ABSTRACT
Human papillomavirus (HPV) is an etiologic factor in head and neck squamous cell carcinoma (HNSCC). HPV(+) cancers respond favorably to therapy potentially due to more robust anti-tumor immune responses. We hypothesized that tumor-derived exosomes (TEX) produced by HPV(+) or HPV(-) HNSCCs differentially modulate anti-tumor immune responses.
Proteomes of exosomes from HPV(+) and HPV(-) HNSCC cell lines were compared in search for proteins putatively involved in the communication with immune system. TEX were isolated from supernatants of HPV(+) (SCC-2, SCC-47, and SCC-90) or HPV(-) (PCI-13 and PCI-30) cells by size exclusion chromatography. A comparison of proteome profiles was performed by high-resolution mass spectrometry. The presence and biological activity of selected immunoregulatory proteins were validated by flow cytometry and co-incubation assays. Exosomes produced by SCC-90 and PCI-30 cells contained 711 proteins, including 80 proteins specific for HPV(+) exosomes and 77 specific for HPV(-) exosomes, associated with similar GO terms such as regulation of cell growth, metabolism, communication, and cellular signaling. Search for proteins localized in the membrane and involved in immune regulation identified a few proteins detected specifically in HPV(+) or HPV(-) exosomes. Only HPV(+) exosomes were enriched in immune effector cell-related CD47 and CD276 antigens; only HPV(-) exosomes contained tumor-protective/growth-promoting antigens, MUC-1 and HLA-DA. Flow cytometry and Western blots confirmed the reciprocal presence/paucity of these proteins in a whole panel of tumor cells and corresponding exosomes. The differential content of protein cargos in HPV(+) and HPV(-) exosomes might contribute to the disparity in immune responses that characterize HPV(+) and HPV(-) HNSCC.
KEYWORDS: Exosomes, head and neck cancer, human papillomavirus, proteomics, immune functions
Introduction
Despite advances in the delivery of surgery and chemo-radiotherapy, HNSCC patients have a poor prognosis, largely because of disease recurrence.1 Five-year survival for HNSCC patients has not improved in 30 years and remains at <50%.2 Established etiologic factors associated with HNSCC include tobacco or alcohol consumption and human papillomavirus (HPV) infection.3–5 HPV16 infection is primarily associated with tumors of the oropharynx and base of the tongue.6,7 The incidence of this site-specific cancer seen largely in young patients has been rapidly increasing.8 Clinical, histopathological and molecular characteristics of HPV(+) and HPV(-) HNSCCs are distinct.9 Patients with HPV(+) tumors respond favorably to initial therapy, but as they age and their tumors recur, the therapeutic options become limited. New therapies based on a better understanding of factors that drive viral oncogenesis are an unmet need for patients with HPV(+) or HPV(-) HNSCCs. Expression of E6 and E7 viral oncoproteins, interacting with and inhibiting p53 and Rb proteins, respectively, is causally linked to malignant transformation of HPV(+) cancers.10 Deregulation of cellular processes involving p53 and Rb, which includes apoptosis and the cell cycle arrest, could be hypothetically associated with differential response of HPV(+) and HPV(-) cancers to DNA damaging factors during oncological treatments. However, cells derived from HPV(+) HNSCC cancers are not more sensitive to cisplatin, radiation or cetuximab, therapies commonly used in HNSCC, than cells derived from HPV(-) cancers, suggesting that the favorable prognosis for patients with HPV(+) cancers is not directly related to an intrinsic sensitivity of these tumors to chemotherapy and/or radiation.11
An alternative hypothesis advanced as a potential explanation for better prognosis of HPV(+) HNSCCs involves the host immune system.12–15 In this paradigm, immune cells activated by the virus are prepared to mediate antitumor activity but due to tumor-induced immune suppression of effector cell functions fail to do so efficiently. Indeed, an increased frequency of immune effector cells specific for tumor-associated antigens (TAAs) has been detected in the circulation of patients with HPV(+) cancers, although these cells appear to be partially dysfunctional.12 Among various tumor-derived factors known to contribute to cross-talk between HNSCC and the immune system, exosomes produced by the tumor (TEX) have been recently recognized as a major contributor to immune cell dysfunction.16
Exosomes are the smallest (30–150 nm in diameter) subset of extracellular vesicles (EVs), which mediate inter-cellular communication.17 TEX convey information from the tumor to various cells in the tumor microenvironment (TME) and by autocrine, juxtacrine or paracrine signaling reprogram cellular functions, converting them into cancer-promoting cells.18 TEX molecular profiles partially recapitulate the content of their parent cells thus serving as potential tumor biomarkers.19 TEX carry a variety of receptors and ligands that are expressed on the parent tumor cell surface and deliver them to recipient cells, inducing functional alterations.20 Immunosuppressive proteins carried by exosomes isolated from plasma of HNSCC patients, such as PD-L1, correlate with the tumor stage, nodal involvement, and disease activity.16 Moreover, exosomes from HPV(+) cancers carry viral proteins and genes in addition to TAA.21 We reported earlier that TEX produced by HPV(+) HNSCC cells promoted human dendritic cell (DC) maturation and enhanced antigen-processing machinery (APM) expression levels in mature DCs, while those produced by HPV(-) cells suppressed DC functions.22 These results support the hypothesis that TEX derived from HPV(+) or HPV(-) HNSCC differentially modulate anti-tumor immune responses and thereby play a role in disease progression and outcome. However, molecular underpinnings of effects that HPV(+) and HPV(-) TEX induce in recipient immune cells are not understood. In this study, we analyzed the proteomes of exosomes produced by HPV(+) and HPV(-) HNSCC cell lines, specifically searching for membrane-associated proteins that could contribute to differential modulation of immune recipient cells by these exosomes.
Results
The general characteristics of exosomes isolated from cell culture supernatants of the HPV(+) or HPV(-) tumor cells were previously described.22 Briefly, vesicles isolated by size exclusion chromatography and eluted in PBS had a diameter ranging from 30 to 150 nm, as determined by TEM and qNANO, and contained endocytic markers TSG101 and ALIX as well as tetraspanins, CD63 and CD81, as determined by Western blots. Moreover, exosomes released by HPV(+) cells (SCC-2, SCC-47, SCC-90), but not by HPV(-) cells (PCI-13 or PCI-30), contained E6/E7 viral proteins and p16 protein.22
Global proteome profiling of exosomes
A shot-gun proteomics approach was used for analysis of exosomes produced by two cell lines SCC-90 and PCI-30, representing HPV(+) and HPV(-) HNSCCs, respectively. This approach identified 711 proteins encoded by unique genes. Among them, 80 proteins were detected only in HPV(+) exosomes produced by SCC-90 cells and 77 proteins were detected only in HPV(-) exosomes produced by PCI-30 cells (Figure 1a). All detected proteins are listed in the Supplementary File (Table S1).
Figure 1.
