Abstract
Sliding clamp proteins encircle duplex DNA and are involved in processive DNA replication and the DNA damage response. Clamp proteins are ring-shaped oligomers (dimers or trimers) and are loaded onto DNA by an ATP-dependent clamp-loader complex that ruptures the interface between two adjacent subunits. Here we measured the solution dynamics of the human clamp protein Proliferating Cell Nuclear Antigen (PCNA) by monitoring the change in the fluorescence of a site-specifically labeled PCNA protein construct. To unravel the origins of clamp subunit interface stability, we carried out comprehensive comparative analysis of the interfaces of seven sliding clamps. We used computational modeling (MD simulations and MM/GBSA binding energy decomposition analyses) to identify conserved networks of hydrophobic residues critical for clamp stability and ring-opening dynamics. The hydrophobic network is shared among clamp proteins and exhibits a “key in a keyhole” pattern where a bulky aromatic residue from one clamp subunit is anchored into a hydrophobic pocket of the opposing subunit. Bioinformatics and dynamic network analyses showed that this oligomeric latch is conserved across DNA sliding clamps from all domains of life and dictates the dynamics of clamp opening and closing.
INTRODUCTION
DNA polymerase holoenzyme is involved in the rapid and accurate replication of genomic DNA during cell division [1]. The polymerase holoenzyme complex is formed by tethering of the polymerase to a sliding clamp – an accessory protein, which encircles primer- template DNA as a closed ring [2, 3]. This topological link to the DNA substrate substantially increases the polymerase processivity [3, 4]. In addition to their critical role in replication, sliding clamps are essential in the DNA damage response (DDR), serving as mobile platforms for the recruitment of DNA repair enzymes and other DDR participants to sites of DNA damage [2, 4–6].
Sliding clamps are functionally conserved from prokaryotes to phages, archaea and higher eukaryotes [3]. In all these organisms, clamp proteins oligomerize to yield remarkably similar toroid shapes (rings), capable of encircling duplex DNA [5, 7]. Most clamps are formed by the oligomerization of two or three subunits, each comprised of two domains connected by an interdomain connector loop (IDCL). This results in an overall clamp architecture (Figure S1) with pseudo six-fold rotational symmetry [4, 8]. One notable exception is the β-clamp, which is a homodimer rather than a trimer [9]. Clamps from T4 phage (gp45), eukaryotes (PCNA) and archaea (PCNA) all feature three equivalent subunits. There are also examples of heterotrimeric clamps: the Rad9-Hus1-Rad1 (9-1-1, checkpoint) clamp and archaeal PCNA from Sulfolobus solfataricus. The large inner diameter of the ring-shaped clamps allows them to slide along duplex DNA without directional bias [4].
Since sliding clamps form stable closed rings on duplex DNA, an activated ATP-driven mechanism must exist for their initial opening and loading onto the primer-template. This function is carried out by the cognate clamp-loader ATPases [2, 5, 10–13]. T4 gp45, PCNA, 9-1-1 and the β clamps are loaded by the following clamp-loader complexes: gp44/62, replication factor C (RFC), Rad17-RFC and γ-complex, respectively [2, 5, 10–13]. These proteins belong to the family of AAA+ ATPases and are composed of one large and four small subunits with the subunit interfaces harboring the ATP binding sites. In the first stage of the clamp-loading cycle the ATP-bound form of the clamp loader associates with the functional proximal face of the sliding clamp [2, 5, 10–13]. In turn, this causes one of the clamp subunit interfaces to open. In the latter stages of the clamp loading cycle, the ATP hydrolysis activity of the clamp loader/clamp complex is triggered upon DNA binding causing a conformational change in the clamp loader/clamp complex [10, 12, 14, 15]. The resulting conformational change induces the closure of the ring-open clamp around double-stranded DNA (dsDNA) followed by the eventual dissociation of the clamp onto DNA from the complex [16].
In this work, the solution dynamics of human PCNA were examined by monitoring the fluorescence signal from a PCNA protein site-specifically labeled at the subunit interface. Previous structural and computational studies of DNA sliding clamps had led to the proposal that ring opening and closing involved the disruption and restoration of a hydrogen bonding network between two antiparallel β strands forming part of the clamps’ subunit interface [17, 18]. Here we report that in addition to this well–known hydrogen-bonding network, a cluster of hydrophobic residues centered on residue Tyr114 also plays a critical role in maintaining the structural integrity of the PCNA clamp ring. We performed comprehensive comparative analysis of the interfaces of seven sliding clamps from all domains of life using computational modeling (MD simulations, MM/GBSA binding energy decomposition analysis, in silico alanine scanning with the Rosetta package and dynamic network analysis). We showed that despite the low overall sequence conservation among clamp proteins, the identified hydrophobic residue network is highly conserved. Next, we determined the energetic contributions from all interfacial residues to identify the most critical contributors to clamp subunit interface stability. We showed that the identified hydrophobic cluster is necessary for clamp oligomerization and for the maintenance of the ring-shaped architecture required for clamp function.
RESULTS
Generation of RFC and PCNA Proteins
Functional hetero-pentameric RFC complex with the full length RFC1 (large) subunit is difficult to purify, involving multiple steps of purification and a very low yield. The yeast RFC protein retained the activity of the wild-type protein when the N-terminal region (residues 1–273) was deleted [14, 19]. Previous analysis of the large p140 (RFC1) subunit of human RFC also revealed that deletion of its N-terminal DNA binding domain did not affect the activity of the wild-type RFC complex [20–22], leading us to generate a truncated RFC1Δ555 construct. This complex composed of RFC1Δ555 and the RFC2,3,4,5 subunits was co-expressed and purified in three-steps in greater yield than the RFC complex (Figure S2b).
Human wild-type PCNA contains six cysteines. Two of the cysteine thiols were determined to be reactive using DTNB assay (Figure S4a). After examination of the crystal structure of human PCNA the two reactive cysteines were assigned to the surface exposed Cys27 and Cys62 (Figure S4b). As these two cysteines are not located at the subunit interface, they are not amenable to labeling to probe the subunit interface dynamics. Cys27 and Cys62 were then mutated to Ser or Met. Of the two C27S/C62S and C27M/C62M mutants generated, the Met mutants gave soluble proteins. The other four cysteines at positions 81, 135, 148, and 162 were converted to Ser. Residues Asn107 or Thr185 was mutated to Cys to introduce fluorescent dye to probe subunit interface dynamics. An AEDANS or fluorescein probe was introduced at the subunit interface at either the C107 or C185 position. Unlike T4 gp45 that carries a native Trp near its subunit interface [15, 23, 24], in human PCNA the native Trp is at position 28 buried in the interior of the protein. However, to remove potential interference, we converted Trp at position 28 to Phe. A Trp residue was then selectively introduced at the subunit interface at either position 185 or 107, to obtain a FRET pair with the AEDANS labeled proteins (Figure S1a, 4b). These mutations are distal to the functional anterior face of the clamp (Figure 1a) and therefore would not interfere with RFC or polymerase binding. The two PCNA mutants then had FRET pair positions (N107W/T185C-AEDANS or N107C-AEDANS/T185W); those without the FRET donor or acceptor served as controls.
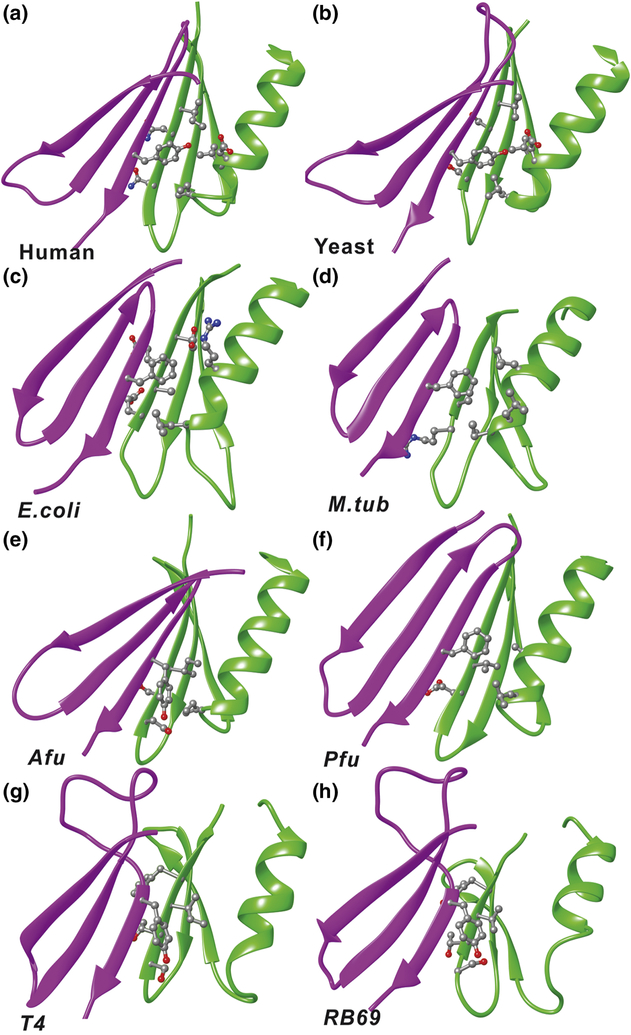
Figure 4. Residue interactions of the anchoring tyrosine (phenylalanine) in the crystal structures.
Side chains of relevant residues are shown in ball and stick representation for: a) Human PCNA; b) Yeast PCNA; c) E. coli β-clamp; d) M.tub clamp; e) Afu PCNA; f) Pfu PCNA; g) T4 gp45 clamp; h) RB69 gp45 clamp. The subunits containing the tyrosine or phenylalanine are shown in purple and the opposing subunits are in green.
Figure 1.
