ABSTRACT
The primordial follicle pool, providing all oocytes available to a female throughout her reproductive life, is established perinatally. The formation of primordial follicle pool is regulated by precise transcriptional and post-transcriptional mechanisms. Recent studies have identified several microRNAs as post-transcriptional regulatory factors in the process of primordial follicle assembly. Here, we showed that miR-92b-3p was significantly upregulated in the stage of primordial follicle assembly in newborn mouse ovaries. Inhibiting miR-92b-3p suppressed the formation of primordial follicles, while overexpression of miR-92b-3p accelerated the processes of cyst breakdown and the following primordial follicle assembly. Accordingly, the expression of follicular development-related genes was reduced upon inhibiting of miR-92b-3p and increased under miR-92b-3p overexpression. Mechanistic studies identified TSC1 as a direct target of miR-92b-3p. miR-92b-3p could activate mTOR/Rps6 signaling through targeting and inhibiting TSC1 expression. In addition, knockdown of TSC1 showed an identical phenotype with that of miR-92b-3p overexpression in accelerating processes of cyst breakdown and primordial follicle formation. Thus, our work demonstrates that miR-92b-3p is a novel regulator of primordial follicle assembly by negatively regulating TSC1 in mTOR/Rps6 signaling.
KEYWORDS: Mir-92b-3p, primordial follicle formation, mTOR, TSC1
Introduction
The entire process of primordial follicle formation involves at least two cell types: the germ cells within the cyst and the surrounding pregranulosa cells [1]. After cyst breakdown perinatally, each oocyte is enclosed by pregranulosa cells to form individual primordial follicle [2]. In mice, the pool of primordial follicles is established in the ovaries during the first 4–5 days after birth [3,4]. They serve as total population of developing follicles and oocytes available to a female for the entire reproductive life [5–7]. Any perturbation of this process can significantly affect the size of primordial follicle pool, resulting in serious reproductive diseases such as ovarian dysgenesis or premature ovarian failure (POF) [8]. However, the mechanisms underlying this process are far from complete.
MicroRNAs (miRNAs) are endogenous noncoding RNAs comprising approximately 20–22 nucleotides, that can partially negatively regulate target gene expression through post-transcriptional degradation or translational repression [9]. They are conserved from worms to mammals and are essential in cell processes such as proliferation, apoptosis, differentiation, and development, cell cycle control, and metabolism [10,11]. Recent studies have shown that some miRNAs are highly enriched in the ovary, suggesting that these RNAs may play vital roles in follicular development [12–14]. For examples, miR-143 inhibits the formation of primordial follicles by suppressing pregranulosa cell proliferation and downregulating the expression of genes related to the cell cycle [15]. MiR-21, miR-125b, let-7b, let-7a, and let-7c share the same expression pattern as that of miR-143 in the follicles at each stage of follicular development [14]. MiR-125b overexpression is also reported to block the process of primordial follicle assembly in cultured newborn mouse ovaries, while its knockdown promotes this process [16]. In contrast, miR-376a has the ability to increase primordial follicle assembly and decrease apoptosis of oocytes by modulating the expression of proliferating cell nuclear antigen (PCNA) in fetal and neonatal mouse ovaries [17].
MiR-92b is highly conserved from Drosophila to human [18]. It was originally identified as an oncogenic miRNA in multiple cancers including glioma, glioblastoma, and nonsmall cell lung cancer [19–21]. As a member of miR-92b cluster, miR-92b-3p was reported to be upregulated in metastatic colorectal carcinoma and targets tumor suppressor F-box and WD-40 domain protein 7 (FBXW7/hCdc4) [22]. However, in pancreatic cancers, it acts as a tumor suppressor by targeting Gabra3 [23], indicating a diverse role of miR-92b-3p in the regulation of different tumor progress. In addition to its roles in tumors, miR-92b-3p also mediates the proliferation and cell cycle progression of pulmonary artery smooth muscles [24], or regulates cardiac hypertrophic growth [25]. Recent clinical studies have identified miR-92b-3p as one of the downregulated miRNAs which play important roles in the etiology and pathophysiology of polycystic ovary syndrome patients [26]. But the role of miR-92b-3p in follicular development is completely lacking. We therefore evaluated the expression of miR-92b-3p during primordial follicle formation, and then investigated its function through microRNA transfection in the newborn mouse ovary culture system. Our results demonstrated for the first time that miR-92b-3p is critical for primordial follicle formation.
Materials and methods
Experimental animals and ovary organ culture
ICR mice were purchased from the Animal Core Facility of Nanjing Medical University. Animal care and experimental procedures were approved by the Animal Care and Use Committee of Nanjing Medical University. Mice were maintained under a 12/12-h dark-light cycle at 22°C with free access to food and water. Adult male and female mice were mated using timed mating, and females were checked for the presence of a vaginal plug in the following morning. The presence of a vaginal plug was denoted as 0.5 day post coitus (dpc). Embryonic ovaries were collected at 16.5 dpc, and neonatal ovaries were collected at 0.5, 2.5, and 6.5 days post parturition (dpp) for immunohistochemistry. Ovaries were isolated from newborn mouse, three to four ovaries were pooled and placed in a 24-well dish and cultured in DMEM:HAM/F12 (invitrogen), which was free of FBS, and incubated at 37°C in a humidified atmosphere of 5% CO2.
