Abstract
Acute respiratory disease caused by influenza viruses is imperfectly mitigated by annual vaccination to select strains. Development of vaccines that elicit lung-resident memory CD8+ T cells (TRM) would offer more universal protection to seasonal and emerging pandemic viruses. Understanding how lung-resident dendritic cells (DCs) regulate TRM differentiation would be an important step in this process. Here, we used CD11c-cre-Irf4f/f (KO) mice, which lack lung-resident IRF4-dependent CD11b+CD24hi DCs and show IRF4 deficiency in other lung cDC subsets, to determine if IRF4-expressing DCs regulate CD8+ memory precursor cells and TRM during influenza A virus (IAV) infection. KO mice showed defective CD8+ T cell memory, stemming from a deficit of T regulatory cells and memory precursor cells with decreased Foxo1 expression. Transfer of wild-type CD11b+CD24hi DCs into KO mice restored CD8+ memory precursor cell numbers to wild-type levels. KO mice recovered from a primary infection harbored reduced numbers of CD8+ TRM and showed deficient expansion of IFNγ+CD8+ T cells and increased lung pathology upon challenge with heterosubtypic IAV. Thus, vaccination strategies that harness the function of IRF4-dependent DCs could promote the differentiation of CD8+ TRM during IAV infection.
Keywords: lung immunity, dendritic cell, T cell memory, influenza
Introduction
Influenza virus infection results in acute respiratory disease and elicits innate immunity, neutralizing antibodies and cytotoxic effector and memory T cells1. Antiviral responses eliminate infected cells but may result in excess inflammation and immune-mediated pathology that prolong illness2. While current influenza vaccines elicit antibodies to strain-specific glycoproteins, vaccination strategies that promote differentiation and retention of memory T cells recognizing conserved internal proteins will enhance universal protection to seasonal and emerging pandemic viruses3,4. Optimal responses of lung-resident dendritic cells (DCs) will be a key component of such vaccine strategies5.
Lung-resident DCs, acting in the lung or after migration to mediastinal lymph nodes (mLN), coordinate immune responses to respiratory viruses. Migratory DCs include CD103+CD11b−CD24hi (CD103+) DCs and two subsets of CD11b+SIRPα+CD103− DCs distinguished as CD24hiCD11b+ (CD11b+P1) or CD24intCD11b+ (CD11b+P2)6,7. The transcription factors IRF8 and BATF3 control CD103+ DC differentiation, while IRF4 is required for the development of CD11b+P1 lung DCs6–9. Analogous subsets of IRF4+CD1c+ and IRF8+CD141+ DCs are present in human respiratory tissue7,10. Monocyte-derived “inflammatory” CD11b+ DCs (iDCs) also accumulate in the lung and mLN after respiratory infection11,12. Functional roles of lung-resident DCs and iDCs during respiratory virus infection have been defined11–18. In Batf3−/− mice, the absence of CD103+ DCs leads to decreased numbers of protective CD8+ T cells during respiratory virus infection including IAV8,19, consistent with their cross-presentation of viral antigens and early production of IL-128,17,20. CD11b+ DCs, including CCR2+ iDCs, express IL-12p70 and CD70 promoting TH1 and CD8+ T cell expansion11,15. However, the difficulty in distinguishing inflammatory and resident CD11b+ DCs with shared markers has hampered definition of how lung-resident CD11b+P1 and CD11b+P2 DCs regulate T cells during IAV infection. CD11c-cre-Irf4f/f mice provide an opportunity to resolve this issue6. In homeostasis, CD11c-cre-Irf4f/f mice lack the IRF4-dependent lung-resident CD11b+P1 DCs, while CD11b+P2 and CD103+ DC subsets are present in normal numbers but lack IRF46. A distinct CD11c-cre-Irf4f/f model, in which a different CD11c-cre construct does not lead to absence of the CD11b+P1 DCs, did not show defects in T cell responses in primary IAV infection, and T memory responses were not studied21.
Viral infection results in circulating CD62LhiCCR7hi central memory T cells (TCM) and CD62LloCCR7lo effector memory T cells (TEM) and extravascular CD69+CD103+ resident memory T cells (TRM) retained in peripheral tissues22, 23. After IAV infection, lung TRM cells contribute to protection against heterosubtypic viruses in mice, although they wane over time in the lower respiratory tract4,24–27. TRM also have been identified in human lungs28, 29. The role of distinct DC subsets in formation of memory T cells during IAV infection is not completely understood. Prior reports showed that CD11b+ DCs support differentiation of CD8+ TCM cells19, while CD103+ DCs promote optimal CD8+ TRM priming but not subsequent TRM differentiation or circulating memory T cells30. Collaboration between functionally unique DC subsets may regulate an optimal balance of CD8+ T effector and memory populations by influencing the differentiation of late effector subsets. In LCMV infection, abundant IL-12 favors IL-7RαloKLRG1+T-BEThi short-lived effector cells (SLECs), and IL-10 promotes IL-7RαhiKLRG1−T-BETlo memory precursor effector cells (MPECs)31. Upon infection resolution, the pool of SLECs contracts, while MPECs convert to memory T cells by upregulating FOXO131–33. T regulatory cell (TREG) production of IL-10 promotes CD8+ memory T cells during LCMV infection34, but the role of DCs in the regulation of induced TREGs or MPECs during IAV infection is less well understood35,36. IRF4 in DCs promotes transcription of Il1021 and inhibits IL-1237, suggesting that IRF4 regulates the balance of IL-12 and IL-10.
Herein, we report that during IAV infection, CD11c-cre-Irf4f/f mice showed increased numbers of IAV-specific IFNγ+CD8+ effector T cells but a marked deficit in FOXP3+ TREGs and CD8+ MPECs. Recovered CD11c-cre-Irf4f/f mice harbored lower numbers of lung CD8+ TEM and TRM cells, and upon exposure to heterosubtypic IAV, showed reduced expansion of IAV-specific IFNγ+TNFα+CD8+ T cells, which correlated with increased lung damage. Thus, IRF4-expressing lung resident DCs promote the differentiation of CD8+ memory T cells during IAV infection.