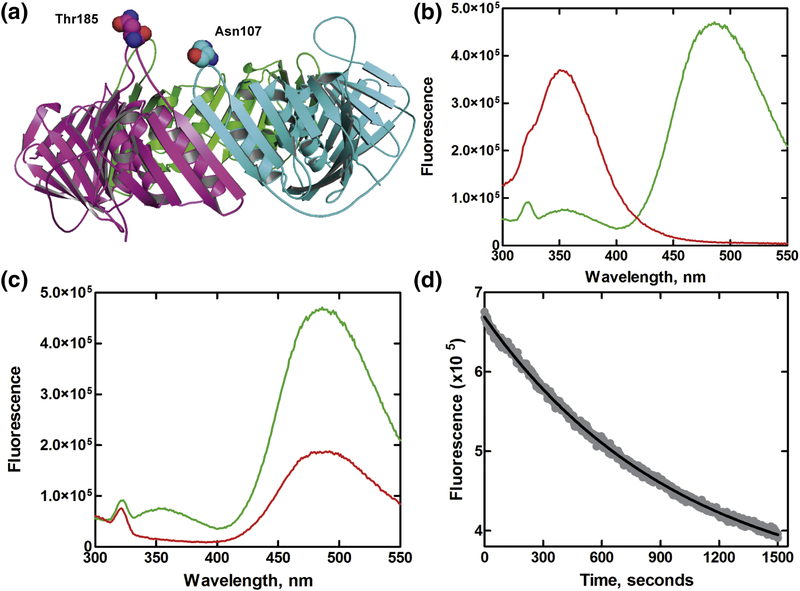
(a) Structure of homotrimeric human PCNA. Side chains of residues Thr185 and Asn107 located at the distal side of the clamp chosen for introducing FRET pair are shown as spheres. (b) Emission spectra of 200 nM PCNA-WC1 mutant with Trp-AEDANS FRET pair (green) and without acceptor (red) excited at 290 nm in reaction buffer (pH 7.8) at 25 °C. (c) Emission spectra of 200 nM PCNA-WC1-AEDANS carrying the FRET pair (green) and without the donor PCNA-C (red) excited at 290 nm in reaction buffer (pH 7.8) at 25 °C. (d) Exchange kinetics of human PCNA subunits by steady state FRET measurements. The exchange reactions of PCNA subunits were followed at 484 nm using PCNA-WC1-AEDANS (200 nM) immediately after the addition of 2μmM 03BCPCNA-W28F in reaction buffer (pH 7.8) at 25 °C. The solid line represents the fit of the experimental data to a single exponential equation.
PCNA proteins were assayed for the stimulation of the ATPase activity of RFC employing the NADH-oxidation coupled enzymatic assay [25]. The ATPase activity of RFC was stimulated by ~10-fold (55.2 ± 6.5 nM/s) by PCNA or its various mutants in the presence of a forked DNA above its basal rate of 6 nM/s (Figure S3). These observations are in good agreement with analogous results for the T4 gp45 [15, 23, 26–28] and yeast clamps [14, 19, 22]. Stimulation of the ATPase activity of RFC by either the labeled or unlabeled PCNA proteins did not differ significantly from the wild-type proteins as evident from their relative rate of ATP hydrolysis (Table S2). The functional integrity of the mutant clamps was confirmed from PCNA-mediated primer extension reactions of Pol δ polymerase holoenzyme reconstituted from Pol δ, RPA, PCNA and RFC complex as previously reported [29], where the wild-type and mutant PCNA proteins are similar in their enhancement of primer extension assay by the Pol δ holoenzyme (Figure S5).
Human PCNA clamp ring is closed in solution
In the crystal structures of the clamp protein, the two residues chosen for FRET pairs (T185 and N107) are separated by 8–12 Å from the backbone Cα atoms [30]. The presence of a FRET donor (W185 or W107) and acceptor (C107- or C185-AEDANS) pair was used to monitor the distance across the PCNA subunit interface in solution. We recorded the FRET energy transfer after excitation at 290 nm from the Trp residue to the dansyl moiety on the adjacent subunit. The presence of a strong FRET signal between Trp and AEDANS was confirmed from the concomitant drop in donor emission and increase in acceptor emission (Figure 1b). A different trend was observed for proteins lacking either the FRET donor (a weak signal seen from direct excitation of dansyl probe) or the acceptor (enhanced fluorescence of Trp) (Figure 1c). The distance between W185 or W107 and C107-or C185-AEDANS was determined for the Trp- AEDANS FRET pair using experimental parameters as described previously for yeast [19] and phage T4 clamps [15, 24], and yielded a Förster distance r of 20–24 Å for either of the FRET pair bearing proteins, consistent with R0 values for a Trp and dansyl pair [31]. This calculated distance agrees with the distance measured between the Cα atoms of residues 185 and 107 in the crystal structure of human PCNA, suggestive of a closed clamp in solution.
The intrinsic stability of the trimeric PCNA ring was determined by examining the subunit dissociation exchange kinetics. We examined the clamp solution dynamics using FRET measurements of PCNA carrying the Trp-AEDANS FRET pair in the presence of a 10-fold excess of PCNA mutant without a Trp (W28F) and AEDANS. This trap protein was added to monitor the exchange of subunits as the FRET signal would decrease upon exchange of the subunits labeled with FRET dyes with the clamp monomers lacking the FRET probes (Figure S6). A distribution of the trimers with varying proportions of FRET pair labels would be obtained upon mixing a clamp with the FRET pair and a clamp without fluorescent donor. The resultant time traces from the clamp exchange were fit to obtain rate constant of subunit exchange, kex = 12.5±3.2 × 10−4 s−1 (Figure 1d). This is ~10-fold slower than the subunit exchange seen for the phage T4 gp45 (9.9± 0.9 × 10−3 s−1) which is partially open in solution [23]. In the presence of RFC and ATP, the rate of subunit exchange showed only a small reduction even after correcting for the contribution of Trp fluorescence from RFC (Table S4) and further addition of end blocked DNA showed no evidence of further reduction as seen in the case of phage T4 [23]. Addition of DNA and polymerase slowed down the rate of exchange slightly suggestive of a stable clamp upon formation of a polymerase holoenzyme complex on DNA.
The specific fluorescence of the PCNA WC1-AEDANS protein at 480 nm upon 290 nm excitation was monitored to evaluate the oligomerization of the clamp protein. There was a dose dependent drop in specific fluorescence with reducing protein concentration (Figure S7). The data were fit to either a cooperative or a non-cooperative model, and the fit could not distinguish between a non-cooperative model with Kd2 = 20 ± 6 nM and Kd1 = 60 ± 14 nM and a cooperative model with Kd2 Kd1=1600±300 nM2A recent report has confirmed that E. coli β-clamp and yeast PCNA clamp oligomerize in a cooperative manner [34]. Our data, in combination with these observations, supports a cooperative model for human PCNA oligomerization.
Mapping the network of interactions at the clamp subunit interface
We performed multiple sequence alignment (Figure S8, Table S3) on four sets of clamp protein sequences: eukaryotes, prokaryotes, archaea and bacteriophages. Eukaryotic clamps exhibited only two highly conserved regions, domain A and domain B that make up the antiparallel β-sheets at the clamp subunit interfaces (Figure S9 & S10). Domain A was composed of residues Lys110, Val112, Ser112, Asp113, Tyr114, Glu115 (KVSDYE). Domain B included residues Gly176, Asn/Thr177, Gly178, Asn179, Ile180, Lys181 (GN/TGNIK). One exception to this conservation pattern was the yeast PCNA clamp with a varying microenvironment generated by semi-conserved residues at the subunit interface, where domain A was reorganized with residues Arg110, Ile111, Ala112, Glu113, Tyr114, Ser115 and domain B with semi-conserved Gly176, Ser177, Gly178, Ser179, Val180, Ile181. Despite the sequence variation in these domains, Tyr114 from domain A and Ile/Val180 from domain B are highly conserved (Figure S8). In addition to the interfacial β-strands, two groups of hydrophobic residues were highly conserved: (i) Phe103, Asp150, Leu151 (indicated by a blue arrow in Figure S8) and, (ii) Ile/Phe154, Ile/Val180. In prokaryotes, the conserved residues that make up the hydrophobic network of the dimeric clamps include Ile78, Phe106, Ile/Leu272 and Leu/Val (Figure S9). In phages Tyr165 plays the role of the large hydrophobic residue at the subunit interface and the hydroxyl group of the side chain of Tyr165 maintains an interaction with Gln125 (Figure S9). While Tyr165 was oriented pointing away from the interface other hydrophobic residues, e.g. Phe91, Leu66 and Leu133 come together to serve an analogous functional role ensuring a stable subunit interface.
Next, we separated the available clamp structures into two groups: trimeric versus dimeric clamps. The trimeric clamps were structurally aligned and overlaid to map out the conserved hydrophobic network of residues at the subunit interfaces. The “anchoring” hydrophobic residue Tyr114 is strongly conserved structurally in H. sapiens, S. cerevisiae and Drosophila melanogaster. In A. fulgidus, H. volcani and P. furiosus the role of the “anchoring” hydrophobic residue is played by a conserved Tyr106, Tyr106 or Phe111, respectively (Figure S9). Importantly, the anchoring residue always faces a pocket of conserved hydrophobic residues across the clamp subunit interface. We found an analogous network of hydrophobic interactions also in the homodimeric β-clamp structures from E. coli and M. tuberculosis (Figure S9). The interaction in E. coli is mediated by residues Phe106, Ile89, Leu273 and Ile272, and by residues Phe116, Ile78, Val290 and Leu259 in M. tuberculosis (Figure S9). In the clamp proteins from phages T4 and RB69, the residue Tyr165 is still conserved. However, its orientation is different from the one found in clamps from other organisms, which may explain the differential stability of phage clamps. In phages, the hydroxyl group of the side chain of Tyr165 maintains an interaction with Gln125 on the same clamp subunit.
Previously we had identified the hydrogen-bonding network across the PCNA subunit interface that ensures assembly of a closed ring and is essential for the clamp’s function [17]. In addition to this network, we found a well-organized cluster of hydrophobic residues on the interior side of the PCNA subunit interface, involving residues Tyr114, Phe103, Ile180, Ile147, Asp150 and Ile154 (Figure S10). Importantly, Tyr114 functions as an anchoring residue for this cluster by inserting itself into a hydrophobic pocket created by residues Ile180, Ile154, Ile147 and Asp150 from the opposing subunit (Figure S10). This “key in a key lock” arrangement, illustrated in Figure S9, appears functionally important for ensuring correct PCNA oligomerization [35]. Furthermore, we showed that the residues forming the Tyr114 hydrophobic cluster are highly conserved despite the overall low sequence similarity among archaeal, eukaryotic and prokaryotic clamps.