Mouse ovary transfection
The miR-92b-3p mimics, inhibitor and corresponding scramble control were designed and synthesized by Gene Pharma (Shanghai, China). The oligonucleotide sequences are listed in Supplementary Table 1. Oligonucleotides were transfected into the mouse 0.5 dpp ovaries with Lipofectamine 2000 Transfection Reagent (Cat# 11,668,019, Invitrogen). Forty-eight hours after transfection, the medium was changed with replacement of half of the complete medium. Ovaries were harvested 96 h after transfection.
Quantitative real-time PCR
Total RNA was isolated from three to four ovaries using the Qiagen RNeasy Mini Kit in combination with on-column DNase treatment (Applied Biosystems, USA). A High Capacity RNA-to-cDNA Kit (Applied Biosystems) was used to synthesize the first strand of cDNA. Reversed transcriptase (RT) reaction was promoted by M-MLV reverse transcriptase (Promega) with purified total RNA (1 μg) as a template and 50 nM RT primer (sequences listed in Supplementary Table 1). Quantitative real-time PCR was performed using the Power SYBR Green PCR Master Mix (Applied Biosystems) with gene-specific primers (sequences listed in Supplementary Table 1). All gene transcripts were normalized to GAPDH, and the relative fold change was calculated using the 2−∆∆CT method.
Immunohistochemistry
Cultured ovaries were collected, fixed in 4% paraformaldehyde for 2 h at room temperature, embedded and continuously sectioned at 5 μm of thickness. After deparaffinization and rehydration, sections were processed for blocking of endogenous peroxidase activity with 3% hydrogen peroxide in methanol for 20 min. Antigen retrieval pretreatment was performed by boiling the sections in 0.01 M citrate buffer, pH 6.0 for 15 min. After blocking with 10% normal goat serum in PBS, sections were incubated with VASA antibody (Ab13840, Abcam) overnight at 4°C. Nonimmunized rabbit serum was used as controls.
Western blotting
Ovaries were lysed with RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS) with 1x protease inhibitor, boiled for 10 min in Laemmli buffer, and then directly loaded onto a 10% SDS-PAGE gel for protein separation. After transferred to a nitrocellulose membrane, blots were blocked with 5% nonfat dry milk in TBS containing 0.5% Tween-20 (TBS-T), washed and incubated with anti-VASA (Ab13840, Abcam) overnight at 4°C. The blots were then washed 3 times in TBS-T and incubated with peroxidase-conjugated goat anti-rabbit IgG secondary antibody (Jackson Immuno Research) for 1 h. The signals were visualized using an Enhanced Chemiluminescence Detection Kit (Pierce Biotechnology, USA) on an ECL Plus Western Blotting Detection System (GE Healthcare, Piscataway, NJ, USA).
Luciferase reporter assay
The 3ʹ-UTR of mouse TSC1 mRNA was cloned and introduced between the KpnI and NheI restriction sites into the modified pGL3 luciferase reporter vector (gift from Dr. Chun Lu from Nanjing Medical University). The firefly luciferase vector was used for internal control. Constructs with mutated 3ʹ-UTR of TSC1 mRNA were used as negative controls. The constructs were confirmed by sequencing. Then a mixture containing 200 ng/mL of the luciferase reporter plasmid and 40 nM miR-92b-3p mimics, miR-92b-3p inhibitors or the corresponding controls was transfected into 293T cells. The luciferase activity was measured by the Dual-luciferase reporter assay system (Cat#: E2920, Promega).
Histological evaluation of follicle numbers
The in vitro cultured mouse ovaries were fixed in 10% buffered formalin for 12 h, embedded in paraffin, serially sectioned at a thickness of 5 μm, and then stained with an antibody against VASA, a specific marker for germ cells. Follicles were counted in every fifth section, and only oocyte with a visible nucleus was counted to avoid duplicate counts. Germ cells not surrounded by pregranulosa cells were scored as unassembled (remaining in cysts). Follicles were scored as primordial follicles if oocytes with visible nuclei were surrounded by pregranulosa cells or a mixture of squamous and cuboidal somatic cells. Follicle populations were expressed as a percentage of the total number of germ cells counted.
Statistical analysis
All experiments were repeated at least three times. All data were analyzed using Student’s t-test. The values were presented as the mean ± SED. Statistical analysis was performed using the t-test or one-way ANOVA. A value of P < 0.05 was considered to indicate statistical significance.
Results
Mir-92b-3p expression is coincident with primordial follicle assembly process in the neonatal mouse ovaries
From an integrated miRNA-seq database YM500v2 [27], we observed a highly expressed miR-92b-3p in human ovaries and brains (Figure 1a). Since miRNA sequences are highly conserved between species, we next validated the online array expression findings in various tissues of adult mice. Results confirmed that miR-92b-3p expression in ovaries and brains was more abundant than in other tissues (Figure 1b). The relatively high expression of miR-92b-3p in adult ovaries promoted us to test whether miR-92b-3p may also express in fetal and neonatal mouse ovaries. Our real-time PCR analysis results showed that miR-92b-3p mRNA expressed at a low level on 16.5 dpc, the stage when the cyst formation is largely ready, then sharply increased and reached the maximum at 0.5dpp, when cyst break down starts and primordial follicles form, and followed by a decrease to a similar level to that of 16.5 dpc on 2.5 dpp, and increased again thereafter (Figure 1c). It was interesting to note that miR-92b-3p expression was much higher in the newborn period, during which primordial follicle assembly occurs, suggesting that miR-92b-3p may play an important role in primordial follicle assembly process.
