ABSTRACT
Besides TopBP1, ETAA1 has been identified more recently as an activator of the ATR-ATRIP complex in human cells. We have examined the role of ETAA1 in the Xenopus egg-extract system, which has been instrumental in the study of ATR-ATRIP. Depletion of ETAA1 from egg extracts did not noticeably reduce the activation of ATR-ATRIP in response to replication stress, as monitored by the ATR-dependent phosphorylation of Chk1 and RPA. Moreover, lack of ETAA1 did not appear to affect DNA replication during an unperturbed S-phase. Significantly, we find that TopBP1 is considerably more abundant than ETAA1 in egg extracts. We proceeded to show that ETAA1 could support the activation of ATR-ATRIP in response to replication stress if we increased its concentration in egg extracts by adding extra full-length recombinant ETAA1. Thus, TopBP1 appears to be the predominant activator of ATR-ATRIP in response to replication stress in this system. We have also explored the biochemical mechanism by which ETAA1 activates ATR-ATRIP. We have developed an in vitro system in which full-length recombinant ETAA1 supports activation of ATR-ATRIP in the presence of defined components. We find that binding of ETAA1 to RPA associated with single-stranded DNA (ssDNA) greatly stimulates its ability to activate ATR-ATRIP. Thus, RPA-coated ssDNA serves as a direct positive effector in the ETAA1-mediated activation of ATR-ATRIP.
KEYWORDS: ETAA1, ATR, ATRIP, TopBP1, Chk1, RPA, Xenopus egg extract
Introduction
Eukaryotic cells must carefully assess the fidelity of the various processes that eventually lead to successful cellular duplication. For example, cells must possess the means to allow faithful replication of the genome and accurate transmission of the duplicated copies to their progeny. Toward this end, cells employ various types of checkpoint-regulatory pathways [1,2]. For example, the kinase ATR and its regulatory partner ATRIP function at the apex of pathways that monitor the fidelity of DNA synthesis during S-phase. ATR-ATRIP also regulates responses to damaged DNA as well as other processes.
The functioning of ATR-ATRIP in checkpoint pathways is subject to stringent regulation. For example, ATR-ATRIP first localizes to potentially problematic regions in the genome by docking with RPA-coated single-stranded DNA (ssDNA), which accumulates at stalled replication forks and other structures [3,4]. However, ATR-ATRIP exhibits minimal kinase activity in the presence of only RPA-ssDNA [5–7]. Hence, other proteins must come into play to activate ATR-ATRIP so that it can phosphorylate downstream target proteins. In a well characterized pathway, binding of TopBP1 to ATR-ATRIP shifts the kinase into its activated conformation [8–10]. TopBP1 achieves this effect by utilizing an ATR-activating domain (AAD), which interacts with both the ATR and ATRIP subunits [8,11].
Other significant aspects of this process are that the association of TopBP1 with checkpoint-inducing structures on chromatin and its subsequent interaction with ATR-ATRIP are also under strict control. For example, TopBP1 docks with the Rad9-Hus1-Rad1 (9-1-1) checkpoint clamp after deposition of this complex onto recessed DNA ends at stalled replication forks by the Rad17-RFC checkpoint clamp loader [12,13]. In addition, the Mre11-Rad50-Nbs1 (MRN) complex regulates the activation of ATR-ATRIP in response to replication stress, at least in part by facilitating the recruitment of TopBP1 to chromatin [14,15].
The role of TopBP1 in the activation of ATR-ATRIP is also conserved in budding yeast. In this system, Dpb11, the yeast homologue of TopBP1, directly activates Mec1-Ddc2, the yeast version of ATR-ATRIP [16]. Significantly, however, additional proteins can also serve as activators of Mec1-Ddc2 in yeast. For example, the C-terminal tail of Ddc1 (the yeast homologue of the Rad9 subunit of the vertebrate 9-1-1 complex) also possesses an AAD [17]. Moreover, the Dna2 protein contains a functional AAD [18]. The diversity of AAD-containing proteins in yeast enables regulation of Mec1-Ddc2 in response to different needs throughout the cell cycle. Such observations raised the question of whether additional activators of ATR might exist in higher eukaryotes.
More recently, several groups identified a novel activator of ATR-ATRIP in human cells called ETAA1 [19–22]. It has been shown that ETAA1 possesses a functional AAD and interacts with RPA through multiple binding motifs. Moreover, ETAA1 is important for the maintenance of genomic stability following various perturbations. However, the exact relationship between ETAA1 and TopBP1 as well as the regulation of ETAA1 are both topics that need further study.
In this report, we have characterized a homologue of ETAA1 in the Xenopus egg-extract system in order to assess its role relative to TopBP1. We have also developed an in vitro system with defined components to reveal that RPA-coated ssDNA plays an important role in the activation of ATR-ATRIP by ETAA1.
Materials and methods
Xenopus egg extracts
Xenopus interphase egg extracts were prepared as described previously [23]. Cycloheximide (50 µg/ml) was added to prevent extracts from entering mitosis. For induction of stalled DNA replication forks, demembranated sperm nuclei (3000/μl) were incubated in extracts with 150 μM (50 μg/ml) aphidicolin, unless indicated otherwise. Chromosomal DNA replication assays were carried out as described previously [23].
Isolation of nuclear and chromatin fractions
For isolation of nuclear fractions, egg extracts were overlaid on a 1 M sucrose cushion (1M sucrose, 80 mM KCl, 2.5 mM K-gluconate, 10 mM Mg-gluconate, and 20 mM HEPES-KOH, pH 7.5) and centrifuged at 6,100 g for 5 min. Nuclear pellets were washed once with 1M sucrose cushion. Nuclear fractions were dissolved in SDS sample buffer for gel loading.
For isolation of chromatin fractions, extracts were mixed with egg lysis buffer (ELB; 10 mM HEPES-KOH, pH 7.7, 250 mM sucrose, 50 mM KCl, and 2.5 mM MgCl2) containing 0.2% Triton X-100. The suspended extracts were layered onto a 0.5 M sucrose cushion (ELB containing an additional 0.25 M sucrose and 0.2% Triton X-100) and centrifuged at 11,700 g for 1 min. The pellet was washed once with ELB containing 0.2% Triton X-100. Chromatin fractions were dissolved in SDS gel sample buffer.
Antibodies
The coding sequence for Xenopus ETAA1 (GenBank: BC076732.1) was amplified from XGC ETAA1 cDNA (Dharmacon) with Q5 High-Fidelity DNA Polymerase (New England Biolabs). To produce a His10-tagged polypeptide containing residues 374–820 of Xenopus ETAA1, we cloned the appropriate DNA sequence into pH10UE [24] and expressed the protein in Escherichia coli Rosetta (DE3)pLysS cells. Anti-Xenopus ETAA1 antibodies were produced in rabbits in a commercial facility (Pocono Rabbit Farm and Laboratory) and affinity purified with the antigen. Affinity-purified rabbit polyclonal antibodies against Xenopus Chk1, ATR, ATRIP, Orc2, RPA70, BLM, Mre11, Cdc45, and TopBP1 were described previously [8,15,25–28]. We purchased the following anti-phosphopeptide antibodies from commercial sources: pSer-345 of human Chk1, equivalent to pSer-344 of Xenopus Chk1 (Cell Signaling Technology); and pSer-33 of human RPA32, equivalent to pThr-36 of Xenopus RPA32 (Bethyl Laboratories). An anti-human Mcm2 mouse monoclonal antibody (BM28) that cross-reacts with Xenopus Mcm2 was obtained from BD Transduction Laboratories. Antiserum against Xenopus RPA32 was kindly provided by Dr. Vincenzo Costanzo (FIRC Institute of Molecular Oncology).
Immunodepletion
For immunodepletion of ETAA1, egg extracts (100 μl) were incubated at 4°C for 1 h with 7.5 μg anti-ETAA1 antibodies bound to protein A Dynabeads (Invitrogen) for two rounds of treatment. Mock depletions were performed with rabbit IgG fraction (Invitrogen). Immunodepletion of TopBP1 was described previously [8].
