Abstract
Spores of several Bacillus species have long history of consumption and safe use as probiotics and a variety of formulations containing these organisms are available in the global market. Considering the difficulties in the identification of Bacillus species and the poor microbiological quality of many probiotic formulations, we used three up-to-date methodological approaches for analyzing the content of ten formulations marketed in Italy and labeled to contain Bacillus spores. We compared the performance of biochemical tests based on the BCL Vitek2 card and MALDI-TOF mass spectrometry, using 16S rDNA sequencing as the reference technique. The BCL card performed well in identifying all Bacillus probiotic strains as well as the Bruker’s MALDI Biotyper. Nevertheless, the MALDI score values were sometimes lower than those indicated by the manufacturer for correct species identification. Contaminant bacteria (Lysinibacillus fusiformis, Acinetobacter baumannii, Bacillus cereus, Brevibacillus choshinensis, Bacillus licheniformis, Bacillus badius) were detected in some formulations. Characterization of the B. cereus contaminant showed the potential pathogenicity of this strain. Microbial enumeration performed by the plate count method revealed that the number of viable cells contained in many of the analyzed products differed from the labeled amount. Overall, our data show that only two of the ten analyzed formulations qualitatively and quantitatively respect what is on the label. Since probiotic properties are most often strain specific, molecular typing of isolates of the two most common Bacillus species, B. clausii and B. coagulans, was also performed. In conclusion, the majority of the analyzed products do not comply with quality requirements, most likely leading to reduced/absent efficacy of the preparation and representing a potential infective risk for consumers.
Introduction
The genus Bacillus is a phenotypically large, heterogeneous collection of Gram-positive or Gram-variable spore-forming, aerobic or facultative anaerobic bacteria that have undergone considerable reclassification following the advances in molecular biology techniques [1, 2]. For their wide range of physiologic characteristics and ability to produce a multitude of enzymes, antibiotics, and metabolites, Bacillus species are used in many medical, pharmaceutical, agricultural, and industrial processes. Different species produce nutraceuticals such as vitamins (e.g., riboflavin, cobalamin, and inositol) and carotenoids and have been used for the synthesis of several health supplements for human consumption [3–5]. In addition, Bacillus spores have a long history of consumption and safe use as probiotics [6], live microorganisms that when administered in adequate amounts confer a health benefit on the host. Probiotic products containing spores for human or animal use are commercialized in several countries, being widespread in Australia, Asia, USA, South America and Europe [6, 7]. Italy has a long story on the use of spore-based probiotics for human consumption, with a Bacillus clausii spore suspension being available since 1958 [8].
The identification of species in the genus Bacillus by classical methods is often difficult, despite still prevailing in many Microbiology laboratories, due to similarities among closely related species that share a pattern of morphological, biochemical, and genetic characteristics. The use of matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) as a diagnostic technique for Bacillus spp. identification has been applied for addressing the challenges associated with the identification of these organisms [9–15]. However, few data are available on the application of MALDI-TOF MS for the identification of probiotic strains belonging to this genus [16–17].
A number of studies have highlighted the poor microbiological quality of many commercial probiotic formulations in terms of identification, viability, or number of microorganisms [16, 18–26], thus potentially precluding the expected health benefit of the preparation and representing a potential infective risk for consumers. On the other hand, limited data are available on the compositional quality of formulations containing Bacillus spores [27, 28].
In this study, we evaluated qualitative and quantitative aspects of ten probiotic formulations marketed in Italy and containing Bacillus spores. We compared the performance of MALDI-TOF MS (Bruker Daltonics, Bremen, Germany) and biochemical tests based on the Vitek2 BCL card (bioMeriéux, Marcy l’Etoile, France), using 16S rDNA sequencing as reference technique for the identification of the probiotic spore-formers. Since the beneficial health effects of probiotics may be strain-specific [29], molecular typing of strains belonging to the most common species isolated from different formulations was also applied.
Materials and methods
Commercial probiotic formulations and bacterial strains
The formulations analyzed in this study are reported in Table 1. All formulations were purchased in pharmacies by the investigators and investigated before the expiration date. Capsules and lyophilized preparations were dissolved in sterile water immediately before the analyses were performed. Liquid formulations were directly used for analyses. The reference strains Bacillus cereus ATCC 14579, Bacillus clausii ATCC 10317, B. clausii ATCC 21536, B. clausii ATCC 21537, B. clausii DSM 8716, and two Bacillus coagulans from our collection (FLtas1 and FP22) were used as control strains.
Table 1. Probiotic products used in this study.
| Formulation n. | Product | Brand | Batch |
|---|---|---|---|
| 1 | Biogermin vials | Union Health S.r.l. | 40239 |
| 2 | Biolactine family bottles | Sella S.r.l. | L096264 |
| 3 | Enterofermenti family vials | SB Pharma C. | L70316 |
| 4 | Enterogermina 2 mld vials | Sanofi S.p.A. | 1739 |
| 5 | Enterolife vials | Paladin Pharma S.p.A. | L100516 |
| 6 | Ferzym Plus capsules | Specchiasol S.r.l. | 02427 |
| 7 | Lactò Più bottles | Recordati OTC S.p.A. | 0134 |
| 8 | Nucleogermina 10 bottles | Pharmaelle S.r.l. | 020415 |
| 9 | Progermila bottles | Chemist’s Research S.r.l. | 171214 |
| 10 | Progermila bambini bottles | Chemist’s Research S.r.l. | 140316 |
Brand and batch of the analyzed formulations.
