ABSTRACT
The expression and function of CD163 in glioma are not fully understood. In this report, we collected totally 1323 glioma samples from the Chinese Glioma Genome Atlas (CGGA) dataset, including 325 RNA-seq data and 301 mRNA microarray data, and 697 glioma samples from The Cancer Genome Atlas (TCGA) dataset to characterize the molecular and clinical features of CD163 in glioma by conducting a large-scale study. We found that CD163 expression was positively associated with the grade of malignancy of glioma. CD163 expression was up-regulated in IDH wild-type glioma and mesenchymal subtype. Gene ontology analysis suggested that CD163-related genes were more involved in immune response and angiogenesis in glioma. Moreover, CD163 showed a positive relationship with stromal and immune cell populations. Kaplan–Meier curves analysis revealed that higher CD163 expression indicated significantly poor survival in glioma and glioblastoma multiforme (GBM). Pearson correlation analysis revealed that CD163 was robustly associated with the immune checkpoints and other macrophage markers. These results demonstrated that CD163 predicts poor prognosis in glioma patients. Additionally, combination of CD163 and immune checkpoints may impair angiogenesis and reverse dysfunctional phenotypes of T cells, which suggest that CD163 may be a promising biomarker and target for immunotherapeutic strategies.
Abbreviations: CGGA: Chinese Glioma Genome Atlas; TCGA: The Cancer Genome Atlas; TAMs: Tumor associated macrophages; IDH: isocitrate dehydrogenase; GBM: glioblastoma
KEYWORDS: CD163, glioma, immunotherapy, prognosis, immune checkpoint
Introduction
Glioma has been classified by the World Health Organization (WHO) into four grades of ascending malignancy according to histological criteria.1 Presenting one of the highest mortality rates, glioblastoma multiforme (GBM, WHO grade IV) is the most aggressive malignancy of the central nervous system with a median survival time of only 12–15 months despite various treatments such as surgical resection, radiotherapy, and chemotherapy.2–4 The failure of conventional oncologic treatment has prompted investigators to look for new and more targeted therapy options.
Increasing evidence indicates that tumor microenvironment plays a critical role in supporting the progression of glioma.5,6 The tumor microenvironment in glioma is composed of various components, including stromal cells, infiltrating immune cells, soluble factors, and extracellular matrix.5,7 The majority of immune cells within brain tumors are macrophages, often comprising up to 30% of the tumor mass.8 As an important element of tumor microenvironment in glioma, tumor-associated macrophages (TAMs) are thought to create a supportive stroma for tumor growth,9which are commonly identified with the expression of CD163, CD204, or CD206.10 In general, in vitro–stimulated macrophages would be categorized as M1 and M2. But in vivo–derived macrophages would be described by multiple markers rather than trying to bin them into an M1 or M2 pool.11,12 M1 macrophages are characterized by the high expression of various pro-inflammatory cytokines and contribute towards immune surveillance; the M1 macrophages are also involved in efficient antigen presentation and pathogen killing. M2 macrophages produce low amounts of pro-inflammatory cytokines and high amounts of the anti-inflammatory cytokines, such as IL-10, IL-6, and TGF-β. They mainly contribute towards immune-suppression and favor tumor promotion.13 Most TAMs are considered to have M2 phenotype.10 Additionally, increased infiltration of TAMs correlate with improved glioma progression and tumor grade, and predict poor prognosis in GBM patients.14,15 This raises the intriguing possibility that targeting TAMs may be a successful therapeutic strategy for intractable glioma and GBM.
CD163 was first identified in 1987 and received its CD number in 1996.16 It is a 130-kDa membrane protein with a short cytoplasmic tail and belongs to the scavenger receptor cysteine-rich (SRCR) family class B domains expressed on subpopulations of mature tissue macrophages. CD163 has been identified as a receptor of Hemoglobin-Haptoglobin (Hb-Hp) complexes with a high affinity. Its functions are involved in clearance and endocytosis of these Hb-Hp complexes by resident macrophages and thereby protect tissues from free Hb-mediated oxidative damage.17,18 In addition, CD163 has been proposed as a scavenger receptor for tumor necrosis factor (TNF)-like weak inducer of apoptosis (TWEAK), a cytokine belonging to TNF superfamily, and mediates angiogenesis.19–21 The binding of CD163 with TWEAK could prevent TWEAK from exerting its biological functions as it is sequestered from the environment.19,22 It has also been suggested that CD163 plays a role as an innate immune sensor for bacteria.23 Expression of CD163 in monocytic cells promote bacteria-induced production of pro-inflammatory cytokines, such as TNF-α. The expression of CD163 is restricted to cells of the monocyte/macrophage lineage. In tissues, CD163 predominantly present on resident tissue macrophages such as red pulp macrophages in the spleen, Kupffer cells in the liver, and interstitial and alveolar macrophages in the lungs. The expression of CD163 is significantly higher in these mature tissue macrophages than in blood monocytes, indicating that CD163 is a differentiation marker of the macrophage lineage with increased expression along the macrophage differentiation pathway.24,25 Especially, in tumor tissues, CD163 is considered as the most specific marker of TAMs.26,27 The number of CD163-positive TAMs is significantly increased in glioma.14 Additionally, CD163 expression was found to be elevated in both male and female GBM patients, and inversely correlated with mean survival times.28 Targeting CD163 on macrophages highly increased the anti-inflammatory potency.29,30 However, the expression and function of CD163 are still obscure in glioma.
Through the analysis of a collection of various human malignancies, we found that GBM patients have a high expression of CD163. Moreover, we reviewed the current evidence and failed to find a single integrative report about CD163 in the whole WHO grade glioma classification. To systematically explore the CD163 expression status in all grades of glioma, we took advantage of the Chinese Glioma Genome Atlas (CGGA) database, including RNA-seq data and microarray data of all the grades of glioma, to undertake an integrative investigation of CD163. We further validated our findings through the RNA-seq data of glioma from The Cancer Genome Atlas (TCGA) database. This is the first comprehensive study characterizing CD163 expression in different grades of glioma, at the molecular and clinical levels through large-scale analysis. Our results reveal that CD163 may be a promising biomarker and therapeutic target for glioma and GBM and potentially for other malignant tumors too.
Results
CD163 expression was positively correlated with the grade of malignancy and enriched in IDH wild-type glioma
To assess the expression of CD163, we analyzed CD163 expression in a panel of 8690 solid tumor tissues ranging from 20 different tumor types obtained from TCGA database. As described in Figure S1, GBM patients had a higher expression of CD163, compared to patients with other carcinomas. To further clarify the prominent molecular heterogeneity across different grades of glioma, we analyzed CD163 expression in the RNA-seq and microarray database of CGGA. In the CGGA database, GBM (WHO IV) showed the highest CD163 expression compared to WHO grade II and grade III glioma (Figure 1(a,b)). This result was further validated in TCGA database (Figure 1(c)), indicating that CD163 expression correlated with the grade of malignancy of glioma in line with other tumors. It is acknowledged that isocitrate dehydrogenase (IDH) mutation status plays a crucial role in glioma progression. Consequently, we investigated the relationship between CD163 expression and IDH mutation status and found that IDH wild-type glioma showed significantly elevated CD163 expression in both CGGA RNA-seq and microarray datasets (Figure 1(d,e)), as well as in the TCGA database (Figure 1(f)). Subsequently, we performed receiver operating characteristic (ROC) analysis to evaluate the diagnostic value of CD163 for IDH wild-type glioma. The analysis in CGGA RNA-seq and microarray database showed that areas under the curve (AUC) were 73.8.0% and 93.7%, respectively. Moreover, in TCGA database, AUC was up to 78.0% (Figures S2(a, b, and c)). Collectively, these data suggest that CD163 was mainly enriched in the wild-type glioma and may serve as a biomarker for IDH wild-type glioma.