Figure 1.
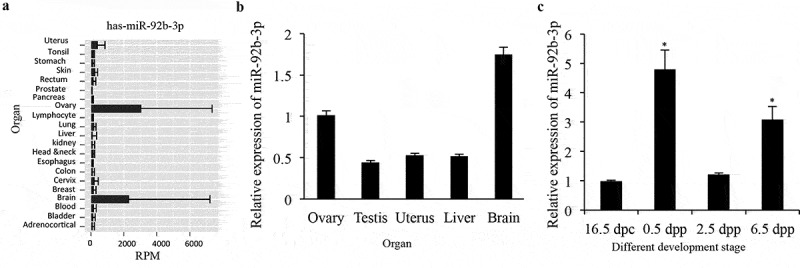
MiR-92b-3p expression in the developing mouse ovary. (a) Expression of miR-92b-3p in human organs from an integrated miRNA-seq database YM500v2. (b) Real-time PCR analysis of miR-92b-3p expression in tissues of adult mice. (c) Real-time PCR analysis of miR-92b-3p expression in developing mouse ovaries. Data were obtained from at least six ovaries and are presented as mean ± SED. MiR-92b-3p was normalized to miR-U6 expression. *p < 0.05 was considered statistically different.
Mir-92b-3p may regulate primordial follicle assembly process in the neonatal mouse ovaries
To assess the effect of miR-92b-3p on the formation of primordial follicles, we overexpressed or silenced miR-92b-3p in 0.5 dpp ovaries through transfection of miR-92b-3p mimics or inhibitor, respectively. It has been demonstrated that there is a high efficiency of microRNA transfection in the newborn mouse ovary culture system [16,17]. To further validate the efficiency of delivery of microRNAs to cultured mouse ovaries, we first transfected 0.5 dpp ovaries with miR-92b-3p labeled with Cy3 (Cy3-miR-92b-3p) and then detected the fluoresce signals under confocal microscopy 96 h post transfection. We observed a homogenous red fluorescence through the whole ovary (Figure 2a). The overexpression or silencing efficiencies in the cultured ovaries were then determined 4 days after transfection by real-time PCR. miR-92b-3p mimics sharply upregulated miR-92b-3p expression, and the inhibitor dramatically downregulated the endogenous miR-92b-3p (Figure 2b and c).
Figure 2.
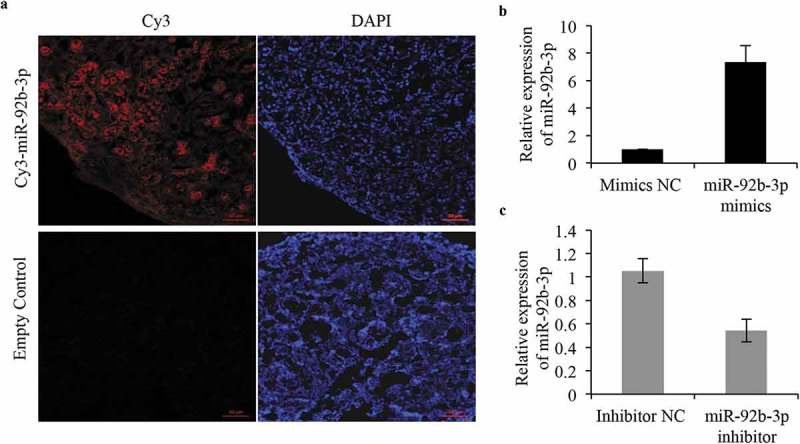
Validation of efficiency of ovarian transfection of microRNA in vitro. (a) Ovaries were collected after incubation with miR-92b-3p labeled with Cy3 and evaluated by confocal microscopy. Strong red fluorescence was observed after 96 h of transfection. DAPI was used for staining nuclei. Bar = 50 μm. (b, c) Real-time PCR analysis of miR-92b-3p expression in ovaries transfected with miR-92b-3p inhibitor, miR-92b-3p mimics, or their respective controls. *p < 0.05.
Then, the cultured ovaries after transfection at 0.5dpp were subjected to histological examination. We counted the oocytes in cultured ovaries every 24 h after miR-92b-3p mimics or inhibitor transfection. The results showed that miR-92b-3p mimics or inhibitor transfection did not affect the total number of oocytes. However, in the miR-92b-3p inhibitor group, ovaries exhibited a significant decreased percentage of germ cells in primordial follicles compared to the controls 96 h after transfection, with a correspondingly significant induction in the percentage of germ cells remaining in cysts (Figure 3a). In contrast, miR-92b-3p mimics-transfected ovaries exhibited more primordial follicles than the mimics controls, with a significant decrease of germ cells in cysts 96 h after transfection (Figure 3b). Increased primordial follicle number in neonatal mouse ovaries has been attributed to the increased number of surviving oocytes during follicle formation [17]. In consistent with the change of oocyte quantification, expression of the genes related to growing oocytes (GDF9, BMP15, ZP3, and Kit) and proliferating granulosa cell (Amhr2) also showed a significant decrease in the miR-92b-3p inhibitor group (Figure 4a), while a dramatic increase in the mimics controls (Figure 4b). These results indicate that miR-92b-3p may promote primordial follicle assembly in neonatal mouse ovaries.