Identification of spore-forming probiotic strains
In order to inactivate bacterial vegetative forms, the microbial suspensions of probiotic organisms were thermally treated by exposing to 80°C for 15 min, serially diluted in PBS and seeded (100 μl per plate) on trypticase soy agar with 5% horse blood (TSH, bioMérieux, Marcy l'Etoile, France). Plates were incubated at 37°C in aerobic atmosphere for 24–48 hours and morphologically different colonies were subjected to identification by biochemical analysis using the BCL card of the Vitek2 system (bioMérieux, Marcy l'Etoile, France). This system expresses the identification profiles as the probability (%) of identity between the tested strain and the database taxa. A probability ranging between 96–99% is associated with an excellent confidence level of identification, between 93–95% with a very good level, between 89–92% with a good level, between 85–88% with an acceptable level, and lower probabilities are considered not discriminative. In parallel, bacteria from single colonies were used for Matrix Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS) analysis in a Microflex LT MALDI Biotyper mass spectrometer (Bruker Daltonics, Bremen, Germany). Bacteria were also subjected to identification by 16S rRNA gene sequencing. Experiments were repeated three times in separate days using different doses of the same batches.
In order to separate the four polyantibiotic-resistant Bacillus strains (OC, NR, SIN, T) contained in formulation 4, aliquots of the product were seeded onto Mueller Hinton agar plates with different antibiotics: chloramphenicol (50 μgml-1), rifampicin (50 μgml-1) plus novobiocin (100 μgml-1), streptomycin (200 μgml-1) plus neomycin (100 μgml-1), and tetracycline (100 μgml-1) [8].
MALDI-TOF MS analysis
A colony was directly spotted on the MALDI plate and treated with 1 μl of ethanol, 1 μl of formic acid and 1 μl of acetonitrile for protein extraction, and then overlaid with 1 μl of saturated α-cyano-4-hydroxycinnamic acid (HCCA) matrix solution and air-dried. Each isolate was tested in duplicate. The loaded plate was then placed in the instrument according to the manufacturer’s instructions. The mass spectra were acquired automatically, within 5–10 min, in the positive linear mode at a laser frequency of 60Hz with an acquisition range from 1.960 to 20.000 Da. The spectra were imported into the integrated MALDI Biotyper software (version 3.1) and analyzed by standard pattern matching with a default database.
16S rRNA gene sequencing
Genomic DNA was extracted and purified as previously described [30]. A sequence ranging from nucleotide 9 to nucleotide 1523 of the 1556 bp 16S rRNA gene (97.4%) was amplified with the universal primers 27F (5’-GAGAGTTTGATCCTGGCTCAG-3’) and 1495R (5’-CTACGGCTACCTTGTTACGA-3’). Amplified fragments were purified and sequenced using the same primers (Eurofins MWG Operon, Germany). Sequences were compared with those contained in the Ribosomal Database Project. Identity scores of 97% and 99% were considered for the identification at genus and species level respectively, according to the Clinical and Laboratory Standards Institute (CLSI) guidelines [31].
Bacterial enumeration
Enumeration of spores and viable bacteria was conducted by the plate count method. Microbial suspensions of probiotic organisms were divided into two aliquots and one aliquot was heat-treated at 80°C for 15 min prior to plating. Thermally treated and untreated suspensions were serially diluted in PBS and seeded on TSH. Plating was performed in triplicate and the experiments were repeated three times in separate days using different doses of the same batches. Plates were incubated at 37°C in aerobic atmosphere for 48 h and the number of CFU was determined. Microbial counts were expressed as the mean ± standard deviation.
Evaluation of B. cereus virulence potential
Analysis of the presence of genes encoding typical B. cereus toxins (sphingomyelinase, sph; enterotoxin T, bcet; enterotoxin FM, entFM; enterotoxin S, entS; phosphatidylinositol-specific phospholipase C, plcA; cytotoxin K, cytK; non-hemolytic enterotoxin complex, nheA, nheB, and nheC; component L2 of hemolysin BL hblC) was performed by specific PCR reactions as previously described [32, 33]. Phosphatidylcholine-specific phospholipase C (PC-PLC) activity in culture supernatants was assayed by an agar-diffusion method using 1.5 mg ml−1 l-α-phosphatidylcholine (Sigma-Aldrich S.r.l., Milano) [32]. Gels were incubated 16 h at 25°C and PC-PLC activity was quantitated by comparison of turbidity areas to those in a standard curve for pure PC-PLC (Sigma-Aldrich S.r.l.).
Molecular typing
Randomly amplified polymorphic DNA (RAPD) fingerprinting of bacterial genomes was performed with the primers RPO2 (5′-GCGATCCCCA-3′), M13 (5′-GAGGGTGGCGGCTCT-3′), and Pro-Up (5′-GCTGCTGGCGGTGG-3′) [8], HLWL85 (5'-ACAACTGCTC-3') [34], OPE02 (5’-GGTGCGGGAA-3’), OPE03 (5’-CCAGATGCAC-3’), OPD02 (5’-GGACCCAACC-3’), and OPD03 (5’-GTCGCCGTCA-3’) [35]. PCR conditions were as follows: 30 cycles consisting of 94°C for 1 min, 36°C for 1 min, and 72°C for 2 min, followed by one cycle consisting of 72°C for 10 min [8]. The reproducibility of RAPD profiles was assessed in at least three separate experiments.
Statistical analysis
All numerical data are expressed as mean plus standard deviations and Student’s t-test has been applied as statistical method.
Results
Identification of the spore formers contained in probiotic formulations
Considering the importance of compositional quality for commercial probiotic products and the difficulties in species identification within the genus Bacillus, we first analyzed the formulations reported in Table 1 searching for heat-resistant bacterial forms (i.e. spores). All morphologically different colonies isolated seeding the thermally-treated probiotic suspensions were subjected to biochemical and MALDI-TOF MS identification. Sequencing of the 16S rRNA gene, the reference technique to identify bacterial isolates, was also applied.