Binding energy contributions to clamp interface stability
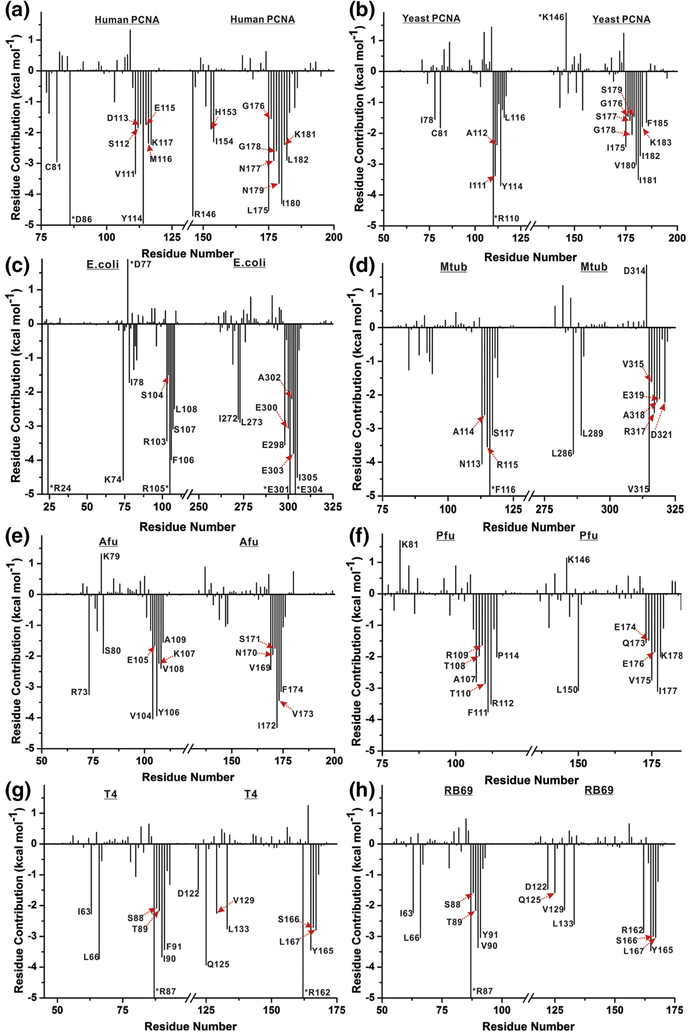
To determine binding energy contributions from interfacial residues to the overall stability of clamp interfaces (in human PCNA, 9-1-1 repair clamp [17], S. cerevisiae PCNA and E. coli β-clamp [18]) we employed MM/GBSA calculations. Like all endpoint free energy methods, MM/GBSA estimates the binding free energy ΔGb of a protein complex by sampling separately the bound and unbound states. The difference in computed van der Waals and electrostatic interactions between the protein partners and the respective solvation free energies of the bound and unbound states serve to estimate ΔGb, providing information on the origins of macromolecular structural stability. Furthermore, the enthalpic contributions to ΔGb can be subdivided into per-residue (one-dimensional (1-D) decomposition) or pairwise residue-residue contributions (two-dimensional (2-D) decomposition). The 1-D decomposition informs how much an individual residue interacts with all other residues across a binding interface and, thus, can be used to identify the hot-spot residues in protein–protein epitopes. The 2-D decomposition shows the contribution of each interfacial residue pair. Thus, 1-D decomposition allows us to quickly identify the important residues contributing the overall interface stability, while the 2-D decomposition provides a more detailed look at the interactions of the most important (top ranked) residues. The 1-D binding energy decomposition results for our clamp proteins highlight the importance of Tyr114 in human and yeast PCNA and the equivalent Phe106 residue in the E. coli β-clamp. Extending these calculations to clamps from M. tub (bacteria), T4 and RB69 (phage), Afu and Pfu (archaea) show similar outcomes. Specifically, the one-dimensional energy decomposition (Figure 2a–h) demonstrated the significance of conserved hydrophobic residues (Tyr or Phe) that align to the Tyr114 residue from the human sequence (Figure S8, Table S3). The dominance of these residues becomes even more obvious if we take into account that implicit representation of the solvent is likely to systematically overestimate charged residue contributions (e.g. from salt bridges) computed with MM/PBSA (MM/GBSA). By contrast, in explicit solvent these interactions would be attenuated leaving the hydrophobic interactions to play a more prominent role. Individual per residue binding energies are sorted (Table 1), showing that Tyr114 (or the equivalent Phe) always appear among the highest ranked residues.
Figure 2. Origins of interface stability for different clamp interfaces from 1-D MM/GBSA decomposition analysis.
Binding energy contributions per residue are shown for a) Human PCNA; b) Yeast PCNA; c) E. coli β-clamp; d) M.tub β-clamp; e) Afu PCNA; f) Pfu PCNA; g) T4 gp45; h) RB69 gp45 clamp. Only residues contributing above a ±1.5 kcal mol−1 threshold in ΔGb are labeled. Energy contributions exceeding the y-axis scale are denoted by an asterisk.
Table 1.
Binding energy contributions from the anchoring hydrophobic residues (Y114 or equivalent) to the clamp interface stability.
| Clamp interface | Residue | Contribution (kcal/mol) | Rank among all interfacial residues | Rank excluding charged residues |
|---|---|---|---|---|
| E. coli | F106 | −3.99 | 7 | 2 |
| M. tuberculosis | F116 | −5.40 | 1 | 1 |
| Human PCNA | Y114 | −4.95 | 2 | 1 |
| Yeast PCNA | Y114 | −3.70 | 2 | 1 |
| T4 phage | Y165 | −3.45 | 6 | 3 |
| RB69 phage | Y165 | −3.47 | 2 | 1 |
| Afu | Y106 | −3.94 | 3 | 3 |
| Pfu | F111 | −3.76 | 1 | 1 |
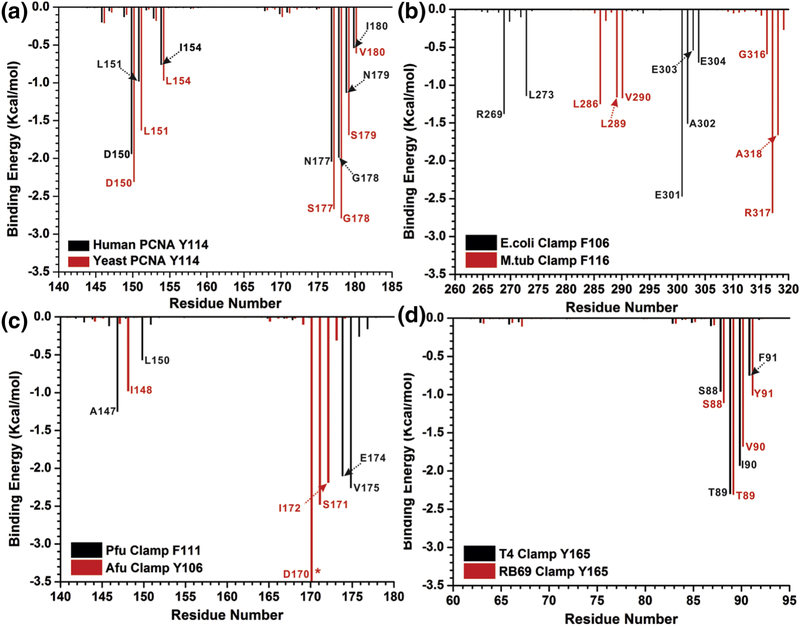
To obtain a more detailed insight into the interactions ensuring clamp interface stability, we employed pairwise MM/GBSA energy decomposition, focusing specifically on the interactions of Tyr114 or equivalent residues. Applying a threshold of −0.5 kcal/mol to the binding energies, we show that seven residues from the opposing subunit form significant interactions with Tyr114 in human and yeast PCNA (Figure 3a). Similarly, in the dimeric E. coli and M. tub β-clamps the Phe106 residue forms five and six interactions, respectively that substantially contribute to the subunit interface stability (Figure 3b). In the archaeal clamps (Afu and Pfu) two phenylalanines (106 and 111) form four critical contact with the opposing subunit (Figure 3c). In bacteriophage clamps (T4 and RB69) the critical Tyr165 residue participates in four contributing pairs (Figure 3d). The binding energy profiles from the pairwise binding energy decomposition appear to be most similar for organisms from the same domain of life (eukaryotes, prokaryotes, archaea and phage). This is especially true for eukaryotic and bacteriophage clamps (Figure 3a,d). The explanation for this lies in the analogous or at the very least similar spatial positioning (Figure 4) of the highly conserved residues in the respective crystal structures.
Figure 3. Residues that contribute to interface stability with the anchoring tyrosine or phenylalanine from pairwise (2-D) MM/GBSA decomposition analysis.
Binding energy contributions for pairs of residues are shown for a) Human PCNA (black) and Yeast PCNA (red); b) E. coli β-clamp (black) and M.tub clamp (red); c) Afu PCNA (black) and Pfu PCNA (red); d) T4 gp45 clamp (black) and RB69 gp45 clamp (red). Only residues contributing above a ±0.5 kcal mol−1 threshold in ΔGb are labeled. Energy contributions exceeding the y-axis scale are denoted by an asterisk.
