Figure 10.
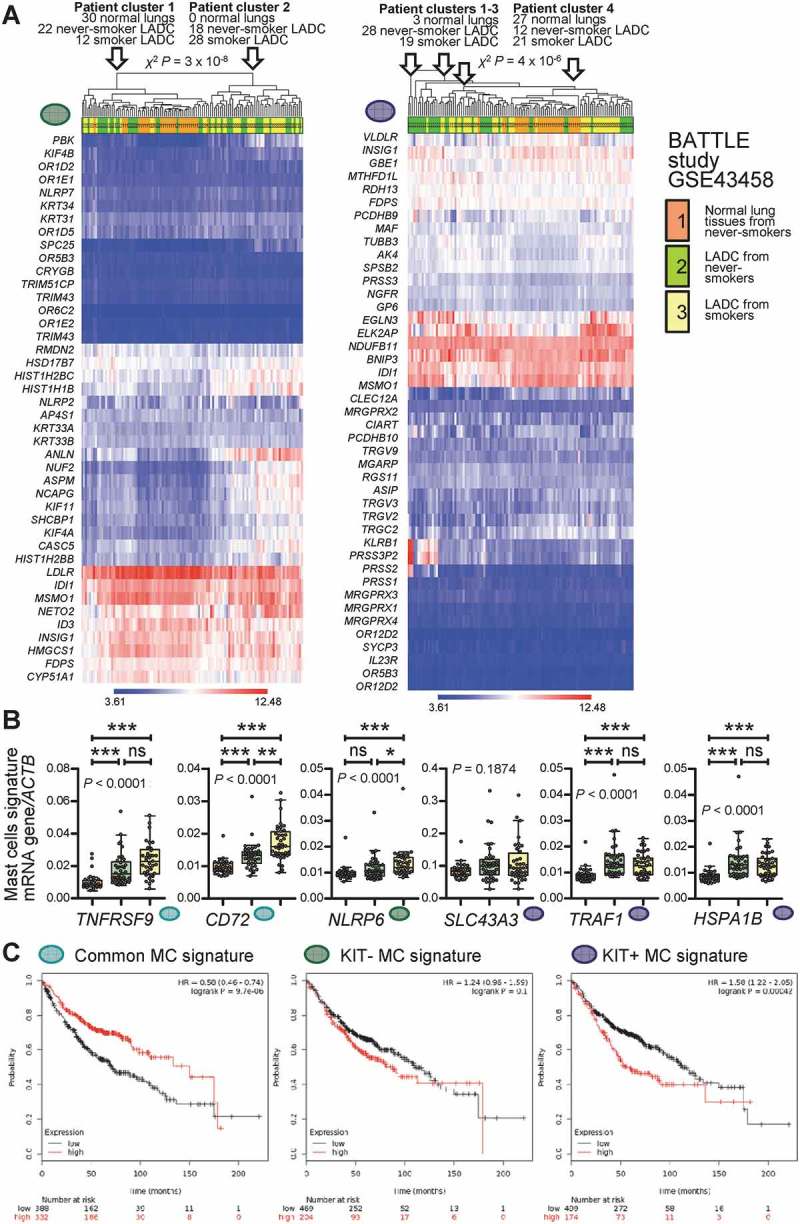
Mast cell signatures in human lung adenocarcinoma.
A Unsupervised clustering of 30 normal lung tissues from never-smokers (orange), 40 lung adenocarcinoma (LADC) tissues from never-smokers (green), and 40 LADC tissues from smokers (yellow) from the Biomarker-integrated Approaches of Targeted Therapy for Lung Cancer Elimination (BATTLE) study [Gene Expression Omnibus (GEO) dataset GSE43458; freely available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE43458]51 by the humanized KIT-independent (left) and KIT-dependent (right) mast cell (MC) signatures from Figure 9D. Both signatures could accurately discriminate normal lung from LADC tissues. P, exact χ2 probability values calculated at http://courses.atlas.illinois.edu/spring2016/STAT/STAT200/pchisq.html. B Data summary of gene expression of the transcripts from Figure 9C normalized to ACTB expression in the BATTLE study validates five of the six genes. Color code is as in Figure 10A. Data are shown as Tukey’s whiskers (boxes: interquartile range; bars: 50% extreme quartiles), raw data points (dots), and Kruskal–Wallis ANOVA P values. ns, *, **, and ***: P> 0.05, P< 0.05, P< 0.01, and P< 0.001, respectively, for the indicated comparisons by Dunn’s post-tests. C Overall survival of patients with LADC stratified by low (black lines) or high (red lines) average expression of MC signatures from Figure 9D. Data from http://kmplot.com/analysis/index.php?p=service&cancer=lung.54 Note that exclusively high expression of the KIT-dependent MC signature correlates with poor survival (A). Shown are Kaplan–Meier survival estimates with hazard ratios (HR) for high compared with low signature expression with their 95% confidence intervals, as well as log-rank test P values.