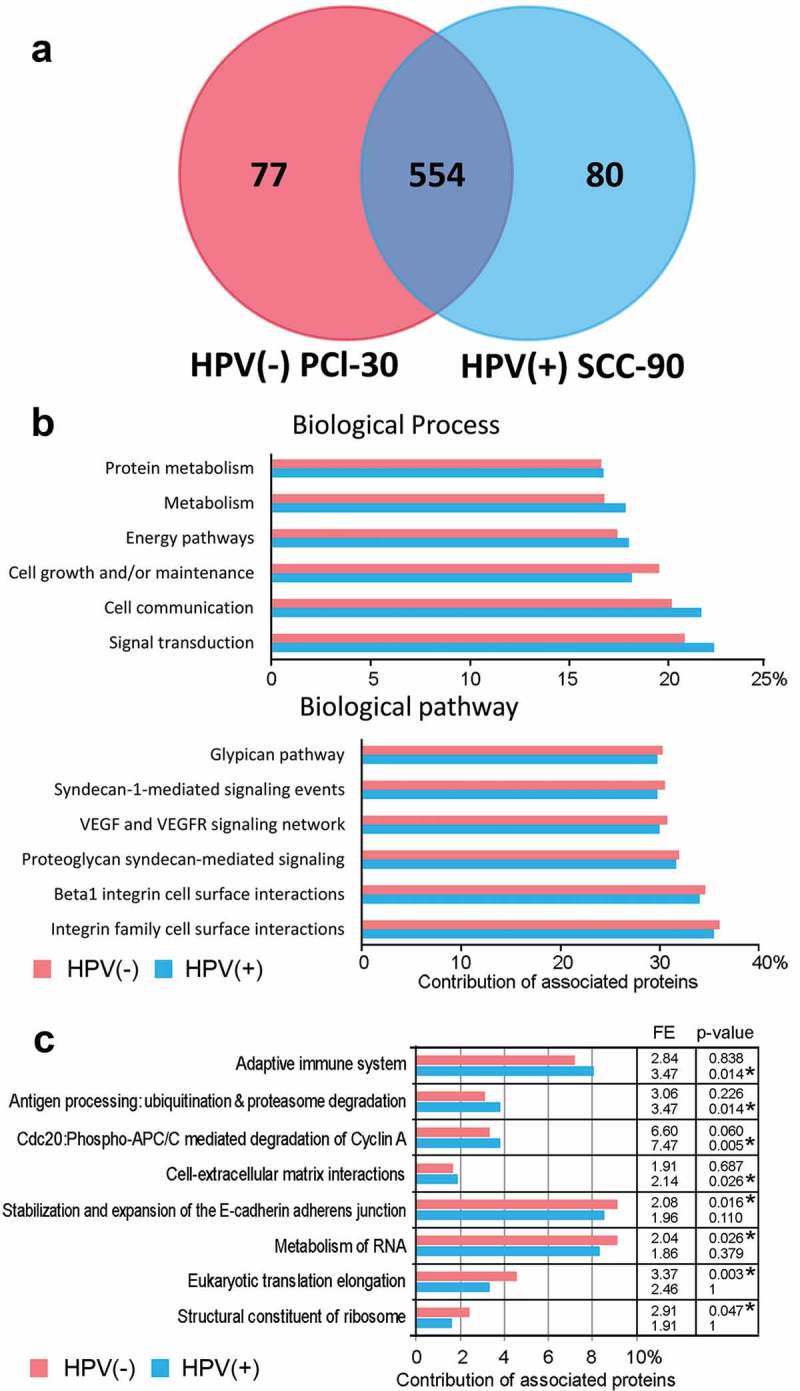
Proteins detected in exosomes released by HNSCC cell lines. Panel a – The Venn diagram illustrating the distribution of proteins in HPV(+) and HPV(-) exosomes. Panel b – Major functional FunRich annotations associated with exosome proteins. Panel c – Functional annotations that showed differences in relative enrichment of proteins in HPV(+) and HPV(-) exosomes; depicted is a relative contribution [%] of proteins associated with a specific pathway in a total set of identified proteins, calculated fold-enrichment (FE) and statistical significance of the overrepresentation (corrected p-value, asterisks mark statistically significant overrepresentation).
Functional gene designations for the identified proteins
Analysis of gene ontology was performed to identify functional pathways associated with the detected proteins. The majority (approximately 2/3) of the detected proteins in exosomes from both HPV(+) and HPV(-) cells were cytoplasmic proteins. They were associated with six major biological functions: (a) regulation of cell growth; (b) cellular metabolism; (c) protein metabolism; (d) energy pathways; (e) cell communication and (f) signal transduction (Figure 1b). Among the proteins detected in both types of exosomes were the integrin family members involved in cell surface interactions and proteins mediating proteoglycan-syndecan signaling as well as the VEGF/VEGFR signaling networks. In general, the majority of biological pathways and processes were similarly enriched in proteins detected in HPV(+) (SCC-90) and HPV(-) (PCI-30) exosomes, as shown in the Supplementary File (Table S2). Several pathways showed statistically significant enrichment (p-value with Bonferroni correction <0.05) for either HPV(+) or HPV(-) exosomes, as shown in Figure 1c. These functional terms included an adaptive immune system or cell-extracellular matrix interactions, which were significantly overrepresented in HPV(+) exosomes, and metabolism of RNA or structural components of ribosomes, which were significantly overrepresented in HPV(-) exosomes. However, no statistically significant differences were detected between these selected sets of proteins in their contribution to the specific pathway.
Based on these results, we concluded that a high level of structural and functional similarity existed between the proteomes of exosomes that were produced by the two HPV(+) and HPV(-) cell lines selected for this study as representative for HNSCC cells.
Analysis of proteins putatively involved in immune signaling
Having previously determined that exosomes produced by HPV(+) and HPV(-) HNSCC cell lines reprogramed functions of immune cells, 22 we were motivated to search for proteins putatively involved in exosome-mediated cross-talk between the tumor and immune cells. By definition, these proteins are localized in the surface membranes of exosomes and are associated with GO terms “intrinsic components of membrane” (0031224) and “immune-related functions” (0002682). As shown in Figure 2a, there were 253 proteins that met the definition of being either intrinsic components of the membrane or serving as regulatory elements in the immune system or both. 172 of these proteins were shared between HPV(+) and HPV(-) exosomes, and 43 were present only in HPV(+) exosomes, while 38 were carried only by HPV(-) exosomes. However, only 19 proteins that associated with both GO terms were identified. These included 4 proteins detected only in exosomes from HPV(+) cells: CD47, CD276 (B7-H3), CALM1 (calmodulin), and CXADR (coxsackie and adenovirus receptor) and 5 proteins identified only in exosomes from HPV(-) cells: C9, MUC-1, HLA-DRA, MUC-4, MUC-16. Ten other proteins were shared by HPV(+) and HPV(-) exosomes. Among the membrane proteins putatively involved in mediating tumor-immune cell interactions, we focused on CD276 and CD47 in HPV(+) exosomes and MUC-1 as well as HLA-DRA in HPV(-) exosomes, based on the well-documented association of these antigens in signaling between immune cells and tumor targets.
Figure 2.