Table 2 summarizes the results obtained with all the identification procedures. On the basis of the sequencing results (Table 2), five (products 1, 3, 4, 5, 7) among the ten tested probiotic formulations resulted to contain spores of the Bacillus species declared on the labels. In four other formulations, spores of contaminant species in addition to the labeled ones were found. In particular, Bacillus licheniformis was recovered from product 2, Bacillus badius from 6, Lysinibacillus fusiformis from 9, and Bacillus cereus and L. fusiformis from 10. Formulation 8 was found to contain B. subtilis spores instead of B. clausii spores. The BCL card, which is produced for the automatic identification of the most significant aerobic endospore-forming species of the family Bacillaceae, was able to correctly identify all the Bacillus isolated strains contained in the probiotic formulations (Table 2). Five strains were identified with a very good probability of correct identification, seven with good probabilities, and one with an acceptable probability. On the other hand, the BCL kit misidentified L. fusiformis as Brevibacillus choshinensis. The Bruker Biotyper expresses the identification of an organism as a score based on pattern matching and considers that a score ≥ 2.00 should be obtained for correct identification at the species level. As shown in Table 2, the B. badius, B. cereus, and B. subtilis isolates were correctly identified with scores always higher than the manufacturer’s cut-off. As regards B. clausii isolates, three of the six probiotic strains gave a ≥ 2.00 score in one of the two replicates. Nevertheless, the other three strains were identified with scores ranging from 1.971–1.772 in concordance with the reference method. A similar performance of MALDI-TOF MS was observed for L. fusiformis (score range 2.073–1.763). B. licheniformis and B. coagulans isolates were never misidentified, but the score values were too low for identification at the species level (global score range 1.847–1.366). The four strains contained in formulation 4 were isolated on selective plates and subjected to MALDI-TOF MS analysis. All strains were identified as B. clausii with following scores: NR 2.121/2.011; OC 1.910/1.661; SIN 1.702/1.702; T 1.797/1.693.
Table 2. Identification of the spore-forming bacteria contained in each probiotic formulation.
| Formulation | Labeled organisms | N. of identified species | Biochemical identification BCL(a) | MALDI-TOF MS identification(b) | 16S rDNA sequencing |
|---|---|---|---|---|---|
| 1 | Bacillus clausii | 1 | B. clausii (94%) | B. clausii (1.953/1.934) | B. clausii |
| 2 |
Bacillus coagulans and other bacteriac |
2 |
B. coagulans (92%) B. licheniformis (90%) |
B. coagulans (1.431/1.366) B. licheniformis (1.847/1.732) |
B. coagulans B. licheniformis |
| 3 | Bacillus clausii | 1 | B. clausii (89%) | B. clausii (1.841/1.772) | B. clausii |
| 4 | Bacillus clausii | 1 | B. clausii (95%) | B. clausii (2.121/1.661) | B. clausii |
| 5 | Bacillus clausii | 1 | B. clausii (93%) | B. clausii (2.120/1.913) | B. clausii |
| 6 | Bacillus coagulans and other bacteriad | 2 |
B. coagulans (92%) B. badius (90%) |
B. coagulans (1.635/1.505) B. badius (2.226/2.095) |
B. coagulans B. badius |
| 7 | Bacillus coagulans | 1 | B. coagulans (94%) | B. coagulans (1.745/1.529) | B. coagulans |
| 8 | Bacillus clausii | 1 | B. subtilis (90%) | B. subtilis (2.216/2.184) | B. subtilis |
| 9 |
Bacillus clausii |
2 |
B. clausii (88%) Brevibacillus choshinensis (96%) |
B. clausii (2.140/1.868) Lysinibacillus fusiformis (1.970/1.867) |
B. clausii L. fusiformis |
| 10 |
Bacillus clausii |
3 |
B. clausii (93%) B. cereus (89%) B. choshinensis (96%) |
B. clausii (1.971/1.815) B. cereus (2.207/2.196) L. fusiformis (2.073/1.763) |
B. clausii B. cereus L. fusiformis |
Labeled organisms and identification by biochemical tests, MALDI-TOF MS, and 16S rDNA sequencing of each bacterial isolate contained in the analyzed formulations.
a Probability of correct identification.
b Identification scores of the two replicates.
c Lactobacillus rhamnosus and Lactobacillus helveticus.
d Lactobacillus acidophilus and Bifidobacterium animalis ssp. lactis.
Enumeration of the organisms contained in probiotic formulations
Table 3 reports the labeled number of bacteria belonging to the Bacillus genus, the total counts (total CFU), and the counts of spores (CFU from spores only) obtained for a unit dose of each product after plate counting on TSH medium. Total CFU were concordant with the labeled number of cells for products 1 and 4. Formulations 2, 3, 5, 6, and 7 produced a lower CFU number per unit dose than that declared by the manufacturers. Total CFU originating from products 8, 9, and 10 were 1–3 log higher than those labeled.
Table 3. Enumeration of the spore formers contained in a unit dose of each probiotic formulation.