Figure 1.
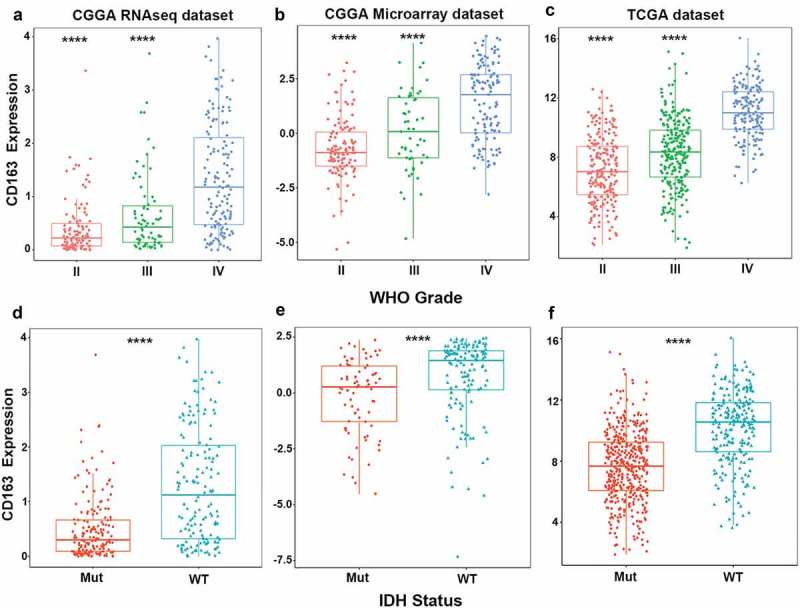
CD163 was significantly enriched in glioblastoma and in IDH wild-type glioma. (a, b, c) CD163 was highly expressed in glioblastoma (WHO IV) at transcription level. (d, e, f) CD163 was significantly up-regulated in IDH wild-type glioma. *** and **** represent p< 0.001 and p< 0.0001, respectively.
CD163 was highly enriched in mesenchymal molecular subtype glioma
We further explored the distribution of CD163 in different molecular subtypes of glioma in both CGGA and TCGA databases. When compared with that in classical, neural, and pro-neural subtypes, CD163 was significantly upregulated in the mesenchymal subtype (Figure 2(a-c)). To further confirm this finding, ROC curves of CD163 expression to obtain diagnostic values of mesenchymal subtype of all grades of glioma were constructed. Interestingly, the AUC was up to 90.1%,93.7%, and 94.2% in the CGGA RNA-seq data, microarray data, and TCGA RNA-seq database, respectively. (Figure 2(d-f)). These results demonstrated that CD163 expression was elevated in mesenchymal subtype and could be indicated as a biomarker for mesenchymal subtype glioma.
Figure 2.
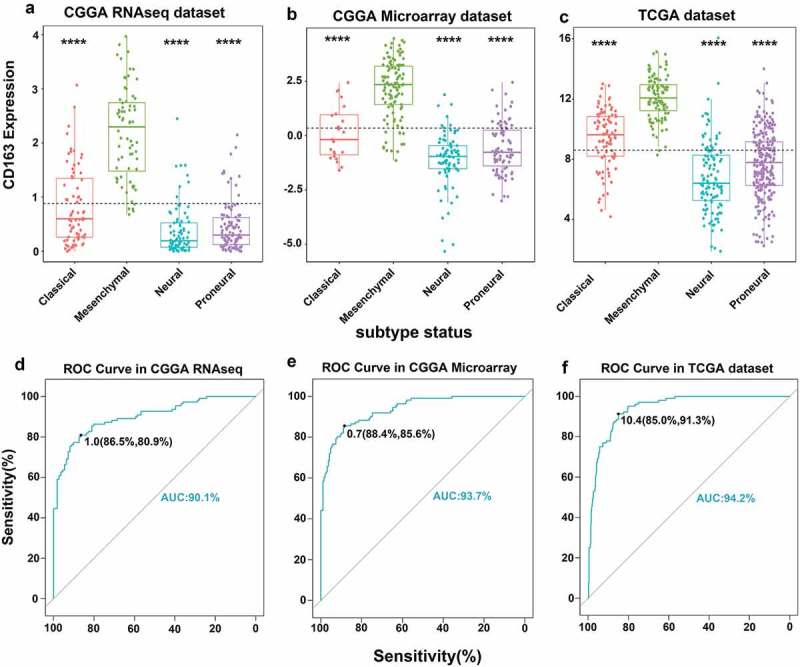
CD163 showed a strong expression pattern in mesenchymal molecular subtype glioma. (a, b, c) CD163 was highly enriched in mesenchymal molecular subtype glioma. (d, e, f) CD163 could serve as a biomarker to predict mesenchymal molecular subtype glioma. *, **, and **** represent p < 0.05, p< 0.01, and p< 0.0001, respectively.
CD163-related biological process
As revealed above, CD163 expression was closely related to the degree of malignancy of glioma. To evaluate the biological features of different CD163 expressions in glioma, the Gene Ontology (GO) analysis with online methods (DAVID, https://david.ncifcrf.gov/) was performed to analyze the significantly related genes (top 600 genes ranked by Pearson |R|) that were shared in the CGGA and TCGA databases. We found the genes that were positively correlated with CD163 expression were more involved in immune system process, immune response, and angiogenesis in both the CGGA and TCGA cohorts (Figure 3(a,b)). To identify the CD163 associated immune signature in glioma, we further analyzed the gene sets related to immune response (http://amigo.geneontology.org/amigo/landing). Results showed that genes positively related to CD163 were significantly enriched in immune response, inflammatory response, and antigen processing and presentation in both the CGGA and TCGA cohorts (Figures S3(a and b)). These results suggested that CD163 may play a crucial role in the regulation of immune system in glioma.
Figure 3.
