ph3072291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dong Y. H., Chang C. H., Wu L. C., Hwang J. S., and Toh S. (2018) Comparative cardiovascular safety of nonsteroidal anti-inflammatory drugs in patients with hypertension: a population-based cohort study. Br. J. Clin. Pharmacol. 84, 1045–1056 10.1111/bcp.13537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brook R. D., Kramer M. B., Blaxall B. C., and Bisognano J. D. (2000) Nonsteroidal anti-inflammatory drugs and hypertension. J. Clin. Hypertens. 2, 319–323 [PubMed] [Google Scholar]
- 48. Muñoz E., Valero R. A., Quintana A., Hoth M., Núñez L., and Villalobos C. (2011) Nonsteroidal anti-inflammatory drugs inhibit vascular smooth muscle cell proliferation by enabling the Ca2+-dependent inactivation of calcium release-activated calcium/orai channels normally prevented by mitochondria. J. Biol. Chem. 286, 16186–16196 10.1074/jbc.M110.198952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Boring M. A., Hootman J. M., Liu Y., Theis K. A., Murphy L. B., Barbour K. E., Helmick C. G., Brady T. J., and Croft J. B. (2017) Prevalence of arthritis and arthritis-attributable activity limitation by urban-rural county classification–United States, 2015. MMWR Morb. Mortal Wkly. Rep. 66, 527–532 10.15585/mmwr.mm6620a2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Crofford L. J. (2013) Use of NSAIDs in treating patients with arthritis. Arthritis Res. Ther. 15, Suppl. 3, S2 10.1186/ar4174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. van Laar M., Pergolizzi J. V. Jr., Mellinghoff H. U., Merchante I. M., Nalamachu S., O'Brien J., Perrot S., and Raffa R. B. (2012) Pain treatment in arthritis-related pain: beyond NSAIDs. Open Rheumatol. J. 6, 320–330 10.2174/1874312901206010320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Atchison J. W., Herndon C. M., and Rusie E. (2013) NSAIDs for musculoskeletal pain management:current perspectives and novel strategies to improve safety. J. Manag. Care Pharm. 19, S3–S19 [PubMed] [Google Scholar]
- 53. Mercadante S. (2001) The use of anti-inflammatory drugs in cancer pain. Cancer Treat. Rev. 27, 51–61 10.1053/ctrv.2000.0192 [DOI] [PubMed] [Google Scholar]
- 54. Lethaby A., Augood C., and Duckitt K. (2000) Nonsteroidal anti-inflammatory drugs for heavy menstrual bleeding. Cochrane Database Syst. Rev. 2000, CD000400 10.1002/14651858.CD000400 [DOI] [PubMed] [Google Scholar]
- 55. Moris D., Kontos M., Spartalis E., and Fentiman I. S. (2016) The role of NSAIDs in breast cancer prevention and relapse: current evidence and future perspectives. Breast Care 11, 339–344 10.1159/000452315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Thun M. J., Henley S. J., and Patrono C. (2002) Nonsteroidal anti-inflammatory drugs as anticancer agents: mechanistic, pharmacologic, and clinical issues. J. Natl. Cancer Inst. 94, 252–266 10.1093/jnci/94.4.252 [DOI] [PubMed] [Google Scholar]
- 57. Pantziarka P., Sukhatme V., Bouche G., Meheus L., and Sukhatme V. P. (2016) Repurposing drugs in oncology (ReDO)–diclofenac as an anti-cancer agent. Ecancermedicalscience 10, 610 10.3332/ecancer.2016.610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pantziarka P., Sukhatme V., Meheus L., Sukhatme V. P., and Bouche G. (2017) Repurposing non-cancer drugs in oncology— how many drugs are out there? bioRxiv. 10.1101/197434 [DOI] [Google Scholar]