| Formulation | Unit dose | Labeled Bacillus no. | Total CFU | CFU from spores only |
| 1 | 1 vial | 2 × 109 | 1.4 ± 1.1 × 109 | 5.9 ± 5.0 × 109 |
| 2 | 1 bottle | 4.55 × 109 | 5.6 ± 0.5 × 104 | 5.5 ± 1.5 × 104 |
| 3 | 1vial | 4 × 109 | 2.8 ± 2.2 × 107 | 1.1 ± 0.5 × 107 |
| 4 | 1 vial | 2 × 109 | 1.2 ± 0.9 × 109 | 1.7 ± 0.7 × 109 |
| 5 | 1 vial | 2 × 109 | 3.7 ± 2.3 × 106 | 3.4 ± 2.1 × 106 |
| 6 | 1 capsule | 5 × 109 | 4.8 ± 3.2 × 104 | 2.0 ± 1.7 × 105 |
| 7 | 1 bottle | 3 × 109 | 1.5 ± 1.5 × 108 | 1.1 ± 5.8 × 107 |
| 8 | 1 bottle | 1 × 1010 | 4.3 ± 2.9 × 1011 | 1.6 ± 1.4 × 1012 |
| 9 | 1 bottle | 1 × 1010 | 7.1 ± 1.5 × 1012 | 9.0 ± 0.5 × 1012 |
| 10 | 1 bottle | 5 × 109 | 4.2 ± 3.1 × 1012 | 1.9 ± 1.1 × 1012 |
| Identified species | ||||
| 2 | 1 bottle |
B. coagulans B. licheniformis |
1.2 ± 0.8 × 104 6.0 ± 3.2 × 103 |
4.3 ± 0.8 × 104 8.5 ± 1.5 × 103 |
| 6 | 1 capsule |
B. coagulans B. badius |
1.5 ± 1.3 × 104 3.5 ± 1.5 × 103 |
2.0 ± 1.7 × 105 9.0 ± 3.7 × 102 |
| 9 | 1 bottle |
B. clausii L. fusiformis |
6.4 ± 0.9 × 1012 7.0 ± 0.7 × 1011 |
6.5 ± 0.4 × 1012 2.5 ± 0.2 × 1012 |
| 10 | 1 bottle |
B. clausii B. cereus L. fusiformis |
1.8 ± 1.1 × 1012 1.0 ± 0.6 × 1010 1.3 ± 0.3 × 1011 |
3.4 ± 1.6 × 1012 2.5 ± 0.5 × 1010 7.0 ± 1.0 × 1011 |
Enumeration of the spore formers on TSH medium. For products containing more than one isolate, separate counts are reported.
The contaminant microorganisms present in products 2, 6, 9, and 10 were recovered at high levels, comparable to the labeled Bacillus species (Table 3, lower part). Interestingly, a Gram-negative organism was also isolated on TSH plates (1.2 ± 0.3 × 1011 total CFU/unit dose) from product 8. The strain was identified as Acinetobacter baumannii by MALDI-TOF MS (scores 2.474/2.302).
Since B. cereus is well known as food pathogen, able to cause diarrheal and emetic syndromes, and exhibits virulence in a strain specific manner, the potential pathogenicity of the B. cereus strain isolated from formulation 10 was evaluated. S1 Fig shows the amplification profile of B. cereus virulence genes from this isolate (S lanes). The B. cereus ATCC 14579 reference strain was used as positive control for PCR amplification (C+ lanes). The sph, entT, entS, plcA, and the three nhe genes were found present in the isolated strain. Moreover, by an agar diffusion assay, we demonstrated that the B. cereus isolate secretes 0.04 Uml-1 of PC-PLC, an amount comparable to a low PC-PLC producer [32]. All together, these data indicate that the B. cereus strain isolated from product 10 is potentially toxic for humans.
Molecular typing of B. clausii and B. coagulans isolates
With the aim of evaluating whether the different probiotic formulations contained an identical B. clausii or B. coagulans strain, molecular typing of the isolates of these species was performed by RAPD-PCR. This kind of whole genome fingerprinting has successfully been used to differentiate strains belonging to the genus Bacillus [8, 35, 36].
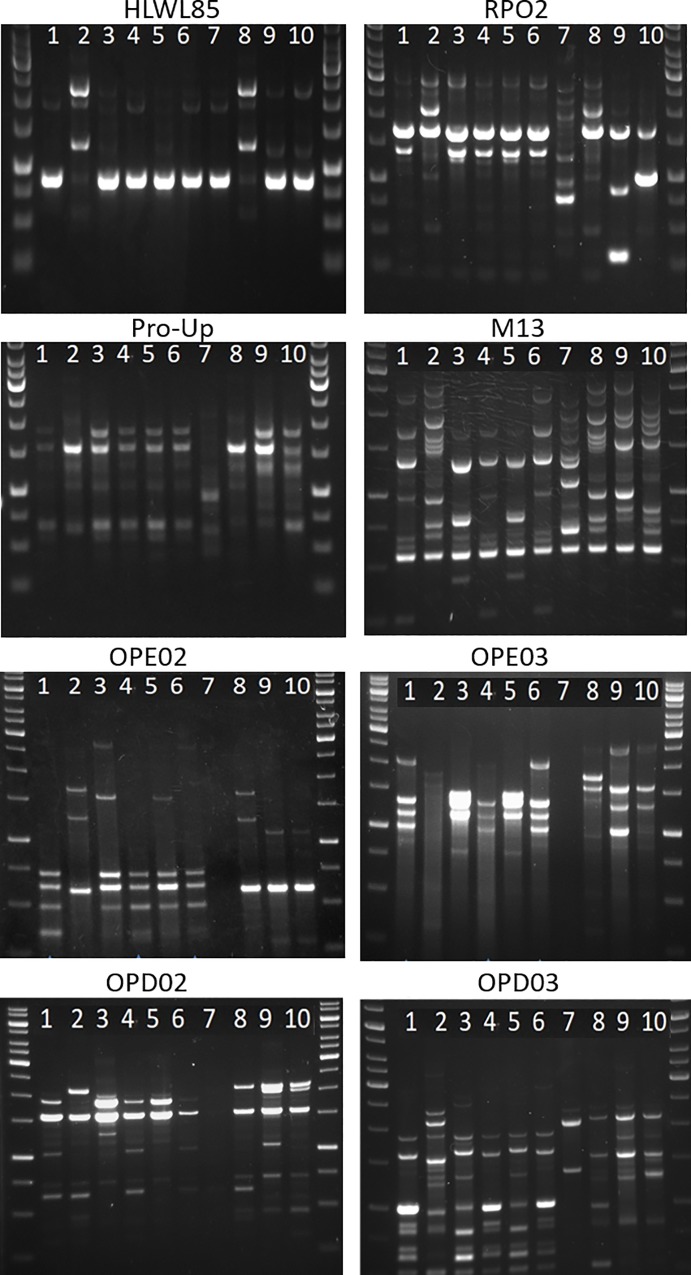
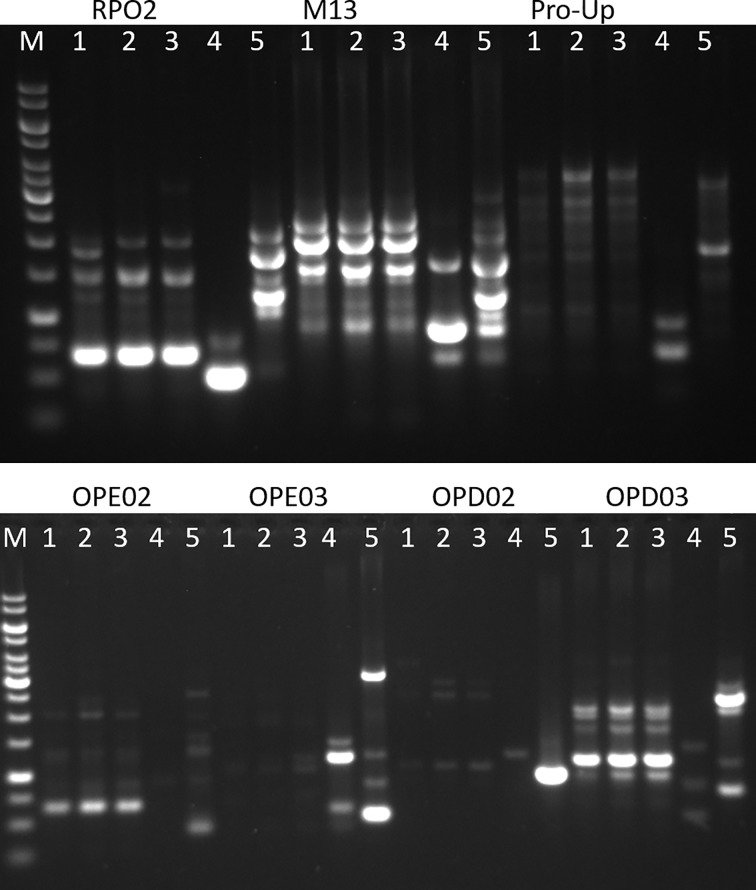
In this study, different primers were used to amplify the genomic DNA of the B. clausii (Fig 1) and B. coagulans isolates (Fig 2). Obtained profiles were compared with those produced from B. clausii (Fig 1) and B. coagulans collection strains (Fig 2). Since previous studies demonstrated the identical profiles obtained by RAPD-PCR amplification of the four B. clausii strains [8], herein we used strain OC as representative of this probiotic formulation.
Fig 1. RAPD-PCR amplification obtained with different primers from B. clausii strains.