Preparation of recombinant proteins
A recombinant baculovirus encoding Xenopus ETAA1 with a His6 tag at the N-terminal end and tandem 3X-FLAG and 2X-Strep tags at the C-terminal end (HFS-ETAA1) was generated with the Bac-to-Bac system (Invitrogen). To prepare recombinant HFS-ETAA1 protein, Sf9 insect cells were infected with the baculovirus and incubated for 2 days. Cell pellets were lysed in 2 ml lysis buffer (10 mM HEPES-KOH, pH 7.5, 300 mM NaCl, 1% NP-40, and 1.5 mM EGTA) containing 1 mM PMSF. Cell lysates were clarified by centrifugation at 16,000 g for 10 min. The supernatants were then diluted to 150 mM NaCl. For purification by means of the His6 tag, diluted supernatants were incubated with nickel agarose beads for 1 h. After the beads were washed, proteins were eluted with 100 μl His6 elution buffer (10 mM HEPES-KOH, pH 7.5, 150 mM NaCl, and 300 mM imidazole). For purification with the FLAG tag, diluted supernatants were incubated with anti-FLAG M2 agarose beads (Sigma) for 2 h. After washing of the beads, proteins were eluted with 100 μl FLAG elution buffer (10 mM HEPES-KOH, pH 7.5, 150 mM NaCl, 0.5 mg/ml 3X-FLAG peptide, 1 mM DTT, 1 mM EDTA, and 10 μg/ml each of pepstatin, chymostatin, and leupeptin). Aliquots of purified proteins were frozen in liquid nitrogen and stored at − 80°C. Procedures for isolation of human p27 and human RPA were previously described [25,29]. The FLAG-tagged polypeptide containing BRCT domains I-III of Xenopus TopBP1 was prepared by published methods [8].
Isolation of ATR-ATRIP complexes from egg extracts
Preparation of ATR-ATRIP complexes in egg extracts with recombinant, FLAG-tagged full-length ATRIP or ΔN222-ATRIP and endogenous ATR was carried out as described previously [8].
Kinase assays
Kinase assays with ATR-ATRIP complexes were performed in the presence of γ-[32P]ATP under the same conditions as before [8]. HFS-ETAA1 and RPA were added at final concentrations of 141 nM and 991 nM, respectively, as indicated. In some cases, a single-stranded DNA containing 138 nucleotides (5'-GAT ATC AGG GCC CTG GAA CAG CAC CTC CAG GGC GCC TTT TTC GAA CTG AGG GTG GCT CCA GGC GCT GCC TCC GGA TCC GCC TCC GGA GCC TCC GCC TTT CTC GAA CTG AGG GTG GCT CCA GCT AGC CAT GGT GGG TCT-3') was included at a final concentration of 188 nM.
Quantitation
For quantitation, immunoblots were analyzed with the Odyssey Imaging System (LI-COR Biosciences). Scanned images were quantified with Fiji (ImageJ). Kinase assay gels were scanned with a Typhoon FLA 9500 phosphorimager and the images were quantified with Fiji (ImageJ).
Results
Characterization of a Xenopus homologue of ETAA1
In order to study ETAA1 in the Xenopus egg-extract system, we produced rabbit polyclonal antibodies against residues 374–820 of a Xenopus homologue of ETAA1. We found that these antibodies recognize a polypeptide of approximately 134 kD in Xenopus egg extracts (Figure 1(a)). In parallel, we also prepared a recombinant form of Xenopus ETAA1 with His6, FLAG, and Twin-Strep tags in baculovirus-infected insect cells. We designated this construct as the HFS-ETAA1 protein. The electrophoretic mobility of the HFS-ETAA1 protein was consistent with the addition of the tags onto the ETAA1 protein (Figure 1(a)).
Figure 1.

Characterization of ETAA1 in Xenopus egg extracts. (a) Interphase egg extracts were immunoblotted with antibodies against Xenopus ETAA1 (lane 1). Recombinant, baculovirus-expressed HFS-ETAA1 protein (lane 2) was electrophoresed on the same gel and immunoblotted with anti-ETAA1 antibodies. (b) Concentration of ETAA1 in egg extracts. The indicated amounts (ng) of antigen used for the production of anti-ETAA1 antibodies (His10-tagged version of residues 374–820) were mixed with egg extracts. Mixtures were subjected to gel electrophoresis and immunoblotted with anti-ETAA1 antibodies. Amounts of egg extract correspond to: 1 μl (lanes 1–4); 0.5 μl (lane 5); and 0.2 μl (lane 6). (c) Interphase egg extracts were incubated without (lane 1) or with sperm chromatin (lanes 2–9) in the absence (lanes 1–5) or presence of APH (lanes 6–9). Chromatin fractions were isolated from the extracts at the indicated times and immunoblotted for ETAA1, RPA70, and Orc2 (loading control). Lane 10 depicts the initial egg extract.
By quantitative immunoblotting, we noted that ETAA1 is present at a concentration of approximately 7.5 nM in egg extracts (Figure 1(b)). This value agrees quite well with the concentration of 8 nM that was obtained in a global proteomic analysis of egg extracts [30]. By comparison, it has been estimated that TopBP1 is present at a concentration of 30 nM in egg extracts [31], which likewise is consistent with the value of 37 nM from the proteomic analysis. Hence, ETAA1 appears to be present at approximately a four- to five-fold lower concentration than TopBP1 in egg extracts.
Xenopus ETAA1 associates with chromatin in a checkpoint-dependent manner
Many regulators of ATR accumulate on chromatin in egg extracts upon replication stress [25,32–34]. By mass spectrometry analysis, Haahr et al. [21] observed enrichment of ETAA1 on chromatin following treatment with the DNA polymerase inhibitor aphidicolin (APH). We utilized our anti-Xenopus ETAA1 antibodies to characterize this process in further detail. For this purpose, we incubated demembranated Xenopus sperm chromatin in egg extracts to form reconstituted nuclei. To elicit the formation of stalled DNA replication forks, we utilized APH [35]. We incubated egg extracts in the absence and presence of APH, isolated reconstituted nuclei at different times, prepared chromatin fractions, and immunoblotted for ETAA1 (Figure 1(c)). We found that only small amounts of ETAA1 bound to chromatin in the absence of APH. By contrast, there was a large increase in the amount of ETAA1 on chromatin from APH-treated extracts (Figure 1(c), compare lanes 2–5 and 6–9). This increase mirrored a similar elevation in the binding of RPA, which we followed by immunoblotting of RPA70, the largest subunit of the RPA complex. Moreover, we noted that a large proportion of the chromatin-bound ETAA1 showed a substantially reduced electrophoretic mobility indicative of a post-translational modification(s). We also observed a lower-molecular weight band of ETAA1, which may represent a breakdown product. Under these conditions, RPA70 and Orc2 exhibited no signs of degradation. Overall, these results indicated that a highly modified form of ETAA1 accumulates on chromatin undergoing replication stress.
Depletion of Xenopus ETAA1 does not appreciably affect the checkpoint-dependent phosphorylation of Chk1 and RPA
To evaluate whether ETAA1 has a role in checkpoint regulation in egg extracts, we set out to remove this protein from these extracts by immunodepletion. We found that our anti-ETAA1 antibodies were very effective for this purpose (Figure 2(a)). After two rounds of treatment with protein A beads that had been coated with anti-ETAA1 antibodies, we could not detect any ETAA1 in egg extracts by immunoblotting. Depletion of ETAA1 did not noticeably affect the levels of TopBP1. Next, we incubated ETAA1-depleted extracts as well as mock-depleted extracts in the absence and presence of APH. Subsequently, we isolated nuclear fractions from the extracts and performed immunoblotting to detect phosphorylation of Chk1 on Ser-344, an indicator for activation of ATR in egg extracts [26,36]. As expected, Chk1 underwent robust phosphorylation in the presence of APH (Figure 2(b)). However, we could not observe any apparent reduction in the APH-dependent phosphorylation of Chk1 in the absence of ETAA1.
Figure 2.