- 59. Handa O., Naito Y., Fukui A., Omatsu T., and Yoshikawa T. (2014) The impact of non-steroidal anti-inflammatory drugs on the small intestinal epithelium. J. Clin. Biochem. Nutr. 54, 2–6 10.3164/jcbn.13-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vallecillo-Hernández J., Barrachina M. D., Ortiz-Masiá D., Coll S., Esplugues J. V., Calatayud S., and Hernández C. (2018) Indomethacin disrupts autophagic flux by inducing lysosomal dysfunction in gastric cancer cells and increases their sensitivity to cytotoxic drugs. Sci. Rep. 8, 3593 10.1038/s41598-018-21455-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Song Z., Chen H., Fiket M., Alexander C., and Chan D. C. (2007) OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J. Cell Biol. 178, 749–755 10.1083/jcb.200704110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Alavi M. V., and Fuhrmann N. (2013) Dominant optic atrophy, OPA1, and mitochondrial quality control: understanding mitochondrial network dynamics. Mol. Neurodegener. 8, 32 10.1186/1750-1326-8-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Otera H., Ishihara N., and Mihara K. (2013) New insights into the function and regulation of mitochondrial fission. Biochim. Biophys. Acta 1833, 1256–1268 10.1016/j.bbamcr.2013.02.002 [DOI] [PubMed] [Google Scholar]
- 64. Liu R., and Chan D. C. (2015) The mitochondrial fission receptor Mff selectively recruits oligomerized Drp1. Mol. Biol. Cell 26, 4466–4477 10.1091/mbc.E15-08-0591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lim S., Smith K. R., Lim S. T., Tian R., Lu J., and Tan M. (2016) Regulation of mitochondrial functions by protein phosphorylation and dephosphorylation. Cell Biosci. 6, 25 10.1186/s13578-016-0089-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Taguchi N., Ishihara N., Jofuku A., Oka T., and Mihara K. (2007) Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J. Biol. Chem. 282, 11521–11529 10.1074/jbc.M607279200 [DOI] [PubMed] [Google Scholar]
- 67. Liesa M., Palacín M., and Zorzano A. (2009) Mitochondrial dynamics in mammalian health and disease. Physiol. Rev. 89, 799–845 10.1152/physrev.00030.2008 [DOI] [PubMed] [Google Scholar]
- 68. Kashatus J. A., Nascimento A., Myers L. J., Sher A., Byrne F. L., Hoehn K. L., Counter C. M., and Kashatus D. F. (2015) Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol. Cell 57, 537–551 10.1016/j.molcel.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kashatus D. F., Lim K. H., Brady D. C., Pershing N. L., Cox A. D., and Counter C. M. (2011) RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat. Cell Biol. 13, 1108–1115 10.1038/ncb2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang Z., Jiang H., Chen S., Du F., and Wang X. (2012) The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell 148, 228–243 10.1016/j.cell.2011.11.030 [DOI] [PubMed] [Google Scholar]
- 71. Campello S., and Scorrano L. (2010) Mitochondrial shape changes: orchestrating cell pathophysiology. EMBO Rep. 11, 678–684 10.1038/embor.2010.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ko A. R., Hyun H. W., Min S. J., and Kim J. E. (2016) The differential DRP1 phosphorylation and mitochondrial dynamics in the regional specific astroglial death induced by status epilepticus. Front. Cell Neurosci. 10, 124 10.3389/fncel.2016.00124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Huang S., Wang Y., Gan X., Fang D., Zhong C., Wu L., Hu G., Sosunov A. A., McKhann G. M., Yu H., and Yan S. S. (2015) Drp1-mediated mitochondrial abnormalities link to synaptic injury in diabetes model. Diabetes 64, 1728–1742 10.2337/db14-0758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Xu S., Pi H., Zhang L., Zhang N., Li Y., Zhang H., Tang J., Li H., Feng M., Deng P., Guo P., Tian L., Xie J., He M., Lu Y., et al. (2016) Melatonin prevents abnormal mitochondrial dynamics resulting from the neurotoxicity of cadmium by blocking calcium-dependent translocation of Drp1 to the mitochondria. J. Pineal Res. 60, 291–302 10.1111/jpi.12310 [DOI] [PubMed] [Google Scholar]