Used primer: HLWL85, RPO2, Pro-Up, M13, OPE02, OPE03, OPD02, OPD03. 1: strain OC from formulation 4; 2: strain from formulation 10; 3: strain from formulation 9; 4: strain from formulation 5; 5: strain from formulation 3; 6: strain from formulation 1; 7: ATCC 10317; 8: DSM 8716; 9: ATCC 21536; 10: ATCC 21537.
Fig 2. RAPD-PCR amplification obtained with different primers from B. coagulans strains.
Used primer: RPO2, M13, Pro-Up, OPE02, OPE03, OPD02, OPD03. 1: strain from formulation 2; 2: strain from formulation 6; 3: strain from formulation 7; 4: FLtas1; 5: FP22.
Global analysis of the generated amplification patterns revealed genomic similarities between i) the strain contained in formulation 10 and B. clausii DSM 8716; ii) the strains contained in the formulations 1, 4, and at less extent 5; and iii) the strains contained in the formulations 9 and 3. As regards B. coagulans strains, the three probiotic isolates showed similar amplification profiles. Nevertheless, the strain isolated from formulation 2 produced characteristic bands when the primers RPO2 and OPD02 were used, thus indicating genomic divergences from the other strains.
Discussion
Quality control of probiotic products is the focus of numerous organizations worldwide, with the European Society for Pediatric Gastroenterology Hepatology and Nutrition recently highlighting the importance of a more stringent control of commercialized probiotic products [37]. As established in the Food and Agriculture Organization (FAO) and the World Health Organization (WHO) guidelines, for being used as probiotic it is essential that the organism is considered as GRAS (generally recognized as safe) [38]. The European Food Safety Authority (EFSA) has developed a list of safe biological agents defined as QPS (qualified presumption of safety) for pre-market safety assessment. In 2018, the Italian Ministry of Health published its guidelines on probiotics in which the assessment of the taxonomic position is considered a crucial point to guarantee safety of the used microorganism [39].
The probiotic market is continuously expanding and new products are constantly developed. Formulations are mainly constituted by lactobacilli, bifidobacteria, spore formers, or yeast. In the global nutraceutical and pharmaceutical market, the spore-containing probiotics based on Bacillus spp. are making a major contribution [6] and, in some countries, have a long tradition of use. The genus Bacillus has undergone considerable taxonomic changes and different genetic approaches and biological assays have been developed for differentiating more than 300 species belonging to this genus [40, 41].
Optimizing the identification procedures for probiotic Bacillus strains is essential for the quality control of preparations containing bacterial spores. Three methods based on different approaches (biochemical/metabolic, proteomic, and genetic) were used in this study to identify Bacillus strains contained in probiotic formulations. Our overall results indicate that the biochemical BCL card test performed well in identifying all the Bacillus probiotic strains. Only the contaminants isolated from the formulations 9 and 10 were identified as B. coshinensis instead of L. fusiformis. The results obtained with MALDI-TOF MS were always concordant with 16S rDNA sequencing. Nevertheless, the obtained scores were sometimes lower than the values suggested by the manufacturer’s guidelines for correct species identification, especially when dealing with B. coagulans strains [16]. However, different replicates always produced correct identification, thus suggesting that even low score values could be considered as accurate for the Bacillus species most commonly present in probiotic formulations.