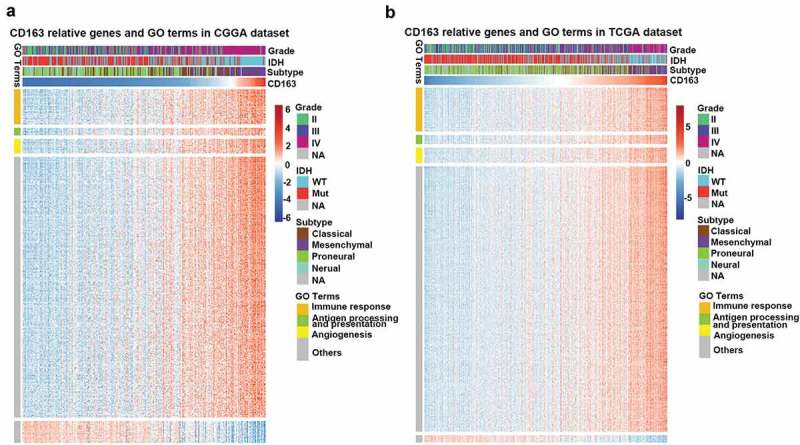
CD163 related immune genes and Gene Ontology (GO) terms in glioma. (a, b) CD163 showed a strong positive correlation with most of the immune genes related to GO terms especially in immune response, antigen process presentation, and angiogenesis.
Relationship between CD163 and infiltrated cells in the tumor microenvironment
Immune cells and stromal cells are important components in the tumor microenvironment. To be thoroughly aware of the relationship between CD163 and the infiltrated cells, ESTIMATE algorithm method described by Yoshihara31 was used. Briefly, this method uses gene expression signatures to infer the fraction of stromal and immune cells in tumor samples. ESTIMATE scores correlate with DNA copy number-based tumor purity across samples from 11 different tumor types, including glioma, profiled on Agilent, Affymetrix platforms or based on RNA sequencing and available through TCGA database. As Figure 4(a,b) (upper panel) indicated, CD163 was positively associated with the immune score and stromal score in both the CGGA RNA-seq and TCGA databases, which suggested that CD163 play an important role in immune regulation and stromal cells regulation. To further evaluate the relationship between CD163 and specific cell populations in the tumor microenvironment of glioma, we used the Microenvironment Cell Populations-counter method.32 This method allows the robust quantification of the absolute abundance of eight immune and two stromal cell populations in heterogeneous tissues from transcriptomic data. The findings showed CD163 had a significantly positive correlation with monocytes (Figure 4(a,b), lower panel). In addition, an intense association between CD163 endothelial cells was observed (Figure 4(a,b), lower panel), which was consistent with the previous reports that CD163 positive macrophage density and vascular density are generally closely correlated in human tumors.33–35 Furthermore, we also found that CD163 was positively associated with fibroblasts (Figure 4(a,b), lower panel), which was consistent with the previous reports that the expression of CD163 was positively associated with fibroblasts.36-38These data suggested that CD163 may participate in the regulation of stromal cells in the tumor microenvironment.
Figure 4.
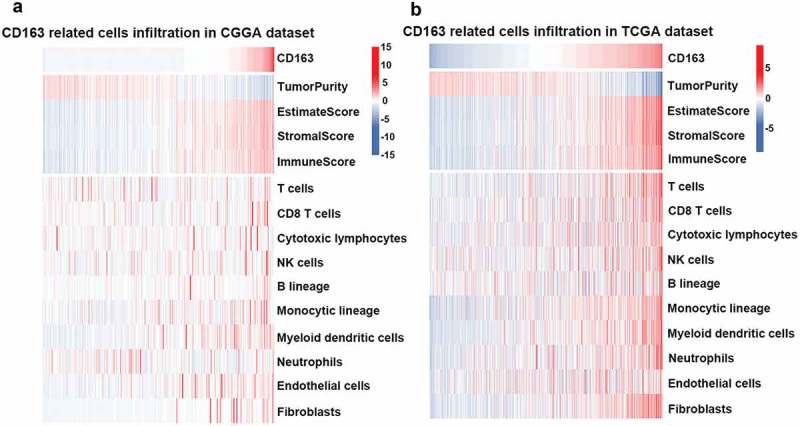
CD163 was tightly associated with immune score and infiltrated cells in tumor microenvironment. (a and b, upper panel) CD163 was positively related with immune score and stromal score. (a and b, lower panel) CD163 was closely related to infiltrated cells in tumor microenvironment.
Relationship between CD163 and inflammatory activities
To further understand CD163-related inflammatory activities, 104 genes were analyzed from seven clusters defined as metagenes,39 representing different types of inflammation and immune response. As shown in Figure 5(a), CD163 expression was positively associated with most of the clusters in CGGA dataset except for IgG, which was mainly associated with activities of B lymphocytes. To validate our findings in the clusters, seven metagenes were generated with results of Gene Sets Variation Analysis (GSVA) of corresponding clusters of genes. Corrgrams derived according to Pearson r value between CD163 and the seven metagenes (Figure 5(b)). CD163 was positively associated with HCK, LCK, and MHC-I, but was negatively associated with IFN-γ and IgG, as observed in Figure 5(a). Similar results were observed in the TCGA database (Figures S4(a and b)). These findings were consistent with the above results indicating that CD163 may participate in the regulation of the inflammatory response.
Figure 5.
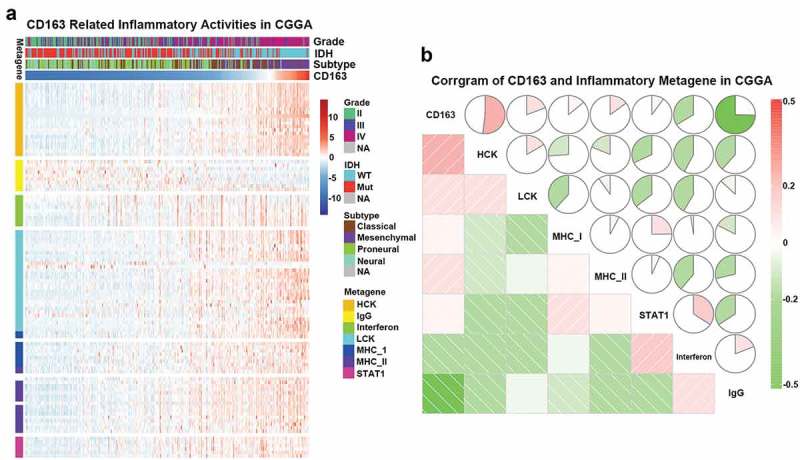
CD163-related inflammatory activities in CGGA database. (a) The heatmap of CD68 related inflammatory metagenes in CGGA cohort. (b) Corrgram of CD68 and inflammatory metagenes in CGGA cohort.
CD163 predicted poor survival in glioma and GBM
To further explore the prognostic value of CD163 in glioma of all grades, Kaplan–Meier curves were generated. As shown in Figure 6(a-c)), patients with higher CD163 expression had a significantly shorter survival than patients with low CD163 expression both in the CGGA and TCGA databases. A similar pattern was observed in the Kaplan–Meier curves of GBM patients (Figure 6(d-f)). These findings indicated that CD163 is a negative prognostic marker in glioma and GBM.
Figure 6.