- 75. Fischer T. D., Hylin M. J., Zhao J., Moore A. N., Waxham M. N., and Dash P. K. (2016) Altered mitochondrial dynamics and TBI pathophysiology. Front Syst. Neurosci. 10, 29 10.3389/fnsys.2016.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Guo C., Sun L., Chen X., and Zhang D. (2013) Oxidative stress, mitochondrial damage and neurodegenerative diseases. Neural Regen. Res. 8, 2003–2014 10.3969/j.issn.1673-5374.2013.21.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Pletjushkina O. Y., Lyamzaev K. G., Popova E. N., Nepryakhina O. K., Ivanova O. Y., Domnina L. V., Chernyak B. V., and Skulachev V. P. (2006) Effect of oxidative stress on dynamics of mitochondrial reticulum. Biochim. Biophys. Acta 1757, 518–524 10.1016/j.bbabio.2006.03.018 [DOI] [PubMed] [Google Scholar]
- 78. Parameyong A., Govitrapong P., and Chetsawang B. (2015) Melatonin attenuates the mitochondrial translocation of mitochondrial fission proteins and Bax, cytosolic calcium overload and cell death in methamphetamine-induced toxicity in neuroblastoma SH-SY5Y cells. Mitochondrion 24, 1–8 10.1016/j.mito.2015.07.004 [DOI] [PubMed] [Google Scholar]
- 79. Wu S., Zhou F., Zhang Z., and Xing D. (2011) Mitochondrial oxidative stress causes mitochondrial fragmentation via differential modulation of mitochondrial fission-fusion proteins. FEBS J. 278, 941–954 10.1111/j.1742-4658.2011.08010.x [DOI] [PubMed] [Google Scholar]
- 80. Thakre-Nighot M., and Blikslager A. T. (2016) Indomethacin induces increase in gastric epithelial tight junction permeability via redistribution of occludin and activation of p38 MAPK in MKN-28 Cells. Tissue Barriers 4, e1187325 10.1080/21688370.2016.1187325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kim T. I., Jin S. H., Kang E. H., Shin S. K., Choi K. Y., and Kim W. H. (2002) The role of mitogen-activated protein kinases and their relationship with NF-κB and PPARγ in indomethacin–Induced apoptosis of colon cancer cells. Ann. N.Y. Acad. Sci. 973, 241–245 10.1111/j.1749-6632.2002.tb04641.x [DOI] [PubMed] [Google Scholar]
- 82. Mansouri A., Ridgway L. D., Korapati A. L., Zhang Q., Tian L., Wang Y., Siddik Z. H., Mills G. B., and Claret F. X. (2003) Sustained activation of JNK/p38 MAPK pathways in response to cisplatin leads to Fas ligand induction and cell death in ovarian carcinoma cells. J. Biol. Chem. 278, 19245–19256 10.1074/jbc.M208134200 [DOI] [PubMed] [Google Scholar]
- 83. Ko S. H., Choi G. E., Oh J. Y., Lee H. J., Kim J. S., Chae C. W., Choi D., and Han H. J. (2017) Succinate promotes stem cell migration through the GPR91-dependent regulation of DRP1-mediated mitochondrial fission. Sci. Rep. 7, 12582 10.1038/s41598-017-12692-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Waldron R. T., Rey O., Zhukova E., and Rozengurt E. (2004) Oxidative stress induces protein kinase C-mediated activation loop phosphorylation and nuclear redistribution of protein kinase D. J. Biol. Chem. 279, 27482–27493 10.1074/jbc.M402875200 [DOI] [PubMed] [Google Scholar]
- 85. Cosentino-Gomes D., Rocco-Machado N., and Meyer-Fernandes J. R. (2012) Cell signaling through protein kinase C oxidation and activation. Int. J. Mol. Sci. 13, 10697–10721 10.3390/ijms130910697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ki Y. W., Park J. H., Lee J. E., Shin I. C., and Koh H. C. (2013) JNK and p38 MAPK regulate oxidative stress and the inflammatory response in chlorpyrifos-induced apoptosis. Toxicol. Lett. 218, 235–245 10.1016/j.toxlet.2013.02.003 [DOI] [PubMed] [Google Scholar]