Results
CD11c-cre-Irf4f/f mice contain greater numbers of influenza antigen-specific CD4+ and CD8+ IFNγ+ T cells in the mediastinal LN after IAV infection
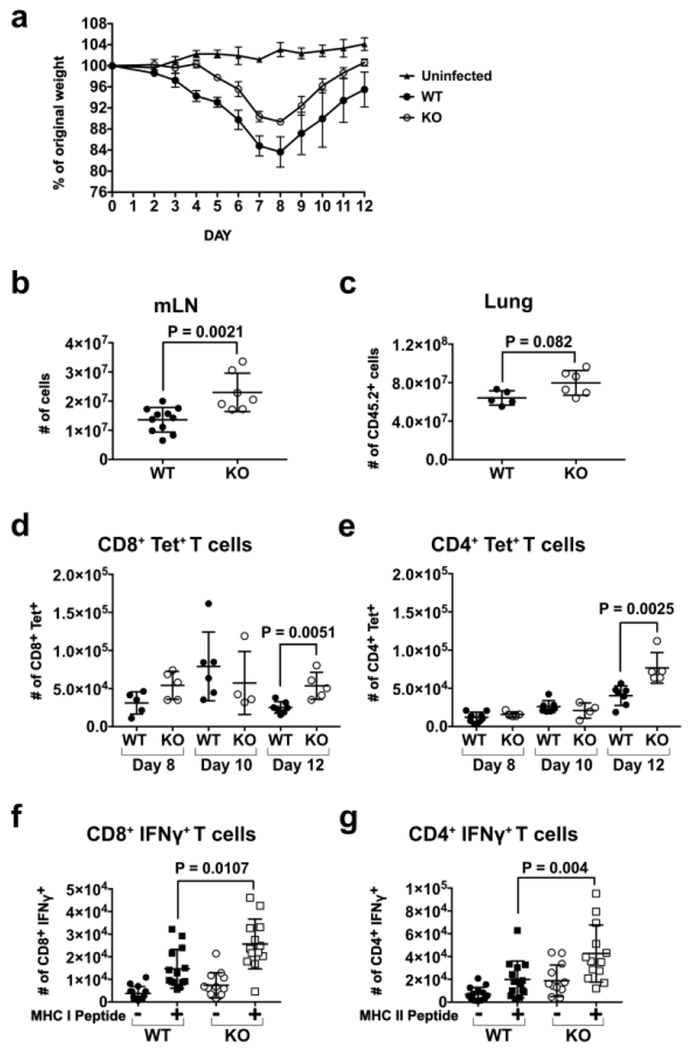
To determine the role of IRF4-dependent DCs during IAV infection, we infected CD11c-cre-Irf4+/+ (WT) and CD11c-cre-Irf4f/f (KO) mice intranasally with a sublethal dose of A/Puerto Rico/8/1934 (PR8) virus. We monitored daily weight loss as a sign of morbidity and the kinetics of virus clearance, but we did not identify significant reproducible differences in these parameters between WT and KO mice (Fig. 1a, Fig. S1a). However, by day 12 post-infection (p.i.), the KO mice showed significantly increased (1.7-fold) numbers of mLN cells (Fig. 1b) and tended to have more CD45.2+ cells in the lung compared to the WT mice (Fig. 1c), suggesting a greater immune response to virus.
Fig. 1. CD11c-cre-Irf4f/f mice harbor higher numbers of antigen-specific CD4+ and CD8+ IFNγ+ T cells in the mLN after IAV infection.
WT and KO mice were infected i.n. with a sublethal dose of PR8 IAV. (a) Weight loss and recovery of WT and KO mice (n=3 each) over 12 days p.i. with PR8 virus. Shown are the mean and SEM. Differences in WT and KO mice were not significant as determined by a multiple t test with the Bonferroni-Sidak method, (b) Total numbers of mLN cells on day 12 p.i. (c) Total numbers of CD45+ cells in the lung on day 12 p.i. (d) Numbers of CD8+ T cells binding H-2Db/NP366-374 tetramers (Tet) and (e) numbers of CD4+ T cells binding I-Ab/NP311-325 tetramers in the mLN on days 8, 10 and 12 p.i. (f) Numbers of CD8+ T cells producing IFNγ after incubation of mLN cells (day 12 p.i.) in vitro with or without NP366-374 peptide. (g) Numbers of CD4+ T cells producing IFNγ after incubation of mLN cells (day 12 p.i.) in vitro with or without NP311-325 peptide. Gating of T cells is shown in Fig. S2a. The data are compiled from 1-2 (a-e) or 4 (f-g) independent experiments, each with 3-4 animals per WT or KO group. Symbols represent individual mice, with the mean and SD indicated. Significance (b-g) was evaluated using Mann-Whitney tests, with p values indicated.
To determine if CD4+ and CD8+ IAV antigen-specific T cells expanded more in KO mice during the infection, we measured numbers of CD4+ and CD8+ nucleoprotein (NP)-specific T cells in the mLN using H-2Db/NP366-374 and I-Ab/NP311-325 tetramers on days 8,10 and 12 p.i. Although numbers of tetramer-positive (Tet+) CD4+ and CD8+ T cells in WT and KO mice were similar on days 8-10 p.i., the numbers of Tet+CD8+ T cells in WT had contracted while the numbers in KO mice remained ~2-fold elevated on 12 p.i. (Fig. 1d). Similarly, on day 12 p.i., numbers of Tet+CD4+ T cells were higher in KO mice (Fig. 1e). To determine if antigen-specific T cells in WT and KO differed in functional capacity, mLN cells were isolated on day 12 p.i. and activated ex vivo with IAV-specific peptides: I-Ab-binding NP311-325 and H-2Db-binding NP366-374 (Fig. S2a). While antigen-specific WT and KO CD4+ and CD8+ T cells produced comparable amounts of IFNγ as judged by mean fluorescence intensity (MFI) (Fig. S1b) and the fraction (frequency of live cells) of mLN CD8+ or CD4+ mLN T cells producing IFNγ was not different (Fig. S1c), KO mice harbored significantly increased (1.7-2-fold) numbers of antigen-specific CD8+ and CD4+ IFNγ+ T cells (Fig. 1f,g). This is consistent with elevated IFNγ in the bronchoalveolar lavage fluid of KO mice compared to WT mice (Fig. S1d). However, numbers of NP-specific CD8+ and CD4+ T cells capable of producing IFNγ in the lung on day 12 p.i. did not differ in WT and KO mice (Fig. S1e). These data show that KO mice expand greater numbers of CD4+ and CD8+ effector T cells in the mLN during IAV infection.