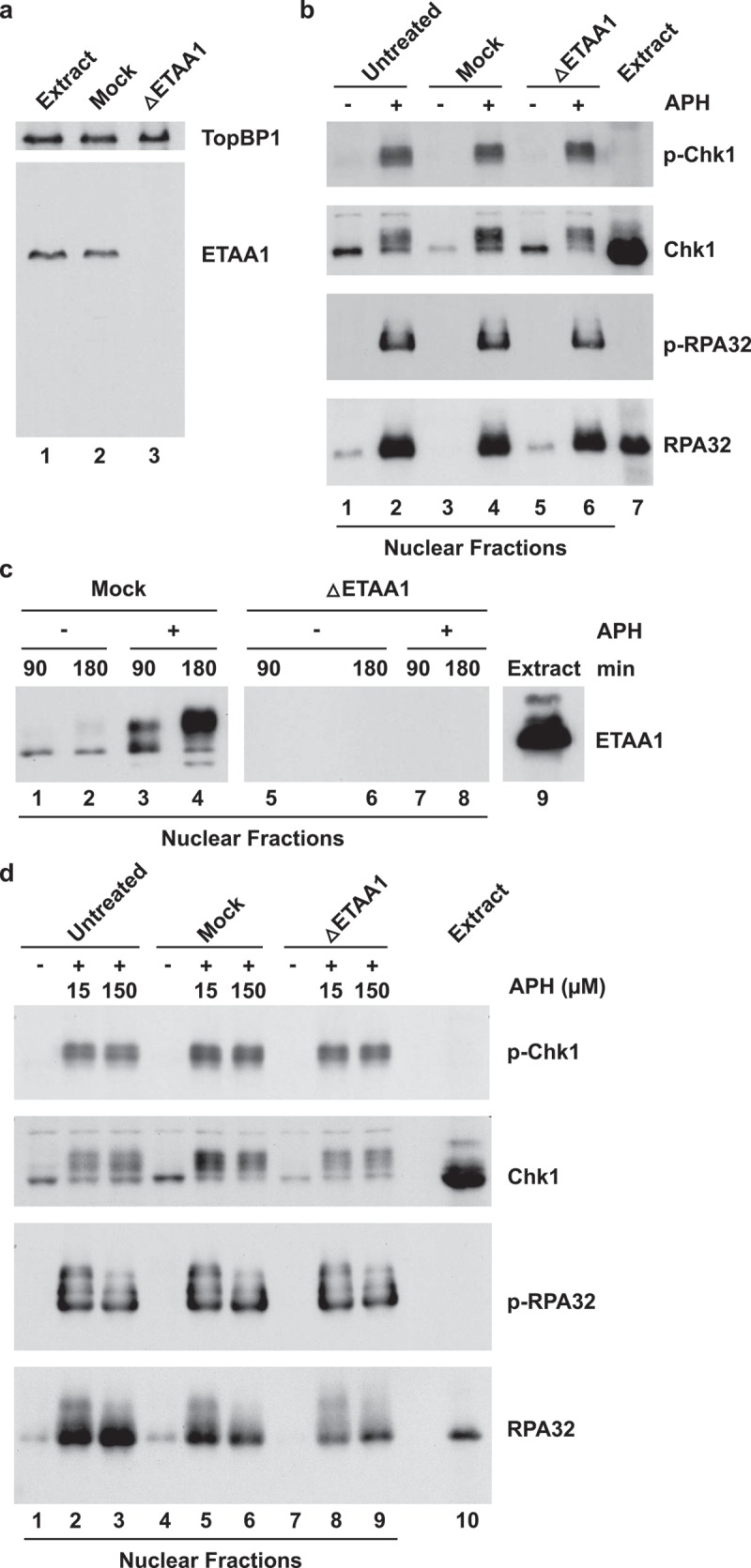
Effect of depletion of ETAA1 on APH-induced phosphorylation of Chk1 and RPA in egg extracts. (a) Egg extracts (lane 1) were mock depleted with control antibodies (lane 2) or immunodepleted with anti-Xenopus ETAA1 antibodies (lane 3) and immunoblotted for ETAA1 and TopBP1, as indicated. (b) Untreated (lanes 1–2), mock-depleted (lanes 3–4), and ETAA1-depleted egg extracts (lanes 5–6) were incubated for 90 min in the absence (lanes 1, 3, and 5) or presence (lanes 2, 4, and 6) of 150 μM APH. Nuclear fractions from the extracts were prepared and immunoblotted with anti-phospho-Chk1 (p-Chk1), anti-Chk1, anti-phospho-RPA32 (p-RPA32), and anti-RPA32 antibodies. Lane 7 depicts the initial egg extract. (c) Mock-depleted (lanes 1–4) and ETAA1-depleted egg extracts (lanes 5–8) were incubated for 90 min or 180 min in the absence (lanes 1, 2, 5, and 6) or presence (lanes 3, 4, 7, and 8) of 150 μM APH. Nuclear fractions were prepared and immunoblotted with anti-ETAA1 antibodies. Note that the space between lanes 5 and 6 is a blank lane. Lane 9 depicts the initial egg extract. (d) Untreated (lanes 1–3), mock-depleted (lanes 4–6), and ETAA1-depleted egg extracts (lanes 7–9) were incubated for 90 min in the absence of APH (lanes 1, 4, and 7) or the presence of either 15 μM (lanes 2, 5, and 8) or 150 μM APH (lanes 3, 6, and 9). Nuclear fractions from the extracts were prepared and immunoblotted with the indicated antibodies. Lane 10 depicts the initial egg extract.
We also probed for phosphorylation of RPA32, the middle subunit of the RPA complex, on Thr-36, the equivalent of Ser-33 in human RPA32 [14] and likewise detected no difference in the absence versus presence of ETAA1. As expected, ETAA1 in nuclear fractions from mock-depleted extracts had become highly modified in response to APH, and there was no ETAA1 detectable in nuclear fractions from ETAA1-depleted extracts (Figure 2(c)). For these experiments, we utilized a relatively high concentration of APH (150 μM), which results in stalled replication forks with relatively short nascent DNA [37,38]. However, we obtained the same results with a lower dose of APH (15 μM), which results in longer stretches of nascent DNA (Figure 2(d)). Similarly, we observed that phosphorylation of Chk1 occurred normally in ETAA1-depleted extracts that we had treated with the restriction endonuclease PflMI to induce double-stranded DNA breaks (not shown). Taken together, these results indicate that ETAA1 plays little, if any role, in the activation of ATR in response to APH-triggered stalled replication forks in the Xenopus egg-extract system. These observations are consistent with the fact that depletion of TopBP1, the first identified activator of ATR, eliminates phosphorylation of Chk1 under essentially the same conditions that we have used in this study [8].
Role of ETAA1 in DNA replication
In the human system, it has been reported that there is increased origin firing in ETAA1-deficient cells [19,21]. A key step in origin firing involves the loading of Cdc45 onto chromatin, which results in activation of the replicative helicase [39–41]. Inhibition of ATR in APH-treated egg extracts results in dysregulated origin firing, which is reflected in elevated loading of Cdc45 onto chromatin [37,38,42,43]. Accordingly, we removed ETAA1 from egg extracts, incubated the extracts in the presence of APH, and finally isolated chromatin to examine the loading of Cdc45 (Figure 3(a)). By immunoblotting, we observed similar amounts of Cdc45 on chromatin in APH-treated extracts in the absence and presence of ETAA1. Under such conditions, addition of caffeine (an inhibitor of ATR and ATM) to APH-treated extracts results in a dramatic increase in the recruitment of Cdc45 to chromatin [37,38,42,43]. Therefore, it appears that ETAA1 does not play a determinative role in the suppression of origin firing in response to APH.
Figure 3.
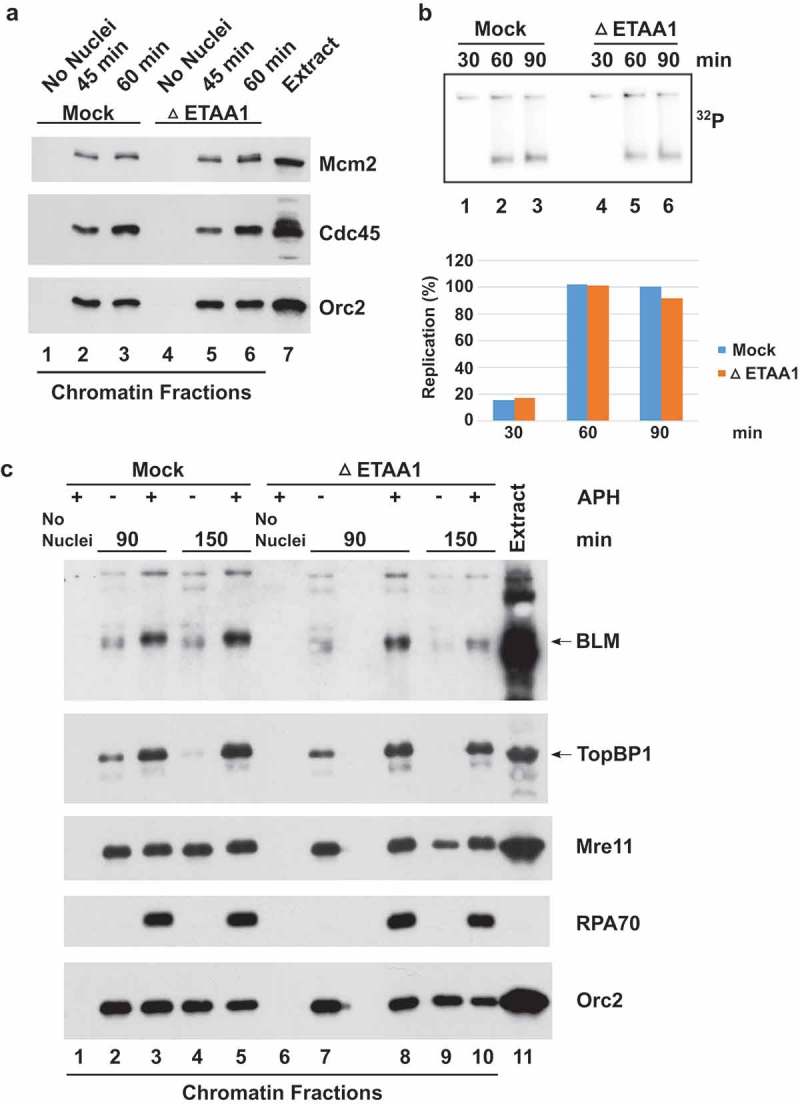
Role of ETAA1 in DNA replication and related processes. (a) Mock-depleted (lanes 1–3) and ETAA1-depleted egg extracts (lanes 4–6) containing APH were incubated in the absence of sperm chromatin for 45 min (lanes 1 and 4) or the presence of sperm chromatin (lanes 2–3 and 5–6) for the indicated times. Incubations were processed for the preparation of chromatin fractions and the resulting samples were immunoblotted for Mcm2, Cdc45, and Orc2. Lane 7 depicts an aliquot of the initial egg extract. (b) Mock-depleted and ETAA1-depleted egg extracts were assayed for chromosomal DNA replication with 32P-labeled dATP at the indicated times as indicated. An agarose gel containing radiolabeled replication products (top) and corresponding quantitation (bottom) are depicted. Values are normalized to the amount of DNA replication at 90 min in mock-depleted extracts. Representative of three experiments. (c) Mock-depleted (lanes 1–5) and ETAA1-depleted egg extracts (lanes 6–10) were incubated in the absence (lanes 1 and 6) or presence of sperm chromatin (lanes 2–5 and 7–10). In addition, extracts either lacked (lanes 2, 4, 7, and 9) or contained APH (lanes 1, 3, 5, 6, 8, and 10). After incubation for the indicated times, extracts were processed for the preparation of chromatin fractions and the resulting samples were immunoblotted for BLM, TopBP1, Mre11, RPA70, and Orc2. Lane 11 depicts an aliquot of the initial egg extract. Note that the space between lanes 7 and 8 is a blank lane.