Figure 3.
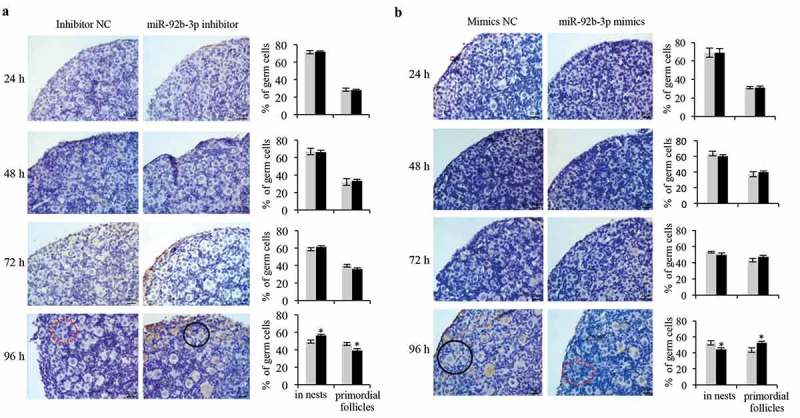
miR-92b-3p promotes primordial follicle assembly in neonatal mouse ovaries. The cultured ovaries after transfection at 0.5 dpp were subjected to immunohistochemistrical analysis with VASA antibody (dark brown) to help visualize the germ cells in the primordial follicles. Follicles were detected and counted in sections of mouse ovaries 1, 2, 3, and 4 days after transfection with (a) Inhibitor NC or miR-92b-3p inhibitor, (b) Mimics NC or miR-92b-3p mimics. Percentage of germ cells in cysts, primordial follicles was shown on the right panel. Data were obtained from at least six ovaries and are presented as mean ± SED. Dashed red circles represent cysts. Black circles represent primordial follicles. *p < 0.05.
Figure 4.
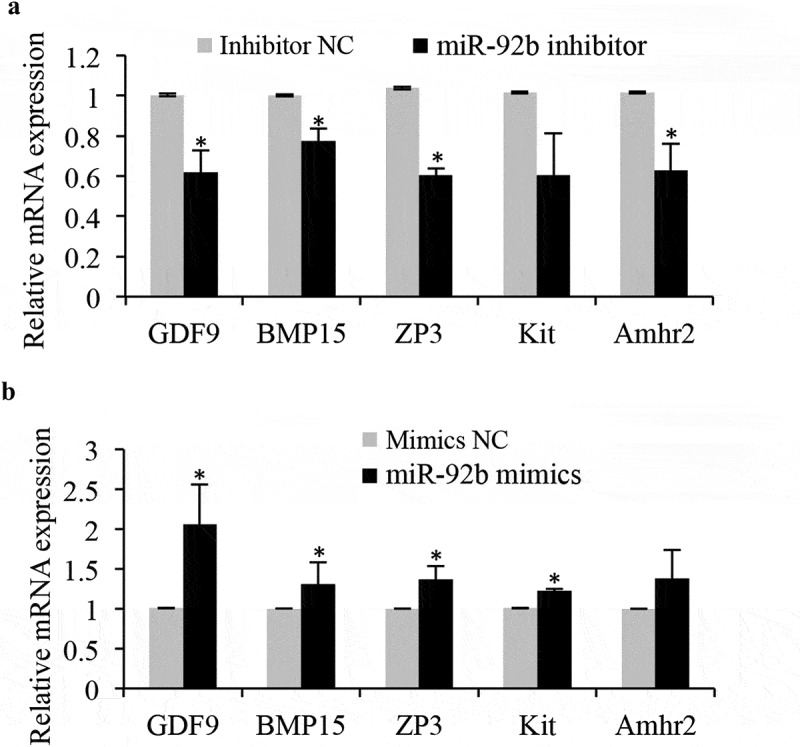
miR-92b-3p activates the expression of the genes related to growing oocytes and granulosa cells. (a) Real-time PCR analysis of genes in control or miR-92b-3p inhibitor-treated ovaries. The levels of mRNA expression in miR-92b-3p inhibitor-treated groups were normalized to the control groups. (b) Real-time PCR analysis of genes in control or miR-92b-3p mimics treated ovaries. The levels of mRNA expression in miR-92b-3p mimics treated groups were normalized to the control groups. *p < 0.05.