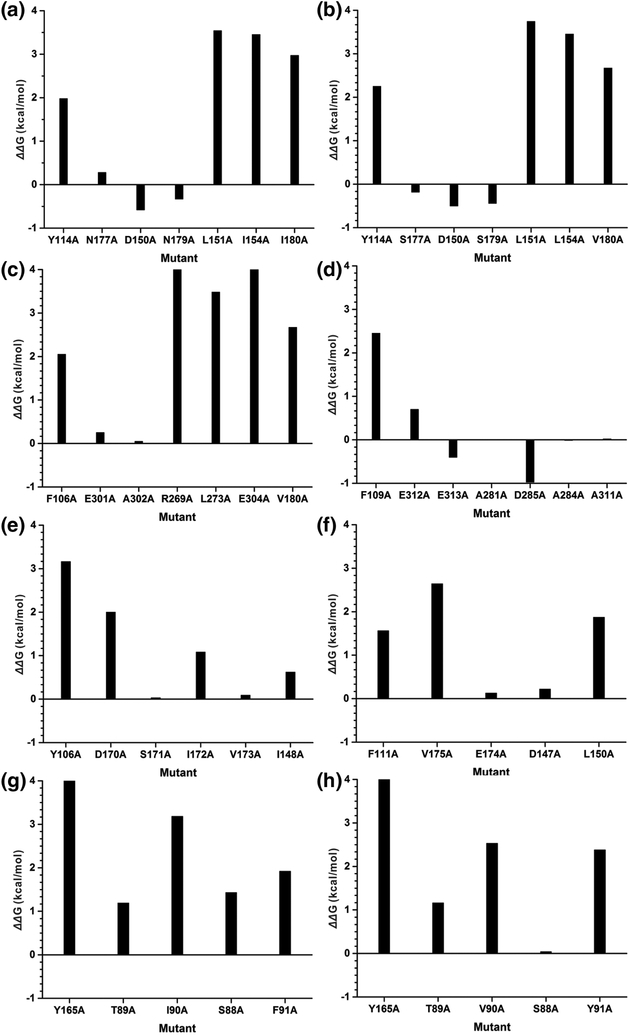
Another way to cross validate the above energy contributions from interfacial residues was to perform computational alanine scanning. We sequentially mutated each residue at the clamp subunit interface using the Rosetta package and evaluated the change in Rosetta score for the protein by applying the default Rosetta scoring function [36]. The difference in Rosetta score was used as a proxy for the free energy change upon alanine mutation (ΔΔG) [37, 38]. For the vast majority of the residues, alanine mutation is highly unfavorable (Figure 5). Notably, the nonpolar residues are more sensitive to the Ala mutation by showing higher free energy penalties than the polar residues. Specifically, mutating Tyr114 or equivalent residues in the series of clamp structures leads to ΔΔG values of from 1.56 to 4.57 kcal/mol and an average of 2.77 kcal/mol, highlighting their important role in maintaining the integrity of the clamp subunit interfaces.
Figure 5. Interfacial binding free energy change upon alanine mutation computed by Rosetta alanine scanning.
a) Human PCNA; b) Yeast PCNA; c) E. coli β-clamp; d) M.tub clamp; e) Afu PCNA; f) Pfu PCNA; g) T4 gp45 clamp; h) RB69 gp45 clamp.
To compare the stability of the wild type human PCNA versus the Y114A, Y114H and Y114L mutants, we adopted the high-precision protocol of Rosetta ddG_monomer application to estimate the change in stability (ΔΔG). The results are shown in Table 2, indicating that the most energetically unfavorable mutation of the anchoring tyrosine residue was to alanine, followed by to leucine, and then by histidine. This is in close agreement with an earlier experimental observation that when a single mutation of Tyr114 to alanine in human PCNA was introduced, the oligomeric structure of the PCNA clamp was completely lost and this mutation also completely abolished the function of the PCNA clamp ring [35].
Table 2.
Change in free energy between mutant and wild type human PCNA. The values in the first column correspond to a single mutation at one of the subunit interfaces. The values in the second column correspond to mutations at all three subunit interfaces.
| Mutation | ΔΔG (kcal/mol) | |
|---|---|---|
| One subunit | Three subunits | |
| Y114A | 7.72 | 20.70 |
| Y114L | 2.06 | 7.96 |
| Y114H | 5.86 | 18.20 |
Network analysis highlights the key anchor residue as a connectivity hotspot
First, we employed Perturbation Response Scanning (PRS) to analyze the sensitivity/influence of residues located at the subunit interfaces of various clamp proteins. PRS analysis is based on a coarse-grained anisotropic network model (ANM) [39] and quantifies the response of a residue to mechanical perturbation at another residue within the ANM network [39]. Each element in the normalized PRS matrix represents the response of residue k to a unit deformation at residue i. Extracting a particular column from the matrix (e.g. for residue Tyr114) highlights how well-connected that residue is within the protein and its likely significance, for example, in communicating allosteric signals to distal parts of the protein. Here, we adapted this method to probe the response of the residues in the clamp proteins upon perturbation on Tyr114 or equivalent residues from the multiple sequence alignment. Similar to the results from the MM/GBSA decomposition, we note the close resemblance among PRS profiles of clamps in organisms from the same life domain (Figure S11). Thus, the PRS method is sensitive to the conservation of the ANM networks in homologous clamp proteins. We identified residues that respond to Tyr114 (or equivalent residue) perturbation with more than half of the maximum response magnitude across the entire protein (Figure S11). The coverage of these highly responsive residues overlaps very well the findings from the 2-D MM/GBSA decomposition (Figure 3). Given the coarse-grain nature of the PRS method, observed prominent response for the cluster of hydrophobic residues anchored by Tyr114 suggests that this cluster is not only energetically important but also occupies a central position within the protein network. The increased connectivity of Tyr114 (or equivalents) is critical for maintaining the integrity of the protein network at the subunit interfaces.
Second, we carried out community network analysis of all clamp proteins systems. Upon dividing the networks into communities, not surprisingly, the dynamic network analysis communities of a clamp protein are mostly defined by its domains, with the residues within a community connected through highly weighted edges (Figure S12). Interestingly, the communities containing the Tyr or Phe anchor residue always extend beyond their subunit reaching across the interface into the adjacent subunit (Figure S13). The strong coupling of the anchor residue to conserved hydrophobic partners from the opposing subunit is evidenced by multiple heavily weighted edges in the network graph and also by the betweenness values presented in Table 3. In the case of either human or yeast clamp, the Tyr114 sits at the boundary of the community that it belongs to, forming a major anchor for the opposing subunit (Figure S13a,b). In the prokaryotic clamps, the Phe is also among the residues that are most heavily connected to the opposing subunit (Figure S13c,d). The connection of the Tyr to the opposing subunit is less prominent in archaeal or bacteriophage clamps (Figure S13e–h), which correlates with their experimentally measured relatively low interface stabilities.
Table 3.
Results from dynamic network analysis: the number of edges connecting the Tyr (Phe) anchor residue to the opposing subunit and the overall betweenness associated with these edges.
| Clamp | Number of Edges | Overall Betweenness |
|---|---|---|
| Human PCNA | 4 | 1.81 |
| Yeast PCNA | 4 | 3.44 |
| E. coli | 5 | 1.95 |
| M. tuberculosis | 3 | 1.92 |
| A.fulgidus | 2 | 1.56 |
| P. furiosus | 2 | 1.68 |
| T4 Phage | 0 | 0.0 |
| RB69 Phage | 1 | 0.48 |
DISCUSSION
Sliding clamp proteins are central players in DNA replication and the DNA damage response. Sliding clamps from different species display various forms in solution. The phage T4 gp45 clamp exists as an open clamp in solution whereas the β-clamp from E. coli and S. cerevisiae PCNA clamp are closed in solution [15, 19, 24]. T4 phage gp45 clamp subunits exchange readily in solution due to its partially open clamp ring structure [23]. We explored the intrinsic stability of human PCNA and clamp ring dynamics using a FRET pair selectively introduced at the subunit interface. Our results indicate that the free human PCNA is closed in solution as seen in the crystal structures. The closed form of the PCNA clamp could be responsible for the improved intrinsic stability of the clamp as the subunits of PCNA exchange at least seven-fold slower than the partially open gp45 clamp. The trimeric form of the human PCNA clamp dissociates into monomers in solution by a cooperative mechanism.
Due to competing active unloading of the clamp from DNA [10, 40], FRET measurements were limited in their ability to calculate the extent of ring opening and closing in the presence of RFC, ATP and polymerase. Unlike phage T4 gp45 sliding clamp whose subunit stability increases in the presence of its cognate clamp loader and polymerase [23], addition of RFC and ATP does not alter the kinetics of PCNA subunit exchange in solution. But addition of DNA polymerase δ shows slight reduction in the subunit exchange rate, confirming inhibition of active unloading of PCNA clamp on DNA by the RFC complex.
We also show that in addition to the well characterized subunit interface H-bonding network, there is also a conserved network of hydrophobic residues that maintains the oligomeric structure and characteristic ring shape of all sliding clamp proteins. This network is comprised of a large aromatic residue (Tyr or Phe) from one subunit, which is “anchored” into a conserved hydrophobic pocket located in the opposing subunit across the clamp interface. The network was clearly identifiable in structures of representative clamps from a number of organisms, including phages, bacteria, archaea and eukaryotes. In addition to the H-bonding network, the hydrophobic residues at the subunit interface contribute significantly to the stability and oligomerization of the clamp ring structures as evidenced by the results from MM/GBSA analysis that delineated energy contributions of individual residues and residue pairs to the interfacial stability in PCNA and related clamp proteins. Furthermore, mutation of the hydrophobic network abolishes both the oligomeric structure and the function of the clamp proteins [35]. Additionally, we show that mutations of the hydrophobic residue Tyr114 in human PCNA or Phe106 in bacterial clamps completely abolish the oligomerization of the subunits. Finally, in the case of the bacterial β-clamp, removal of the “docking” site aromatic cavity through the introduction of a double mutation, was found to completely prevent dimerization [41]. In phage T4, the conserved large hydrophobic “anchor” Tyr residue is pointing inwards in the same subunit interacting with the side chain of a conserved Asn maintaining a hydrogen bond between Asn and Tyr side chains. This lack of a direct hydrophobic interaction between the subunits could explain the fast subunit exchange in solution as well as the partially opened structure of the clamp unlike the β-clamp and PCNA. This is also corroborated by observations of a highly dynamic gp45 clamp protein as compared to β-clamp and PCNA [10, 42, 43]. Recently clamp interface dynamics was systematically examined using hydrogen-deuterium exchange in eight clamp proteins from different species, observing a widely varying dynamic behavior despite the common overall protein architecture [43]. For example, the subunit interface in yeast PCNA was less dynamic than human PCNA. Our computational results rationalize these observed differences among clamps in dynamic subunit association by providing a detailed account of important residue interactions. Our findings suggests that clamps from different species have highly varying microenvironments at the subunit interfaces resulting from different hydrophobic contacts, hydrogen-bonding patterns and salt-bridge interactions.