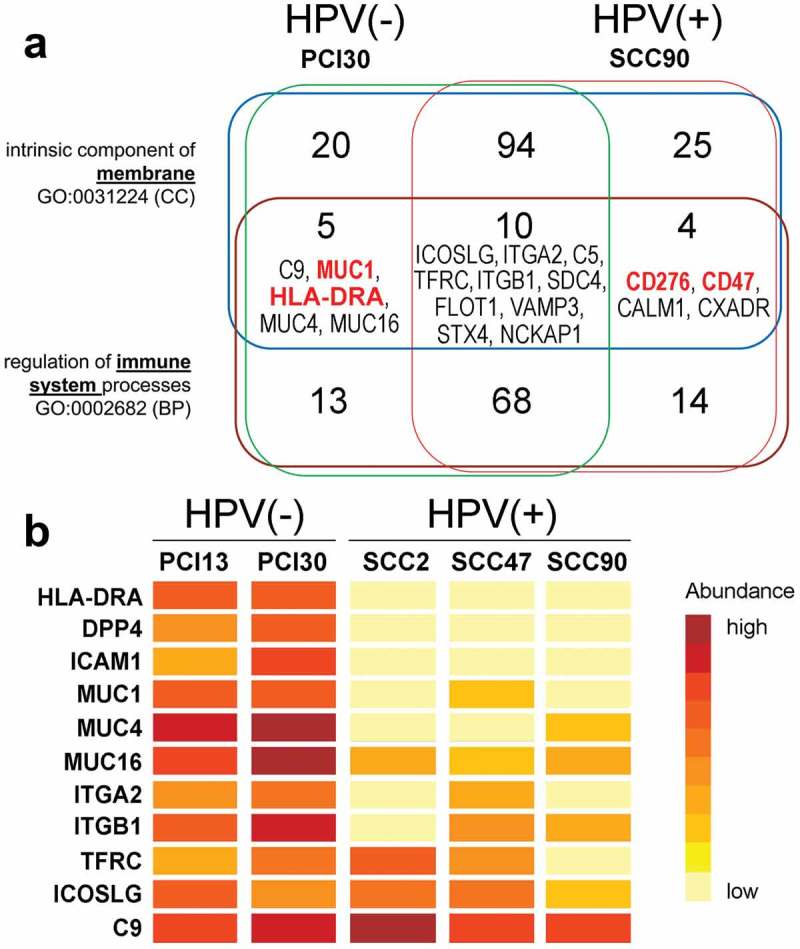
Selected immune-related proteins present in exosomes produced by HPV(+) and HPV(-) tumor cells. Panel a – Number of proteins related to immune functions [GO:0002682] and membrane localization [GO:0031224] identified in exosomes produced by PCI30 or SCC90 cells. Panel b – Relative abundance of the selected immune-related proteins in exosomes produced by the 5 different HNSCC cell lines that we have analyzed and quantified by MS/MS (abundances of selected proteins are sorted regarding deciles of all normalized signals).
To further analyze immune-related proteins in exosomes produced by two HPV(-) (PCI-13 and PCI-30) and 3 HPV(+) cell lines (SCC-2, SCC-47, SCC-90), a label-free MS/MS-based protein quantitation was performed. This analysis revealed a relative abundance of the identified proteins but gave a weaker coverage of exosome proteomes (only 217 proteins were identified). Nevertheless, 11 proteins with immune-related functions [GO:0002682] and membrane localization [GO:0031224] were quantified, as shown in Figure 2b. In agreement with the results presented above, a markedly higher abundance of HLA-DRA, MUC1, MUC4, and MUC16 in HPV(-) exosomes than in HPV(+) exosomes was observed. Moreover, HPV(-) exosomes showed a higher abundance of DDP4, ICAM1, and ITGA2. The relative abundance of ITGB1, TFRC, ICOSLG, and C9 was similar between both sub-panels of exosomes. Surprisingly, proteins previously detected only in SCC90-derived exosomes (Figure 2a) i.e. CD276, CD47, CALM1, and CXADR, were not identified in this quantitative analysis. It is likely that variably low abundance of these proteins in exosomes from different cell lines prevented their recognition in the quantitative MS/MS.
Validation of CD47 and CD276 presence on exosomes
To validate the presence of the proteins identified on exosomes by LC-MS/MS on the surface of parent cells, flow cytometry was performed using antibodies specific for the identified antigens. All five cells lines [three HPV(+) and two HPV(-)], as well as exosomes isolated from supernatants of these cells, were studied. Figure 3a shows that only HPV(+) tumor cells were positive for CD47, while HPV(-) tumor cells were negative. As expected based on the LC-MS/MS data, exosomes produced by HPV(+) cells carried CD47, while exosomes produced by HPV(-) cell did not. In Figure 3b, expression of CD276 on HPV(+) tumor cells and on HPV(-) tumor cells is evident, although the expression levels are higher on HPV(+) tumor cells than HPV(-) tumor cells. Likewise, HPV(+) exosomes carry higher levels of CD276 than do HPV(-) exosomes. Thus, in case of CD276, low levels of this protein were detected on the HPV(-) parent cells and on HPV(-) exosomes, indicating that Ab-based flow cytometry had greater sensitivity relative to LC-MS/MS for detection of low abundance proteins on cells or exosomes. Nevertheless, flow cytometry-based analysis confirmed a markedly higher level of both CD47 and CD276 antigens in exosomes released by HPV(+) HNSCC cell lines and their paucity, respectively, in HPV(-) exosomes.
Figure 3.
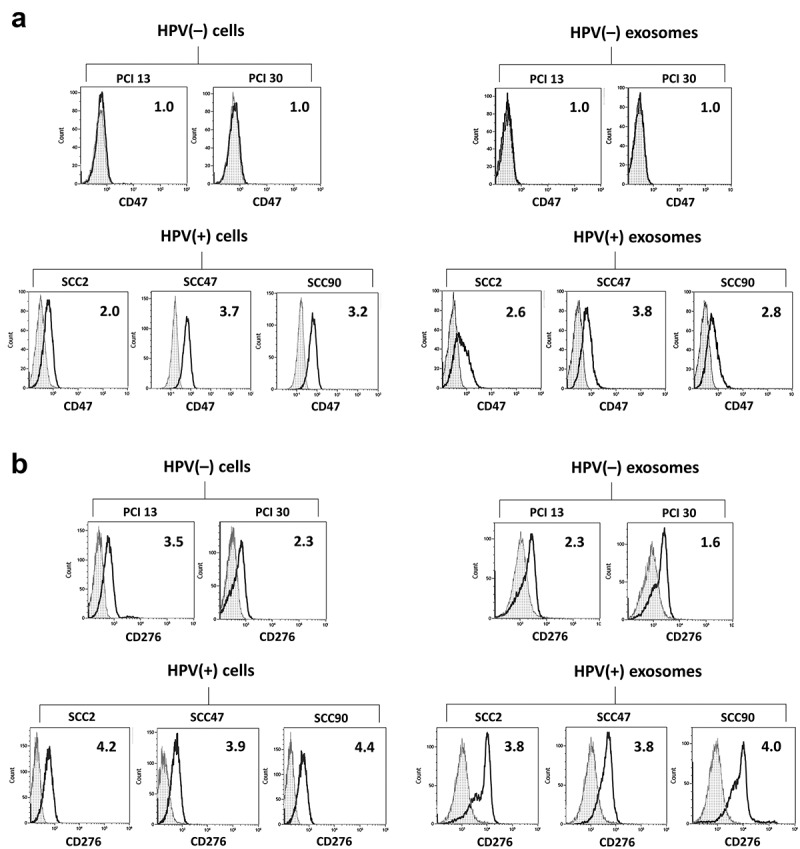
Expression of CD47 (Panel a) and CD276 (Panel b) antigens on the membranes of HPV(+) and HPV(-) tumor cells and exosomes released by these cells analyzed by flow cytometry. On-bead flow cytometry for exosomes was performed as described in methods. The numbers indicate RFI values.