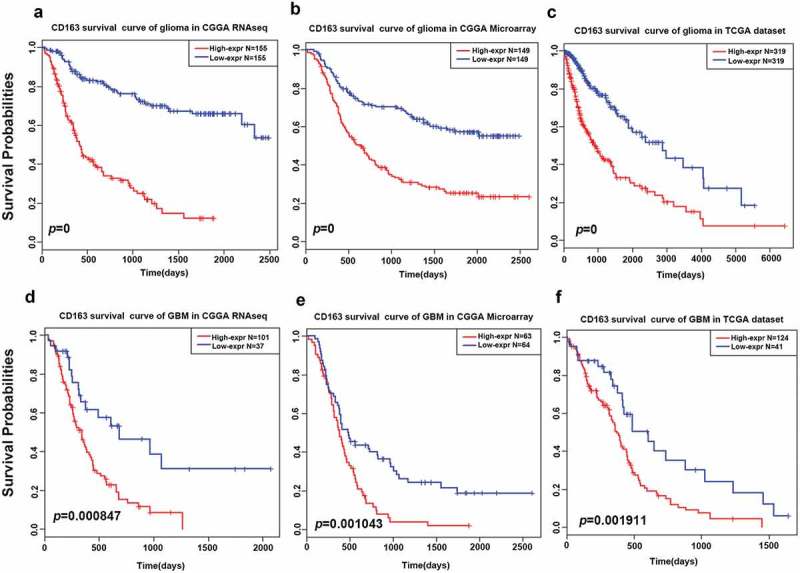
Survival analysis of CD163 in glioma and glioblastoma. (a, b, and c) Survival analysis of CD163 in whole grade glioma. (d, e, and f) Survival analysis of CD163 in glioblastoma.
CD163 was synergistic with checkpoint members and markers of macrophages in glioma and GBM
We identified CD163 as a potential immunotherapeutic target in our previous results and this led us to analyze the correlation between CD163 and immune checkpoints. Pearson correlation was performed in both the CGGA and TCGA databases. Figures S5 A and B showed that CD163 had a high concordance with PD-1, PD-L1, TIM-3, LAG3, B7H3, and B7H4 in glioma. The heterogeneity across different grades of glioma led us to investigate the relationship between these immune checkpoints in GBM. As indicated in Figures S5(c and d), these immune checkpoints showed an even higher correlation with CD163. These findings indicated that the combination therapy of CD163 and immune checkpoints may be useful in overcoming limitations associated to the use of immune checkpoints alone.
Furthermore, Macrophage Scavenger Receptor1 (MSR1), also known as CD204 has additionally been identified as a specific marker for TAMs.40,41 It was also reported that CD14, CD68, CD11B are macrophage markers.15 To fully understand the relationship between CD163 and other macrophage markers, Pearson correlation analysis was performed. As Figures S6A and B indicated, CD163 showed a high correlation with the macrophage marker members both in the CGGA RNA-seq and microarray databases. In line with the CGGA database, the correlation between CD163 and other macrophage markers was also robustly positive in the TCGA database (Figure S6c). Similar results were observed in Figures S6(d-f). Taken together, these results point to the rising MSR1, CD11B, or CD68 levels in cases when glioma patients acquire resistance to therapy of targeting CD163.
CD163+macrophages impaired T cell function in glioma
To further investigate the role of CD163 involved in the immune regulation, CD163+ and CD163- cells were sorted from the tumor tissues of glioma patients by fluorescence activating cell sorter (FACS). After sorting, the purity of CD163+ or CD163- macrophages was no less than 95% (Figures S7(a)). The mRNA expression of anti-inflammatory factors (Arginase-1, IL-10, IL-6, and TGF-β) in CD163+ macrophages was significantly higher than those in the CD163- macrophages, whereas pro-inflammatory factors of TNF-α and iNOS expression in CD163+ macrophages were lower than those in the CD163- macrophages (Figures S7(b)). In addition, we investigated the effect of CD163+ macrophages on the function of CD8+ T cells derived from the peripheral blood of the glioma patients in vitro. When T cells were co-cultured with CD163+ and CD163- macrophages, the percentage of PD-1 and TIM-3 within CD8+ T cells was significantly higher than that within CD163- macrophages. However, the percentage of IFN-γ and Granzyme B in CD8+ T cells was significantly decreased (Figures S7(f)). To further evaluate the function of CD163 in immune response, we performed the reversion of M2 to M1. We have reported that pseudomonas aeruginosa-mannose-sensitive hemagglutinin (PA-MSHA) could re-educate M2 to M1 in malignant pleural effusion through TLR4-mediated pathway. Thus, we used PA-MSHA to convert M2 to M1 to analyze the effect on CD8+ T cells. After treatment with PA-MSHA, CD163+ macrophages obtained from glioma tissues became smaller and turned round, shapes of which resembled M1 macrophages rather than M2 macrophages (Figures S7(c)). Also, the frequency of purified CD163+ macrophages was decreased after treatment with PA-MSHA in vitro (Figures S7(d)). In addition, we found that the mRNA expression of anti-inflammatory factors (Arginase-1, IL-10, IL-6, and TGF-β) in CD163+ macrophages treated with PA-MSHA was lower than that in cells untreated with PA-MSHA. However, expression of pro-inflammatory factors (TNF-α and iNOS) in these cells was increased after treatment with PA-MSHA (Figures S7(e)). Subsequently, we investigated the effect of treatment with PA-MSHA on CD8+ T cells. After treatment with PA-MSHA, the percentages of PD-1 and TIM-3 within CD8+ T cell populations co-cultured with CD163+ macrophages were decreased compared with those of the untreated group, whereas the percentages of IFN-γ and Granzyme B were increased (Figures S7(f)). Taken together, these results indicate that the switch of M2 to M1 reversed the impaired cytotoxicity of CD8+ T cells in glioma, indicating that CD163+ macrophages play an important role in T cell immune regulation in gliomas. Thus, high expression of M2-related cytokines in glioma tissues-derived CD163+ macrophages reveals that CD163+ TAMs are defined as M2 macrophages in gliomas and are involved in immune regulation.
Discussion
Given the extremely poor prognosis and high resistance to conventional therapies in glioma, it is worth exploring further mechanisms which could contribute to glioma development and therapy resistance.42,43
In general, as many as 30–50% of the cells in tumor microenvironment are tumor-associated macrophages (TAMs) in glioma. Clinically, TAMs within the brain tend to be linked with a poor outcome and their accumulation is associated with higher tumor grades. Furthermore, TAMs promote numerous tumor-promoting activities such as angiogenesis, enhanced tumor cell migration and invasiveness, and suppression of adaptive antitumor immunity.44,45 Thus, targeting the infiltrated TAMs may represent an attractive strategy to improve survival of glioma patients.