As regards the quality of the analyzed formulations, all the labeled Bacillus species were recovered, with the only exception of product 8 that includes B. subtilis instead of B. clausii. However, contaminant microorganisms were frequently observed. Formulations 2, 6, 9, and 8 contained one contaminant organism and even two additional species were found in product 10. These results raise some concern about quality control in the procedures during preparation of these formulations.
It appears of relevance the finding that most of the isolated contaminants may behave as human pathogens. B. licheniformis is increasingly recognized as cause of serious diseases such as bacteremia, peritonitis, food poisoning and eye infections mainly in immunocompromised patients [42–45]. L. fusiformis, typically isolated from different environments, has occasionally been reported as opportunistic pathogen [46]. A. baumannii is an opportunistic nosocomial pathogen responsible for a vast array of infections with high mortality rate and one of the six most important multidrug-resistant microorganisms in hospitals worldwide. Notably, gut colonization with A. baumannii has been demonstrated to frequently precede bacteremia in critically ill patients [47]. B. cereus is well known to cause foodborne intoxications as well as local and systemic infections in humans [42, 48]. The pathogenic potential of this bacterium is related to the secretion of several virulence proteins such as hemolysins, phospholipases, trimeric toxins (HBL and NHE), and CytK [48–50]. The finding that the B. cereus strain contained in formulation 10 has the potential to produce many virulence factors highlights that the presence of this food pathogen in a probiotic formulation is far for being of negligible importance.
The number of viable cells contained into a probiotic formulation is one of the qualifications that the FAO and the WHO document have recommended [38]. The Italian Ministry of Health guidelines indicate that the minimal amount of a probiotic to be active is 1 × 109 CFU per day [39] since lower numbers of microorganisms could preclude an effective health benefit. In our study, the number of viable cells of the species declared to be contained in the analyzed formulations most often do not comply with the label and the Italian guideline requirements.
The confirmed identity of the microorganism, not only at the species level, but also at the strain level is a prerequisite to ensure that a commercial product will deliver the claimed beneficial health effect. Molecular typing of the most common Bacillus species contained in the tested probiotic Italian products (B. clausii and B. coagulans) indicates the presence of different strains. Therefore, the demonstrated efficacy for one product cannot be automatically translated to another product containing the same Bacillus species.
In conclusion, only two of the ten analyzed formulations (1 and 4) qualitatively and quantitatively respect what is on the label, thus suggesting that probiotic products should have a more stringent quality control process that ensures contents match what is on the label. By the way, only these two formulations are registered as medicinal products and therefore subjected to more rigorous quality controls. As regards formulations containing Bacillus spores, quality controls require trained personnel able to morphologically recognize different bacterial colonies and modern technologies for the identification of these bacteria.
Supporting information
S lanes: B. cereus strain isolated from formulation 10. C+ lanes: B. cereus ATCC 14579 reference strain.
(TIF)
Acknowledgments
The authors pay tribute to the late Professor Mario Campa for his long-standing inspiration.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work received financial support from Sanofi S.p.A. in the form of research materials. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Maughan H, Van der Auwera G. Bacillus taxonomy in the genomic era finds phenotypes to be essential though often misleading. Infect Genet Evol. 2011; 11(5):789–97. 10.1016/j.meegid.2011.02.001 [DOI] [PubMed] [Google Scholar]
- 2.Beveridge TJ. Mechanism of Gram variability in select bacteria. J Bacteriol. 1990; 172(3): 1609–1620. 10.1128/jb.172.3.1609-1620.1990 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohammed Y, Lee B, Kang Z, Du G. Development of a two-step cultivation strategy for the production of vitamin B12 by Bacillus megaterium. Micr Cell Fact. 2014; 13:102 10.1186/s12934-014-0102-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka K, Takanaka S, Yoshida K. A second-generation Bacillus cell factory for rare inositol production. Bioengineered. 2014; 5(5):331–4. 10.4161/bioe.29897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takano H. The regulatory mechanism underlying light-inducible production of carotenoids in non-phototrophic bacteria. Biosci Biotechnol Biochem. 2016; 80:1264–73. 10.1080/09168451.2016.1156478 . [DOI] [PubMed] [Google Scholar]
- 6.Elshaghabee FMF, Rokana N, Gulhane RD, Sharma C, Panwar H. Bacillus as potential probiotics: status, concerns, and future perspectives. Front Microbiol. 2017; 8:1490 10.3389/fmicb.2017.01490 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baccigalupi L, Ricca E and Ghelardi E. Non-LAB Probiotics: Spore Formers From: Venema K and do Carmo AP editors. Probiotics and Prebiotics: Current Research and Future Trends. Caister Academic Press, U.K; 2015. Pages: 93–104. [Google Scholar]