The same flow cytometry-based approach was used for validation of MUC-1 and HLA-DRA protein levels on the HPV(-) tumor parent cells and exosomes. As Figure 4 shows, HPV(-) cells and exosomes produced by these cells contain high levels of both proteins. In contrast, HPV(+) cells and HPV(+) exosomes do not express/carry these proteins.
Figure 4.
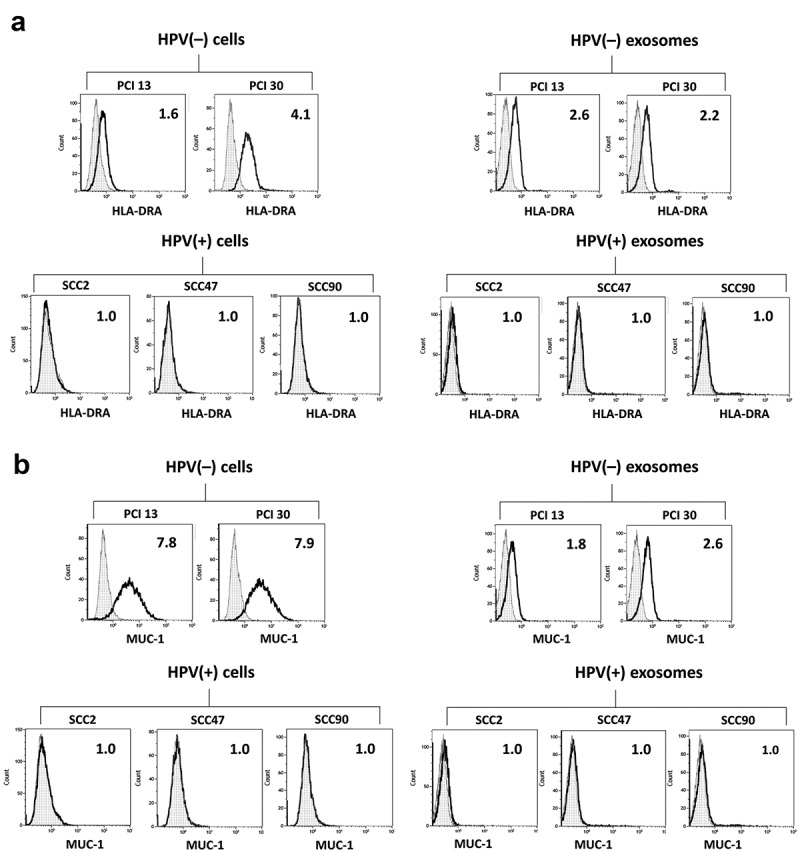
Expression of HLA-DRA (Panel a) and MUC-1 (Panel b) antigens on the membranes of HPV(+) and HPV(-) tumor cells and exosomes released by these cells analyzed by flow cytometry. On-bead flow cytometry for exosomes was performed as described in methods. The numbers indicate RFI values.
Functional analysis of CD47 and MUC-1 carried by HPV(+) or HPV(-) exosomes
In vitro experiments were performed to show that HPV(+) exosomes enriched in CD47 were phagocytosed less efficiently by human activated monocytes than HPV(-) exosomes with lower CD47 levels in the membrane. To this end, monocytes isolated from human PBMC were co-incubated with exosomes that were released by PCI30 or SSCC90 cells and labeled with PKH26. Following optimization of the monocyte/exosome ratio and co-incubation time, the efficiency of phagocytosis of HPV(+) exosomes (SCC90) and of HPV(-) exosomes (PCI30) by monocytes was compared by flow cytometry. We found that the mean fluorescence intensity (MFI) ratio for PCI30/SCC90 exosomes (i.e. CD47−/low/CD47+ exosomes) was about 1.6 (the Supplementary File Table S3); Figure 5b shows a representative flow cytometry experiment.
Figure 5.
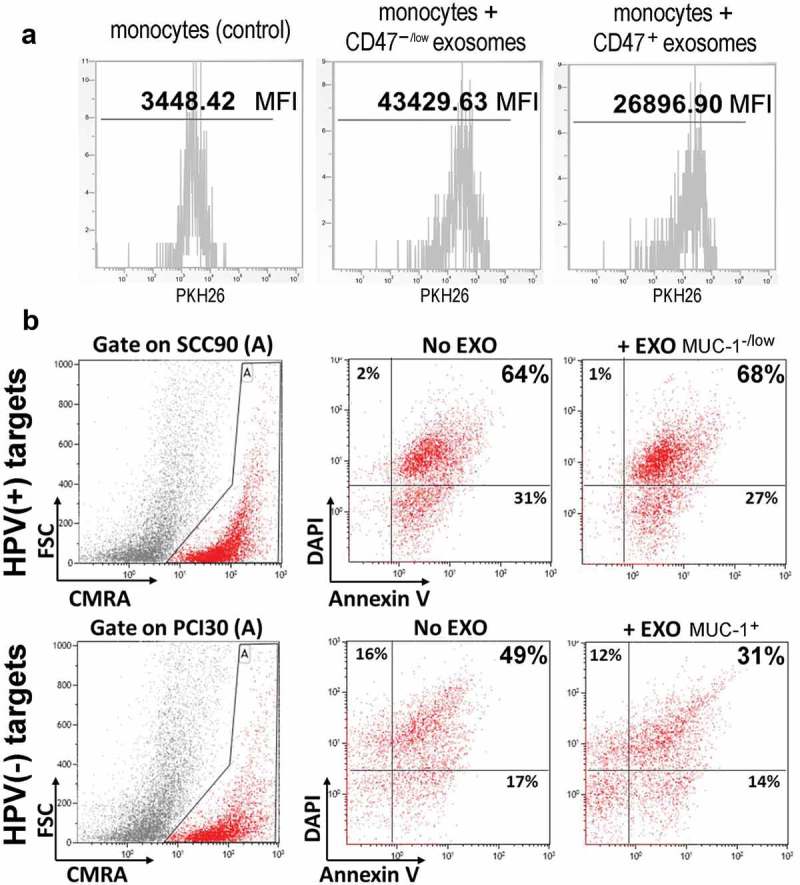
Functional importance of CD47 and MUC-1. Panel a – Flow cytometry assessment of human CD14+ monocytes co-incubated with PKH26-labeled exosomes. Uptake/phagocytosis of CD47+ and CD47−/low exosomes isolated from SSC90 or PCI30 cells, respectively, was measured. Co-incubations were performed for 15 minutes with the 1 × 105 monocytes/10 µg exosome protein ratio; denoted is the mean fluorescence intensity (MFI). Panel b – NK cell-induced apoptosis of tumor cells pre-incubated with autologous MUC-1+ and MUC-1−/low exosomes. Target SCC90 and PCI30 cells (HPV(+) and HPV(-), respectively) labeled with the CMRA cell tracker were co-incubated with exosomes (10 µg protein). Exosome-treated (+EXO) and not treated (No EXO) target cells were then co-incubated with activated effector NK cells (at 1:5 target:effector ratio) for 2 h and apoptosis of target cells was measured by flow cytometry using the Annexin V and PI staining (with the gate set on target cells); the percentages of apoptotic target cells are indicated in each quadrant.