In order to evaluate whether ETAA1 has any role in DNA replication during an unperturbed cell cycle, we examined incorporation of 32P-lableled dAMP (from a radiolabeled deoxynucleoside triphosphate precursor) into chromosomal DNA as a function of time. We observed very similar time-courses for DNA replication in the presence and absence of ETAA1 (Figure 3(b)). Therefore, absence of ETAA1 does not appear to affect the overall progression of DNA replication in an obvious manner.
Bass et al. [19] also observed that ETAA1 associates with other DNA damage-response proteins, including the BLM/Top3α/RMI1/RMI2 complex, that are involved in the repair of stalled replication forks. We examined whether depletion of ETAA1 might affect the recruitment of BLM to chromatin. In parallel, we also examined TopBP1 and Mre11, a component of the Mre11-Rad50-Nbs1 (MRN) complex, and RPA. We observed that lack of ETAA1 did not typically impair the binding of BLM, TopBP1, Mre11, or RPA to chromatin (Figure 3(c)). Taken together, these results suggest that absence of ETAA1 does not dramatically affect replication and some replication-related processes in egg extracts.
ETAA1 can substitute for TopBP1 when present at a sufficiently high concentration
As described above, ETAA1 is present at significantly lower concentrations (four- to five-fold) than TopBP1 in egg extracts. Accordingly, we wished to examine whether ETAA1 could substitute for TopBP1 to support the APH-triggered activation of ATR if present at a higher concentration. To address this issue clearly, we decided to remove TopBP1 from egg extracts by immunodepletion and then add extra recombinant ETAA1 to the extracts (Figure 4). A consideration for this approach is that TopBP1 is necessary for chromosomal DNA replication [31,44]. Thus, addition of APH to TopBP1-depleted extracts does not result in the formation of stalled replication forks, which are a prerequisite for activation of ATR-ATRIP and the ensuing phosphorylation of Chk1.
Figure 4.

ETAA1 can substitute for TopBP1 when present at a sufficiently high concentration. (a) Egg extracts (lane 1) were mock-depleted with control antibodies (lane 2) or immunodepleted with anti-TopBP1 antibodies (lane 3) and immunoblotted with anti-TopBP1 (top) or anti-ETAA1 antibodies (bottom). (b) Mock-depleted (lanes 1 and 2) and TopBP1-depleted extracts (lanes 3–5) were incubated in the absence (lane 1) or presence of APH (lane 3–5). In addition, some extracts were supplemented with recombinant BRCT I-III domain of TopBP1 (lanes 4 and 5) and recombinant HFS-ETAA1 protein (lane 5). Nuclear fractions from the extracts were immunoblotted with the indicated antibodies. (c) Mock-depleted extracts (lanes 1–4) were incubated in the absence (lanes 1 and 3) or presence of APH (lanes 2 and 4). Some extracts were supplemented with recombinant HFS-ETAA1 protein (lanes 3 and 4). Nuclear fractions from the extracts were immunoblotted with the indicated antibodies.
Certain truncation mutants of TopBP1 are able to support initiation of replication but cannot promote activation of ATR [9,10,23]. For example, a truncated form of TopBP1 containing BRCT domains I-III can support normal levels of DNA replication in egg extracts [23]. This polypeptide lacks the AAD, which resides between BRCT domains VI and VII, and thus cannot activate ATR. Accordingly, we depleted egg extracts of their endogenous TopBP1 and then added a recombinant TopBP1 BRCT I-III fragment back to the extracts in order to support initiation of DNA replication (Figure 4(a and b)). Parenthetically, we consistently noticed that there was a partial reduction in the amount of ETAA1 in TopBP1-depleted extracts. We could detect some ETAA1 in anti-TopBP1 immunoprecipitates. However, this binding was sensitive to washing with 0.1% NP-40 (not shown). Therefore, TopBP1 and ETAA1 may form a weak complex in egg extracts.
Next, we proceeded to add recombinant ETAA1 to the extracts at a concentration of approximately 50 nM, which is slightly higher than the concentration of endogenous TopBP1 (30–37 nM). Finally, we incubated the extracts in the absence and presence of APH and then monitored the phosphorylation of Chk1 and RPA. As expected, there was no phosphorylation of Chk1 or RPA in APH-treated, TopBP1-depleted extracts (Figure 4(b)). Addition of the TopBP1 BRCT I-III fragment did not restore phosphorylation of these proteins. On the other hand, further addition of recombinant HFS-ETAA1 to such extracts resulted in efficient phosphorylation of Chk1 and RPA in the presence of APH (Figure 4(b)). In control experiments, we showed that added recombinant ETAA1 could not induce phosphorylation of Chk1 in mock-depleted extracts (Figure 4(c)). Taken together, these results suggest that ETAA1 can substitute for TopBP1 in egg extracts if present at a sufficiently high concentration.
RPA-ssDNA promotes the ETAA1-dependent activation of ATR-ATRIP in a defined biochemical system
An important question involves the issue of what cellular factor(s) might regulate the ability of ETAA1 to activate ATR. For example, interaction of TopBP1 with the 9-1-1 and MRN complexes regulates its ability to trigger the activation of ATR [12–15]. To address this question, we attempted to set up a defined in vitro system in which the activation of ATR-ATRIP would be dependent upon addition of recombinant full-length ETAA1 protein (Figure 5). For this purpose, we utilized the recombinant HFS-ETAA1 protein. As the source of ATR-ATRIP, we prepared a complex of ATR and FLAG-tagged ATRIP by using Xenopus egg extracts, as described previously (Figure 5(a)) [8]. For most experiments, we utilized full-length ATRIP. Initially, we mixed HFS-ETAA1 and ATR-ATRIP in the presence of model ATR substrates, e.g., PHAS-I and a GST-tagged fusion protein containing residues 62–122 of Xenopus Mcm2 [8]. However, we could not observe significant phosphorylation of these substrates above background levels with full-length HFS-ETAA1 (not shown).
Figure 5.