Based on our results, we propose a revised model for the oligomeric clamp stability and clamp ring opening and closing. This model incorporates two structural elements: (1) a hydrogen bonding network between two anti-parallel β-strands and (2) the hydrophobic aggregation of residues from adjacent subunits across the subunit interface. Association with the clamp loader unzips this H-bonding network and also releases the aromatic “anchor” residue. Conversely, during clamp assembly on DNA, ring closure is mediated by re-zipping the H-bonding interactions as well as reorganization of the hydrophobic interaction across the subunit interface of the clamp. The C-terminal tail of gp43 polymerase is composed of amino acids with aromatic side chains. This tail binds to the partially open subunit interface of the clamp to form a DNA polymerase holoenzyme with a dramatically increased subunit stability mediated by a clamp ring that is now closed tightly on DNA. By extension, the implication of our observations is that small molecule inhibitors that could bind to the hydrophobic pocket of open clamps will prevent the clamp ring from generating a productive DNA polymerase complex. This approach would complement the current strategies of identifying molecules that inhibit DNA replication by blocking clamp ring assembly targeting the sites of the interdomain loop in the clamp ring structure [44–50]. Thus, inhibition of holoenzyme DNA polymerase assembly by blocking clamp ring closure could be exploited as a strategy to slow down rapid DNA synthesis observed in cancer and other disease states.
MATERIAL AND METHODS
The sequences of oligonucleotides used in the loading assays were template DNA: 5′-ACACAG ACGTACTATCATGACGCATCAGACAACGTGCGTCAGCTATTACGTGCGGAAGGAGT-3′, primer DNA: 5′-biotin-ACTCCTTCCGCACGTAATAGCTG-3′ and flap DNA: 5′-TGGGT GGGAGGGAGTGGGATGATAGTACGTCTGTGT-3′. For PCNA clamp ring closing and assembly assays, a forked-DNA substrate was employed obtained from annealing the oligonucleotides by heating to 92°C and cooling to room temperature over 2h. The oligonucleotides (HPLC purified and Biotin labeled) were purchased from IDT Technologies, Inc. ATP and streptavidin were purchased from Sigma-Aldrich. IAEDANS was purchased from Molecular probes. NiNTA was from Qiagen. HiPrep Q HP, Heparin and Sephadex 16/60 columns were all from GE Healthcare. The pET23 vector carrying the human wild-type PCNA with a C-terminal His6-tag was a gift from Ulrich Hubscher. The plasmids containing the five subunits of human RFC p36-p37-p38-p40-pET-Duet-1(AmpR) and pCDF-1b-p140 (StrepR) were generously provided to us by Paul Modrich, Duke University Medical School [51].
Purification of wild-type and mutant human PCNA
The wild-type PCNA was expressed and purified as described with the following modifications [35, 52]. Briefly, the pET23 vector carrying the gene for PCNA with a C-terminal His6-tag was transformed into BL21 (DE3) and selected on agar plates containing 50 μg/ml ampicillin. Two to three colonies were selected from the agar plate and grown overnight at 37 °C in LB containing ampicillin (50 μg/ml). A 10 ml of the overnight inoculum was diluted into 1 L of terrific broth with 50 μg/ml ampicillin (8 × 1L). The cells were grown at 37° C until the OD600 nm reached ~0.6. The temperature of the culture was dropped to 18 °C and protein expression was induced by the addition of 0.3 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG). The cells were grown for an additional 10–16 h at 18° C after which the cells were pelleted by centrifugation in a Beckman centrifugation device at 5000 rpm at 4° C for 15 min. The cells were resuspended in 5 ml of 20 mM TrisHCl (pH 7.8), 0.8 M NaCl and 5 mM imidazole (pH 7.5) buffer for every gram of the cell pellet in the presence of an EDTA-free protease inhibitor tablet. The re-suspended cells were then lysed using sonication, and centrifuged at 15000 rpm for 30 min at 4 °C. The lysate was then loaded onto a NiNTA His-bind resin column (10 ml bed volume), washed with 10 column volumes of lysis buffer, followed by 10 column volumes of 20 mM TrisHCl (pH 7.8), 0.8 M NaCl and 10 mM imidazole and 10 column volumes of 20 mM TrisHCl (pH 7.8), 0.2 M NaCl and 20 mM imidazole. The protein was eluted with a buffer containing 20 mM TrisHCl (pH 7.8), 100 mM NaCl, 100 mM imidazole (pH 7.5). The proteins were collected in 10 ml fractions. The fractions were analyzed on a 12% SDS-PAGE and the fractions containing the protein were pooled together. The eluted protein was diluted two-fold with a buffer containing 20 mM Tris-HCl (pH 8.0), 5% glycerol and loaded onto a 5 ml Q HP (GE) column. The column was washed with 2 column volumes of a buffer containing 20 mM TrisHCl (pH 8.0), 100 mM NaCl before eluting the protein with a linear gradient of 0.1 – 0.7 M NaCl over 60 min. The fractions were analyzed on a 12% SDS-PAGE and the fractions containing PCNA were pooled together and concentrated using an Amicon centrifugation device with a 10 kD cut-off and buffer exchanged into the storage buffer. The concentrated protein was then flash-frozen in liquid N2 in 10 μl aliquots and stored at −80°C. Protein concentration was calculated based on an extinction coefficient of 15010 M−1 cm−1.
Mutagenesis of PCNA
The pET23 vector carrying the wild-type PCNA gene was used as a template for generating the mutants. The plasmids encoding the mutant PCNA-WC1 and PCNA-WC2 carrying the mutations T185W-N107C-C27M-W28F-C62M-C81S-C148S-C135S-C162S and N107W-T185C-C27M-W28F-C62M-C81S-C148S-C135S-C162S, respectively, were obtained by multisite direct mutagenesis using the mutagenic primers (see Table S1). The PCNA mutants were expressed and purified to homogeneity using procedures identical to the wild-type protein. The PCNA-W28F mutant was expressed and purified identically to the wild-type PCNA. The PCNA-C1 mutant carrying N107C-C27M-W28F-C62M-C81S-C148S-C135S-C162S mutation was generated by mutagenic PCR and the mutant protein was expressed and purified by procedures identical to the wild-type and other PCNA mutant proteins.
Cloning, expression and purification of human RFC
The gene for the mutant RFCΔ1555 lacking the N-terminal domain was PCR amplified from the plasmid containing the wild-type RFC1 gene [51] using the forward primer - 5’-GCTTACCATGGACAAAGAACAGGTTGCCGAAG-3’ and reverse primer - 5’-CTGAGGATCCTTATTTTTTCGAGCTTTTACC-3’ and then inserted into the NcoI and BamHI sites of pCDF-1b vector. The identity of the gene was confirmed by dideoxy sequencing. The pCDF-1b vector carrying the RFC1Δ555 gene and the pET-Duet1 vector carrying RFC2–3-4–5 were transformed into BLR (BL21 (DE3)/recA−) competent cells by electroporation and selected on agar plates containing 50 μg/ml streptomycin and 50 μg/ml ampicillin. Colonies were selected from the agar plate and grown overnight at 37 °C in LB containing 50 μg/ml streptomycin and 50 μg/ml ampicillin. A 10 ml of the overnight inoculum was diluted into 1 L of terrific broth at 37 °C supplemented with 50 μg/ml streptomycin and 100 μg/ml ampicillin (8 × 1 L). The cells were grown at 37 °C until the OD600 nm reached ~0.6. The temperature of the culture was dropped to 15 °C and the protein expression was induced by the addition of 0.5 mM IPTG. The cells were then grown for an additional 10–16 h at 15 °C after which the cells were pelleted by centrifugation in a Beckman centrifugation device at 5000 rpm at 4 °C for 20 min. The cells were re-suspended in 10 ml of 20 mM Tris-HCl, (pH 7.5) and 150 mM NaCl buffer for every gram of the cell pellet and then collected by centrifugation at 5000 rpm at 4 °C for 20 min. The cells were re-suspended in 5 ml of 20 mM HEPES (pH 7.5), 2 mM EDTA, 5% glycerol, 0.01% NP40, 50 mM NaCl, 2 mM DTT for every gram of the cell pellet in the presence of EDTA-free protease inhibitor cocktail (3 tablets) from Roche Diagnostics. The re-suspended cells were then lysed using sonication and centrifuged at 15000 rpm for 30 min at 4 °C and was repeated to obtain a clear supernatant. The supernatant was loaded onto a 25 ml column generated with SP Sepharose FF resin and equilibrated with a buffer containing 25 mM HEPES (pH 7.5), 0.1 mM EDTA, 5% glycerol, 0.01% NP40, 200 mM NaCl. After a 25 ml wash with equilibration buffer, the column was developed with a 100 ml linear gradient of NaCl (0.2–0.8 M) in equilibration buffer. The fractions were analyzed on a 10% SDS PAGE and the peak protein complex fractions were pooled together and diluted with an equal volume of 100 mM potassium phosphate (pH 7.5). A 5 ml CHT2-I (BioRad) equilibrated with a buffer solution containing 50 mM potassium phosphate (pH 7.5), 5% glycerol, 0.01% NP40, 100 mM NaCl was charged with the protein solution at 1 ml/min. After 15 ml wash with equilibration buffer, the column was developed with 15 ml of Solution HA (5% glycerol, 0.01% NP40, 100 mM NaCl) containing 140 mM potassium phosphate (pH 7.5), 15 ml of Solution HA containing 185 mM phosphate (pH 7.5), 15 ml of solution HA containing 230 mM phosphate (pH 7.5), 15 ml of solution HA containing 275 mM phosphate (pH 7.5), and 15 ml of solution HA containing 350 mM phosphate (pH 7.5). The fractions were analyzed on a 10% SDS PAGE and the protein complex peak fractions that eluted between 185 mM and 275 mM phosphate were pooled together. The protein solution was then diluted with a buffer containing 25 mM HEPES (pH 7.5), 0.1 mM EDTA, 5% glycerol, 0.01% NP40 (Buffer Q) to a salt concentration of 100 mM NaCl and loaded at 1 ml/min onto a 5 ml Q FF column equilibrated with Buffer Q. The protein was eluted with a 100-ml linear gradient of NaCl (0.1–0.5 M) in Buffer Q. The fractions were analyzed on a 10% SDS-PAGE and the fractions containing the pure truncated RFC complex fractions were pooled, flash frozen in liquid N2 in 10 μl aliquots containing 20% glycerol, and stored at −80°C. The protein concentration was calculated based on an extinction coefficient of 136650 M−1 cm−1.