TSC1 is a direct target of mir-92b-3p during primordial follicle assembly
In order to find out the potential miR-92b-3p targeted genes, the targeted mRNAs were predicted by both the computational prediction algorithms miRDB (http://www.mirdb.org/) and Targetscan (http://www.targetscan.org/mamm_31/), separately. There were 258 targeted mRNAs overlapped from these two online databases for miRNA target prediction and functional annotations (Figure 5a). These target genes were then submitted to the FunRich database (http://www.funrich.org/) [28] for functional enrichment and interaction network analysis. Among these 258 putative miR-92b-3p targets, Tuberous Sclerosis Complex 1 (TSC1, highlighted in dashed red circle) appeared to be particularly important (Figure 5b). The minimum free energy of hybridization between the predicted target gene and miR-92b-3p by RNAhybrid also supported the possibility that 3ʹUTR of TSC1 contains the potential miR-92b-3p binding seed sequences (Figure 5c). To further confirm the prediction that TSC1 is a direct target of miR-92b-3p, the wild-type (WT) or mutated (Mut) 3ʹUTR sequences of TSC1 were cloned into pGL3 luciferase reporter vector and co-transfected with miR-92b-3p mimics or negative control into 293T cells. The following luciferase reporter assay showed that miR-92b-3p mimics significantly inhibited luciferase activity of WT-TSC1-3ʹUTR but not that of Mut-TSC1-3ʹUTR (Figure 5d), indicating the authentic binding between miR-92b-3p and TSC1-3ʹUTR. Therefore, we determined TSC1 expression during primordial follicle assembly in cultured ovaries 96 h after transfection with miR-92b-3p inhibitor or miR-92b-3p mimics by qRT-PCR and western blot. Results showed that the application of miR-92b-3p inhibitor promoted both the mRNA and protein levels of TSC1 while exogenous expression of miR-92b-3p mimics inhibited TSC1 expression (Figure 5e and f).
Figure 5.
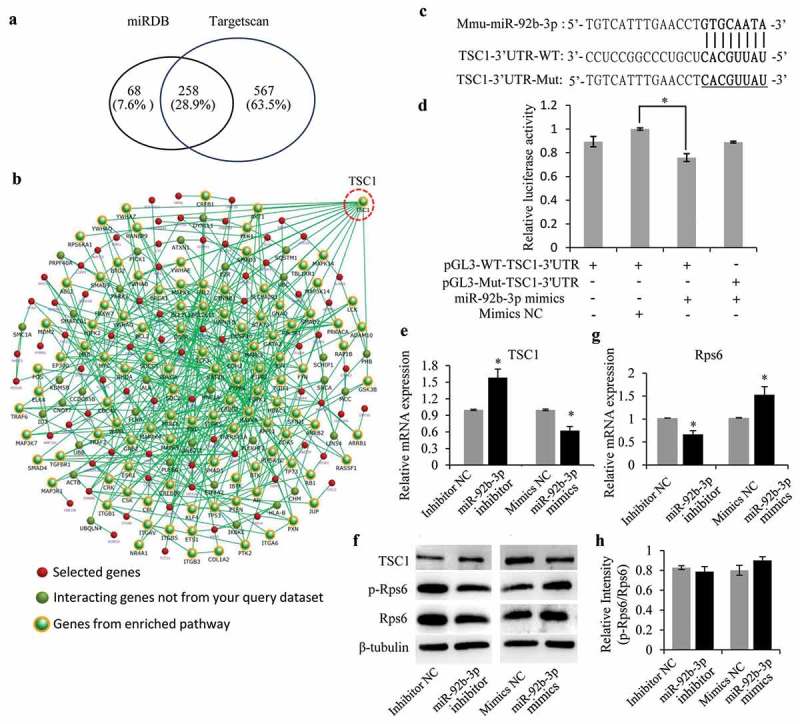
miR-92b-3p targets TSC1 to activate Rps6 during primordial follicle assembly. (a) Vein diagram showing the potential 258 miR-92b-3p targeted mRNAs from two computational prediction algorithms miRDB and Targetscan. (b) FunRich database analysis of the 258 miR-92b-3p targets showing TSC1 to be an important target. (c) The sequence alignment of mouse miR-92b-3p and the 3′-UTR of TSC1. Mutations of the 3′-UTR of TSC1 (underlined CACGUUAU) was used to create the mutant luciferase reporter construct. The WT and Mut-TSC1 3ʹUTR were separately cloned into the modified pGL3 luciferase reporter vector. (d) Luciferase reporter assay showing that the activity of WT-TSC1 3ʹUTR, but not Mut-TSC1 3ʹUTR, was repressed by miR-92b-3p overexpression. (e) TSC1 mRNA expression was detected by Real-time PCR analysis after miR-92b-3p inhibitor or miR-92b-3p mimics transfection. (f) TSC1 protein expression was detected by western blotting after miR-92b-3p inhibitor or miR-92b-3p mimics transfection. P-Rps6 and total Rps6 were also tested. β-tubulin was used as a loading control. (g) Rps6 mRNA expression was detected by Real-time PCR analysis after miR-92b-3p inhibitor or miR-92b-3p mimics transfection. (h) The relative expression levels of p-Rps6 and total Rps6 proteins were analyzed by ImageJ software. *P < 0.05.
Mir-92b-3p activates mTOR/Rps6 signaling during primordial follicle assembly
TSC1 is an important upstream negative regulator of the mTORC1 in mTOR/Rps6 signaling pathway [29,30]. Given that miR-92b-3p downregulated TSC1 protein expression during primordial follicle formation, it is possible that miR-92b-3p may regulate primordial follicle assembly process in the neonatal mouse ovaries by altering TSC1/mTOR/Rps6 signaling. To test this hypothesis, we transfected the 0.5dpp ovaries with miR-92b-3p mimics or inhibitor, followed by immunoblotting analysis. Results showed that the activation of p-Rps6 (235/236) was inhibited after 96 h of treatment with miR-92b-3p inhibitor, while miR-92b-3p mimics promoted phosphorylation of Rps6 (Figure 5f). Of note, this result also correlated with decreased expression of both protein and mRNA of Rps6 by miR-92b-3p inhibitor treatment, or enhanced Rps6 expression under miR-92b-3p mimics (Figure 5f and g). The relatively consistent p-Rps6/Rps6 protein intensity ratio between each treatment and control groups (Figure 5h) further indicated that alteration of Rps6 activation may be caused by changes of Rps6 expression. Together, miR-92b-3p could target and inhibit TSC1 expression, thus further enhancing Rps6 expression and activity during primordial follicle assembly.