- 8.Senesi S, Celandroni F, Tavanti A, Ghelardi E. Molecular characterization and identification of Bacillus clausii strains marketed for use in oral bacteriotherapy. Appl Environ Microbiol. 2001; 67(2):834–9. 10.1128/AEM.67.2.834-839.2001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.AlMasoud N, Xu Y, Nicolaou N, Goodacre R. Optimization of matrix assisted desorption/ionization time of flight mass spectrometry (MALDI-TOF-MS) for the characterization of Bacillus and Brevibacillus species. Anal Chim Acta. 2014; 840:49–57. 10.1016/j.aca.2014.06.032 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Böhme K, Fernández-No IC, Barros-Velázquez J, Gallardo JM, Cañas B, Calo-Mata P. Rapid species identification of seafood spoilage and pathogenic Gram-positive bacteria by MALDI-TOF mass fingerprinting. Electrophoresis. 2011; 32(21):2951–65. 10.1002/elps.201100217 . [DOI] [PubMed] [Google Scholar]
- 11.Farfour E, Leto J, Barritault M, Barberis C, Meyer J, Dauphin B, et al. Evaluation of the Andromas Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry System for identification of aerobically growing Gram-positive Bacilli. J Clin Microbiol. 2012; 50(8):2702–7. 10.1128/JCM.00368-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Starostin KV, Demidov EA, Bryanskaya AV, Efimov VM, Rozanov AS, Peltek SE. Identification of Bacillus strains by MALDI TOF MS using geometric approach. Scient Rep. 2015; 5:16989 10.1038/srep16989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernández-No IC, Böhme K, Díaz-Bao M, Cepeda A, Barros-Velázquez J, Calo-Mata P. Characterisation and profiling of Bacillus subtilis, Bacillus cereus and Bacillus licheniformis by MALDI-TOF mass fingerprinting. Food Microbiol. 2013; 33(2):235–42. 10.1016/j.fm.2012.09.022 . [DOI] [PubMed] [Google Scholar]
- 14.Hotta Y, Sato J, Sato H, Hosoda A, Tamura H. Classification of the genus Bacillus based on MALDI-TOF MS analysis of ribosomal proteins coded in S10 and spc operons. J Agric Food Chem. 2011; 59(10):5222–30. 10.1021/jf2004095 . [DOI] [PubMed] [Google Scholar]
- 15.Shu L-J & Yang Y-L. Bacillus classification based on matrix-assisted laser desorption ionization time-of-flight mass spectrometry—effects of culture conditions. Scient Rep. 2017; 7(1):15546 10.1038/s41598-017-15808-5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vecchione A, Celandroni F, Mazzantini D, Senesi S, Lupetti A, Ghelardi E. Compositional quality and potential gastrointestinal behavior of probiotic products commercialized in Italy. Front Med. 2018; 5:59 10.3389/fmed.2018.00059 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yadav P, Sharma P, Arora R. Content analysis of commercially available probiotics. Indian Pediatr. 2018; 55(4):344–5. . [PubMed] [Google Scholar]
- 18.Hamilton-Miller JM, Shah S. Deficiencies in microbiological quality and labelling of probiotic supplements. Int J Food Microbiol. 2002; 72:175–6. . [DOI] [PubMed] [Google Scholar]
- 19.Fasoli S, Marzotto M, Rizzotti L, Rossi F, Dellaglio F, Torriani S. Bacterial composition of commercial probiotic products as evaluated by PCR-DGGE analysis. Int J Food Microbiol. 2003; 82:59–70. . [DOI] [PubMed] [Google Scholar]
- 20.Temmerman R, Pot B, Huys G, Swings J. Identification and antibiotic susceptibility of bacterial isolates from probiotic products. Int J Food Microbiol. 2003; 81:1–10. . [DOI] [PubMed] [Google Scholar]
- 21.Drago L, De Vecchi E, Nicola L, Colombo A, Gismondo MR. Microbiological evaluation of commercial probiotic products available in Italy. J Chemother. 2004; 16:463–7. . [PubMed] [Google Scholar]
- 22.Aureli P, Fiore A, Scalfaro C, Casale M, Franciosa G. National survey outcomes on commercial probiotic food supplements in Italy. Int J Food Microbiol. 2010; 137(2–3):265–73. 10.1016/j.ijfoodmicro.2009.12.016 . [DOI] [PubMed] [Google Scholar]
- 23.Brink M, Senekal M, Dicks LMT. Market and product assessment of probiotic/prebiotic containing functional food and supplements manufactured in South Africa. S Afr Med J. 2005; 95:114–9. . [PubMed] [Google Scholar]
- 24.Elliot E, Teversham K. An evaluation of nine probiotics available in South Africa, 2003 August. S Afr Med J. 2004; 94(2):121–4. . [PubMed] [Google Scholar]
- 25.Lin WH, Hwang CF, Chen LW, Tsen HY. Viable counts, characteristic evaluation for commercial lactic acid bacteria products. Food Microbiol. 2006; 23:74–8. 10.1016/j.fm.2005.01.013 . [DOI] [PubMed] [Google Scholar]
- 26.Drisko J, Bischoff B, Giles C, Adelson ME, Rao Raja-Venkitesh S, Mc Callum R. Evaluation of five probiotic products for label claims by DNA extraction and polymerase chain reaction analysis. Dig Dis Sci. 2005; 50:1113–7. . [PubMed] [Google Scholar]
- 27.Patrone V, Molinari P, Morelli L. Microbiological and molecular characterization of commercially available probiotics containing Bacillus clausii from India and Pakistan. Int J Food Microbiol. 2016; 237:92–7. 10.1016/j.ijfoodmicro.2016.08.012 . [DOI] [PubMed] [Google Scholar]
- 28.Hoa NT, Baccigalupi L, Huxham A, Smertenko A, Van PH, Ammendola S, et al. Characterization of Bacillus species used for oral bacteriotherapy and bacterioprophylaxis of gastrointestinal disorders. Appl Environ Microbiol. 2000; 66(12):5241–7. 10.1128/aem.66.12.5241-5247.2000 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venugopalan V, Shriner KA, Wong-Beringer A. Regulatory Oversight and Safety of Probiotic Use. Emerg Infect Dis. 2010; 16:1661–5. 10.3201/eid1611.100574 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Celandroni F, Ghelardi E, Pastore M, Lupetti A, Kolstø AB, Senesi S. Characterization of the chemotaxis fliY and cheA genes in Bacillus cereus. FEMS Microbiol Lett. 2000; 90(2):247–53. 10.1111/j.1574-6968.2000.tb09294.x . [DOI] [PubMed] [Google Scholar]