CD11c+ lung DCs in CD11c-cre-Irf4f/f mice show decreased IL-10 and increased IL-12a RNA levels during IAV infection
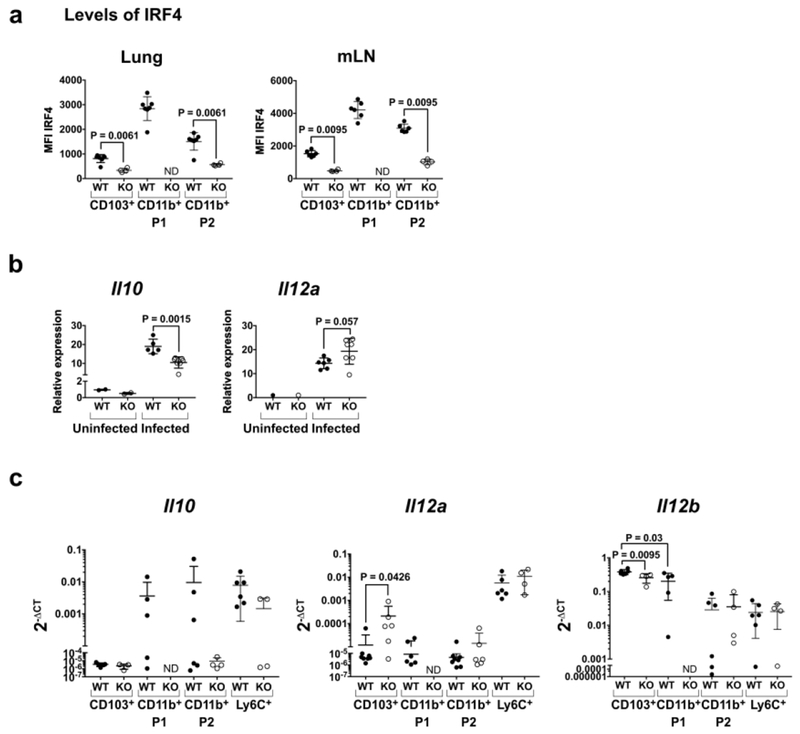
Upon IAV infection, the lung-resident CD103+ and CD11b+ DCs acquire and present viral antigens, produce cytokines including IL-12 and IL-10 and migrate to the draining mLN where they elicit an adaptive immune T cell response leading to production of IFNγ1. Migratory CD103+ DCs are a significant source of IL-12 at early time points post-infection by intracellular pathogens, while IRF4-expressing CD11b+ DCs may be a primary source of IL-1020,21. In infected WT mice, the CD11b+ and CD103+ lung-resident migratory DC subsets express distinct levels of IRF4 in the lung and mLN, with the highest level in CD11b+P1 DCs (Fig. 2a; Fig. S2b). To determine if IRF4 deficiency might alter the balance of DC-produced IL-12 and IL-10, total CD11c+ cells from the lung were isolated day 3 p.i. and assayed for Il10 and Il12a RNA by qPCR. In the KO mice, CD11c+ lung cells expressed significantly lower (1.8-fold) levels of Il10 and tended to express higher levels of Il12a (Fig. 2b).
Fig. 2. CD11c+ lung DCs in CD11c-cre-Irf4f/f mice show decreased Il10 and increased Il12a RNA upon IAV infection.
(a) Expression of IRF4 determined by intracellular staining of the indicated DC subsets in lung and mLN of WT and KO mice on day 5 p.i. Gating of DC subsets is shown in Fig. S2b. (b) Relative levels of Il10 and Il12a RNA in total CD11c+ lung cells on days 0 and 3 p.i. determined by qPCR. (c) Levels of Il10, Il12a and Il12b RNA relative to Gapdh RNA in sorted lung DC subsets on day 5 p.i. determined by qPCR and compiled from 2 independent experiments. In all panels, symbols represent individual mice, with the mean and SD indicated. ND, not determined due to absence of the CD11b+P1 subset in KO mice. Significance was evaluated using a t test (panel b) or one-way ANOVA with a Tukey’s multiple comparison test (panels a-c), with p values indicated.
To identify the DC subsets synthesizing Il10, Il12a and Il12b RNA, we sorted lung DC subsets on day 5 p.i. CD11c+MHCII+Ly6C−CD88− lung-resident DCs were sorted as CD103+CD11b−(CD103+), CD103−CD11b+SIRPα+CD24hi (CD11b+P1) and CD103-CD11b+SIRPα+CD24int (CD11b+P2); all three subsets were CD64− (Fig. S2b). We also sorted CD11b+Ly6Chi iDCs that accumulate by day 5 p.i.; for KO mice, we sorted the ~50% of iDCs that were GFP+, indicating Irf4 deletion. WT CD11b+ P1 and P2 DCs contained Il-10 RNA (Fig. 2c), although this RNA was not detectable in all WT mice, perhaps due to the kinetics of infection varying in individual mice. At early time points, some DCs have migrated from the lung to the mLN, and the number of activated (IL-10 or IL-12a producing) DCs remaining in each lung may vary. Ly6Chi iDCs also contained 11-10 RNA, and these levels were reduced in IRF4−/− GFP+ iDCs in some KO mice (Fig. 2c). Among WT DCs, Il12a RNA was detectable in Ly6Chi iDCs, while all DC subsets contained Il12b RNA (Fig. 2c). Notably, Il12a RNA was elevated ~17-fold in KO CD103+ DCs (Fig. 2c). Thus, the absence of the CD11b+P1 DCs and Irf4-deficiency in other DC subsets led to decreased Il10 and increased Il12a RNA in DCs.