- 87. Sui X., Kong N., Ye L., Han W., Zhou J., Zhang Q., He C., and Pan H. (2014) p38 and JNK MAPK pathways control the balance of apoptosis and autophagy in response to chemotherapeutic agents. Cancer Lett. 344, 174–179 10.1016/j.canlet.2013.11.019 [DOI] [PubMed] [Google Scholar]
- 88. Mebratu Y., and Tesfaigzi Y. (2009) How ERK1/2 activation controls cell proliferation and cell death: Is subcellular localization the answer? Cell Cycle 8, 1168–1175 10.4161/cc.8.8.8147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kubli D. A., and Gustafsson Å. B. (2012) Mitochondria and mitophagy: the yin and yang of cell death control. Circ. Res. 111, 1208–1221 10.1161/CIRCRESAHA.112.265819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ashrafi G., and Schwarz T. L. (2013) The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 20, 31–42 10.1038/cdd.2012.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Shimura T., Kobayashi J., Komatsu K., and Kunugita N. (2016) Severe mitochondrial damage associated with low-dose radiation sensitivity in ATM- and NBS1-deficient cells. Cell Cycle 15, 1099–1107 10.1080/15384101.2016.1156276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Luo C., Li Y., Wang H., Feng Z., Li Y., Long J., and Liu J. (2013) Mitochondrial accumulation under oxidative stress is due to defects in autophagy. J. Cell Biochem. 114, 212–219 10.1002/jcb.24356 [DOI] [PubMed] [Google Scholar]
- 93. Smith G., and Gallo G. (2017) To mdivi-1 or not to mdivi-1: is that the question? Dev. Neurobiol. 77, 1260–1268 10.1002/dneu.22519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Tang W. X., Wu W. H., Qiu H. Y., Bo H., and Huang S. M. (2013) Amelioration of rhabdomyolysis-induced renal mitochondrial injury and apoptosis through suppression of Drp-1 translocation. J. Nephrol. 26, 1073–1082 10.5301/jn.5000268 [DOI] [PubMed] [Google Scholar]
- 95. Gharanei M., Hussain A., Janneh O., and Maddock H. (2013) Attenuation of doxorubicin-induced cardiotoxicity by mdivi-1: a mitochondrial division/mitophagy inhibitor. PLoS ONE 8, e77713 10.1371/journal.pone.0077713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zhang N., Wang S., Li Y., Che L., and Zhao Q. (2013) A selective inhibitor of Drp1, mdivi-1, acts against cerebral ischemia/reperfusion injury via an anti-apoptotic pathway in rats. Neurosci. Lett. 535, 104–109 10.1016/j.neulet.2012.12.049 [DOI] [PubMed] [Google Scholar]
- 97. Manczak M., Kandimalla R., Yin X., and Reddy P. H. (2019) Mitochondrial division inhibitor 1 reduces dynamin-related protein 1 and mitochondrial fission activity. Hum. Mol. Genet. 28, 177–199 10.1093/hmg/ddy335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Wang J., Hansen K., Edwards R., Van Houten B., and Qian W. (2015) Mitochondrial division inhibitor 1 (mdivi-1) enhances death receptor-mediated apoptosis in human ovarian cancer cells. Biochem. Biophys. Res. Commun. 456, 7–12 10.1016/j.bbrc.2014.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Akita M., Suzuki-Karasaki M., Fujiwara K., Nakagawa C., Soma M., Yoshida Y., Ochiai T., Tokuhashi Y., and Suzuki-Karasaki Y. (2014) Mitochondrial division inhibitor-1 induces mitochondrial hyperfusion and sensitizes human cancer cells to TRAIL-induced apoptosis. Int. J. Oncol. 45, 1901–1912 10.3892/ijo.2014.2608 [DOI] [PubMed] [Google Scholar]