In another series of experiments, “tumor protective” functions of MUC-1+ HPV(-) exosomes was investigated. HPV(-) exosomes were co-incubated with autologous tumor targets (PCI30) labeled with the CMRA cell tracker dye for 15 min prior to the addition of IL2-activated NK cells. In parallel, MUC-1−/low HPV(+) exosomes were similarly co-incubated with the labeled autologous tumor targets (SSC90) and then with IL-2 activated NK cells. The abilities of NK effector cells to lyse these exosome-pretreated tumor targets were compared. As shown in Figure 5b, MUC-1+ exosomes from HPV(-) cells (PCI30) were more effective in protecting tumor cells from lysis by activated NK cells.
Both experiments with tumor cells co-incubated with HPV(+) CD47+ or HPV(-) MUC-1+ exosomes indicate that these proteins retain their respective biological activities, as expected. Thus, CD47+ exosomes were phagocytosed less efficiently by monocytes presumably due to “do-not-eat-me” signaling by CD47, and MUC-1+ exosomes protected tumor cells from lysis mediated by activated NK cells.
Discussion
In this study, proteomic profiles of exosomes produced by HPV(+) and HPV(-) HNSCC cell lines were compared with a special emphasis on surface membrane-associated proteins potentially mediating the tumor-immune cell cross-talk. TEX have been shown by us and others to suppress functions of immune cells, largely due to numerous immunoinhibitory proteins they deliver to these recipient cells 20,23 While the global proteomes of HPV(+) and HPV(-) exosomes were structurally and functionally similar, we identified two small subsets of membrane-associated proteins that were carried exclusively by HPV(+) or HPV(-) exosomes and were not shared. These membrane-associated proteins are of special interest, because of their potential involvement in interactions of tumor-derived exosomes with immune recipient cells. The presence or absence of these proteins in exosomes originating from HPV(+) or HPV(-) tumor cells is likely to have an impact on interactions of TEX with immune recipient cells that result in reprogramming of their functions.
We have previously shown that TEX produced by HPV(+) and HPV(-) tumor cells and co-incubated with primary human immune cells reprogrammed functions of recipient cells.22 These exosomes inhibited activation and proliferation of activated CD8 + T cells or induced their apoptosis; however, only HPV(+) exosomes enhanced expression of antigen-processing machinery (APM) components in dendritic cells (DCs), thus promoting their immune activity.22 This finding suggested that HPV(+) and HPV(-) exosomes differentially modulate interactions with DCs. TEX-driven reprogramming of recipient cells is initiated by exosomes making contact with components of the surface membrane prior to exosome internalization by the recipient cell.24 The cargo TEX carry and to a certain extent also the nature of the recipient cell determine the mechanism responsible for exosome internalization by this cell.24 As protein-protein interactions have been shown to mediate exosome attachment and uptake into cells, the presence of proteins such as CD47 and CD267 in the membrane proteome of HPV(+) exosomes and their absence/paucity in HPV(-) exosomes is highly significant. CD47, highly expressed on most tumor cells, is a ligand of an inhibitory receptor SIRPα (signal regulatory protein alpha).25 It transduces inhibitory signals via SIRPα on macrophages and/or other myeloid cells and inhibits phagocytosis.25 CD47 is referred to as a “do-not-eat-me” molecule, and its role in protecting tumor as well as hematopoietic cells from removal by the RES is well established.26 It has been reported that the presence of CD47 in exosomes limited their clearance by circulating monocytes, and enhanced immune cell uptake and therapeutic efficacy of exosomes in a mouse model of pancreatic cancer.27 The presence of CD47 on all three HPV(+) HNSCC cells as well as HPV(+) exosomes indicates that they are similarly protected from phagocytosis, allowing for prolonged and potentially more immunogenic interactions of tumor antigens these exosomes carry with immune cells. In contrast, the absence of CD47 on HPV(-) tumor cells and HPV(-) exosomes favors their rapid phagocytosis. Remarkably, simple in vitro co-incubation of CD47+ exosomes released by HPV(+) cancer cells showed that the CD47 protein on exosomes had biological activity and, as expected, inhibited exosome uptake/phagocytosis by human monocytes.
CD276 (B7-H3) is a member of the B7 co-stimulatory family with prominent expression in HNSCC28 and other tumor types.29 While its function as a T-cell co-stimulatory protein has been controversial, and it has been viewed by some as a co-inhibitory rather than co-stimulatory molecule,30 its role in modulating immune responses has been well documented.31 Strong expression of CD276 on HPV(+) tumor cells as well as HPV(+) exosomes and its relative paucity on HPV(-) tumor cells and exosomes argues for the existence of more effective immune reactivity in the former. Furthermore, calmodulin detected by MS on HPV(+) exosomes regulates calcium binding and thus immune cell activation.32 Coxsackie/adenovirus receptor (CXADR) is an adhesion molecule facilitating exosome binding to recipient cells.33 The presence of these proteins supportive of immune activities in HPV(+) cells and their exosomes fits well with the hypothesis of more robust, virus-primed anti-tumor immunity in HPV(+) malignancies.
A repertoire of the protein cluster unique for HPV(-) tumors and for TEX they produce is also provocative. Identified proteins play a key role in anti-tumor defense, mostly as negative regulators of immune responses. The MHC class II molecule, HLA-DRA, is a ligand for the T-cell receptor, and its signaling promotes Treg generation.34 Finally, MUC-1, MUC-4, and MUC-16 glycoproteins play a major role in protecting tumor cells from pathogens or adverse events, including chemotherapy or immune interventions, and/or promote tumor growth and migration.35,36 Using in vitro co-incubation of MUC-1+ exosomes we isolated from supernatants of HPV(-) tumor cells we showed in preliminary experiments that these exosomes decreased the sensitivity of tumor cell targets to activated NK cell-mediated lysis, while the HPV(+) and MUC-1−/low exosomes did not protect target tumor cells from lysis. Our data support the conclusion that proteins detected only in HPV(-) exosomes are engaged in the protection of tumor cells either from activated immune cells, pathogens or other forms of injury. As TEX are known to be engaged in autocrine signaling and functional reprogramming of their own activities, the proteomic findings we report here provide a potential explanation for greater resistance of HPV(-) HNSCC to therapy and for poor outcome of this cancer.9,37 Our previous study also showed that the molecular content of TEX, especially the presence of immunoinhibitory proteins, such as PD-L1, correlated with the disease stage and activity in HNSCC.16 TEX have been implicated in mediating resistance to chemotherapies by mechanisms that are not well defined.38 The unique protein content of the TEX cargo, as indicated by our study, might provide a mechanistic underpinning for the resistance of tumors to immune cells or therapies that is mediated by exosomes.