CD11c-cre-Irf4f/f mice show quantitative and qualitative defects in CD8+ memory precursor T cells during the resolution phase of infection
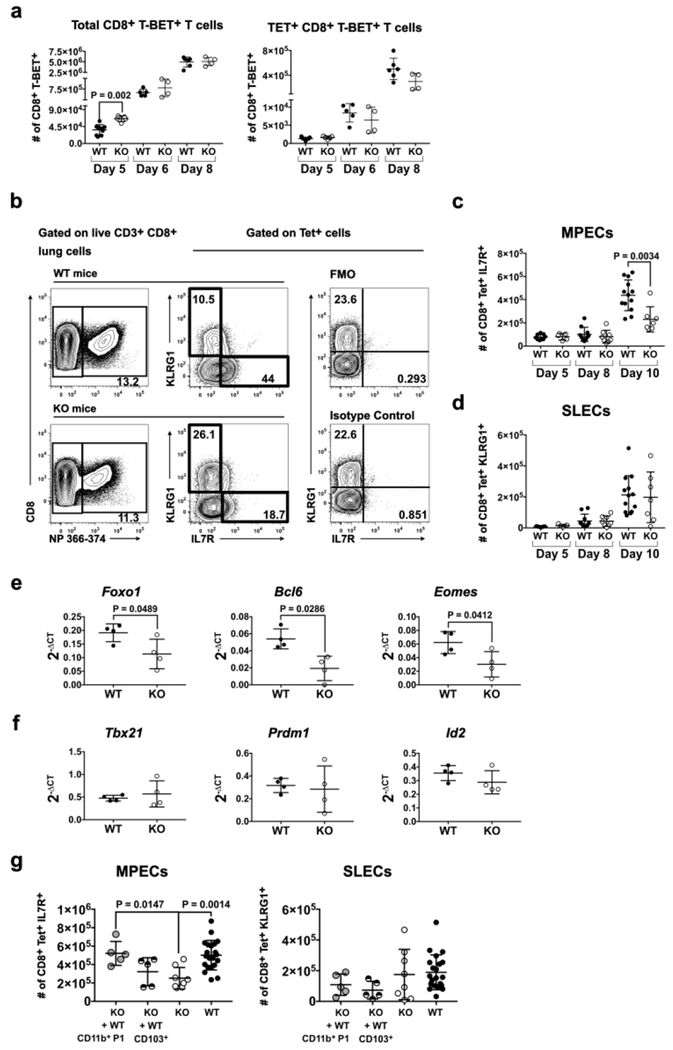
IL-12 induces the expression of the transcription factor T-BET in T cells thus promoting the differentiation of IFNγ-producing effector T cells31. Consistent with elevated Il12a and reduced Il10 RNA in KO DCs, KO mice have increased (~1.8-fold) numbers of CD44hi CD8+ and CD4+ T cells expressing T-BET compared to WT mice in the lung on day 5 p.i. (Fig. 3a; Fig. S3a,b). On day 5 p.i., we detected very few Tet+ T cells, and it was difficult to determine if they had higher levels of T-BET in KO mice. T-BET+ T cell numbers (either Tet+ or the total pool) were similar in WT and KO mice days 6 and 8 p.i. (Fig. 3a), suggesting distinct kinetics in WT and KO mice. Depending on the expression level of T-BET, CD8+ T cells differentiate into SLEC and MPEC subsets of late effectors as determined by IL-7Rα and KLRG1 expression (Fig. 3b; Fig. S3c)31. By day 10 p.i., KO mice showed a significant reduction (1.9-fold) in numbers of antigen-specific (Tet+) IL-7Rα+KLRG1− MPECs in the lung (Fig. 3c; Fig. S3c). However, numbers of Tet+IL-7Rα− KLRG1+ SLECS did not differ in WT and KO lungs (Fig. 3d).
Fig. 3. CD11c-cre-Irf4f/f mice show quantitative and qualitative defects in CD8+ memory precursor T cells during the resolution phase of infection.
Lung cells were harvested from infected WT and KO mice on day 5, 6, 8 and 10 p.i. Surface and intracellular flow cytometry identified subsets of CD4+ and CD8+ late effector T cells, gated as in Fig. S3b,c. (a) Numbers of CD8+CD44hiT-BET+ and CD8+CD44hiT-BET+ Tet(NP366-374)+ T cells, (b) Gating of Tet(NP366-374)+CD8+KLRG1−IL-7Rα+ MPECs and CD8+Tet+KLRG1+IL-7Rα− SLECs. (c) Numbers of MPECs on day 5, 8 and 10 p.i. (d) Numbers of SLECs on day 5, 8 and 10 p.i. (e) Expression of Foxo1, Bcl6 and Eomes RNA in CD8+Tet+KLRG1−IL-7Rα+ MPECs sorted on day 10 p.i. determined by qPCR. (f) Expression of Tbx21, Prdm1 and Id2 genes related to T cell effector function in MPECs sorted on day 10 p.i. determined by qPCR. Data in panels a-f are from 1-3 independent experiments each with 4-5 animals per WT or KO group. (g) Numbers of MPECs and SLECs on day 10 p.i. after WT CD11b+ P1 or CD103+ DC transfer into KO mice at the time of IAV infection. Data sets showing DC transfer in panel g are from 3 independent experiments, with each experiment involving transfer of either DC subset into 1-2 KO recipients; KO and WT alone data sets include mice analyzed in the DC transfer experiments and the data shown in panels c and d. In all panels, symbols represent individual mice, with the mean and SD indicated. Significance was evaluated using Mann-Whitney (panels a, c-d), unpaired t (panels e-f), and one-way ANOVA with Tukey’s multiple comparison test (panel g), with p values indicated.
To determine if the MPECs in KO mice show an altered transcriptional program, we sorted CD8+Tet+MPECs (GFP−) from WT and KO lungs on day 10 p.i. and assayed expression of selected genes important for memory T cell differentiation (Fig. 3e,f). KO MPECS contained reduced (1.7-fold) levels of Foxo1 RNA, a transcription factor that regulates the program of memory T cells (Fig. 3e)38,39. RNA levels of the FOXO1-regulated transcription factors Bcl6 and Eomes, which promote the development of MPECs to memory CD8+ T cells, also were reduced (2-2.8-fold) in KO MPECs (Fig. 3e). RNA levels of genes involved in CD8+ T cell effector function (Tbx21, Prdm1, Id2) and survival (Bcl2, Bcl2l11, Noxa), tissue residence of TRM (Klf2, S1pr1), or regulation of memory (Foxo3) were not different in WT and KO MPECs (Fig. 3f; Fig. S3d–f). Taken together, these data suggest that the absence of the CD11b+P1 DC subset and changes in the balance of DC-produced IL-12 and IL-10 in infected KO mice result in decreased numbers of CD8+ MPECs with reduced capacity to complete memory T cell differentiation.