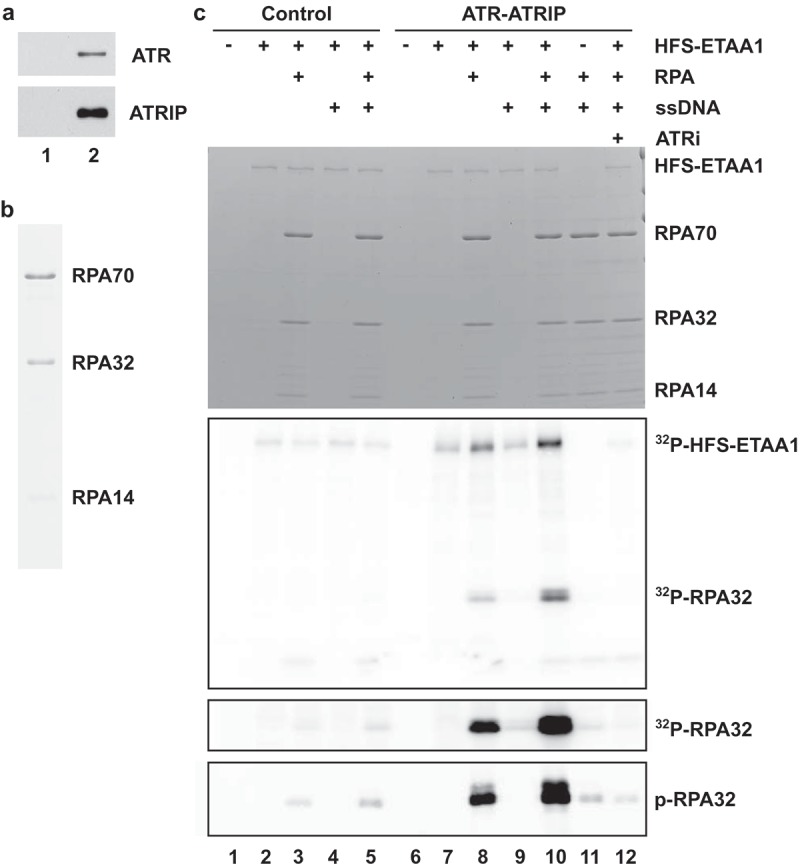
Full-length ETAA1 activates ATR-ATRIP in a defined biochemical system. (a) Isolation of ATR-ATRIP complexes. Control buffer (lane 1) and full-length ATRIP-FLAG (lane 2) were added to egg extracts containing anti-FLAG M2 antibody beads. After incubation, the beads were reisolated, washed, and incubated with 3X-FLAG peptide. The eluates were immunoblotted with anti-ATR (top) and anti-FLAG antibodies (bottom). (b) Purification of RPA. Recombinant human RPA was purified as described in Materials and Methods and stained with Coomassie blue. (c) Control eluates (lanes 1–5) and eluates containing ATR-ATRIP complex (lanes 6–12) from panel A were incubated in kinase buffer containing γ-[32P]ATP in the absence (lanes 1, 6, and 11) or presence HFS-ETAA1 protein (lanes 2–5, 7–10, and 12). Some incubations also contained RPA (lanes 3, 5, 8, and 10–12), ssDNA (lanes 4–5, and 9–12), and 50 μM ATRi (lane 12). Reactions were subjected to SDS gel electrophoresis. The gel was stained with Coomassie blue (top panel). Incorporation of 32P into proteins was detected by phosphorimaging (second panel from top). For the third panel from the top, we have depicted a longer exposure of the section of the gel containing 32P-labeled RPA32. For the bottom panel, we electrophoresed aliquots of the same samples in a different gel and performed immunoblotting with anti-phospho-RPA32 antibodies.
Previous studies have indicated that human ETAA1 exhibits highly specific binding to RPA [19–22]. Hence, we asked whether RPA could promote the ability of ETAA1 to activate ATR-ATRIP. For this purpose, we utilized recombinant human RPA that had been purified from bacteria (Figure 5(b)) [29]. Since RPA functions through binding to ssDNA, we also included an ssDNA template in some incubations. For these experiments, we utilized an oligonucleotide comprised of a 138-mer of ssDNA. A DNA molecule of this length would be able to accommodate multiple RPA molecules, approximately four [45,46].
We proceeded to incubate HFS-ETAA1 and ATR-ATRIP in the presence of RPA or ssDNA or both (Figure 5(c)). Although we could observe some phosphorylation of the middle subunit of RPA (RPA32) in the absence of DNA, the addition of ssDNA greatly stimulated the phosphorylation of RPA32 (Figure 5(c), compare lanes 8 and 10). In particular, inclusion of ssDNA resulted in a 4.9 ± 0.6-fold increase (mean ± SEM, n = 4 experiments) in the phosphorylation of RPA32. This reaction appears to be specific for RPA32 because we did not observe any phosphorylation of RPA70 or RPA14 in these experiments. As a control, we performed the reactions in the presence of ssDNA but in the absence of RPA in order to confirm that the phosphorylated band corresponds to RPA32 (Figure 5(c), lane 9). By using anti-phosphopeptide antibodies, we established that at least one phosphorylation site in RPA32 corresponds to Ser-33. This residue is a well-documented target of ATR [47,48].
There was no phosphorylation of RPA32 in the presence of only ATR-ATRIP and RPA-ssDNA (Figure 5(c), lane 11). Furthermore, no phosphorylation of RPA32 occurred in the presence of control preparations that lacked ATR-ATRIP (Figure 5(c), lanes 1–5). We also noted that in the complete reactions the ETAA1 protein itself underwent phosphorylation (Figure 5(c), lane 10). As was the case for phosphorylation of RPA, phosphorylation of ETAA1 was highest in the presence of both RPA and ssDNA. Therefore, ETAA1 itself also appears to be a substrate of ATR-ATRIP.
In order to establish that the phosphorylation of RPA is indeed due to ATR-ATRIP, we made use of a specific ATR kinase inhibitor called ETP-46464 (ATRi) [49]. We observed that addition of ATRi completely inhibited the phosphorylation of RPA in the cell-free reaction (Figure 5(c), lane 12). This inhibitor also blocked the phosphorylation of ETAA1.
ETAA1 carries out the activation of ATR by utilizing an ATR-activating domain (AAD), which was initially found in TopBP1 [8]. This domain is well conserved in ETAA1 [19,21,22]. All known AADs contain a critical tryptophan residue. Mutation of this critical tryptophan (W) to arginine (R) in both human and Xenopus TopBP1 abolishes the ATR-activating function [8]. This residue corresponds to W107 of human ETAA1 [19,21,22]. In Xenopus ETAA1, this position is equivalent to W63 (Figure 6(a)). We produced a W63R mutant of Xenopus HFS-ETAA1 in baculovirus-infected insect cells and tested this protein for its ability to promote the phosphorylation of RPA by ATR-ATRIP (Figure 6(b)). We observed that the HFS-ETAA1-W63R protein exhibited no activity in the kinase assay (Figure 6(c)). Thus, the phosphorylation of RPA that we have observed in our experiments requires an intact AAD in ETAA1.
Figure 6.
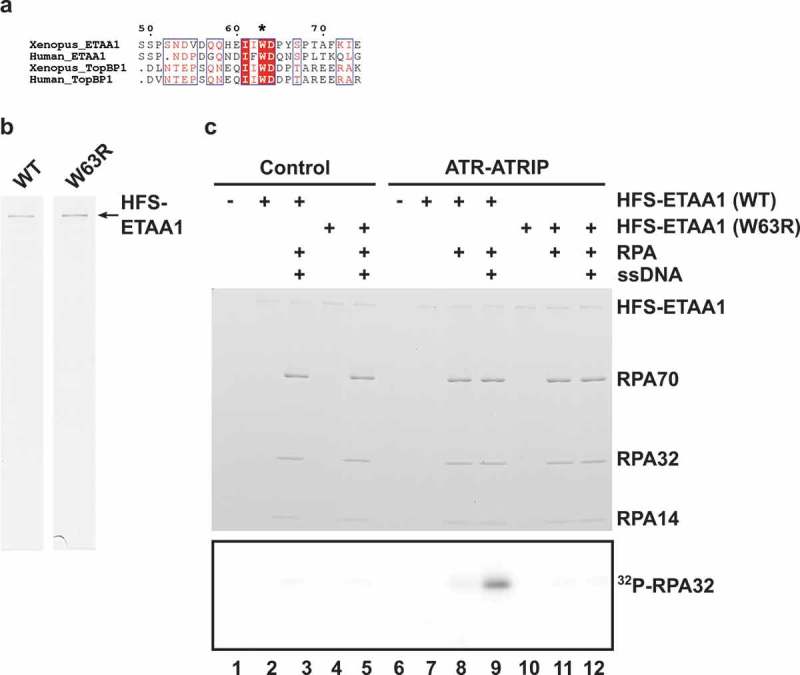
ETAA1-dependent phosphorylation of RPA requires an intact AAD. (a) Alignment of AADs around the critical tryptophan residues in ETAA1 and TopBP1 from Xenopus laevis and humans. The conserved tryptophan (W63) in Xenopus ETAA1 is denoted with an asterisk. (b) Wild-type (WT) and mutant W63R HFS-ETAA1 proteins were purified with anti-FLAG antibodies as described in Materials and Methods and stained with Coomassie blue. (c) Control eluates (lanes 1–5) and eluates containing ATR-ATRIP complexes (lanes 6–12) were incubated in kinase buffer containing γ-[32P]ATP in the absence of recombinant HFS-ETAA1 (lanes 1 and 6) or the presence of either WT (lanes 2–3 and 7–9) or W63R HFS-ETAA1 protein (lanes 4–5 and10-12). Some incubations also contained RPA (lanes 3, 5, 8–9, and 11–12) and ssDNA (lanes 3, 5, 9, and 12). Reactions were subjected to SDS gel electrophoresis. The gel was stained with Coomassie blue (top panel). Incorporation of 32P into proteins was detected by phosphorimaging (bottom panel). The portion of the image containing 32P-RPA32 is depicted.