In conclusion, proteomic analysis of TEX from HPV(+) and HPV(-) HNSCC cells identified differential enrichment in membrane-associated TEX proteins that are biologically active and are able to promote immune interactions [HPV(+) exosomes] or protect tumor cells from adverse immune or drug effects [HPV(-) exosomes].
Materials and methods
Cell lines
Three HPV-positive cell lines (UM-SCC-2, UM-SCC-47, UPCI-SCC-90, all gifts from Robert L. Ferris, UPCI) and two HPV-negative cell lines (PCI-13, PCI-30) established, characterized and maintained in the Whiteside laboratory39 were cultured in 150 cm2 cell culture flasks and 25 ml DMEM supplemented with 1% (v/v) penicillin and streptomycin and 10% (v/v) exosome-depleted fetal bovine serum (Gibco, A2720801) at 37°C and an atmosphere of 5% CO2 in air. All cultures were authenticated just prior to the described experiments. The cell expansion range varied from 40% to 80% confluency. After 48–72 h of incubation, supernatants were collected and used for exosome isolation.
Exosome isolation from cell culture supernatants
Culture supernatants were centrifuged at 2,000 x g for 10 min at room temperature (RT) and at 10,000 x g for 30 min at 4°C followed by filtration on 0.22 µm syringe-filters (Millipore, SLGP033RB). Pre-conditioned supernatants were concentrated from 50 ml to 1 ml on Vivacell 100 filter units (Sartorius, VC1042). Aliquots (1 mL) of concentrated supernatants were loaded on mini-SEC columns,40 and exosomes were eluted with PBS. Exosomes were collected in the void volume fraction #4 (1 mL). For some experiments, particularly Western blots, #4 fractions were concentrated using 100,000 MWCO Vivaspin 500 Centrifugal Concentrators (Sartorius, VS0142) by centrifugation at 2,000 x g for 10–15 min. To determine protein concentration in the isolated exosome fraction #4, Pierce BCA protein assay kit (Thermo Scientific, 23227) was used according to the manufacturer’s instructions.
Proteomics analysis
Exosome samples were prepared for proteomic analysis according to a modified FASP41 protocol, as previously reported42 – version with trypsin digestion only. As a consequence, each sample was divided into two peptide fractions eluted at pH 5 and 2, respectively, and analyzed individually. The analysis was performed with the use of Dionex UltiMate 3000 RSLC nanoLC System connected to Q Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific).
Peptides from each fraction (0.5 µg) were separated on a reverse phase Acclaim PepMap RSLC nanoViper C18 column (75 µm × 25 cm, 2 µm granulation) using acetonitrile gradient (from 4 to 60%, in 0.1% formic acid) at 30°C and a flow rate of 300 nL/min (for 230 min). The spectrometer was operated in data-dependent MS/MS mode with survey scans acquired at a resolution of 70,000 at m/z 200 in MS mode, and 17,500 at m/z 200 in MS2 mode. Spectra were recorded in the scanning range of 300–2000 m/z in the positive ion mode. Higher energy collisional dissociation (HCD) ion fragmentation was performed with normalized collision energies set to 25. Protein identification was performed using Swiss-Prot human database with a precision tolerance 10 ppm for peptide masses and 0.08 Da for fragment ion masses.
All raw data obtained for each dataset were imported into Protein Discoverer v.1.4 (Thermo Fisher Scientific) <Thermo raw files> for protein identification and quantification. Fractions derived from each sample (accordingly related to pH 5 and pH 2) were combined before analysis, then Mascot and Sequest engines were used for database searches. Protein was considered as positively identified if at least two peptides per protein were found by both search engines, and a peptide score reached the significance threshold FDR = 0.01 (assessed by the Percolator algorithm); a protein was further considered as “present” if detected in at least one sample of a given type. Abundances of identified proteins were estimated in Proteome Discoverer using Precursor Ions Area detector node, which calculates the abundance of a given protein based on the average intensity of three most intensive distinct peptides for this protein, with further normalization to the total ion current.
Functional analysis of proteomic data based on gene ontology
A list of genes corresponding to identified proteins was annotated at Gene Ontology (GO) using FunRich 3.1.3 software and FunRich internal database.43
Uptake/phagocytosis of exosomes by monocytes
Peripheral venous blood specimens were obtained from normal human volunteers who signed the IRB approved consent form (IRBB #0506140). CD14+monocytes were isolated using AutoMACS and were evaluated for purity by flow cytometry (enrichment >90%). Monocytes (1x105) were co-incubated with different concentrations (2.5, 5, 10 or 20 µg protein) of PKH26-labeled exosomes for 5, 15 or 30 min to establish the optimal time and exosome concentration for uptake and processing by monocytes. Monocytes co-incubated with PBS were used as controls for all experiments. Optimized conditions (10 µg of labeled exosomes, 15 min uptake) were used to compare CD47+ HPV(+) exosomes with CD47− HPV(-) exosomes using flow cytometry. MIF values were compared to those seen with PBS-treated monocytes.
Protection of tumor cells from NK cell-mediated lysis by exosomes
Human NK cells isolated from PBMC by AutoMACS were cultured in IL-2 supplemented medium as previously described.44 The adherent target tumor cells (SCC90 and PCI30) were detached from plastic and were labeled with the CMRA cell tracker dye (Invitrogen). Tumor cell aliquots (5 x 104 cells) were pre-incubated with autologous exosomes (10, 20 or 30 µg protein) for 15 min. Next, effector aNK cells were added to obtain the T:E ratios of 1:0, 1:1 1: 5 and 1:10. Target and effector cells were co-incubated for 2 h at 37°C and then were stained with Annexin V and PI using reagents from a commercial kit. Flow cytometry was performed as described below with the gate set on labeled target cells. Heat-treated tumor cells were used as a positive control for apoptosis. NK cell-mediated apoptosis of HPV(+) and HPV(-) tumor cells co-incubated with autologous exosomes was compared.
Flow cytometry
Cells growing in culture were detached from the flask using TrypLE solution (Gibco, 12604013) and centrifuged at 400 g for 5 min. Supernatants were discarded, and the resuspended cells were washed twice with PBS, counted and dispensed into polystyrene round-bottom 12 × 75 mm2 tubes 200x103 in 200 µL of flow cytometry staining buffer/tube (eBioscience, 00-4222-26). Pre-titered fluorochrome-labeled mAbs (see below for Ab specificity) were added to each tube, and the cells were stained for 30 min in the dark at RT. Next, cells were washed 2X with PBS and suspended in a final volume (200 µL) of PBS. Isotype control Abs were used in all experiments. The data were acquired using GalliosTM flow cytometer (Beckman Coulter) and analyzed with Kaluza 1.5 software.