To determine which DC subset regulates CD8+ Tet+ MPEC numbers, we performed adoptive intranasal transfer of sorted WT CD11b+P1 or CD103+ DC subsets into KO mice, followed immediately by PR8 virus infection. By day 10 p.i., KO mice that received WT CD11b+P1 DCs showed a 2-fold increase in numbers of MPECs, compared to KO not receiving transferred DCs (Fig. 3g). Transfer of WT CD103+ DCs did not lead to a significant increase in MPECs (Fig. 3g). Numbers of SLECs did not change upon DC transfer (Fig. 3g). These data show that CD11b+P1 DCs preferentially promote MPEC numbers in the primary infection.
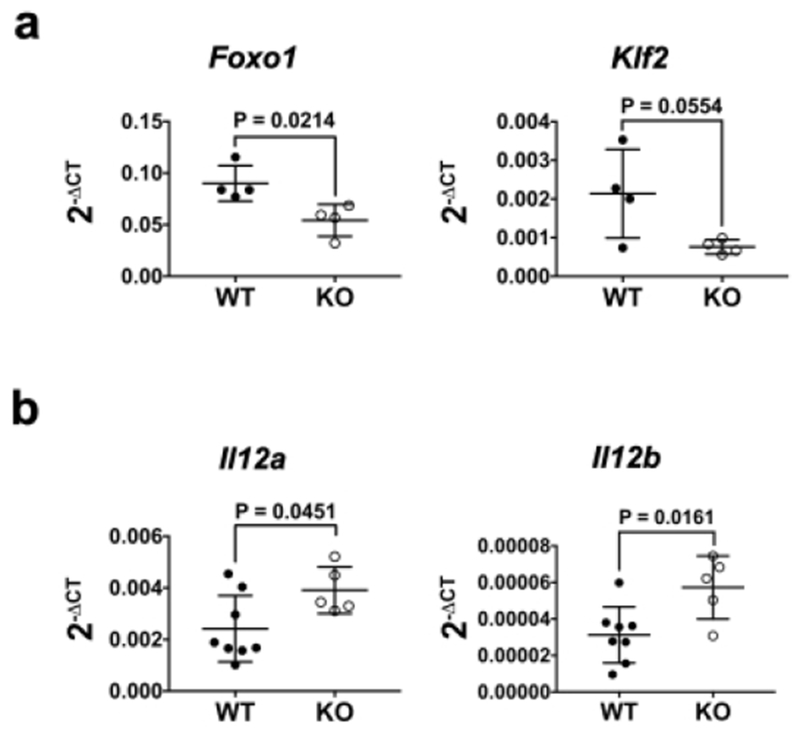
Tet+CD8+ T cells in the mLN of KO mice on day 10 p.i. also contained reduced (1.7-2.8-fold) amounts of Foxo1 and Klf2 RNA (Fig. 4a). This was consistent with increased (1.6-1.9-fold) amounts of Il12a and Il12b RNA in the mLN of KO mice on day 8 p.i. (Fig. 4b). Since FOXO1 opposes CD8+ T cell effector programming and KLF2 regulates LN egress38,40, the reduction of Foxo1 RNA may promote retention of T cell effectors in the mLN. Indeed, we observed a greater number of Tet+CD8+ T cells in the mLN of KO mice on day 12 p.i. (Fig. 1d).
Fig. 4. The mLN of CD11c-cre-Irf4f/f mice contains elevated Il12 RNA and antigen-specific CD8+ T cells with reduced expression of Foxo1 RNA late in infection.
(a) CD8+ Tet+ T cells (gated as in Fig. 3b) were sorted from the mLN on day 10 p.i. and assessed for expression of Foxo1 and Klf2 RNA by qPCR. (b) Total mLN RNA was assessed for expression of Il12a and Il12b RNA by qPCR. Symbols represent individual mice, with the mean and SD indicated. Significance was evaluated using unpaired t tests, with p values indicated.
CD11c-cre-Irf4f/f mice show reduced numbers of CD4+ T regulatory cells in the mLN and lung after infection
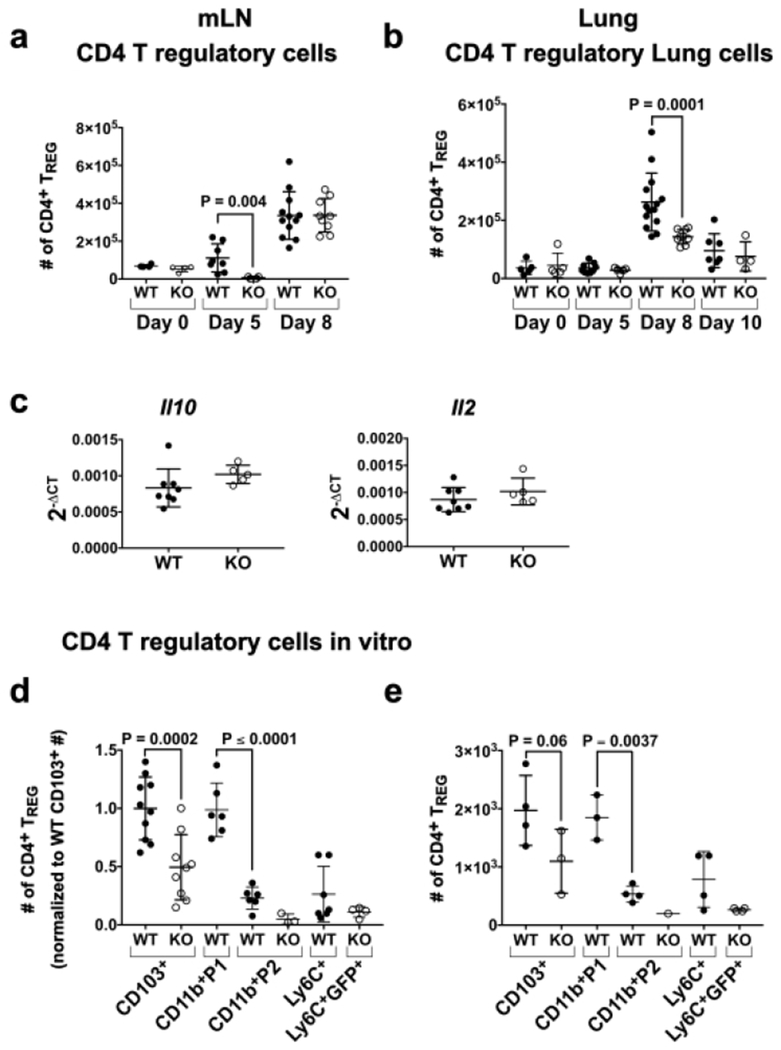
IL-10 produced by CD4+ TREGs is required for the transition of MPECs to memory T cells in LCMV infection34. Thus, we hypothesized that decreased numbers of induced TREGs may underlie the MPEC defect in KO mice. In homeostasis, the low numbers of CD4+CD25+FOXP3+ TREGs in the mLN and lung did not differ in WT and KO mice (Fig. 5a,b; Fig. S4a). While the numbers of TREGs in the mLN of WT were increased by day 5 p.i., no increase in TREGs was observed in KO mice (16-fold difference) (Fig. 5a). By day 8 p.i., numbers of TREGs in the mLN were comparable in WT and KO mice, suggesting a delay in their differentiation or accumulation in KO mice. Consistent with this, RNA levels of the TREG promoting cytokines Il10 and 112 in the mLN did not differ in WT and KO mice on day 8 p.i. (Fig. 5c). Numbers of TREGs peaked in the lung on day 8 p.i. in WT mice, but notably this expansion was markedly attenuated (1.8-fold) in KO mice and did not occur at later time points (Fig. 5b).