- 100. Chuang Y. C., Lin T. K., Yang D. I., Yang J. L., Liou C. W., and Chen S. D. (2016) Peroxisome proliferator-activated receptor-γ dependent pathway reduces the phosphorylation of dynamin-related protein 1 and ameliorates hippocampal injury induced by global ischemia in rats. J. Biomed. Sci. 23, 44 10.1186/s12929-016-0262-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Yu T., Sheu S. S., Robotham J. L., and Yoon Y. (2008) Mitochondrial fission mediates high glucose-induced cell death through elevated production of reactive oxygen species. Cardiovasc. Res. 79, 341–351 10.1093/cvr/cvn104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Anderson E. J., Katunga L. A., and Willis M. S. (2012) Mitochondria as a source and target of lipid peroxidation products in healthy and diseased heart. Clin. Exp. Pharmacol. Physiol. 39, 179–193 10.1111/j.1440-1681.2011.05641.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Traverso N., Menini S., Maineri E. P., Patriarca S., Odetti P., Cottalasso D., Marinari U. M., and Pronzato M. A. (2004) Malondialdehyde, a lipoperoxidation-derived aldehyde, can bring about secondary oxidative damage to proteins. J. Gerontol. A Biol. Sci. Med. Sci. 59, B890–B895 10.1093/gerona/59.9.B890 [DOI] [PubMed] [Google Scholar]
- 104. Łuczaj W., and Skrzydlewska E. (2003) DNA damage caused by lipid peroxidation products. Cell Mol. Biol. Lett. 8, 391–413 [PubMed] [Google Scholar]
- 105. Blair I. A. (2008) DNA adducts with lipid peroxidation products. J. Biol. Chem. 283, 15545–15549 10.1074/jbc.R700051200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kagan V. E., Tyurin V. A., Jiang J., Tyurina Y. Y., Ritov V. B., Amoscato A. A., Osipov A. N., Belikova N. A., Kapralov A. A., Kini V., Vlasova I. I., Zhao Q., Zou M., Di P., Svistunenko D. A., et al. (2005) Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat. Chem. Biol. 1, 223–232 10.1038/nchembio727 [DOI] [PubMed] [Google Scholar]
- 107. Ott M., Gogvadze V., Orrenius S., and Zhivotovsky B. (2007) Mitochondria, oxidative stress and cell death. Apoptosis 12, 913–922 10.1007/s10495-007-0756-2 [DOI] [PubMed] [Google Scholar]
- 108. Wigdal S. S., Kirkland R. A., Franklin J. L., and Haak-Frendscho M. (2002) Cytochrome c release precedes mitochondrial membrane potential loss in cerebellar granule neuron apoptosis: lack of mitochondrial swelling. J. Neurochem. 82, 1029–1038 [DOI] [PubMed] [Google Scholar]
- 109. Dey S., Bindu S., Goyal M., Pal C., Alam A., Iqbal M. S., Kumar R., Sarkar S., and Bandyopadhyay U. (2012) Impact of intravascular hemolysis in malaria on liver dysfunction: involvement of hepatic free heme overload, NF-κB activation, and neutrophil infiltration. J. Biol. Chem. 287, 26630–26646 10.1074/jbc.M112.341255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Shorrock C. J., and Rees W. D. (1989) Mechanisms of gastric damage by non-steroidal anti-inflammatory drugs. Scand. J. Rheumatol. Suppl. 78, 5–11; discussion 30–2 [DOI] [PubMed] [Google Scholar]
- 111. Ivey K. J. (1988) Mechanisms of nonsteroidal anti-inflammatory drug-induced gastric damage. Actions of therapeutic agents. Am. J. Med. 84, 41–48 10.1016/0002-9343(88)90253-7 [DOI] [PubMed] [Google Scholar]
- 112. Fromm D. (1987) How do non-steroidal anti-inflammatory drugs affect gastric mucosal defenses? Clin. Invest. Med. 10, 251–258 [PubMed] [Google Scholar]
- 113. Wallace J. L. (2008) Prostaglandins, NSAIDs, and gastric mucosal protection: why doesn't the stomach digest itself? Physiol. Rev. 88, 1547–1565 10.1152/physrev.00004.2008 [DOI] [PubMed] [Google Scholar]