Preparation of fluorescent dye labeled PCNA clamp proteins
The mutant PCNA proteins PCNA-WC1, PCNA-WC2 and PCNA-C1 were labeled selectively at N107C, T185C and N107C, respectively, with IAEDANS to generate a fluorescent label at the subunit interface as follows: a DMF solution of IAEDANS was added in aliquots to the PCNA protein in a buffer containing 20 mM HEPES (pH 7.8), 50 mM NaCl, 1 mM EDTA and 10% glycerol over 30 min to a final concentration of 10 mM with end-over-end mixing at 4 °C. The reaction was then allowed to proceed at 4 °C for an additional 8 h. The reaction was quenched with 10 mM β-ME and dialyzed against 20 mM HEPES (pH 7.8), 50 mM NaCl, 1 mM β-ME, 1 mM EDTA and 10% glycerol. The dialyzed protein solution was further purified on a sephadex G50 column (70 × 1 cm) and the fractions containing the protein were concentrated and stored at −80 °C. The purified protein was stored in a buffer containing 20 mM Tris HCl (pH 7.8), 1 mM β-mercaptoethanol (β-ME), 0.5 mM EDTA, 100 mM NaCl and 20 % glycerol at −80 °C. The labeling efficiency was determined to carry approximately three moles of the fluorophore for every mole of the trimeric PCNA clamp.
PCNA-C1 mutant protein carrying fluorescein was obtained by labeling the protein with fluorescein dye at N107C position using thiol chemistry under identical labeling and purification procedures.
Assaying ATPase activity of RFC clamp loader by PCNA
PCNA proteins were assayed for stimulation of the ATPase activity of the RFC clamp loader complex in the presence of DNA using a coupled enzymatic assay as described previously. The ATPase assays were performed in a Varian spectrophotometer using an NADH-oxidation coupled enzymatic system in reaction buffer (25 mM Tris (pH 7.8), 150 mM potassium acetate, 10 mM magnesium acetate) containing 1 mM adenosine 5′-triphosphate (ATP), 2 mM phosphoenol pyruvate, 0.2 mM β-reduced nicotinamide adenine dinucleotide (NADH), 5 U/ml lactate dehydrogenase and 10 U/ml pyruvate kinase monitoring the reaction at 340 nm. ATP hydrolysis rates were followed by monitoring the oxidation of NADH to NAD+ at 340 nm and the rates obtained in AU/min were converted to M/min by dividing the rates by 6220 M−1 cm−1, the extinction coefficient of NADH at 340 nm [25].
Steady state FRET measurements
Steady state fluorescence measurements were carried out in a FluoroMax 4 spectrofluorometer from HORIBA at 25°C. FRET emission spectra for the PCNA clamp carrying fluorescent probes were obtained by exciting at 290 nm. Corrections for the fluorescence intensity from dilution and inner filter effects were made by using the formula F= Fobs antilog (Aex/2), where Aex is the absorbance at the excitation wavelength of 290 nm. Emission spectra for the PCNA clamp ring opening and closing both in the absence and presence of RFC clamp loader were obtained by exciting the Trp at the subunit interface at 290 nm.
Kinetics of the PCNA clamp subunit exchange
The PCNA-WC1-AEDANS or PCNA-WC2-AEDANS protein (200 nM) was mixed with 2 μM PCNA-W28F protein at 25 °C to measure the subunit exchange. The fluorescence emission from 290 nm excitation was monitored at 484 nm with a 4 nm slit. The dissociation of the PCNA clamp subunits was monitored from the decrease in FRET signal due to the exchange of the subunits of labeled PCNA clamp with the unlabeled PCNA trap protein. The dissociation kinetics data were fit to a single-exponential equation.
Model for PCNA clamp subunit dissociation
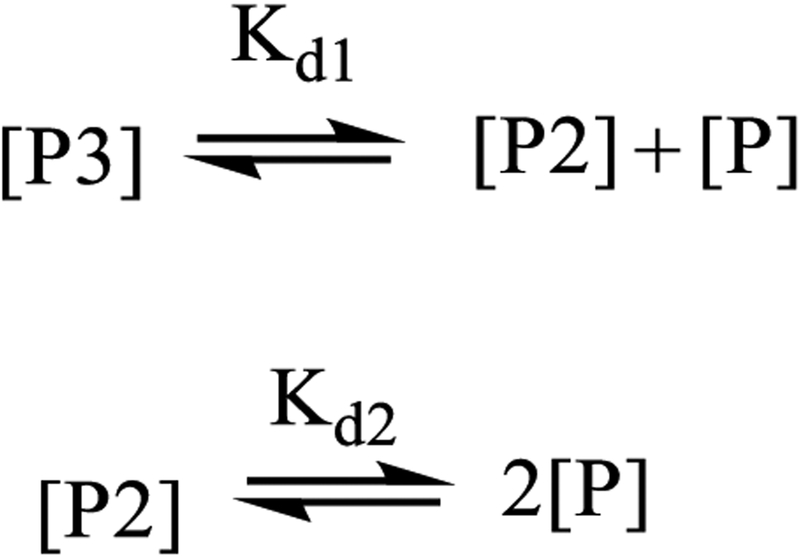
The protein was excited at 290 nm and the emission at 480 nm was monitored over various concentrations ranging from 2 nM to 500 nM. The fluorescence dilution data thus obtained were fit to the following model using Dynafit 3 [53].
Here P3, P2 and P represent the trimer, dimer and monomer of PCNA, respectively. Additionally, the data were fit to a cooperative model and a non-cooperative model. In the cooperative model, Kd2>>Kd1, whereas Kd1=3Kd2 in a non-cooperative model.
Solution structure of trimeric PCNA clamp ring
The structure of the PCNA clamp ring in solution was examined by following fluorescence energy transfer between the FRET pair Trp and AEDANS at the subunit interface. The Forster radius (R0) of Trp and AEDANS in PCNA, energy transfer efficiency and FRET-based distance between Trp and AEDANS at the PCNA subunit interface were calculated as previously described [15, 19, 31].
Molecular Dynamics Simulation
Molecular dynamics (MD) simulations were carried out on the following clamp structures: human PCNA (1VYM) [30], yeast PCNA (1PLQ) [54], E. coli β-clamp (2POL) [55], Mtu β-clamp (3RB9) [56], Afu PCNA (1RWZ) [57], Pfu PCNA (1GE8) [11], T4 phage gp45 clamp (3U6O) [58], RB69 phage gp45 clamp (1B8H) [59]. These simulations were used for analysis of the sliding clamp subunit interfaces through the molecular mechanics Generalized Born solvent accessible surface area (MM/GBSA) method and MM/GBSA energy decomposition [60, 61]. The simulation systems were set up using the tleap module of AMBER 9 [62, 63]. Hydrogen atoms were added and the protonation states for ionizable residues were set at pH 7.0 using the WHATIF webserver [64]. Each system was then solvated with TIP3P water molecules [65] with a minimum distance of 10.0 Å from the protein surface to the edge of the periodic simulation box. Counter ions were added to neutralize the net protein charge and additional Na+ and Cl− ions introduced (100 mM NaCl concentration) to mimic physiological conditions. Energy minimization was then conducted for 5000 steps with fixed backbone atoms followed by 5000 steps of minimization with harmonic restraints on the protein backbone atoms (25 kcal/mol force constant). The temperature of the simulated systems was then gradually raised to 300 K over 200-ps dynamics in the NVT ensemble, keeping the protein backbone restrained. The equilibration was continued for 1 ns in the NPT ensemble while the harmonic restraints were gradually released. 20-ns production runs for each system were carried out in the isothermal isobaric ensemble (1 atm and 300 K). Long-range electrostatic interactions were computed using the smooth particle mesh Ewald (SPME) algorithm [66]. A non-bonded cutoff distance of 10 Å with a switching function at 8.5 Å was used. The integration time step was 2 fs and all bonds between hydrogen and heavy atoms were fixed to eliminate the highest frequency oscillatory motions. The r-RESPA multiple time step method [32] was adopted with a 2-fs time step for bonded, 2-fs time step for short-range non-bonded interactions and 4-fs time step for long-range electrostatic interactions. All simulations were performed using the NAMD 2.9 code [33, 67] with the AMBER Parm99SB force field [68]. Data were analyzed using the PTRAJ utility in AMBER [63] and custom VMD TCL scripts [69].
MM/GBSA Binding Energy Decomposition
To perform the MM/GBSA calculations we selected snapshots at 4 ps intervals from the latter half of each MD trajectory. Free energy of binding (ΔGb) could then be computed as follows:
where ΔEMM represents gas-phase molecular mechanics binding energy (van der Waals and electrostatics); ΔGsol is the change in solvation free energy and ΔS is the gas-phase entropy change upon binding. The electrostatic solvation energy contribution to ΔGsol is computed by analytically solving the Born equation [70, 71]. While the general MM/GBSA expression contains an entropic term TΔS, entropy is a collective property and cannot be decomposed on a per residue basis. Therefore, the MM/GBSA decomposition (either 1-D or 2-D) accounted only for the enthalpic contribution to ΔGb. The dielectric constants for protein and water were set to 1.0 and 78.0, respectively. The non-polar contribution to ΔGsol was estimated from the solvent-accessible surface area: ΔGb. = γA + b. The solvation parameters γ and b were set to 0.0072 kcal mol−1 Å−2 and 0 kcal mol−1 and the surface area A calculated using MolSurf in AMBER 9 [63]. The probe radius of the solvent was 1.4 Å. The optimized set of atomic radii in AMBER 9 was used, and the atomic charges of the protein were taken from the ff99SB force field. MM/GBSA energy decomposition [60, 61] was carried out to evaluate the contributions to subunit interface stability in all clamps that were subject to MD simulations. We performed two types of decomposition: one-dimensional and two-dimensional. The first type quantifies the energetic contribution of each residue to the interface stability. The second type further decomposes the energetic contribution from all residue pairs within a distance from the subunit interface.