Fig. 5. CD11c-cre-Irf4f/f mice show reduced numbers of CD4+ FOXP3+ TREGs in the mLN and lung after IAV infection.
(a-b) Lung and mLN cells were harvested from infected mice on days 0, 5, 8 and 10 p.i. Surface and intracellular staining identified TREGs (Fig. S4a). Shown are numbers of CD4+CD25+FOXP3+ TREGs in the (a) mLN and (b) lung, (c) Expression of Il10 and Il2 RNA in total mLN cells on day 8 p.i. determined by qPCR. (d) Numbers of FOXP3+ TREGs in cultures normalized to the average of WT CD103+mice (gated as in Fig. S4b) after in vitro incubation of naïve CD4+ OT-II T cells with OVA323-339 peptide and isolated lung DC subsets (1000 per well) sorted on day 3 p.i. (e) Numbers of FOXP3+ TREGs in cultures from one representative experiment. Symbols represent individual mice, with the mean and SD indicated. The data are compiled from 1-3 independent experiments each with 3-5 animals per WT or KO group. Significance was evaluated using a Mann-Whitney test (panels a-c) or one-way ANOVA with Tukey’s multiple comparison test (panel d-e), with p values indicated.
Differentiation of naïve CD4+ T cells to FOXP3+ TREGs is promoted by DC production of IL-10 and TGFβ41. To compare the ability of the lung-resident DCs in IAV-infected mice to promote TREG differentiation, we sorted lung DC subsets on day 3 p.i. and incubated them with naïve CD4+CD25− T cells from OT-II mice (bearing an OVA-specific TOR transgene) and OVA 323-339 peptide. After 6 days, numbers of CD4+CD44+CD25+FOXP3+ T cells were quantified; shown are a compilation of 3 experiments, in which data were normalized, and the TREG numbers in one representative experiment (Fig. 5d,e; Fig. S4b). WT CD103+ DCs induced TREG differentiation, and this capacity was diminished ~2-fold in KO CD103+ DCs. While WT CD11b+P1 DCs also induced significant TREG numbers, TREG induction by the other CD11b+ DCs (P2 and Ly6Chi iDCs) was minimal. Taken together, these data suggest that both Irf4 deficiency in CD103+ DCs and the absence of CD11b+P1 DCs decreased TREG differentiation in KO mice during IAV infection.
CD11c-cre-Irf4f/f mice recovered from infection show reduced numbers of CD8+ resident memory T cell populations in the lung.
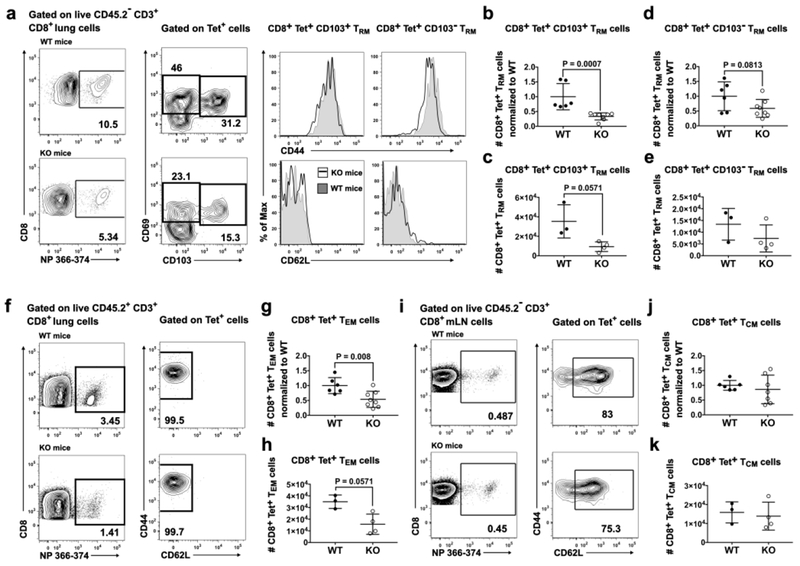
In view of the deficit in CD8+ MPECs and CD4+ TREGs in KO mice, we next investigated the T cell memory populations that remained in the lung and mLN on day 42 p.i. Memory T cells in the lung were identified as extravascular (resident memory, TRM) or vascular (effector memory, TEM) after i.v. injection of a labeled anti-CD45 mAb shortly before euthanasia (Fig. 6a,f; Fig. S5a,b). KO mice showed a 3-fold reduction in numbers of CD8+Tet+CD69+CD44+CD62L−CD103+ TRM, and a trend to reduced numbers of CD8+Tet+CD69+CD44+CD62LȡCD103ȡ TRM (Fig. 6b–e). Shown are a compilation of two experiments with numbers normalized to the average value in WT mice (Fig. 6b,d) and cell numbers from one representative experiment (Fig. 6c,e). Numbers of CD8+Tet+CD44+CD62L−CD69−CD103− TEM also were reduced (1.9-fold) in KO mice (Fig. 6g,h; Fig. S5b). In contrast, numbers of CD8+Tet+CD44+CD62L+ TCM in the mLN were not different in WT and KO mice (Fig. 6i–k; Fig. S5c). Numbers of CD4+Tet+ TRM, TEM and TCM did not differ in WT and KO mice (Fig. S5d). Thus, low numbers of MPECs in KO mice at the late stage of infection correlated with reduced numbers of lung CD8+ TRM and TEM in mice fully recovered from infection.