- 114. Musumba C., Pritchard D. M., and Pirmohamed M. (2009) Review article: cellular and molecular mechanisms of NSAID-induced peptic ulcers. Aliment. Pharmacol. Ther. 30, 517–531 10.1111/j.1365-2036.2009.04086.x [DOI] [PubMed] [Google Scholar]
- 115. Zhang J. M., and An J. (2007) Cytokines, inflammation, and pain. Int. Anesthesiol. Clin. 45, 27–37 10.1097/AIA.0b013e318034194e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Pradère J. P., Hernandez C., Koppe C., Friedman R. A., Luedde T., and Schwabe R. F. (2016) Negative regulation of NF-κB p65 activity by serine 536 phosphorylation. Sci. Signal. 9, ra85 10.1126/scisignal.aab2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Solt L. A., and May M. J. (2008) The IκB kinase complex: master regulator of NF-κB signaling. Immunol. Res. 42, 3–18 10.1007/s12026-008-8025-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Scholz C. C., Cavadas M. A., Tambuwala M. M., Hams E., Rodríguez J., von Kriegsheim A., Cotter P., Bruning U., Fallon P. G., Cheong A., Cummins E. P., and Taylor C. T. (2013) Regulation of IL-1β–induced NF-κB by hydroxylases links key hypoxic and inflammatory signaling pathways. Proc. Natl. Acad. Sci. U.S.A. 110, 18490–18495 10.1073/pnas.1309718110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Zhang H., Chen J., Liu X., Awar L., McMickle A., Bai F., Nagarajan S., and Yu S. (2013) IL-17 induces expression of vascular cell adhesion molecule through signalling pathway of NF-κB, but not Akt1 and TAK1 in vascular smooth muscle cells. Scand. J. Immunol. 77, 230–237 10.1111/sji.12030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Xie S., Li J., Wang J. H., Wu Q., Yang P., Hsu H. C., Smythies L. E., and Mountz J. D. (2010) IL-17 activates the canonical NF-κB signaling pathway in autoimmune B cells of BXD2 mice to upregulate the expression of regulators of G-protein signaling 16. J. Immunol. 184, 2289–2296 10.4049/jimmunol.0903133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Vincenti M. P., and Brinckerhoff C. E. (2002) Transcriptional regulation of collagenase (MMP-1, MMP-13) genes in arthritis: integration of complex signaling pathways for the recruitment of gene-specific transcription factors. Arthritis Res. 4, 157–164 10.1186/ar401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Li Y. F., Xu X. B., Chen X. H., Wei G., He B., and Wang J. D. (2012) The nuclear factor-κB pathway is involved in matrix metalloproteinase-9 expression in RU486-induced endometrium breakdown in mice. Hum. Reprod. 27, 2096–2106 10.1093/humrep/des110 [DOI] [PubMed] [Google Scholar]
- 123. Burke S. J., Lu D., Sparer T. E., Masi T., Goff M. R., Karlstad M. D., and Collier J. J. (2014) NF-κB and STAT1 control CXCL1 and CXCL2 gene transcription. Am. J. Physiol. Endocrinol. Metab. 306, E131–E149 10.1152/ajpendo.00347.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Wang L., Wang L., Dai Z., Wu P., Shi H., and Zhao S. (2017) Lack of mitochondrial ferritin aggravated neurological deficits via enhancing oxidative stress in a traumatic brain injury murine model. Biosci. Rep. 37, BSR20170942 10.1042/BSR20170942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Isaya G. (2014) Mitochondrial iron-sulfur cluster dysfunction in neurodegenerative disease. Front. Pharmacol. 5, 29 10.3389/fphar.2014.