In principle, the enhanced phosphorylation of RPA by ATR-ATRIP in the presence of ETAA1 and ssDNA could be due to the co-localization of ATR-ATRIP, ETAA1, and RPA on the DNA. Alternatively, RPA-ssDNA might act directly on ETAA1 to stimulate its ability to activate ATR-ATRIP, perhaps by eliciting a conformational change in ETAA1. These possibilities are not necessarily mutually exclusive. It is also possible that RPA32 bound to ssDNA could serve as a better substrate for ATR-ATRIP, but we likewise observe enhanced phosphorylation of ETAA1 on RPA-ssDNA. In order to explore these possibilities, we made use of a mutant Xenopus ATRIP called ΔN222 [50]. This mutant lacks the N-terminal RPA-binding domain of ATRIP and thus cannot associate effectively with ssDNA through RPA. An analogous mutant was first described in the case of human ATRIP [51].
Accordingly, we prepared complexes of ATR-ATRIP containing either wild-type (WT) or ΔN222 ATRIP and compared these complexes side-by-side for their activity in the ETAA1-dependent phosphorylation of RPA bound to ssDNA (Figure 7(a and b)). We observed that the complex containing ΔN222 ATRIP also exhibited a similar strong enhancement as the WT complex in kinase activity in the presence of RPA and ssDNA, even though ΔN222 ATRIP cannot associate with RPA. These experiments indicate that increased phosphorylation of RPA in the presence of ssDNA does not simply depend on the tethering of ATR-ATRIP and ETAA1 in close proximity on ssDNA. Overall, these experiments indicate that association of ETAA1 with RPA-ssDNA strongly enhances its ability to activate ATR-ATRIP, which we have monitored through phosphorylation of RPA. This in vitro system should be valuable for further characterization of the ETAA1-mediated activation of ATR-ATRIP.
Figure 7.
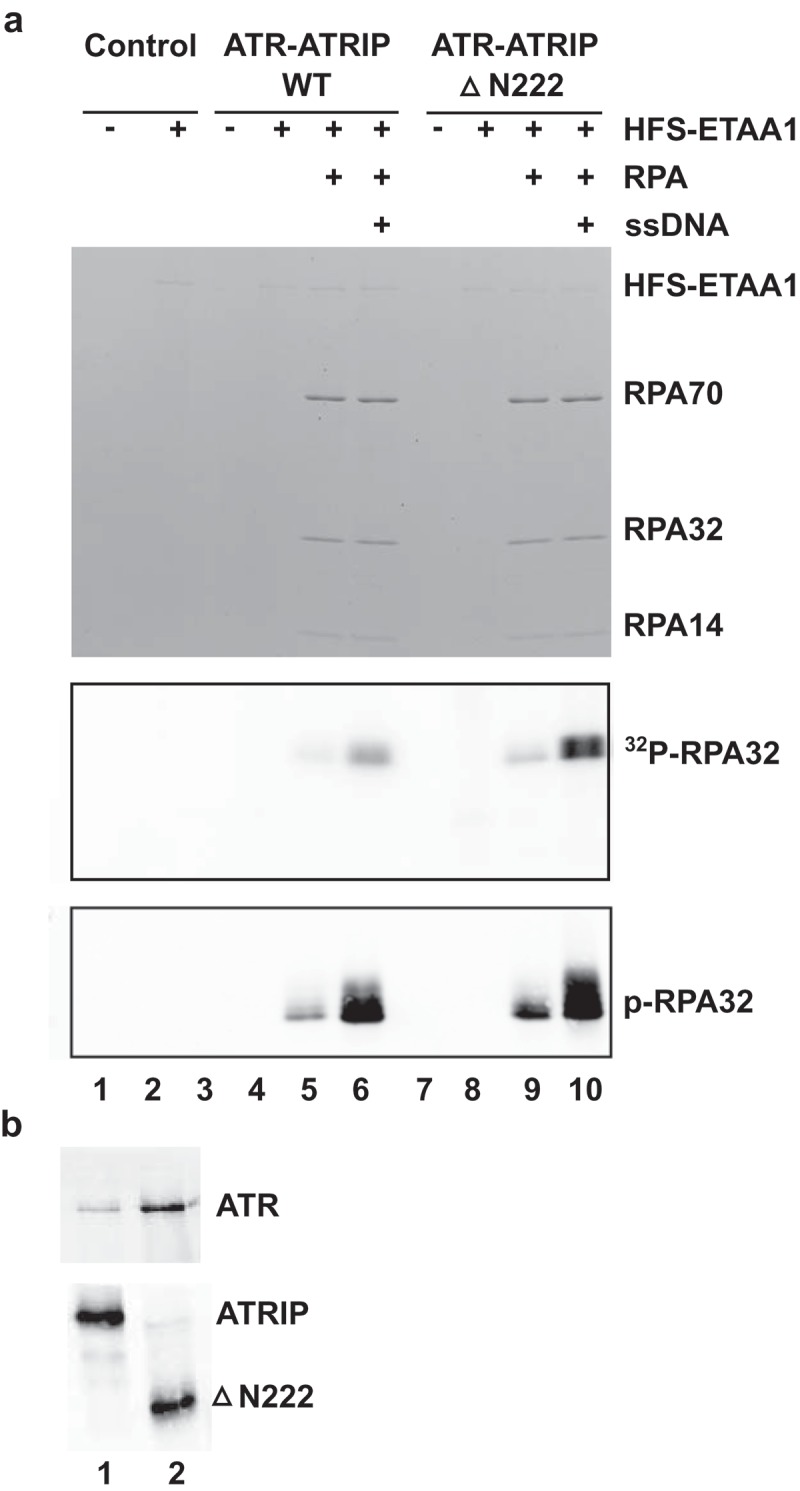
RPA-ssDNA can still promote activation of a mutant ATR-ATRIP complex that lacks the RPA-binding domain in ATRIP. (a) Control eluates (lanes 1–2) and eluates containing ATR in a complex with either full-length ATRIP (lanes 3–6) or ΔN222 ATRIP (lanes 7–10) were incubated in kinase buffer containing γ-[32P]ATP in the absence (lanes 1, 3, and 7) or presence HFS-ETAA1 protein (lanes 2, 4–6, and 8–10). Some incubations also contained RPA (lanes 5–6 and 9–10) and ssDNA (lanes 6 and 10). Reactions were subjected to SDS gel electrophoresis. The gel was stained with Coomassie blue (top panel). Incorporation of 32P into proteins was detected by phosphorimaging (middle panel). The portion of the image containing 32P-RPA32 is depicted. For the bottom panel, we electrophoresed aliquots of the same samples in a different gel and performed immunoblotting with anti-phospho-RPA32 antibodies. (b) ATR-ATRIP complexes. Full-length (lane 1) and ΔN222 versions of ATRIP-FLAG (lane 2) were added to egg extracts containing anti-FLAG M2 antibody beads. After incubation, the beads were reisolated, washed, and eluted with 3X-FLAG peptide. The eluates were immunoblotted with anti-ATR (top) and anti-ATRIP antibodies (bottom).
Discussion
The Xenopus egg-extract system was previously utilized for the demonstration that TopBP1 serves as a direct activator of the ATR-ATRIP complex [8]. In these studies, removal of TopBP1 from egg extracts resulted in the virtual elimination of the activation of ATR-ATRIP in response to the addition of APH. In view of the recent discovery of ETAA1, another activator of ATR, it was important to assess the role of this protein in the egg-extract system. In this report, we have confirmed that TopBP1 is the predominant, if not exclusive, activator of ATR for phosphorylation of Chk1 in response to stalled replication forks (as well as double-stranded DNA breaks) in egg extracts.
We observed that TopBP1 is significantly more abundant than ETAA1 in egg extracts. Therefore, we examined whether the addition of extra recombinant ETAA1 could support phosphorylation of Chk1 in extracts that had been depleted of their endogenous TopBP1. As described above, in order to carry out these experiments, we needed to supplement the TopBP1-depleted extracts with a fragment of TopBP1 that could sustain DNA replication but not activation of ATR. Under these conditions, we observed that extra recombinant ETAA1 could support ATR-dependent phosphorylation of Chk1 and RPA in APH-treated extracts.