Fig. 6. CD11c-cre-Irf4f/f mice recovered from primary IAV infection show reduced numbers of CD8+ TRM and TEM populations in the lung.
On day 42 p.i., CD45.2+ cells in blood were marked by i.v. injection of an a-CD45 mAb before euthanasia and identification of memory T cell subsets using flow cytometry (Fig. S5). (a) Gating of extravascular (i.v. CD45~) CD8+Tet+CD69hiCD44hiCD62L“CD103+ or CD103− TRM cells in the lung, (b) Numbers of CD8+Tet+CD69hiCD44hiCD62l−CD103+TRM normalized to the average of WT mice, (c) Numbers of CD8+Tet+CD69hiCD44hiCD62l−CD103+TRM cells from one representative experiment, (d) Numbers of CD8+Tet+CD69hiCD44hiCD62L−CD103-TRM normalized to the average of WT mice, (e) Numbers of CD8+Tet+CD69hiCD44hiCD62L“CD103− TRM cells from one representative experiment, (f) Gating of vascular (i.v. CD45+) CD8+Tet+CD62L-CD44+ TEM cells, (g) Numbers of CD8+Tet+CD44+CD62L−CD69−CD103- TEM normalized to the average of WT mice, (h) Numbers of CD8+Tet+CD44+CD62I_-CD69-CD103-TEM cells from one representative experiment, (i) Gating of CD8+Tet+CD62L+CD44+ TCm in the mLN. (j) Numbers of CD8+Tet+CD62L+CD44+ TCm normalized to the average of WT mice, (k) Numbers of CD8+Tet+CD62L+CD44+ TCm from one representative experiment. Symbols represent individual mice, with the mean and SD indicated. Normalized data are compiled from 2 independent experiments with 3–4 animals per WT and KO group. Significance was evaluated using a Mann-Whitney test, with p values indicated.
CD11c-cre-Irf4f/f mice have a decreased CD8+ IFNγ+ T cell response upon secondary infection with a heterosubtypic virus
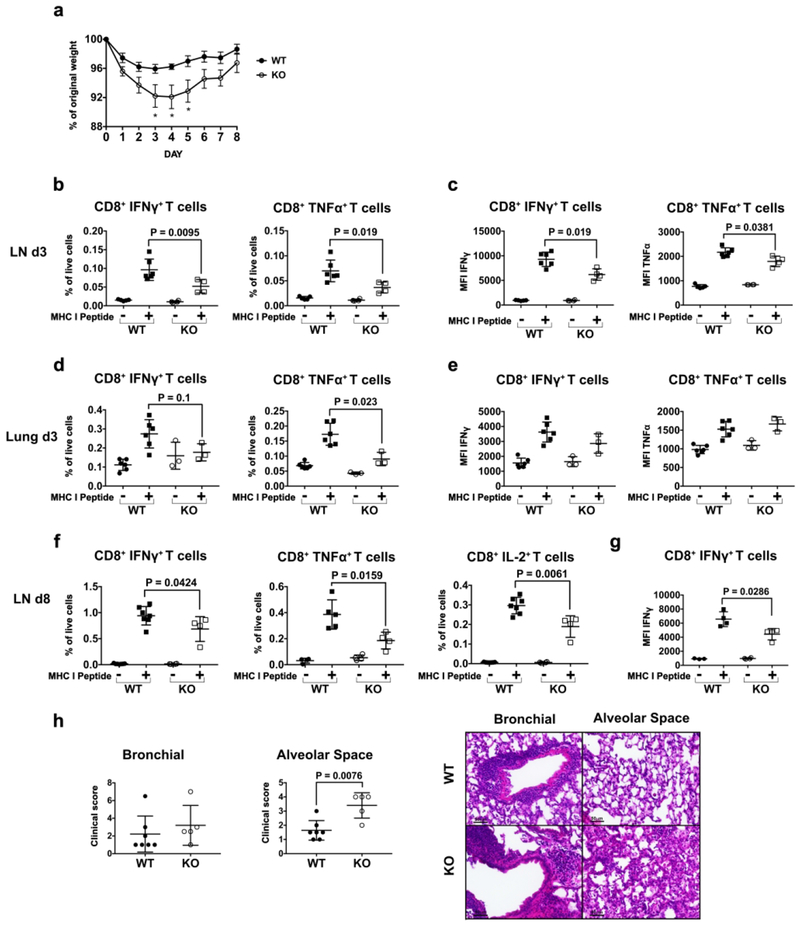
To determine the functional consequences of the CD8+ T memory defect in KO mice, sequential primary and secondary infections were done with the related X31 (H3N2) and PR8 (H1N1) viruses that share conserved NP T cell epitopes but differ in the HA and NA proteins that are targets of neutralizing antibodies42. WT and KO mice were infected with a sublethal dose of the less virulent X31, and after 42 days, the mice were infected with a lethal dose of the more virulent PR8. During the secondary infection with PR8, the KO mice lost more weight than WT mice, suggesting increased morbidity (Fig. 7a; Fig. S6a). The viral load in WT and KO mice was not different on day 3 p.i. and is not detectable by day 8 p.i. (not shown). On days 3 and 8 p.i., T cells in the mLN and lung were activated ex vivo with the I-Ab-binding NP311-325 and Db-binding NP366-374 peptides. On day 3 p.i., KO mice showed a significant decrease (~1.9-fold) in the fraction (% of live cells) of NP-specific CD8+ T cells producing IFNγ or TNFα in the mLN (Fig. 7b), but differences in absolute numbers of these cells did not reach statistical significance (Fig. S6b). The amount of IFNγ and TNFα per cell was also decreased 1.5-1.9-fold in KO mice, as judged by MFI (Fig. 7c). In the lung, KO mice showed a 1.9-fold decrease in the fraction (% of live cells) of NP-specific CD8+ T cells producing TNFα (Fig. 7d) and 1.9-fold decreased numbers of antigen-specific CD8+ TNFα+ or IFNγ+ cells (Fig. S6c), but no differences in the amount of IFNγ and TNFα per cell (Fig. 7e). On day 8 p.i. in the mLN, the fraction (% of live cells) of NP-specific CD8+ T cells producing IFNγ, TNFα or IL-2 and the amount of IFNγ per CD8+ T cell also were reduced (1.5-2-fold) in KO mice (Fig. 7f,g), although differences in absolute numbers of these cells did not reach statistical significance (Fig. S6d). In contrast, KO mice showed no difference in the fraction of NP-specific CD4+IFNγ+ T cells in the mLN compared to WT mice on day 3 or day 8 p.i. (Fig. S6e,f). H&E staining of lung tissue sections on day 8 p.i. revealed that while peri-bronchial inflammation was similar in WT and KO mice, the KO mice showed a 2-fold increase in parenchymal damage characterized by increased alveolar inflammation and thickening of the alveolar wall (Fig. 7h). Extensive lung injury due to viral infection in KO mice is also visible as inflammatory exudate in alveolar spaces and desquamated alveolar epithelial cells upon Periodic Acid-Schiff staining (Fig. S6h).