TSC1 negatively regulates primordial follicle assembly
Given that miR-92b-3p could downregulate TSC1 protein expression and promote primordial follicle assembly in neonatal mouse ovaries, we wondered whether depletion of TSC1 could also activate primordial follicle assembly, as does miR-92b-3p during primordial follicle formation. To test this hypothesis, we first transfected 0.5dpp ovaries with siTSC1, followed by qRT-PCR and immunoblotting analyses 96 h after transfection. Results showed that both mRNA and protein levels of TSC1 were dramatically inhibited by TSC1 siRNA transfection (Figure 6a and b). Ovary histological examination showed that the percentage of germ cells in primordial follicles was significantly elevated in siTSC1 group than that in the controls, with a correspondingly significant reduction in the percentage of germ cells remaining in cysts (Figure 6c and d). Real-time PCR analysis for the expression of the genes related to growing oocytes showed that siTSC1 also increased these gene expressions (Figure 6e). Together, the results suggested that depletion of TSC1 increased cyst breakdown and promoted primordial follicle formation.
Figure 6.
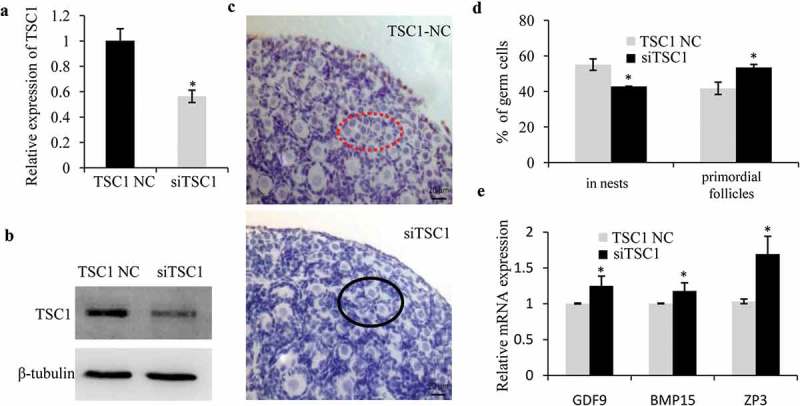
TSC1 negatively regulates primordial follicle assembly in the mouse ovary. Real-time PCR (a) and western blot (b) analyses of TSC1 in 0.5dpp ovaries transfected with TSC1 siRNA or respective controls and cultured for 4 days. (c, d) The 0.5dpp ovaries were transfected with TSC1 NC or TSC1 siRNA and then collected 4 days later to count the germ cells in cyst (dashed red circle) and primordial follicles (black circle). The representative images were shown in (c), and percentage of germ cells in cysts, primordial follicles was shown in (d). (e) Real-time PCR analysis of growth-related genes in TSC1 NC or TSC1 siRNA treated ovaries. The levels of mRNA expression in TSC1 siRNA treated group were normalized to the control groups. * p < 0.05.
Discussion
As a highly conserved serine/threonine kinase, mTORC1, and mTORC2 represent two large physically and functionally distinct signaling complexes, with Raptor (regulatory-associated protein of mTOR) and Rictor (rapamycin-insensitive companion of mTOR) for mTORC1 and mTORC2, respectively [31,32]. mTORC1 is sensitive to rapamycin and promotes cell growth largely through regulation of the activity of its downstream targets S6 kinase 1 (S6K1) and the translational regulators eukaryotic translation initiation factor 4E-BP1 [33], which are then responsible for phosphorylation and activation of Rps6 and eIF4E [34,35]. mTORC1 is known to modulate many cellular processes such as protein synthesis, ribosome biogenesis and autophagy that ultimately determine cell growth and proliferation in response to growth factors and nutrients [33,36,37]. In cells, mTORC1 activity is negatively regulated by a heterodimeric complex of tuberous sclerosis complex 1 (TSC1 or hamartin) and TSC2 (or tuberin) [38]. The suppressive effect of TSC1/TSC2 complex on the activation of mTORC1 is through a GTPase activating protein domain located in TSC2, and the function of TSC1 is to stabilize TSC2 and protect it from ubiquitination and degradation [39,40]. Therefore, inhibition or depletion of TSC1 would release the activity of mTORC1 from TSC1/TSC2 complex, which further activated mTOR/Rps6 signaling. Our study fits well with this hypothesis by showing that miR-92b-3p could activate mTOR/Rps6 signaling through targeting and inhibiting TSC1 expression during primordial follicle assembly.