Along similar lines, the concentration of ETAA1 varies substantially in different human cell lines [21]. For example, U2OS cells express considerably lower levels of ETAA1 than HeLa cells. Thus, knockdown of TopBP1 abolished hydroxyurea-induced phosphorylation of Chk1 in U2OS cells but not in HeLa cells [21]. Consistent with the results presented here, overexpression of GFP-ETAA1 restored phosphorylation of Chk1 in hydroxyurea-treated, TopBP1-depleted U2OS cells. Nonetheless, there appear to be significant qualitative differences between the pathways containing TopBP1 and ETAA1. For example, even though depletion of ETAA1 did not affect phosphorylation of Chk1 in U2OS cells, there was a reduction in phosphorylation of RPA [19].
It was recently shown in human cells that ETAA1 promotes sustained activity of ATR during an unperturbed S-phase to suppress a mitotic transcription factor [52]. Since there is minimal transcription and no G2 phase in early Xenopus development, this type of regulation would not need to be operable at this stage [53]. We also have not been able to detect any effect on replication during an unperturbed S-phase in ETAA1-depleted extracts, but there is also a rapid S-phase characteristic of early embryogenesis in egg extracts. After submission of this manuscript, Bass and Cortez [54] reported that removal of ETAA1 from human cells weakens the spindle checkpoint response. This checkpoint is absent in dividing early Xenopus embryos [55]. It will be interesting to evaluate the role that ETAA1 plays at later stages of Xenopus embryonic development.
It is important that AAD-containing proteins such as TopBP1 and ETAA1 carry out the activation of ATR only under the appropriate circumstances. Accordingly, the interaction of TopBP1 with ATR-ATRIP is regulated by a number of other proteins, such as the 9-1-1 and MRN complexes. To explore this issue for ETAA1, we set out to develop an in vitro system with defined components. Since we could not observe good phosphorylation of model substrates in the presence of only recombinant ETAA1 and ATR-ATRIP, we tested the addition of other components. RPA was a clear candidate because ETAA1 exhibits a highly specific interaction with this protein [19–22]. We could detect phosphorylation of RPA in the presence of ETAA1 and ATR-ATRIP. However, when we performed these reactions in the additional presence of ssDNA, which would efficiently recruit RPA, we observed a dramatic increase in the phosphorylation of the RPA32 subunit of the complex. Thus, RPA-ssDNA acts as strong positive effector in the ETAA1-mediated activation of ATR-ATRIP.
These observations suggest that docking of ETAA1 onto RPA-ssDNA in living cells would result in a significant increase in the kinase activity of ATR-ATRIP. From our in vitro studies, it is not possible to discern whether this form of ATR-ATRIP would correspond to a fully activated in vivo version. Nonetheless, it seems likely that the substantial amount of kinase activity that we observe in the presence of only ETAA1, ATR-ATRIP, and RPA-ssDNA would be physiologically impactful. By analogy with TopBP1, it seems plausible, even likely, that additional factors might contribute to the stabilization or further stimulation or both of the ETAA1-activated form ATR-ATRIP complex in living cells.
Previous studies have indicated that RPA is a significant ultimate target of the ETAA1-dependent pathway [19–22]. Our studies have reinforced this concept by showing explicitly that RPA is a direct in vitro substrate of ETAA1-activated ATR-ATRIP, as would be anticipated from the in vivo studies. Within the human RPA complex, we observed phosphorylation of RPA32, but not RPA70 and RPA14. Using anti-phosphopeptide antibodies, we have found that Ser-33 of RPA32 is well phosphorylated in our biochemical reactions. This site has been documented as a physiologically important target of ATR [47,48].
In closing, we have utilized the Xenopus egg-extract system to illuminate significant features of the role and regulation of ETAA1. The use of diverse experimental systems enhances understanding of key cellular regulatory molecules.
Funding Statement
This work was supported by NIH grants GM070891 and GM043974 to W.G.D.
Acknowledgments
We are grateful to Kanomi Sasaki-Capela for technical assistance. Dr. Marc Wold (University of Iowa) kindly provided the construct for expression of human RPA in bacteria. We also thank Dr. Oscar Fernandez-Capetillo (Karolinska Institute) for providing ETP46464. Anti-Xenopus RPA32 serum was the kind gift of Dr. Vincenzo Costanzo (FIRC Institute of Molecular Oncology).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Blackford AN, Jackson SP.. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol Cell. 2017. June 15;66(6):801–817. PubMed PMID: 28622525. [DOI] [PubMed] [Google Scholar]
- [2].Saldivar JC, Cortez D, Cimprich KA. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat Rev Mol Cell Biol. 2017. October;18(10):622–636. PubMed PMID: 28811666; PubMed Central PMCID: PMCPMC5796526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cortez D, Guntuku S, Qin J, et al. ATR and ATRIP: partners in checkpoint signaling. Science. 2001. November 23;294(5547):1713–1716. PubMed PMID: 11721054 [DOI] [PubMed] [Google Scholar]
- [4].Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003. June 6;300(5625):1542–1548. PubMed PMID: 12791985. [DOI] [PubMed] [Google Scholar]
- [5].Choi JH, Lindsey-Boltz LA, Kemp M, et al. Reconstitution of RPA-covered single-stranded DNA-activated ATR-Chk1 signaling. Proc Nat Acad Sci USA. 2010. August 3;107(31):13660–13665. PubMed PMID: 20616048; PubMed Central PMCID: PMC2922256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kumagai A, Dunphy WG. How cells activate ATR. Cell Cycle. 2006. June;5(12):1265–1268. PubMed PMID: 16760665. [DOI] [PubMed] [Google Scholar]
- [7].Ball HL, Ehrhardt MR, Mordes DA, et al. Function of a conserved checkpoint recruitment domain in ATRIP proteins. Mol Cell Biol. 2007. May;27(9):3367–3377. PubMed PMID: 17339343; PubMed Central PMCID: PMCPMC1899971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kumagai A, Lee J, Yoo HY, et al. TopBP1 activates the ATR-ATRIP complex. Cell. 2006. March 10;124(5):943–955. PubMed PMID: 16530042. [DOI] [PubMed] [Google Scholar]
- [9].Yan S, Lindsay HD, Michael WM. Direct requirement for Xmus101 in ATR-mediated phosphorylation of Claspin bound Chk1 during checkpoint signaling. J Cell Biol. 2006. April 24;173(2):181–186. PubMed PMID: 16618813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hashimoto Y, Tsujimura T, Sugino A, et al. The phosphorylated C-terminal domain of Xenopus Cut5 directly mediates ATR-dependent activation of Chk1. Genes Cells. 2006. September;11(9):993–1007. PubMed PMID: 16923121. [DOI] [PubMed] [Google Scholar]
- [11].Mordes DA, Glick GG, Zhao R, et al. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008. June 1;22(11):1478–1489. PubMed PMID: 18519640; PubMed Central PMCID: PMCPMC2418584. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Delacroix S, Wagner JM, Kobayashi M, et al. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007. June 15;21(12):1472–1477. PubMed PMID: 17575048; PubMed Central PMCID: PMCPMC1891424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lee J, Kumagai A, Dunphy WG. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J Biol Chem. 2007. September 21;282(38):28036–28044. PubMed PMID: 17636252. [DOI] [PubMed] [Google Scholar]
- [14].Duursma AM, Driscoll R, Elias JE, et al. A role for the MRN complex in ATR activation via TOPBP1 recruitment. Mol Cell. 2013. April 11;50(1):116–122. PubMed PMID: 23582259; PubMed Central PMCID: PMCPMC3669687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lee J, Dunphy WG. The Mre11-Rad50-Nbs1 (MRN) complex has a specific role in the activation of Chk1 in response to stalled replication forks. Mol Biol Cell. 2013. May;24(9):1343–1353. PubMed PMID: 23468519; PubMed Central PMCID: PMCPMC3639046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Navadgi-Patil VM, Burgers PM. Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. J Biol Chem. 2008. December 19;283(51):35853–35859. PubMed PMID: 18922789; PubMed Central PMCID: PMCPMC2602893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Majka J, Niedziela-Majka A, Burgers PM. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol Cell. 2006. December 28;24(6):891–901. PubMed PMID: 17189191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kumar S, Burgers PM. Lagging strand maturation factor Dna2 is a component of the replication checkpoint initiation machinery. Genes Dev. 2013. February 1;27(3):313–321. PubMed PMID: 23355394; PubMed Central PMCID: PMC3576516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bass TE, Luzwick JW, Kavanaugh G, et al. ETAA1 acts at stalled replication forks to maintain genome integrity. Nat Cell Biol. 2016. November;18(11):1185–1195. PubMed PMID: 27723720; PubMed Central PMCID: PMCPMC5245861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Feng S, Zhao Y, Xu Y, et al. Ewing Tumor-associated Antigen 1 interacts with Replication Protein A to promote restart of stalled replication forks. J Biol Chem. 2016. October 14;291(42):21956–21962. PubMed PMID: 27601467; PubMed Central PMCID: PMCPMC5063979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Haahr P, Hoffmann S, Tollenaere MA, et al. Activation of the ATR kinase by the RPA-binding protein ETAA1. Nat Cell Biol. 2016. November;18(11):1196–1207. PubMed PMID: 27723717. [DOI] [PubMed] [Google Scholar]