The first-identified function of CD163 is related to its capacity to bind and internalize Hb-Hp complexes and as such appears to participate in the clearance of free Hb-Hp complexes, thus preventing oxidative tissue damage. Later on, its functional repertoire was expanded, with the identification of CD163 as a receptor for TWEAK, which is a TNF superfamily member and mediates angiogenesis. Furthermore, a recent report showed that CD163 expression is associated with angiogenesis.46 Additionally, in tumor tissue, CD163 represents the most classical and most specific marker of TAMs and is also a prognostic factor in glioma and other types of cancer.15,28 Han reported that presence of CD163+ TAMs exacerbated T cell immunosuppression, since depletion of CD163+ TAMs significantly improved T cell proliferation and pro-inflammatory cytokine production.47 Keigo also reported that CD163+ TAMs promote T-cell apoptosis and immunosuppression via IL-10 and PD-L1 and show unfavorable prognosis in oral squamous cell carcinoma patients.48Therefore, targeting CD163 could be one of the potential ways to eliminate the TAMs and markedly inhibit the immune escape, thus leading to the retardation of glioma progression.49,50 However, a comprehensive report about CD163 expression and its functions in glioma were not available. In our study, we firstly performed the largest and most comprehensive research to identify the expression pattern and distribution of CD163 in 1323 glioma samples using the CGGA and TCGA databases. We found that GBM patients had the most up-regulated CD163 expression, which is consistent with previous reports.14,15 Moreover, we also noted that a high expression of CD163 was associated with a high enrichment of phenotypes of known malignant molecules, such as IDH wild-type state, mesenchymal phenotype. All these findings suggested that CD163 may play an important role in the biological processes of glioma.
Additionally, through the analysis of the relationship between CD163 and biological processes, we discovered that CD163 was positively associated with angiogenesis and inflammatory activities. A previous report has demonstrated that the CD163+ TAMs density positively correlated with tumor microvessel density and CD163+ TAMs actively contribute to the process of vasculogenesis,44 which is consistent with our findings. Anti-angiogenic agents have been hypothesized to be beneficial in treating solid tumors, due to the important role that angiogenesis plays in tumor growth, invasion, and metastasis.51 Disappointingly, these agents have not been successful in trials, which may be because increased levels of TAMs may still be present after anti-angiogenesis therapies; the number of CD163+ TAMs significantly correlated with an overall decreased survival among patients with recurrent GBM.14 Recently, Wei Lu52 demonstrated that overexpressed CD163 in bone marrow stromal cells (BMSCs) elevated protein levels of endothelial-associated markers such as CD31, Flk-1, eNOS, and VE-cadherin, significantly promoting angiogenesis. They also found that CD163 was involved in Hmbox1/CD163/FGF-2 signal pathway in BMSC differentiation into vascular endothelial-like cells, which suggested that CD163 was a key regulator in angiogenesis and provided a novel target for further investigating the gene control of BMSC differentiation into vascular endothelial-like cells. Therefore, the combined targeting of CD163 and angiogenesis may be an effective therapy for treating patients with glioma.
We then undertook an in-depth analysis of the relationship between CD163 and inflammatory-related clusters. The findings revealed that CD163 was positively correlated with immune response and antigen presentation. Furthermore, Fabriek53 also reported that the CD163 positive perivascular macrophages in the human brain express several molecules such as MHC-II, B7-1, B7-2, and CD40 involved in antigen recognition, presentation, and costimulation, which supported our results. It has been extensively reported that the expression of CD163 can be regulated by various factors in vitro and in vivo. With the treatment of glucocorticoids, the percentage of CD163 positive monocytes increased markedly, from 10–30% to 90%.54 The other anti-inflammatory mediators such as IL-10 and IL-6 also promote CD163 expression. Inversely, pro-inflammatory factors, such as LPS, IFN-γ, and TNF-α suppress the mRNA and protein levels of CD163. We also found that the expression of CD163 was indirectly proportional to the expression of IFN-γ, consistent with the previous studies. The regulation of CD163 by pro- and anti-inflammatory mediators demonstrated a relationship between CD163 and the resolution of inflammation.
Through analyzing the relationship between CD163 expression and T cell-specific function, the immunosuppressive role of CD163 was further confirmed. As tumor progression occurs, T cell activation is facilitated to suppress tumor growth. However, CD163 expression is promoted in TAMs, thus inhibiting T cell activation and leading to the formation of the immunosuppressive microenvironment in glioma, and lastly leading to immune escape. Collectively, CD163 can have dual targeting functions; on the one hand, CD163 acts as a regulator of immune response and targeting CD163 may inhibit immune escape and recover the function of dysfunctional T cells. On the other hand, CD163 acts as a non-immunological regulator and targeting CD163 may intervene in TAMs-regulated tumor angiogenesis and decrease tumor growth.
In addition, we also found that higher CD163 expression level predicted significantly worse survival rate in glioma and GBM patients. Similarly, as our results suggested, CD163 expression was higher within glioma specimens than in the surrounding peripheral areas in both male and female patients, and was inversely correlated with mean survival times.28 Furthermore, higher numbers of CD163 positive TAMs infiltration were associated with a poor prognosis in various types of solid tumors.40,55,56 In our previous report, we observed that CD163 positive TAMs contribute to the progression of tumor growth, and the increased infiltration presents a poor prognosis in lung cancer.49,50 This discovery provided a reliable foundation for our current research.
Recently, an increasing research interest has focused on studying a combination of immune checkpoint blockades, which may be a promising therapy for treating glioma. The first phase III trial of nivolumab, the FDA approved checkpoint inhibitor, targeting PD-1 for unresectable or metastatic melanomas and non-small cell lung cancer (NSCLCs) are ongoing in patients with GBM (ClinicalTrials.gov identifier: NCT02017717).57 Immune checkpoint blockades have shown great progress for the treatment of cancer, however, both endogenous and exogenous T cells are inhibited by the immunosuppressive tumor microenvironment. TAMs in the microenvironment play pivotal and complex roles in tumor development and progression. Inhibition of CSF-1R can lead to preferential TAMs depletion and modulate the remaining myeloid cells toward a more anti-tumor phenotype and away from pro-tumor properties.58–60 The combination of anti-CSF-1R and anti-PD-1 antibodies promotes antitumor activity in pre-clinical models, and this combination is actively investigated in several clinical trials for many types of tumors.59,61–63 It is encouraging that one clinical trial has already reported that dual PD-1 and CSF-1R blockade may provide durable clinical benefit in patients with malignant glioma (ClinicalTrials.gov identifier: NCT02526017). Recently, the concept of targeting CD163 therapy has also been established in quite a number of studies.29,64 However, the concept of using a combination therapy of CD163 and immune checkpoints has not been envisaged till date. In our study, we initially analyzed the correlation between CD163 and immune checkpoints, and found that CD163 has a high concordance with immune checkpoints, such as PD-L1, PD-1, TIM-3, LAG-3, B7-H3, and B7-H4. These findings provide promising opportunities for the combined treatment of glioma.
In conclusion, this is the first study exploring the clinical expression and biological processes, including inflammatory and immune response of CD163, using glioma samples in the CGGA and TCGA databases on a large scale. Our results elucidated the involvement of the CD163 pathway in immune responses and tumor progression that may lead to the development of CD163 targeting therapy strategies for assessing the efficacy and receptiveness in glioma treatment.