- 31.CLSI. Interpretative Criteria for Identification of Bacteria and Fungi by DNA Target Sequencing Approved Standard MM18-A. 2008. CLSI, Wayne, PA [Google Scholar]
- 32.Ghelardi E, Celandroni F, Salvetti S, Barsotti C, Baggiani A, Senesi S. Identification and characterization of toxigenic Bacillus cereus isolates responsible for two food-poisoning outbreaks. FEMS Microbiol Lett. 2002; 208(1):129–34. 10.1111/j.1574-6968.2002.tb11072.x . [DOI] [PubMed] [Google Scholar]
- 33.Thaenthanee S, Wong AC, Panbangred W. Phenotypic and genotypic comparisons reveal a broad distribution and heterogeneity of hemolysin BL genes among Bacillus cereus isolates. Int J Food Microbiol. 2005; 105(2):203–12. 10.1016/j.ijfoodmicro.2005.04.003 . [DOI] [PubMed] [Google Scholar]
- 34.Aguado V & García-Jaloń I. Random Amplified Polymorphic DNA typing applied to the study of cross-contamination by Listeria monocytogenes in processed food products. J Food Prot. 2001; 64:716–20. . [DOI] [PubMed] [Google Scholar]
- 35.Khowal S, Siddiqui MZ, Ali S, Khan MT, Khan MA, Naqvi SH, et al. A report on extensive lateral genetic reciprocation between arsenic resistant Bacillus subtilis and Bacillus pumilus strains analyzed using RAPD-PCR. Mol Phylogen Evol. 2017; 107:443–54. 10.1016/j.ympev.2016.12.010 . [DOI] [PubMed] [Google Scholar]
- 36.Daffonchio D, Borin S, Frova G, Gallo R, Mori E, Fani R, et al. Sorlini C. A randomly amplified polymorphic DNA marker specific for the Bacillus cereus group is diagnostic for Bacillus anthracis. Appl Environ Microbiol. 1999; 65(3):1298–303. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kolaček S, Hojsak I, Berni Canani R, Guarino A, Indrio F, Orel R, et al. ESPGHAN working group for probiotics and prebiotics. Commercial probiotic products: a call for improved quality control. A position paper by the ESPGHAN working group for probiotics and prebiotics. J Pediatr Gastroenterol Nutr. 2017; 65(1):117–24. 10.1097/MPG.0000000000001603 . [DOI] [PubMed] [Google Scholar]
- 38.Food and Agriculture Organization/World Health Organization. Guidelines for the Evaluation of Probiotics in Food. London, ON: Food and Agriculture Organization of the United Nations/World Health Organization; (2002). [Google Scholar]
- 39.Ministero della Salute, Direzione Generale per l’Igiene e la Sicurezza degli Alimenti e la Nutrizione. Linee Guida su Probiotici e Prebiotici, Revisione marzo 2018.
- 40.Alcaraz LD, Moreno-Hagelsieb G, Eguiarte LE, Souza V, Herrera-Estrella L, Olmedo G. Understanding the evolutionary relationships and major traits of Bacillus through comparative genomics. BMC Genomics. 2010; 11:332 10.1186/1471-2164-11-332 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y, Lai Q, Dong C, Sun F, Wang L, Li G, et al. Phylogenetic diversity of the Bacillus pumilus group and the marine ecotype revealed by multilocus sequence analysis. PLoS ONE. 2013; 8(11):e80097 10.1371/journal.pone.0080097 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Celandroni F, Salvetti S, Gueye SA, Mazzantini D, Lupetti A, Senesi S, et al. Identification and pathogenic potential of clinical Bacillus and Paenibacillus isolates. PLoS One. 2016; 11(3):e0152831 10.1371/journal.pone.0152831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeon YL, Yang JJ, Kim MJ, Lim G, Cho SY, Park TS, et al. Combined Bacillus licheniformis and Bacillus subtilis infection in a patient with esophageal perforation. J Med Microbiol. 2012; 61(Pt 12):1766–9 10.1099/jmm.0.042275-0 [DOI] [PubMed] [Google Scholar]
- 44.Park DJ, Yun JC, Baek JE, Jung EY, Lee DW, Kim MA, et al. Relapsing Bacillus licheniformis peritonitis in a continuous ambulatory peritoneal dialysis patient. Nephrology. 2006; 11:21–2. 10.1111/j.1440-1797.2006.00539.x . [DOI] [PubMed] [Google Scholar]
- 45.Padhi TR, Sharma S, Das S, Das T. Bacillus licheniformis as a cause of delayed-onset recurrent pseudophakic endophthalmitis-a rare case report. Retin Cases Brief Rep. 2012; 6(1):43–5. 10.1097/ICB.0b013e3182051df9 . [DOI] [PubMed] [Google Scholar]
- 46.Wenzler E, Kamboj K, Balada-Llasat JM. Severe Sepsis Secondary to Persistent Lysinibacillus sphaericus, Lysinibacillus fusiformis and Paenibacillus amylolyticus Bacteremia. Int J Infect Dis. 2015; 35:93–5. 10.1016/j.ijid.2015.04.016 . [DOI] [PubMed] [Google Scholar]
- 47.Thom KA, Hsiao WWL, Harris AD, Stine OC, Rasko DA, Johnson JK. Patients with Acinetobacter baumannii bloodstream infections are colonized in the gastrointestinal tract with identical strains. American journal of infection control. 2010; 38(9):751–3. 10.1016/j.ajic.2010.03.005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeßberger N, Krey VM, Rademacher C, Böhm ME, Mohr A-K, Ehling-Schulz M, et al. From genome to toxicity: a combinatory approach highlights the complexity of enterotoxin production in Bacillus cereus. Front Microbiol. 2015; 6:560 10.3389/fmicb.2015.00560 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Senesi S, Ghelardi E. Production, secretion and biological activity of Bacillus cereus enterotoxins. Toxins (Basel) 2010; 2(7):1690–703. 10.3390/toxins2071690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramarao N, Sanchis V. The pore-forming haemolysins of Bacillus cereus: a review. Toxins. 2013. June 5(6):1119–39. 10.3390/toxins5061119 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S lanes: B. cereus strain isolated from formulation 10. C+ lanes: B. cereus ATCC 14579 reference strain.
(TIF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.