On-bead flow cytometry was used for exosome staining. First, exosomes were placed on avidin-labeled beads and biotinylated anti-CD63 Ab was added as previously described.17 For flow cytometry-based detection of antigens carried by exosomes coupled to beads, the method described by Morales-Kastresana45 was modified as previously described by us.16
Funding Statement
This work was supported by the Ministerstwo Nauki i Szkolnictwa Wyższego [6839/IA/SN/2018];National Institutes of Health [RO-1CA168628]; National Institutes of Health [R21CA205644]; National Science Centre Poland [2013/11/B/NZ7/01512].
Abbreviations
| EVs | extracellular vesicles |
| HNSCC | head and neck squamous cell carcinoma |
| HPV | human papillomavirus |
| TEX | tumor-derived exosomes |
Acknowledgments
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. This work was supported by the National Science Centre, Poland, Grant 2013/11/B/NZ7/01512 (to MP). In addition, the support was provided by NIH Grants RO-1CA168628 and R21CA205644 (to TLW). SL was supported in part by funds from the Pittsburgh-Essen Partnership.
The authors gratefully acknowledge the contribution by Dr. Chan-Sook Hong, who performed the flow cytometry-based aNK cell assays with exosome-treated tumor cell targets.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Cooper JS, Pajak TF, Forastiere AA, Jacobs J, Campbell BH, Saxman SB, Kish JA, Kim HE, Cmelak AJ, Rotman M, et al. Postoperative concurrent radiotherapy and chemotherapy for high-risk squamous-cell carcinoma of the head and neck. N Engl J Med. 2004;350(19):1937–1944. doi: 10.1056/NEJMoa032646. [DOI] [PubMed] [Google Scholar]
- 2.Argiris A, Li Y, Forastiere A.. Prognostic factors and long-term survivorship in patients with recurrent or metastatic carcinoma of the head and neck. Cancer. 2004;101(10):2222–2229. doi: 10.1002/cncr.20640. [DOI] [PubMed] [Google Scholar]
- 3.Muscat JE, Richie JP Jr., Thompson S, Wynder EL.. Gender differences in smoking and risk for oral cancer. Cancer Res. 1996;56:5192–5197. [PubMed] [Google Scholar]
- 4.Applebaum KM, Furniss CS, Zeka A, Posner MR, Smith JF, Bryan J, Eisen EA, Peters ES, McClean MD, Kelsey KT. Lack of association of alcohol and tobacco with HPV16-associated head and neck cancer. J Natl Cancer Inst. 2007;99(23):1801–1810. doi: 10.1093/jnci/djm233. [DOI] [PubMed] [Google Scholar]
- 5.Mork J, Lie AK, Glattre E, Hallmans G, Jellum E, Koskela P, Møller B, Pukkala E, Schiller JT, Youngman L, et al. Human papillomavirus infection as a risk factor for squamous-cell carcinoma of the head and neck. N Engl J Med. 2001;344(15):1125–1131. doi: 10.1056/NEJM200104123441503. [DOI] [PubMed] [Google Scholar]
- 6.Gillison ML, Koch WM, Capone RB, Spafford M, Westra WH, Wu L, Zahurak ML, Daniel RW, Viglione M, Symer DE, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92(9):709–720. [DOI] [PubMed] [Google Scholar]
- 7.van Houten VM, Snijders PJ, van Den Brekel MW, Kummer JA, Meijer CJ, van Leeuwen B, Denkers F, Smeele LE, Snow GB, Brakenhoff RH. Biological evidence that human papillomaviruses are etiologically involved in a subgroup of head and neck squamous cell carcinomas. Int J Cancer. 2001;93(2):232–235. doi: 10.1002/ijc.1313. [DOI] [PubMed] [Google Scholar]
- 8.Curado MP, Hashibe M. Recent changes in the epidemiology of head and neck cancer. Curr Opin Oncol. 2009;21(3):194–200. doi: 10.1097/CCO.0b013e32832a68ca. [DOI] [PubMed] [Google Scholar]
- 9.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, Westra WH, Chung CH, Jordan RC, Lu C, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363(1):24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leemans CR, Snijders PJF, Brakenhoff RH. The molecular landscape of head and neck cancer. Nat Rev Cancer. 2018;18(5):269–282. doi: 10.1038/nrc.2018.11. [DOI] [PubMed] [Google Scholar]
- 11.Nagel R, Martens-de Kemp SR, Buijze M, Jacobs G, Braakhuis BJ, Brakenhoff RH. Treatment response of HPV-positive and HPV-negative head and neck squamous cell carcinoma cell lines. Oral Oncol. 2013;49(6):560–566. doi: 10.1016/j.oraloncology.2013.03.446. [DOI] [PubMed] [Google Scholar]
- 12.Welters MJP, Ma W, Santegoets SJ, Goedemans R, Ehsan I, Jordanova ES, van Ham VJ, van Unen V, Koning F, van Egmond SI, et al. Intratumoral HPV16-specific T-cells constitute a type 1 oriented tumor microenvironment to improve survival in HPV16-driven oropharyngeal cancer. Clin Cancer Res. 2017. doi: 10.1158/1078-0432.CCR-17-2140. [DOI] [PubMed] [Google Scholar]
- 13.Krishna S, Ulrich P, Wilson E, Parikh F, Narang P, Yang S, Read AK, Kim-Schulze S, Park JG, Posner M, et al. Human papilloma virus specific immunogenicity and dysfunction of CD8+ T cells in head and neck cancer. Cancer Res. 2018;78(21):6159–6170. doi: 10.1158/0008-5472.CAN-18-0163. [DOI] [PubMed] [Google Scholar]
- 14.Nordfors C, Grun N, Tertipis N, Ahrlund-Richter A, Haeggblom L, Sivars L, Du J, Nyberg T, Marklund L, Munck-Wikland E, et al. CD8+ and CD4+ tumour infiltrating lymphocytes in relation to human papillomavirus status and clinical outcome in tonsillar and base of tongue squamous cell carcinoma. Eur J Cancer. 2013;49(11):2522–2530. doi: 10.1016/j.ejca.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 15.Laban S, Hoffmann TK. Human papillomavirus immunity in oropharyngeal cancer: time to change the game? Clin Cancer Res. 2018;24(3):505–507. doi: 10.1158/1078-0432.CCR-17-2991. [DOI] [PubMed] [Google Scholar]
- 16.Theodoraki MN, Yerneni SS, Hoffmann TK, Gooding WE, Whiteside TL. Clinical significance of PD-L1(+) exosomes in plasma of head and neck cancer patients. Clin Cancer Res. 2018;24(4):896–905. doi: 10.1158/1078-0432.CCR-17-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang HG, Grizzle WE. Exosomes: a novel pathway of local and distant intercellular communication that facilitates the growth and metastasis of neoplastic lesions. Am J Pathol. 2014;184(1):28–41. doi: 10.1016/j.ajpath.2013.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whiteside TL. Exosome and mesenchymal stem cell cross-talk in the tumor microenvironment. Semin Immunol. 2017. doi: 10.1016/j.smim.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruivo CF, Adem B, Silva M, Melo SA. The biology of cancer exosomes: insights and new perspectives. Cancer Res. 2017;77(23):6480–6488. doi: 10.1158/0008-5472.CAN-17-0994. [DOI] [PubMed] [Google Scholar]