Methods
Sample and data collection
We obtained the CD163 transcriptional data from CGGA (http://www.cgga.org.cn/) and TCGA (http://cancergenome.nih.gov/). A total of 325 samples of RNA-seq data and 301 samples of mRNA microarray data ranging from WHO grade II to grade IV gliomas were examined in this study. To validate the CGGA dataset findings, we analyzed 697 glioma samples of all grades from the TCGA dataset. We analyzed the data from 121, 122 and 271 females in the CGGA microarray, CGGA RNA-seq and TCGA databases. The details of patients’ information from these three datasets are described in supplementary Table 1.
Statistical analysis
R language was used as the main tool for the statistical analysis and generating figures. Kaplan–Meier plots were generated by the survival package, and the log‐rank test was used to compare survival curves between high and low CD163 expression patients. In the survival analysis, patients with no prognostic information were excluded. The Estimation of STromal and Immune cells in Malignant Tumours using Expression data (ESTIMATE) package was used to evaluate the infiltrated cells and Gene Sets Variation Analysis (GSVA) package was used to calculate the inflammatory activities in glioma microenvironment. Other figures were generated by several R packages, such as pheatmap, pROC, circlize, and corrgram. Gaussian test was performed before data analysis that required Gaussian distribution. GraphPad Prism version 7.0 (GraphPad Software Inc., USA) was used to perform the statistical analyses for in vitro experiments. Student’s t-test and one-way ANOVA were conducted to compare the differences between variables, as appropriate. The data are presented as the means ± SDs of three independent experiments. A p-value less than 0.05 is considered to be statistically significant. All statistical tests were two-sided.
Funding Statement
This study was supported by the National Key Research and Development Program of China (NO. 2016YFC1303501) and the Major Science and Technology Projects of Henan Province (NO. 22130001).
Acknowledgments
We thank Lifeng Li, Guohui Qin and Zhibo Shen for the technical assistance. We also thank Ling Cao and Qun Gao for their comments and advice.
Disclosure of potential conflicts of interest
Authors declare no conflicts of interest.
Authors’ contributions
SL, CZ, and YZ conceived and designed the project. SL, CZ, and ZZ collected and performed the data analysis. SL and CZ drafted the manuscript. NRM, YP, and LH prepared figures and reviewed the paper. LY reviewed the revised paper and performed experiments. All authors read and approved the final manuscript.
Availability of data and materials
CD163 expression data of gliomas was obtained from CGGA (http://www.cgga.org.cn/) and TCGA (http://cancergenome.nih.gov/).
Supplementary data
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW.. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Burton EC, Prados MD.. Malignant gliomas. Curr Treat Options Oncol. 2000;1:459–468. [DOI] [PubMed] [Google Scholar]
- 3.Buckner JC, Brown PD, O‘Neill BP, Meyer FB, Wetmore CJ, Uhm JH. Central nervous system tumors. Mayo Clin Proc. 2007;82(10):1271–1286. doi: 10.4065/82.10.1271. [DOI] [PubMed] [Google Scholar]
- 4.Stupp R, Hegi ME, Mason WP, van Den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. doi: 10.1016/s1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 5.Rape A, Ananthanarayanan B, Kumar S. Engineering strategies to mimic the glioblastoma microenvironment. Adv Drug Deliv Rev. 2014;79-80:172–183. doi: 10.1016/j.addr.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nusblat LM, Carroll MJ, Roth CM. Crosstalk between M2 macrophages and glioma stem cells. Cell Oncol (Dordr). 2017;40(5):471–482. doi: 10.1007/s13402-017-0337-5. [DOI] [PubMed] [Google Scholar]
- 7.Shi Y, Ping YF, Zhou W, He ZC, Chen C, Bs B, Zhang L, Chen L, Lan X, Zhang XC, et al. Tumour-associated macrophages secrete pleiotrophin to promote PTPRZ1 signalling in glioblastoma stem cells for tumour growth. Nat Commun. 2017;8:15080. doi: 10.1038/ncomms15080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graeber MB, Scheithauer BW, Kreutzberg GW. Microglia in brain tumors. Glia. 2002;40(2):252–259. doi: 10.1002/glia.10147. [DOI] [PubMed] [Google Scholar]
- 9.Garris C, Pittet MJ. Therapeutically reeducating macrophages to treat GBM. Nat Med. 2013;19(10):1207–1208. doi: 10.1038/nm.3355. [DOI] [PubMed] [Google Scholar]
- 10.Yang L, Zhang Y. Tumor-associated macrophages: from basic research to clinical application. J Hematol Oncol. 2017;10(1). 58. doi: 10.1186/s13045-017-0430-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41(1):14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ginhoux F, Schultze JL, Murray PJ, Ochando J, Biswas SK. New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat Immunol. 2016;17(1):34–40. doi: 10.1038/ni.3324. [DOI] [PubMed] [Google Scholar]
- 13.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu-Emerson C, Snuderl M, Kirkpatrick ND, Goveia J, Davidson C, Huang Y, Riedemann L, Taylor J, Ivy P, Duda DG, et al. Increase in tumor-associated macrophages after antiangiogenic therapy is associated with poor survival among patients with recurrent glioblastoma. Neuro Oncol. 2013;15(8):1079–1087. doi: 10.1093/neuonc/not082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prosniak M, Harshyne LA, Andrews DW, Kenyon LC, Bedelbaeva K, Apanasovich TV, Heber-Katz E, Curtis MT, Cotzia P, Hooper DC. Glioma grade is associated with the accumulation and activity of cells bearing M2 monocyte markers. Clin Cancer Res. 2013;19(14):3776–3786. doi: 10.1158/1078-0432.CCR-12-1940. [DOI] [PubMed] [Google Scholar]
- 16.Zwadlo G, Voegeli R, Schulze Osthoff K, Sorg C. A monoclonal antibody to a novel differentiation antigen on human macrophages associated with the down-regulatory phase of the inflammatory process. Exp Cell Biol. 1987;55:295–304. [DOI] [PubMed] [Google Scholar]
- 17.Law SK, Micklem KJ, Shaw JM, Zhang XP, Dong Y, Willis AC, Mason DY. A new macrophage differentiation antigen which is a member of the scavenger receptor superfamily. Eur J Immunol. 1993;23(9):2320–2325. doi: 10.1002/eji.1830230940. [DOI] [PubMed] [Google Scholar]
- 18.Kristiansen M, Graversen JH, Jacobsen C, Sonne O, Hoffman HJ, Law SK, Moestrup SK. Identification of the haemoglobin scavenger receptor. Nature. 2001;409(6817):198–201. doi: 10.1038/35051594. [DOI] [PubMed] [Google Scholar]