- [22].Lee YC, Zhou Q, Chen J, et al. RPA-binding protein ETAA1 is an ATR activator involved in DNA replication stress response. Curr Biol. 2016. December 19;26(24):3257–3268. PubMed PMID: 27818175; PubMed Central PMCID: PMCPMC5173396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kumagai A, Shevchenko A, Shevchenko A, et al. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell. 2010. February 5;140(3):349–359. PubMed PMID: 20116089; PubMed Central PMCID: PMCPMC2857569. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hu RG, Wang H, Xia Z, et al. The N-end rule pathway is a sensor of heme. Proc Nat Acad Sci USA. 2008. January 08;105(1):76–81. PubMed PMID: 18162538; PubMed Central PMCID: PMCPMC2224235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lee J, Kumagai A, Dunphy WG. Claspin, a Chk1-regulatory protein, monitors DNA replication on chromatin independently of RPA, ATR, and Rad17. Mol Cell. 2003. February;11(2):329–340. PubMed PMID: 12620222. [DOI] [PubMed] [Google Scholar]
- [26].Kumagai A, Guo Z, Emami KH, et al. The Xenopus Chk1 protein kinase mediates a caffeine-sensitive pathway of checkpoint control in cell-free extracts. J Cell Biol. 1998;142(6):1559–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Carpenter PB, Mueller PR, Dunphy WG. Role for a Xenopus Orc2-related protein in controlling DNA replication. Nature. 1996;379(6563):357–360. [DOI] [PubMed] [Google Scholar]
- [28].Li W, Kim SM, Lee J, et al. Absence of BLM leads to accumulation of chromosomal DNA breaks during both unperturbed and disrupted S phases. J Cell Biol. 2004. June 21;165(6):801–812. PubMed PMID: 15197177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A: expression, complex formation, and functional characterization. J Biol Chem. 1994. April 15;269(15):11121–11132. PubMed PMID: 8157639. [PubMed] [Google Scholar]
- [30].Wuhr M, Freeman RM Jr., Presler M, et al. Deep proteomics of the Xenopus laevis egg using an mRNA-derived reference database. Curr Biol. 2014. July 07;24(13):1467–1475. PubMed PMID: 24954049; PubMed Central PMCID: PMCPMC4090281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hashimoto Y, Takisawa H. Xenopus Cut5 is essential for a CDK-dependent process in the initiation of DNA replication. Embo J. 2003. May 15;22(10):2526–2535. PubMed PMID: 12743046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hekmat-Nejad M, You Z, Yee M, et al. Xenopus ATR is a replication-dependent chromatin-binding protein required for the DNA replication checkpoint. Curr Biol. 2000;10(24):1565–1573. [DOI] [PubMed] [Google Scholar]
- [33].You Z, Kong L, Newport J. The role of single-stranded DNA and polymerase alpha in establishing the ATR, Hus1 DNA replication checkpoint. J Biol Chem. 2002. July 26;277(30):27088–27093. PubMed PMID: 12015327. [DOI] [PubMed] [Google Scholar]
- [34].Hoogenboom WS, Klein Douwel D, Knipscheer P. Xenopus egg extract: A powerful tool to study genome maintenance mechanisms. Dev Biol. 2017. August 15;428(2):300–309. PubMed PMID: 28427716. [DOI] [PubMed] [Google Scholar]
- [35].Dasso M, Newport JW. Completion of DNA replication is monitored by a feedback system that controls the initiation of mitosis in vitro: studies in Xenopus. Cell. 1990;61(5):811–823. [DOI] [PubMed] [Google Scholar]
- [36].Guo Z, Kumagai A, Wang SX, et al. Requirement for ATR in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 2000;14(21):2745–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Luciani MG, Oehlmann M, Blow JJ. Characterization of a novel ATR-dependent, Chk1-independent, intra-S-phase checkpoint that suppresses initiation of replication in Xenopus. J Cell Sci. 2004. December 1;117(Pt 25):6019–6030. PubMed PMID: 15536124; PubMed Central PMCID: PMC2701543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yanow SK, Gold DA, Yoo HY, et al. Xenopus Drf1, a regulator of Cdc7, displays checkpoint-dependent accumulation on chromatin during an S-phase arrest. J Biol Chem. 2003. October 17;278(42):41083–41092. PubMed PMID: 12897072. [DOI] [PubMed] [Google Scholar]
- [39].Walter J, Newport J. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol Cell. 2000. April;5(4):617–627. PubMed PMID: 10882098. [DOI] [PubMed] [Google Scholar]
- [40].Mimura S, Masuda T, Matsui T, et al. Central role for Cdc45 in establishing an initiation complex of DNA replication in Xenopus egg extracts. Genes Cells. 2000. June;5(6):439–452. PubMed PMID: 10886370. [DOI] [PubMed] [Google Scholar]
- [41].Mimura S, Takisawa H. Xenopus Cdc45-dependent loading of DNA polymerase alpha onto chromatin under the control of S-phase Cdk. Embo J. 1998. October 1;17(19):5699–5707. PubMed PMID: 9755170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Shechter D, Costanzo V, Gautier J. ATR and ATM regulate the timing of DNA replication origin firing. Nat Cell Biol. 2004. July;6(7):648–655. PubMed PMID: 15220931. [DOI] [PubMed] [Google Scholar]
- [43].Marheineke K, Hyrien O. Control of replication origin density and firing time in Xenopus egg extracts: role of a caffeine-sensitive, ATR-dependent checkpoint. J Biol Chem. 2004. July 2;279(27):28071–28081. PubMed PMID: 15123715. [DOI] [PubMed] [Google Scholar]
- [44].Van Hatten RA, Tutter AV, Holway AH, et al. The Xenopus Xmus101 protein is required for the recruitment of Cdc45 to origins of DNA replication. J Cell Biol. 2002. November 25;159(4):541–547. PubMed PMID: 12438414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66: 61–92. PubMed PMID: 9242902. [DOI] [PubMed] [Google Scholar]
- [46].Chen R, Wold MS. Replication protein A: single-stranded DNA‘s first responder. Bioessays. 2014. December;36(12):1156–1161. PubMed PMID: 25171654; PubMed Central PMCID: PMCPMC4629251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Byrne BM, Oakley GG. Replication protein A, the laxative that keeps DNA regular: the importance of RPA phosphorylation in maintaining genome stability. Semin Cell Dev Biol. 2018. April 20 PubMed PMID: 29665433 DOI: 10.1016/j.semcdb.2018.04.005 [DOI] [PubMed] [Google Scholar]
- [48].Marechal A, Zou L. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 2015. January;25(1):9–23. PubMed PMID: 25403473; PubMed Central PMCID: PMCPMC4650586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Toledo LI, Murga M, Zur R, et al. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol. 2011. June;18(6):721–727. PubMed PMID: 21552262; PubMed Central PMCID: PMCPMC4869831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kim S-M, Kumagai A, Lee J, et al. Phosphorylation of Chk1 by ATM- and Rad3-related (ATR) in Xenopus egg extracts requires binding of ATRIP to ATR but not the stable DNA-binding or coiled-coil domains of ATRIP. J Biol Chem. 2005. November 18;280(46):38355–38364. PubMed PMID: 16186122. [DOI] [PubMed] [Google Scholar]
- [51].Ball HL, Myers JS, Cortez D. ATRIP binding to replication protein A-single-stranded DNA promotes ATR-ATRIP localization but is dispensable for Chk1 phosphorylation. Mol Biol Cell. 2005. May;16(5):2372–2381. PubMed PMID: 15743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Saldivar JC, Hamperl S, Bocek MJ, et al. An intrinsic S/G2 checkpoint enforced by ATR. Science. 2018. August 24;361(6404):806–810. PubMed PMID: 30139873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982. October;30(3):675–686. PubMed PMID: 6183003. [DOI] [PubMed] [Google Scholar]
- [54].Bass TE, Cortez D. Quantitative phosphoproteomics reveals mitotic function of the ATR activator ETAA1. J Cell Biol. 2019. February 12 PubMed PMID: 30755469 DOI: 10.1083/jcb.201810058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Minshull J, Sun H, Tonks NK, et al. A MAP kinase-dependent spindle assembly checkpoint in Xenopus egg extracts. Cell. 1994. November 4;79(3):475–486. PubMed PMID: 7954813. [DOI] [PubMed] [Google Scholar]