The roles of the mTOR signaling pathway in follicular development have been extensively studied in recent years. It has been demonstrated that somatic mTORC1 signals are essential to control the activation of primordial follicles and the developmental fates of dormant oocytes [41]. It is also known that inhibiting mTOR activation results in a delayed process of cyst breakdown and the retarded subsequent follicle development [42]. Furthermore, deletion of TSC1 or TSC2 in oocytes could lead to premature activation of the entire pool of primordial follicles and subsequent POF due to the enhanced mTORC1 signaling in oocytes, suggesting that TSC1/mTOR signaling is essential to suppress the activation of primordial follicles [30,39,43]. However, the role of TSC1 during primordial follicle formation remains largely unknown. We found that knockdown of TSC1 in the 0.5dpp ovaries accelerated the breakdown of cysts and assembly of primordial follicles, suggesting that TSC1 may be a key upstream modulator in mTOR signaling to control not only the activation but also the assembly of the primordial follicles in neonatal mouse ovaries. Regarding the molecular mechanisms, we showed that TSC1 is a direct target of miR-92b-3p, and miR-92b-3p could activate mTOR/Rps6 signaling through targeting and inhibiting TSC1 expression. In addition, overexpression of miR-92b-3p showed an identical phenotype with that of siTSC1 in accelerating processes of cyst breakdown and primordial follicle assembly, while inhibition of miR-92b-3p delayed these processes, suggesting that miR-92b-3p negatively regulates TSC1 in mTOR/Rps6 signaling to increase the cyst breakdown and promote primordial follicle formation.
In conclusion, we identified miR-92b-3p as one of the upstream regulators for TSC1 in mTOR/Rps6 signaling pathway, which plays important roles in the process of cyst breakdown and primordial follicle assembly. Our work further extends the knowledge of a regulatory role of miR-92b-3p during early folliculogenesis in mammalian ovaries, and offers new insights into ovarian physiology and pathology.
Funding Statement
This work was supported by National Key Research and Development Program of China [2018YFC1003703, 2018YFC1004203], National Natural Science Foundation of China [81501797 to XQL], the Key Project of Science and Technology Innovation of Nanjing Medical University [2017NJMUCX007], and Jiangsu Six Talent Peaks Project.
Acknowledgments
We thank Dr. Chun Lu from Nanjing Medical University for kindly providing us the modified pGL3 luciferase reporter vector.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplementary data for this article can be accessed here.
References
- [1].Pepling ME. From primordial germ cell to primordial follicle: mammalian femalegerm cell development. Genesis. 2006;44(12):622–632. [DOI] [PubMed] [Google Scholar]
- [2].Pepling ME, Sundman EA, Patterson NL, et al. Differences in oocyte development and estradiol sensitivity among mouse strains. Reproduction. 2010;139(2):349–357. [DOI] [PubMed] [Google Scholar]
- [3].Pepling ME, Spradling AC. Mouse ovarian germ cell cysts undergo programmed breakdown to form primordial follicles. Dev Biol. 2001;234(2):339–351. [DOI] [PubMed] [Google Scholar]
- [4].Bristol-Gould SK, Kreeger PK, Selkirk CG, et al. Postnatal regulation of germ cells by activin: the establishment of the initial follicle pool. Dev Biol. 2006;298(1):132–148. [DOI] [PubMed] [Google Scholar]
- [5].Zhang H, Adhikari D, Zheng W, et al. Combating ovarian aging depends on the use of existing ovarian follicles, not on putative oogonial stem cells. Reproduction. 2013;146(6):R229–233. [DOI] [PubMed] [Google Scholar]
- [6].McGee EA, Hsueh AJ. Initial and cyclic recruitment of ovarian follicles. Endocr Rev. 2000;21(2):200–214. [DOI] [PubMed] [Google Scholar]
- [7].Wang C, Zhou B, Xia G. Mechanisms controlling germline cyst breakdown and primordial follicle formation. Cell Mol Life Sci. 2017;74(14):2547–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Guigon CJ, Magre S. Contribution of germ cells to the differentiation and maturation of the ovary: insights from models of germ cell depletion. Biol Reprod. 2006;74(3):450–458. [DOI] [PubMed] [Google Scholar]
- [9].Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. [DOI] [PubMed] [Google Scholar]
- [10].Gregory RI, Chendrimada TP, Cooch N, et al. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123(4):631–640. [DOI] [PubMed] [Google Scholar]
- [11].Cheng AM, Byrom MW, Shelton J, et al. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33(4):1290–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Watanabe T, Takeda A, Tsukiyama T, et al. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20(13):1732–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ro S, Song R, Park C, et al. Cloning and expression profiling of small RNAs expressed in the mouse ovary. RNA. 2007;13(12):2366–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yao N, Lu CL, Zhao JJ, et al. A network of miRNAs expressed in the ovary are regulated by FSH. Front Biosci(Landmark Ed). 2009;14:3239–3245. [DOI] [PubMed] [Google Scholar]
- [15].Zhang J, Ji X, Zhou D, et al. miR-143 is critical for the formation of primordial follicles in mice. Front Biosci (Landmark Ed). 2013;18:588–597. [DOI] [PubMed] [Google Scholar]