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Ježek J., Cooper K. F., and Strich R. (2018) Reactive oxygen species and mitochondrial dynamics: the yin and yang of mitochondrial dysfunction and cancer progression. Antioxidants 7, E13 10.3390/antiox7010013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Zhou X., Wang H. Y., Wu B., Cheng C. Y., Xiao W., Wang Z. Z., Yang Y. Y., Li P., and Yang H. (2017) Ginkgolide K attenuates neuronal injury after ischemic stroke by inhibiting mitochondrial fission and GSK-3β-dependent increases in mitochondrial membrane permeability. Oncotarget 8, 44682–44693 10.18632/oncotarget.17967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Hung C. H., Cheng S. S., Cheung Y. T., Wuwongse S., Zhang N. Q., Ho Y. S., Lee S. M., and Chang R. C. (2018) A reciprocal relationship between reactive oxygen species and mitochondrial dynamics in neurodegeneration. Redox Biol. 14, 7–19 10.1016/j.redox.2017.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Mazumder S., De R., Sarkar S., Siddiqui A. A., Saha S. J., Banerjee C., Iqbal M. S., Nag S., Debsharma S., and Bandyopadhyay U. (2016) Selective scavenging of intra-mitochondrial superoxide corrects diclofenac-induced mitochondrial dysfunction and gastric injury: a novel gastroprotective mechanism independent of gastric acid suppression. Biochem. Pharmacol. 121, 33–51 10.1016/j.bcp.2016.09.027 [DOI] [PubMed] [Google Scholar]
- 130. Wieckowski M. R., Giorgi C., Lebiedzinska M., Duszynski J., and Pinton P. (2009) Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat. Protoc. 4, 1582–1590 10.1038/nprot.2009.151 [DOI] [PubMed] [Google Scholar]
- 131. Xu S., Pi H., Chen Y., Zhang N., Guo P., Lu Y., He M., Xie J., Zhong M., Zhang Y., Yu Z., and Zhou Z. (2013) Cadmium induced Drp1-dependent mitochondrial fragmentation by disturbing calcium homeostasis in its hepatotoxicity. Cell Death Dis. 4, e540 10.1038/cddis.2013.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Losón O. C., Song Z., Chen H., and Chan D. C. (2013) Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell 24, 659–667 10.1091/mbc.e12-10-0721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Dey S., Guha M., Alam A., Goyal M., Bindu S., Pal C., Maity P., Mitra K., and Bandyopadhyay U. (2009) Malarial infection develops mitochondrial pathology and mitochondrial oxidative stress to promote hepatocyte apoptosis. Free Radic. Biol. Med. 46, 271–281 10.1016/j.freeradbiomed.2008.10.032 [DOI] [PubMed] [Google Scholar]
- 134. Dey S., Mazumder S., Siddiqui A. A., Iqbal M. S., Banerjee C., Sarkar S., De R., Goyal M., Bindu S., and Bandyopadhyay U. (2014) Association of heme oxygenase 1 with the restoration of liver function after damage in murine malaria by Plasmodium yoelii. Infect. Immun. 82, 3113–3126 10.1128/IAI.01598-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Chowdhury S. R., Ray U., Chatterjee B. P., and Roy S. S. (2017) Targeted apoptosis in ovarian cancer cells through mitochondrial dysfunction in response to Sambucus nigra agglutinin. Cell Death Dis. 8, e2762 10.1038/cddis.2017.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Biswas D., Ghosh M., Kumar S., and Chakrabarti P. (2016) PPARα-ATGL pathway improves muscle mitochondrial metabolism: implication in aging. FASEB J. 30, 3822–3834 10.1096/fj.201600571RR [DOI] [PubMed] [Google Scholar]
- 137. Carrasco-Pozo C., Gotteland M., and Speisky H. (2011) Apple peel polyphenol extract protects against indomethacin-induced damage in Caco-2 cells by preventing mitochondrial complex I inhibition. J. Agric. Food Chem. 59, 11501–11508 10.1021/jf202621d [DOI] [PubMed] [Google Scholar]
- 138. Spinazzi M., Casarin A., Pertegato V., Salviati L., and Angelini C. (2012) Assessment of mitochondrial respiratory chain enzymatic activities on tissues and cultured cells. Nat. Protoc. 7, 1235–1246 10.1038/nprot.2012.058 [DOI] [PubMed] [Google Scholar]