- 20.Whiteside TL. Exosomes carrying immunoinhibitory proteins and their role in cancer. Clin Exp Immunol. 2017;189(3):259–267. doi: 10.1111/cei.12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honegger A, Leitz J, Bulkescher J, Hoppe-Seyler K, Hoppe-Seyler F. Silencing of human papillomavirus (HPV) E6/E7 oncogene expression affects both the contents and the amounts of extracellular microvesicles released from HPV-positive cancer cells. Int J Cancer. 2013;133(7):1631–1642. doi: 10.1002/ijc.28164. [DOI] [PubMed] [Google Scholar]
- 22.Ludwig S, Sharma P, Theodoraki MN, Pietrowska M, Yerneni SS, Lang S, Ferrone S, Whiteside TL. Molecular and functional profiles of exosomes from HPV(+) and HPV(−) head and neck cancer cell lines. Front Oncol. 2018;8:445. doi: 10.3389/fonc.2018.00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hellwinkel JE, Redzic JS, Harland TA, Gunaydin D, Anchordoquy TJ, Graner MW. Glioma-derived extracellular vesicles selectively suppress immune responses. Neuro Oncol. 2016;18(4):497–506. doi: 10.1093/neuonc/nov170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. 2014;3:1. doi: 10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barclay AN, Brown MH. The SIRP family of receptors and immune regulation. Nat Rev Immunol. 2006;6(6):457–464. doi: 10.1038/nri1859. [DOI] [PubMed] [Google Scholar]
- 26.Weiskopf K. Cancer immunotherapy targeting the CD47/SIRPalpha axis. Eur J Cancer. 2017;76:100–109. doi: 10.1016/j.ejca.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 27.Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA, Lee JJ, Kalluri R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546(7659):498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L, Cao NN, Wang S, Man HW, Li PF, Shan BE. Roles of coinhibitory molecules B7-H3 and B7-H4 in esophageal squamous cell carcinoma. Tumour Biol. 2016;37(3):2961–2971. doi: 10.1007/s13277-015-4132-5. [DOI] [PubMed] [Google Scholar]
- 29.Kang FB, Wang L, Jia HC, Li D, Li HJ, Zhang YG, Sun DX. B7-H3 promotes aggression and invasion of hepatocellular carcinoma by targeting epithelial-to-mesenchymal transition via JAK2/STAT3/Slug signaling pathway. Cancer Cell Int. 2015;15:45. doi: 10.1186/s12935-015-0195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zang X, Thompson RH, Al-Ahmadie HA, Serio AM, Reuter VE, Eastham JA, Scardino PT, Sharma P, Allison JP. B7-H3 and B7x are highly expressed in human prostate cancer and associated with disease spread and poor outcome. Proc Natl Acad Sci U S A. 2007;104(49):19458–19463. doi: 10.1073/pnas.0709802104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Kang FB, Shan BE. B7-H3-mediated tumor immunology: friend or foe? Int J Cancer. 2014;134(12):2764–2771. doi: 10.1002/ijc.28474. [DOI] [PubMed] [Google Scholar]
- 32.Chin D, Means AR. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 2000;10:322–328. [DOI] [PubMed] [Google Scholar]
- 33.Loustalot F, Kremer EJ, Salinas S. Membrane dynamics and signaling of the coxsackievirus and adenovirus receptor. Int Rev Cell Mol Biol. 2016;322:331–362. doi: 10.1016/bs.ircmb.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 34.Costantini F, Barbieri G. The HLA-DR mediated signalling increases the migration and invasion of melanoma cells, the expression and lipid raft recruitment of adhesion receptors, PD-L1 and signal transduction proteins. Cell Signal. 2017;36:189–203. doi: 10.1016/j.cellsig.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 35.Nath S, Mukherjee P. MUC1: a multifaceted oncoprotein with a key role in cancer progression. Trends Mol Med. 2014;20(6):332–342. doi: 10.1016/j.molmed.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li W, Wu C, Yao Y, Dong B, Wei Z, Lv X, Zhang J, Xu Y. MUC4 modulates human glioblastoma cell proliferation and invasion by upregulating EGFR expression. Neurosci Lett. 2014;566:82–87. doi: 10.1016/j.neulet.2014.02.033. [DOI] [PubMed] [Google Scholar]
- 37.Fakhry C, Westra WH, Li S, Cmelak A, Ridge JA, Pinto H, Forastiere A, Gillison ML. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100(4):261–269. doi: 10.1093/jnci/djn011. [DOI] [PubMed] [Google Scholar]
- 38.Guenat D, Hermetet F, Pretet JL, Mougin C. Exosomes and other extracellular vesicles in HPV transmission and carcinogenesis. Viruses. 2017;9(8):E211. doi: 10.3390/v9080211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heo DS, Snyderman C, Gollin SM, Pan S, Walker E, Deka R, Barnes EL, Johnson JT, Herberman RB, Whiteside TL. Biology, cytogenetics, and sensitivity to immunological effector cells of new head and neck squamous cell carcinoma lines. Cancer Res. 1989;49:5167–5175. [PubMed] [Google Scholar]
- 40.Hong CS, Funk S, Muller L, Boyiadzis M, Whiteside TL. Isolation of biologically active and morphologically intact exosomes from plasma of patients with cancer. J Extracell Vesicles. 2016;5:29289. doi: 10.3402/jev.v5.29289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiśniewski JR, Zougman A, Mann M. Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J Proteome Res. 2009;8:5674–5678. doi: 10.1021/pr900748n. [DOI] [PubMed] [Google Scholar]
- 42.Gawin M, Wojakowska A, Pietrowska M, Marczak Ł, Chekan M, Jelonek K, Lange D, Jaksik R, Gruca A, Widłak P. Proteome profiles of different types of thyroid cancers. Mol Cell Endocrinol. 2018;472:68–79. doi: 10.1016/j.mce.2017.11.020. [DOI] [PubMed] [Google Scholar]
- 43.Pathan M, Keerthikumar S, Chisanga D, Alessandro R, Ang CS, Askenase P, Batagov AO, Benito-Martin A, Camussi G, Clayton A, et al. A novel community driven software for functional enrichment analysis of extracellular vesicles data. J Extracell Vesicles. 2017;6(1):1321455. doi: 10.1080/20013078.2017.1321455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hong CS, Sharma P, Yerneni SS, Simms P, Jackson EK, Whiteside TL, Boyiadzis M. Circulating exosomes carrying an immunosuppressive cargo interfere with cellular immunotherapy in acute myelogenous leukemia. Sci Rep. 2017;7(1):14684. doi: 10.1038/s41598-017-14661-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morales-Kastresana A, Jones JC. Flow cytometric analysis of extracellular vesicles. Methods Mol Biol. 2017;1545:215–225. doi: 10.1007/978-1-4939-6728-5_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