- [16].Wang S, Liu J, Li X, et al. MiR-125b regulates primordial follicle assembly by targeting activin receptor Type 2a in neonatal mouse ovary. Biol Reprod. 2016;94(4):83. [DOI] [PubMed] [Google Scholar]
- [17].Zhang H, Jiang X, Zhang Y, et al. microRNA 376a regulates follicle assembly by targeting Pcna in fetal and neonatal mouse ovaries. Reproduction. 2014;148(1):43–54. [DOI] [PubMed] [Google Scholar]
- [18].Chen Z, Liang S, Zhao Y, et al. miR-92b regulates Mef2 levels through a negative-feedback circuit during Drosophila muscle development. Development. 2012;139(19):3543–3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wu ZB, Cai L, Lin SJ, et al. The miR-92b functions as a potential oncogene by targeting on Smad3 in glioblastomas. Brain Res. 2013;1529:16–25. [DOI] [PubMed] [Google Scholar]
- [20].Wang K, Wang X, Zou J, et al. miR-92b controls glioma proliferation and invasion through regulating Wnt/beta-catenin signaling via Nemo-like kinase. Neuro Oncol. 2013;15(5):578–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lei L, Huang Y, Gong W. Inhibition of miR-92b suppresses nonsmall cell lung cancer cells growth and motility by targeting RECK. Mol Cell Biochem. 2014;387(1–2):171–176. [DOI] [PubMed] [Google Scholar]
- [22].Gong L, Ren M, Lv Z, et al. miR-92b-3p promotes colorectal carcinoma cell proliferation, invasion, and migration by inhibiting FBXW7 In vitro and in vivo. DNA Cell Biol. 2018;37(5):501–511. [DOI] [PubMed] [Google Scholar]
- [23].Long M, Zhan M, Xu S, et al. miR-92b-3p acts as a tumor suppressor by targeting Gabra3 in pancreatic cancer. Mol Cancer. 2017;16(1):167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hao X, Ma C, Chen S, et al. Reverse the down regulation of miR-92b-3p by hypoxia can suppress the proliferation of pulmonary artery smooth muscle cells by targeting USP28. Biochem Biophys Res Commun. 2018;503(4):3064–3077. [DOI] [PubMed] [Google Scholar]
- [25].Hu ZQ, Luo JF, Yu XJ, et al. Targeting myocyte-specific enhancer factor 2D contributes to the suppression of cardiac hypertrophic growth by miR-92b-3p in mice. Oncotarget. 2017;8(54):92079–92089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Xu B, Zhang YW, Tong XH, et al. Characterization of microRNA profile in human cumulus granulosa cells: identification of microRNAs that regulate Notch signaling and are associated with PCOS. Mol Cell Endocrinol. 2015;404:26–36. [DOI] [PubMed] [Google Scholar]
- [27].Cheng WC, Chung IF, Tsai CF, et al. YM500v2: a small RNA sequencing (smRNA-seq) database for human cancer miRNome research. Nucleic Acids Res. 2015;43(Database issue):D862–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Pathan M, Keerthikumar S, Ang CS, et al. FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15(15):2597–2601. [DOI] [PubMed] [Google Scholar]
- [29].Fingar DC, Blenis J. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene. 2004;23(18):3151–3171. [DOI] [PubMed] [Google Scholar]
- [30].Gorre N, Adhikari D, Lindkvist R, et al. mTORC1 Signaling in oocytes is dispensable for the survival of primordial follicles and for female fertility. PLoS One. 2014;9(10):e110491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Jacinto E, Loewith R, Schmidt A, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6(11):1122–1128. [DOI] [PubMed] [Google Scholar]
- [32].Sarbassov DD, Ali SM, Kim DH, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14(14):1296–1302. [DOI] [PubMed] [Google Scholar]
- [33].Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124(3):471–484. [DOI] [PubMed] [Google Scholar]
- [35].Inoki K, Corradetti MN, Guan KL. Dysregulation of the TSC-mTOR pathway in human disease. Nat Genet. 2005;37(1):19–24. [DOI] [PubMed] [Google Scholar]
- [36].Sarbassov DD, Guertin DA, Ali SM, et al. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307(5712):1098–1101. [DOI] [PubMed] [Google Scholar]
- [37].Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12(1):21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Crino PB, Nathanson KL, Henske EP. The tuberous sclerosis complex. N Engl J Med. 2006;355(13):1345–1356. [DOI] [PubMed] [Google Scholar]
- [39].Adhikari D, Zheng W, Shen Y, et al. Tsc/mTORC1 signaling in oocytes governs the quiescence and activation of primordial follicles. Hum Mol Genet. 2010;19(3):397–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chong-Kopera H, Inoki K, Li Y, et al. TSC1 stabilizes TSC2 by inhibiting the interaction between TSC2 and the HERC1 ubiquitin ligase. J Biol Chem. 2006;281(13):8313–8316. [DOI] [PubMed] [Google Scholar]
- [41].Zhang H, Risal S, Gorre N, et al. Somatic cells initiate primordial follicle activation and govern the development of dormant oocytes in mice. Curr Biol. 2014;24(21):2501–2508. [DOI] [PubMed] [Google Scholar]
- [42].Zhang J, Liu W, Sun X, et al. Inhibition of mTOR signaling pathway delays follicle formation in mice. J Cell Physiol. 2017;232(3):585–595. [DOI] [PubMed] [Google Scholar]
- [43].Adhikari D, Flohr G, Gorre N, et al. Disruption of Tsc2 in oocytes leads to overactivation of the entire pool of primordial follicles. Mol Hum Reprod. 2009;15(12):765–770. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