- 19.Bover LC, Cardo-Vila M, Kuniyasu A, Sun J, Rangel R, Takeya M, Aggarwal BB, Arap W, Pasqualini R. A previously unrecognized protein-protein interaction between TWEAK and CD163: potential biological implications. J Immunol. 2007;178:8183–8194. [DOI] [PubMed] [Google Scholar]
- 20.Kaplan MJ, Lewis EE, Shelden EA, Somers E, Pavlic R, McCune WJ, Richardson BC. The apoptotic ligands TRAIL, TWEAK, and Fas ligand mediate monocyte death induced by autologous lupus T cells. J Immunol. 2002;169:6020–6029. [DOI] [PubMed] [Google Scholar]
- 21.Wiley SR, Cassiano L, Lofton T, Davis-Smith T, Winkles JA, Lindner V, Liu H, Daniel TO, Smith CA, Fanslow WC. A novel TNF receptor family member binds TWEAK and is implicated in angiogenesis. Immunity. 2001;15:837–846. [DOI] [PubMed] [Google Scholar]
- 22.Moreno JA, Munoz-Garcia B, Martin-Ventura JL, Madrigal-Matute J, Orbe J, Paramo JA, Ortega L, Egido J, Blanco-Colio LM. The CD163-expressing macrophages recognize and internalize TWEAK: potential consequences in atherosclerosis. Atherosclerosis. 2009;207(1):103–110. doi: 10.1016/j.atherosclerosis.2009.04.033. [DOI] [PubMed] [Google Scholar]
- 23.Fabriek BO, van Bruggen R, Deng DM, Ligtenberg AJ, Nazmi K, Schornagel K, Vloet RP, Dijkstra CD, van Den Berg TK. The macrophage scavenger receptor CD163 functions as an innate immune sensor for bacteria. Blood. 2009;113(4):887–892. doi: 10.1182/blood-2008-07-167064. [DOI] [PubMed] [Google Scholar]
- 24.Backe E, Schwarting R, Gerdes J, Ernst M, Stein H. Ber-MAC3: new monoclonal antibody that defines human monocyte/macrophage differentiation antigen. J Clin Pathol. 1991;44:936–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Gorp H, Delputte PL, Nauwynck HJ. Scavenger receptor CD163, a Jack-of-all-trades and potential target for cell-directed therapy. Mol Immunol. 2010;47(7–8):1650–1660. doi: 10.1016/j.molimm.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 26.Franco R, Fernandez-Suarez D. Alternatively activated microglia and macrophages in the central nervous system. Prog Neurobiol. 2015;131:65–86. doi: 10.1016/j.pneurobio.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 27.Colin S, Chinetti-Gbaguidi G, Staels B. Macrophage phenotypes in atherosclerosis. Immunol Rev. 2014;262(1):153–166. doi: 10.1111/imr.12218. [DOI] [PubMed] [Google Scholar]
- 28.Lisi L, Ciotti GM, Braun D, Kalinin S, Curro D, Dello Russo C, Coli A, Mangiola A, Anile C, Feinstein DL, et al. Expression of iNOS, CD163 and ARG-1 taken as M1 and M2 markers of microglial polarization in human glioblastoma and the surrounding normal parenchyma. Neurosci Lett. 2017;645:106–112. doi: 10.1016/j.neulet.2017.02.076. [DOI] [PubMed] [Google Scholar]
- 29.Graversen JH, Svendsen P, Dagnaes-Hansen F, Dal J, Anton G, Etzerodt A, Petersen MD, Christensen PA, Moller HJ, Moestrup SK. Targeting the hemoglobin scavenger receptor CD163 in macrophages highly increases the anti-inflammatory potency of dexamethasone. Mol Ther. 2012;20(8):1550–1558. doi: 10.1038/mt.2012.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komohara Y, Hirahara J, Horikawa T, Kawamura K, Kiyota E, Sakashita N, Araki N, Takeya M. AM-3K, an anti-macrophage antibody, recognizes CD163, a molecule associated with an anti-inflammatory macrophage phenotype. J histochem cytochem. 2006;54(7):763–771. doi: 10.1369/jhc.5A6871.2006. [DOI] [PubMed] [Google Scholar]
- 31.Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, Trevino V, Shen H, Laird PW, Levine DA, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, Selves J, Laurent-Puig P, Sautes-Fridman C, Fridman WH, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17(1):218. doi: 10.1186/s13059-016-1070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454(7203):436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 34.Leek RD, Lewis CE, Whitehouse R, Greenall M, Clarke J, Harris AL. Association of macrophage infiltration with angiogenesis and prognosis in invasive breast carcinoma. Cancer Res. 1996;56:4625–4629. [PubMed] [Google Scholar]
- 35.Murdoch C, Muthana M, Coffelt SB, Lewis CE. The role of myeloid cells in the promotion of tumour angiogenesis. Nat Rev Cancer. 2008;8(8):618–631. doi: 10.1038/nrc2444. [DOI] [PubMed] [Google Scholar]
- 36.Cheng Z, Zhang D, Gong B, Wang P, Liu F. CD163 as a novel target gene of STAT3 is a potential therapeutic target for gastric cancer. Oncotarget. 2017;8(50):87244–87262. doi: 10.18632/oncotarget.20244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou J, Wang XH, Zhao YX, Chen C, Xu XY, Sun Q, Wu HY, Chen M, Sang JF, Su L, et al. Cancer-associated fibroblasts correlate with tumor-associated macrophages infiltration and lymphatic metastasis in triple negative breast cancer patients. J Cancer. 2018;9(24):4635–4641. doi: 10.7150/jca.28583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang A, Qian Y, Ye Z, Chen H, Xie H, Zhou L, Shen Y, Zheng S. Cancer-associated fibroblasts promote M2 polarization of macrophages in pancreatic ductal adenocarcinoma. Cancer Med. 2017;6(2):463–470. doi: 10.1002/cam4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rody A, Holtrich U, Pusztai L, Liedtke C, Gaetje R, Ruckhaeberle E, Solbach C, Hanker L, Ahr A, Metzler D, et al. T-cell metagene predicts a favorable prognosis in estrogen receptor-negative and HER2-positive breast cancers. Breast Cancer Res. 2009;11(2):R15. doi: 10.1186/bcr2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reinartz S, Schumann T, Finkernagel F, Wortmann A, Jansen JM, Meissner W, Krause M, Schworer AM, Wagner U, Muller-Brusselbach S, et al. Mixed-polarization phenotype of ascites-associated macrophages in human ovarian carcinoma: correlation of CD163 expression, cytokine levels and early relapse. Int J Cancer. 2014;134(1):32–42. doi: 10.1002/ijc.28335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komohara Y, Ohnishi K, Kuratsu J, Takeya M. Possible involvement of the M2 anti-inflammatory macrophage phenotype in growth of human gliomas. J Pathol. 2008;216(1):15–24. doi: 10.1002/path.2370. [DOI] [PubMed] [Google Scholar]
- 42.Yang MY, Zetler PM, Prins RM, Khan-Farooqi H, Liau LM. Immunotherapy for patients with malignant glioma: from theoretical principles to clinical applications. Expert Rev Neurother. 2006;6(10):1481–1494. doi: 10.1586/14737175.6.10.1481. [DOI] [PubMed] [Google Scholar]