Computed changes in subunit interface stability upon point mutation
We also computed the change in free energy (ΔΔG) for three PCNA mutants, Y114A, Y114H and Y114L relative to wild type human PCNA, using the high-level precision protocol of Rosetta ddG_monomer [37, 72]. In this application, the crystal structure of human PCNA (1VYM) was first minimized with the Rosetta scoring function, the flexibility of both sidechains and backbone were then considered to generate 50 structures for the wild-type variant and each of the mutants. The difference between the mean of the top three scored wild type structures and the corresponding mean of the mutant structures were considered as an estimate of ΔΔG [72].
Protein Network Analysis
To discover residue networks encompassing the clamp’s hydrophobic anchor (Y114 or equivalent) and contributing to clamp interface stability, we relied on two methods (i) perturbation response scanning and (ii) community network analysis. In PRS [73], a protein is treated as an anisotropic network model (ANM) [74], in which the nodes (centered on the Cα atoms) represent all protein residues, and the edges (harmonic springs) represent the inter-residue interactions within a cutoff of 12 Å. Based on linear response theory [73, 75], the collective dynamics of the network of nodes and edges is described by the Hessian matrix of the ANM potential [76]. We applied a perturbation force to the nodes at the clamps’ subunit interfaces one residue at a time while monitoring the collective displacement of the ANM network in response to this perturbation, representing the outcome as a N × N heat map. Row and column averages reveal the propensity of a given residue (e.g. Y114) to act as either a sensor or effector of the perturbation, providing a quantitative assessment of its influence/sensitivity within the network.
Community network analysis was performed with the NetworkView plugin in VMD [77, 78]. Network graphs were created with nodes representing protein residues and edges connecting pairs of nodes within a 5 Å cutoff distance and more than 75% occupancy over the MD simulation. The edges between nodes i and jwere weighted by wi,j = −ln(|cij|) where Cij denotes the respective cross correlation coefficient. After computing the network graphs, we subdivided the nodes into communities of highly intra-connected nodes using the Girvan-Newman algorithm [77]. Subsequently, critical nodes were located at the interfaces of neighboring communities, with probabilities of communication between nodes represented by the weights of the connecting edges. This analysis allows us to identify residue clusters that are strongly connected and moving together as a unit during molecular dynamics. These are precisely the clusters of residues that play a critical role in maintaining the integrity of the clamp subunit interfaces.
Supplementary Material
Scheme 1.
Model of PCNA sliding clamp subunit dissociation
Highlights.
A cluster of hydrophobic residues in DNA clamp proteins maintains their structural integrity
The identified network of hydrophobic residues is conserved in clamps from all domains of life
We provide a revised model for clamp protein oligomerization, opening and DNA-loading
ACKNOWLEDGEMENTS
We thank Dr. Ignacio General for sharing the Matlab scripts for PRS analysis. This work was supported by National Institutes of Health grants [GM013306 to S.J.B; GM110387 to I.I.]. Computational resources were provided in part by allocations from the National Science Foundation XSEDE program [CHE110042 to I.I.] and the National Energy Research Scientific Computing Center supported by the DOE Office of Science [contract DE-AC02–05CH11231].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
No conflict of interest declared.
References
- [1].Benkovic SJ, Valentine AM, Salinas F Replisome-mediated DNA replication. Annu. Rev. Biochem 2001, 70, 181–208. [DOI] [PubMed] [Google Scholar]
- [2].Bloom LB Loading clamps for DNA replication and repair. DNA Repair. 2009, 8, 570–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Davey MJ, Jeruzalmi D, Kuriyan J, O’Donnell M Motors and switches: AAA+ machines within the replisome. Nat. Rev. Mol. Cell. Biol 2002, 3, 826–835. [DOI] [PubMed] [Google Scholar]
- [4].Jeruzalmi D, O’Donnell M, Kuriyan J Clamp loaders and sliding clamps. Curr. Opin. Struct. Biol 2002, 12, 217–224. [DOI] [PubMed] [Google Scholar]
- [5].Hedglin M, Kumar R, Benkovic SJ Replication clamps and clamp loaders. Cold Spring Harb. Perspect. Biol 2013, 5, a010165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Stoimenov I, Helleday T PCNA on the crossroad of cancer. Biochem. Soc. Trans 2009, 37, 605–613. [DOI] [PubMed] [Google Scholar]
- [7].Ivanov I, Chapados BR, McCammon JA, Tainer JA Proliferating cell nuclear antigen loaded onto double-stranded DNA: dynamics, minor groove interactions and functional implications. Nucleic Acids Res. 2006, 34, 6023–6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kuriyan J, O’Donnell M Sliding clamps of DNA polymerases. J. Mol. Biol 1993, 234, 915–925. [DOI] [PubMed] [Google Scholar]
- [9].Bowman GD, O’Donnell M, Kuriyan J Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature. 2004, 429, 724–730. [DOI] [PubMed] [Google Scholar]
- [10].Yao N, Turner J, Kelman Z, Stukenberg PT, Dean F, Shechter D, et al. Clamp loading, unloading and intrinsic stability of the PCNA, beta and gp45 sliding clamps of human, E. coli and T4 replicases. Genes Cells. 1996, 1, 101–113. [DOI] [PubMed] [Google Scholar]
- [11].Matsumiya S, Ishino Y, Morikawa K Crystal structure of an archaeal DNA sliding clamp: proliferating cell nuclear antigen from Pyrococcus furiosus. Protein Sci. 2001, 10, 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhuang Z, Berdis AJ, Benkovic SJ An alternative clamp loading pathway via the T4 clamp loader gp44/62-DNA complex. Biochemistry. 2006, 45, 7976–7989. [DOI] [PubMed] [Google Scholar]
- [13].Kelch BA, Makino DL, O’Donnell M, Kuriyan J Clamp loader ATPases and the evolution of DNA replication machinery. BMC Biol. 2012, 10, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kumar R, Nashine VC, Mishra PP, Benkovic SJ, Lee TH Stepwise loading of yeast clamp revealed by ensemble and single-molecule studies. Proc. Natl. Acad. Sci. U. S. A 2010, 107, 19736–19741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Alley SC, Abel-Santos E, Benkovic SJ Tracking sliding clamp opening and closing during bacteriophage T4 DNA polymerase holoenzyme assembly. Biochemistry. 2000, 39, 3076–3090. [DOI] [PubMed] [Google Scholar]
- [16].Tainer JA, McCammon JA, Ivanov I Recognition of the ring-opened state of proliferating cell nuclear antigen by replication factor C promotes eukaryotic clamp-loading. J. Am. Chem. Soc 2010, 132, 7372–7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Xu X, Guardiani C, Yan C, Ivanov I Opening pathways of the DNA clamps proliferating cell nuclear antigen and Rad9-Rad1-Hus1. Nucleic Acids Res. 2013, 41, 10020–10031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang L, Xu X, Kumar R, Maiti B, Liu CT, Ivanov I, et al. Probing DNA clamps with single-molecule force spectroscopy. Nucleic Acids Res. 2013, 41, 7804–7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhuang Z, Yoder BL, Burgers PM, Benkovic SJ The structure of a ring-opened proliferating cell nuclear antigen-replication factor C complex revealed by fluorescence energy transfer. Proc. Natl. Acad. Sci. U. S. A 2006, 103, 2546–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Uhlmann F, Cai J, Gibbs E, O’Donnell M, Hurwitz J Deletion analysis of the large subunit p140 in human replication factor C reveals regions required for complex formation and replication activities. J. Biol. Chem 1997, 272, 10058–10064. [DOI] [PubMed] [Google Scholar]
- [21].Podust VN, Podust LM, Goubin F, Ducommun B, Hubscher U Mechanism of inhibition of proliferating cell nuclear antigen-dependent DNA synthesis by the cyclin-dependent kinase inhibitor p21. Biochemistry. 1995, 34, 8869–8875. [DOI] [PubMed] [Google Scholar]
- [22].Gomes XV, Gary SL, Burgers PM Overproduction in Escherichia coli and characterization of yeast replication factor C lacking the ligase homology domain. J. Biol. Chem 2000, 275, 14541–14549. [DOI] [PubMed] [Google Scholar]
- [23].Soumillion P, Sexton DJ, Benkovic SJ Clamp subunit dissociation dictates bacteriophage T4 DNA polymerase holoenzyme disassembly. Biochemistry. 1998, 37, 1819–1827. [DOI] [PubMed] [Google Scholar]
- [24].Alley SC, Shier VK, Abel-Santos E, Sexton DJ, Soumillion P, Benkovic SJ Sliding clamp of the bacteriophage T4 polymerase has open and closed subunit interfaces in solution. Biochemistry. 1999, 38, 7696–7709. [DOI] [PubMed] [Google Scholar]
- [25].Gilbert SP, Mackey AT Kinetics: a tool to study molecular motors. Methods. 2000, 22, 337–354. [DOI] [PubMed] [Google Scholar]
- [26].Capson TL, Benkovic SJ, Nossal NG Protein-DNA cross-linking demonstrates stepwise ATP-dependent assembly of T4 DNA polymerase and its accessory proteins on the primer-template. Cell. 1991, 65, 249–258. [DOI] [PubMed] [Google Scholar]
- [27].Capson TL, Peliska JA, Kaboord BF, Frey MW, Lively C, Dahlberg M, et al. Kinetic characterization of the polymerase and exonuclease activities of the gene 43 protein of bacteriophage T4. Biochemistry. 1992, 31, 10984–10994. [DOI] [PubMed] [Google Scholar]
- [28].Carver TE Jr., Sexton DJ, Benkovic SJ Dissociation of bacteriophage T4 DNA polymerase and its processivity clamp after completion of Okazaki fragment synthesis. Biochemistry. 1997, 36, 14409–14417. [DOI] [PubMed] [Google Scholar]