- 43.Mitchell DA, Fecci PE, Sampson JH. Immunotherapy of malignant brain tumors. Immunol Rev. 2008;222:70–100. doi: 10.1111/j.1600-065X.2008.00603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen JJ, Lin YC, Yao PL, Yuan A, Chen HY, Shun CT, Tsai MF, Chen CH, Yang PC. Tumor-associated macrophages: the double-edged sword in cancer progression. J Clin Oncol. 2005;23(5):953–964. doi: 10.1200/JCO.2005.12.172. [DOI] [PubMed] [Google Scholar]
- 45.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141(1):39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koh YW, Park CS, Yoon DH, Suh C, Huh J. CD163 expression was associated with angiogenesis and shortened survival in patients with uniformly treated classical Hodgkin lymphoma. PLoS One. 2014;9(1):e87066. doi: 10.1371/journal.pone.0087066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han Q, Shi H, Liu F. CD163(+) M2-type tumor-associated macrophage support the suppression of tumor-infiltrating T cells in osteosarcoma. Int Immunopharmacol. 2016;34:101–106. doi: 10.1016/j.intimp.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 48.Kubota K, Moriyama M, Furukawa S, Rafiul H, Maruse Y, Jinno T, Tanaka A, Ohta M, Ishiguro N, Yamauchi M, et al. CD163(+)CD204(+) tumor-associated macrophages contribute to T cell regulation via interleukin-10 and PD-L1 production in oral squamous cell carcinoma. Sci Rep. 2017;7(1):1755. doi: 10.1038/s41598-017-01661-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang F, Yang L, Gao Q, Huang L, Wang L, Wang J, Wang S, Zhang B, Zhang Y. CD163+CD14+ macrophages, a potential immune biomarker for malignant pleural effusion. Cancer Immunol Immunother. 2015;64(8):965–976. doi: 10.1007/s00262-015-1701-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang L, Wang F, Wang L, Huang L, Wang J, Zhang B, Zhang Y. CD163+ tumor-associated macrophage is a prognostic biomarker and is associated with therapeutic effect on malignant pleural effusion of lung cancer patients. Oncotarget. 2015;6(12):10592–10603. doi: 10.18632/oncotarget.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu C, Kros JM, Cheng C, Mustafa D. The contribution of tumor-associated macrophages in glioma neo-angiogenesis and implications for anti-angiogenic strategies. Neuro Oncol. 2017;19(11):1435–1446. doi: 10.1093/neuonc/nox081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu W, Su L, Yu Z, Zhang S, Miao J. The new role of CD163 in the differentiation of bone marrow stromal cells into vascular endothelial-like cells. Stem Cells Int. 2016; 2016:2539781.doi: 10.1155/2016/2539781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fabriek BO, Van Haastert ES, Galea I, Polfliet MM, Dopp ED, Van Den Heuvel MM, Van Den Berg TK, De Groot CJ, Van Der Valk P, Dijkstra CD. CD163-positive perivascular macrophages in the human CNS express molecules for antigen recognition and presentation. Glia. 2005;51(4):297–305. doi: 10.1002/glia.20208. [DOI] [PubMed] [Google Scholar]
- 54.Wenzel I, Roth J, Sorg C. Identification of a novel surface molecule, RM3/1, that contributes to the adhesion of glucocorticoid-induced human monocytes to endothelial cells. Eur J Immunol. 1996;26(11):2758–2763. doi: 10.1002/eji.1830261131. [DOI] [PubMed] [Google Scholar]
- 55.Bingle L, Brown NJ, Lewis CE. The role of tumour-associated macrophages in tumour progression: implications for new anticancer therapies. J Pathol. 2002;196(3):254–265. doi: 10.1002/path.1027. [DOI] [PubMed] [Google Scholar]
- 56.Marchesi F, Cirillo M, Bianchi A, Gately M, Olimpieri OM, Cerchiara E, Renzi D, Micera A, Balzamino BO, Bonini S, et al. High density of CD68+/CD163+ tumour-associated macrophages (M2-TAM) at diagnosis is significantly correlated to unfavorable prognostic factors and to poor clinical outcomes in patients with diffuse large B-cell lymphoma. Hematol Oncol. 2015;33(2):110–112. doi: 10.1002/hon.2142. [DOI] [PubMed] [Google Scholar]
- 57.Maxwell R, Jackson CM, Lim M. Clinical trials investigating immune checkpoint blockade in glioblastoma. Curr Treat Options Oncol. 2017;18(8):51. doi: 10.1007/s11864-017-0492-y. [DOI] [PubMed] [Google Scholar]
- 58.Ries CH, Cannarile MA, Hoves S, Benz J, Wartha K, Runza V, Rey-Giraud F, Pradel LP, Feuerhake F, Klaman I, et al. Targeting tumor-associated macrophages with anti-CSF-1R antibody reveals a strategy for cancer therapy. Cancer Cell. 2014;25(6):846–859. doi: 10.1016/j.ccr.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 59.Zhu Y, Knolhoff BL, Meyer MA, Nywening TM, West BL, Luo J, Wang-Gillam A, Goedegebuure SP, Linehan DC, DeNardo DG. CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer Res. 2014;74(18):5057–5069. doi: 10.1158/0008-5472.can-13-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Candido JB, Morton JP, Bailey P, Campbell AD, Karim SA, Jamieson T, Lapienyte L, Gopinathan A, Clark W, McGhee EJ, et al. CSF1R(+) macrophages sustain pancreatic tumor growth through T cell suppression and maintenance of key gene programs that define the squamous subtype. Cell Rep. 2018;23(5):1448–1460. doi: 10.1016/j.celrep.2018.03.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peranzoni E, Lemoine J, Vimeux L, Feuillet V, Barrin S, Kantari-Mimoun C, Bercovici N, Guerin M, Biton J, Ouakrim H, et al. Macrophages impede CD8 T cells from reaching tumor cells and limit the efficacy of anti-PD-1 treatment. Proc Natl Acad Sci U S A. 2018;115(17):E4041–e4050. doi: 10.1073/pnas.1720948115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eissler N, Mao Y, Brodin D, Reutersward P, Andersson Svahn H, Johnsen JI, Kiessling R, Kogner P. Regulation of myeloid cells by activated T cells determines the efficacy of PD-1 blockade. Oncoimmunology. 2016;5(12):e1232222. doi: 10.1080/2162402x.2016.1232222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cannarile MA, Weisser M, Jacob W, Jegg AM, Ries CH, Ruttinger D. Colony-stimulating factor 1 receptor (CSF1R) inhibitors in cancer therapy. J Immunother Can. 2017;5(1):53. doi: 10.1186/s40425-017-0257-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Greisen SR, Moller HJ, Stengaard-Pedersen K, Hetland ML, Horslev-Petersen K, Jorgensen A, Hvid M, Deleuran B. Soluble macrophage-derived CD163 is a marker of disease activity and progression in early rheumatoid arthritis. Clin Exp Rheumatol. 2011;29:689–692. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
CD163 expression data of gliomas was obtained from CGGA (http://www.cgga.org.cn/) and TCGA (http://cancergenome.nih.gov/).