- [29].Hu Z, Perumal SK, Yue H, Benkovic SJ The human lagging strand DNA polymerase delta holoenzyme is distributive. J. Biol. Chem 2012, 287, 38442–38448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kontopidis G, Wu SY, Zheleva DI, Taylor P, McInnes C, Lane DP, et al. Structural and biochemical studies of human proliferating cell nuclear antigen complexes provide a rationale for cyclin association and inhibitor design. Proc. Natl. Acad. Sci. U. S. A 2005, 102, 1871–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lakowicz JR Principles of Fluorescence Spectroscopy. 2nd ed. New York, Boston, Dordrecht, London, Moscow: Kluwer Academic/Plenum Publishers; 1999. p. 54. [Google Scholar]
- [32].Tuckerman M, Berne BJ, Martyna GJ Reversible Multiple Time Scale Molecular-Dynamics. J. Chem. Phys 1992, 97, 1990–2001. [Google Scholar]
- [33].Kale L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, et al. NAMD2: Greater scalability for parallel molecular dynamics. J. Comput. Phys 1999, 151, 283–312. [Google Scholar]
- [34].Binder JK, Douma LG, Ranjit S, Kanno DM, Chakraborty M, Bloom LB, et al. Intrinsic stability and oligomerization dynamics of DNA processivity clamps. Nucleic Acids Res. 2014, 42, 6476–6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Jonsson ZO, Podust VN, Podust LM, Hubscher U Tyrosine 114 is essential for the trimeric structure and the functional activities of human proliferating cell nuclear antigen. EMBO J. 1995, 14, 5745–5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].O’Meara MJ, Leaver-Fay A, Tyka MD, Stein A, Houlihan K, DiMaio F, et al. Combined covalent-electrostatic model of hydrogen bonding improves structure prediction with Rosetta. J. Chem. Theory Comput 2015, 11, 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kortemme T, Baker D A simple physical model for binding energy hot spots in protein-protein complexes. Proc. Natl. Acad. Sci. U. S. A 2002, 99, 14116–14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kortemme T, Kim DE, Baker D Computational alanine scanning of protein-protein interfaces. Sci. STKE 2004, 219, pl2. [DOI] [PubMed] [Google Scholar]
- [39].General IJ, Liu Y, Blackburn ME, Mao WZ, Gierasch LM, Bahar I ATPase Subdomain IA Is a Mediator of Interdomain Allostery in Hsp70 Molecular Chaperones. PLOS Comput. Biol 2014,10, e1003624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hedglin M, Perumal SK, Hu Z, Benkovic S Stepwise assembly of the human replicative polymerase holoenzyme. eLife. 2013, 2, e00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hingorani MM, O’Donnell M ATP binding to the Escherichia coli clamp loader powers opening of the ring-shaped clamp of DNA polymerase III holoenzyme. J. Biol. Chem 1998, 273, 24550–24563. [DOI] [PubMed] [Google Scholar]
- [42].Gadkari VV, Harvey SR, Raper AT, Chu WT, Wang J, Wysocki VH, et al. Investigation of sliding DNA clamp dynamics by single-molecule fluorescence, mass spectrometry and structure-based modeling. Nucleic Acids Res. 2018, 46, 3103–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Fang J, Nevin P, Kairys V, Venclovas C, Engen JR, Beuning PJ Conformational analysis of processivity clamps in solution demonstrates that tertiary structure does not correlate with protein dynamics. Structure. 2014, 22, 572–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Smith SJ, Gu L, Phipps EA, Dobrolecki LE, Mabrey KS, Gulley P, et al. A Peptide mimicking a region in proliferating cell nuclear antigen specific to key protein interactions is cytotoxic to breast cancer. Mol. Pharmacol 2015, 87, 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Gu L, Smith S, Li C, Hickey RJ, Stark JM, Fields GB, et al. A PCNA-derived cell permeable peptide selectively inhibits neuroblastoma cell growth. PLOS One. 2014, 9, e94773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lingeman RG, Hickey RJ, Malkas LH Expression of a novel peptide derived from PCNA damages DNA and reverses cisplatin resistance. Cancer Chemother. Pharmacol 2014, 74, 981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Tan Z, Wortman M, Dillehay KL, Seibel WL, Evelyn CR, Smith SJ, et al. Small-molecule targeting of proliferating cell nuclear antigen chromatin association inhibits tumor cell growth. Mol. Pharmacol 2012, 81, 811–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Actis M, Inoue A, Evison B, Perry S, Punchihewa C, Fujii N Small molecule inhibitors of PCNA/PIP-box interaction suppress translesion DNA synthesis. Bioorg. Med. Chem 2013, 21, 1972–1977. [DOI] [PubMed] [Google Scholar]
- [49].Dillehay KL, Lu S, Dong Z Antitumor effects of a novel small molecule targeting PCNA chromatin association in prostate cancer. Mol. Cancer. Ther 2014, 13, 2817–2826. [DOI] [PubMed] [Google Scholar]
- [50].Dillehay KL, Seibel WL, Zhao D, Lu S, Dong Z Target validation and structure-activity analysis of a series of novel PCNA inhibitors. Pharmacol. Res. Perspect 2015, 3, e00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kadyrov FA, Genschel J, Fang Y, Penland E, Edelmann W, Modrich P A possible mechanism for exonuclease 1-independent eukaryotic mismatch repair. Proc. Natl. Acad. Sci. U. S. A 2009, 106, 8495–8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Jonsson ZO, Hindges R, Hubscher U Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen. EMBO J. 1998, 17, 2412–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kuzmic P Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal. Biochem 1996, 237, 260–273. [DOI] [PubMed] [Google Scholar]
- [54].Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994, 79, 1233–1243. [DOI] [PubMed] [Google Scholar]
- [55].Kong XP, Onrust R, O’Donnell M, Kuriyan J Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992, 69, 425–437. [DOI] [PubMed] [Google Scholar]
- [56].Kukshal V, Khanam T, Chopra D, Singh N, Sanyal S, Ramachandran RM tuberculosis Sliding β-Clamp Does Not Interact Directly with the NAD+ - Dependent DNA Ligase. PLOS One. 2012, 7, e35702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Chapados BR, Hosfield DJ, Han S, Qiu J, Yelent B, Shen B, et al. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 2004, 116, 39–50. [DOI] [PubMed] [Google Scholar]
- [58].Kelch BA, Makino DL, O’Donnell M, Kuriyan J How a DNA polymerase clamp loader opens a sliding clamp. Science. 2011, 334, 1675–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Shamoo Y, Steitz TA Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell. 1999, 99, 155–166. [DOI] [PubMed] [Google Scholar]
- [60].Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, et al. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res 2000, 33, 889–897. [DOI] [PubMed] [Google Scholar]
- [61].Gohlke H, Kiel C, Case DA Insights into Protein–Protein Binding by Binding Free Energy Calculation and Free Energy Decomposition for the Ras–Raf and Ras–RalGDS Complexes. Journal of Molecular Biology. 2003, 330, 891–913. [DOI] [PubMed] [Google Scholar]
- [62].Case DA, Betz RM, Cerutti DS, Cheatham TE III, Darden TA, Duke RE, Giese TJ, Gohlke H, Goetz AW, Homeyer N, Izadi S, Janowski P, Kaus J, Kovalenko A, Lee TS, LeGrand S, Li P, Lin C, Luchko T, Luo R, Madej B, Mermelstein D, Merz KM, Monard G, Nguyen H, Nguyen HT, Omelyan I, Onufriev A, Roe DR, Roitberg A, Sagui C, Simmerling CL, Botello-Smith WM, Swails J, Walker RC, Wang J, Wolf RM, Wu X, Xiao L and Kollman PA (2016), AMBER 2016, University of California, San Francisco. [Google Scholar]
- [63].Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, et al. The Amber biomolecular simulation programs. J. Comput. Chem 2005, 26, 1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Vriend G WHAT IF: a molecular modeling and drug design program. J. Mol. Graph 1990, 8, 52–56. [DOI] [PubMed] [Google Scholar]
- [65].Jorgensen WL, Chandrasekhar J, Madura1 JD, Impey WL, Klein ML Comparison of simple potential functions for simulating liquid water. J. Chem. Phys 1983, 79, 926–935. [Google Scholar]
- [66].Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG A smooth particle mesh Ewald method. J. Chem. Phys 1995, 103, 8577–8593. [Google Scholar]
- [67].Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, et al. Scalable molecular dynamics with NAMD. J. Comput. Chem 2005, 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins-Structure Funct. Bioinform 2006, 65, 712–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph Model 1996, 14, 33–38. [DOI] [PubMed] [Google Scholar]
- [70].Still WC, Tempczyk A, Hawley RC, Hendrickson T Semianalytical treatment of solvation for molecular mechanics and dynamics. Journal of the American Chemical Society. 1990, 112, 6127–6129. [Google Scholar]
- [71].Constanciel R, Contreras R Self consistent field theory of solvent effects representation by continuum models: Introduction of desolvation contribution. Theoret. Chim. Acta 1984, 65, 1–11. [Google Scholar]
- [72].Kellogg EH, Leaver-Fay A, Baker D Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins. 2011, 79, 830–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Atilgan C, Atilgan AR Perturbation-response scanning reveals ligand entry-exit mechanisms of ferric binding protein. PLOS Comput. Biol 2009, 5, e1000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Eyal E, Yang LW, Bahar I Anisotropic network model: systematic evaluation and a new web interface. Bioinformatics. 2006, 22, 2619–2627. [DOI] [PubMed] [Google Scholar]
- [75].Ikeguchi M, Ueno J, Sato M, Kidera A Protein Structural Change Upon Ligand Binding: Linear Response Theory. Phys. Rev. Lett 2005, 94, 078102. [DOI] [PubMed] [Google Scholar]
- [76].Atilgan AR, Durell SR, Jernigan RL, Demirel MC, Keskin O, Bahar I Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys. J 2001, 80, 505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Sethi A, Eargle J, Black AA, Luthey-Schulten Z Dynamical networks in tRNA:protein complexes. Proc. Natl. Acad. Sci. U. S. A 2009, 106, 6620–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Eargle J, Luthey-Schulten Z NetworkView: 3D display and analysis of protein.RNA interaction networks. Bioinformatics. 2012, 28, 3000–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.