Fig. 7. CD11c-cre-Irf4f/f mice show a decreased CD8+ effector T cell response upon secondary infection with a heterosubtypic IAV.
WT and KO mice were infected with X31 virus and then infected with a lethal dose of PR8 virus 41 days later, (a) Weight loss over 8 days p.i. Shown are the mean and SEM, n=5-7. Data were analyzed using a multiple t test with the Bonferroni-Sidak method. Significant results (*) correspond to an adjusted p<0.05. (b) On day 3 p.i., the fraction (frequency of live cells) of CD8+ T cells producing IFNγ or TNFα (gated as in Fig. S2a) after incubation of mLN cells in vitro with or without NP366–374 peptide, (c) The MFI of IFNγ or TNFα expression in NP-specific mLN T cells on day 3 p.i. (d) On day 3 p.i., the fraction (frequency of live cells) of CD8+ T cells producing IFNγ or TNFα after incubation of lung cells in vitro with or without NP366-374 peptide, (e) The MFI of IFNγ or TNFα expression in NP-specific lung T cells on day 3 p.i. (f) On day 8 p.i., the fraction (frequency of live cells) of CD8+ T cells producing IFNγ, TNFα or IL-2 after incubation of mLN cells in vitro with or without NP366–374 peptide, (g) The MFI of IFNγ expression in NP-specific mLN T cells on day 8 p.i. (h) Lung sections (day 8 p.i.) stained with H&E were scored for inflammation and tissue damage in the bronchial and alveolar space. In panels b-h, symbols represent individual mice, with the mean and SD indicated. The data shown are from one of two independent experiments with 4-6 animals each per WT and KO group. Significance was evaluated using Mann-Whitney tests, with p values indicated.
Similarly, upon primary infection with a sublethal dose of the more virulent PR8 virus followed by secondary infection with X31 virus, KO mice showed decreased numbers of antigen-specific CD8+IFNγ+ and CD8+IL-2+ T cells but no difference in the numbers of antigen-specific CD4+IFNγ+ T cells compared to WT mice (Fig. S6g). These data show that defects in the CD8+ TRM and TEM subsets after a primary influenza infection in KO mice led to a weaker CD8+ T cell effector response upon infection with a heterosubtypic virus, correlating with increased morbidity and lung pathology.
Discussion
In the context of a heterogeneous CD11b+ population encompassing lung-resident DCs and iDCs, the role of the resident CD11b+CD24hi DC subset in effector and memory T cell differentiation during IAV infection remained unclear. To address this, we used CD11c-cre-Irf4f/f (KO) mice bearing Irf4 deficiency in CD11c+ cells, which in homeostasis lack the IRF4-dependent lung-resident CD11b+CD24hi P1 DCs, while CD11b+CD24int P2 and CD103+ DC subsets are present in normal numbers but lack IRF46. In a sublethal IAV infection, these DC defects resulted in increased numbers of IFNγ+CD8+ T cell effectors yet reduced numbers of MPECs. The remaining MPECs in KO mice expressed lower levels of Foxo1 and its downstream targets Bcl6 and Eomes important for solidification of the memory CD8+ T cell transcriptional program. These quantitative and qualitative defects in the MPEC population led to decreased numbers of CD8+ TRM and TEM cells in the memory T cell pool of recovered mice. Upon infection with a heterosubtypic IAV, NP-specific CD8+ T cell effector responses were significantly reduced in the KO mice and correlated with increased lung parenchyma pathology consistent with injury caused by viral infection. Thus during IAV infection, IRF4-expressing DCs act to attenuate the CD8+ T cell effector response while promoting a population of IAV-specific CD8+ memory precursor cells that ultimately leads to a robust population of tissue-localized TRM and circulating TEM cells.
While the molecular mechanisms of memory T cell differentiation have been well defined32,33, our understanding of the possibly distinct roles of DC subsets in CD8+ memory T cell differentiation during IAV infection was incomplete. Early cross-presentation of viral antigens by CD103+ DCs is important for priming of TRM, but CD103+ DCs are dispensable for full TRM differentiation or generation of circulating memory T cells30. The role of CD11b+P1 DC responses in IAV infection were previously queried with different CD11c-cre-Irf4f/f mice bearing a “late-acting” CD11c-cre that does not delete Irf4 in pre-cDCs, leaving normal numbers of the CD11b+P1 subset in KO mice21. These KO mice did not show any differences in the fraction of IAV-specific CD4+ or CD8+ IFNγ or TNFα T cells on day 8 p.i. This suggested that IRF4 expression in CD11b+P1 and other lung cDCs was not required for IAV-specific T cell responses. However, differentiation of effector CD8+ T cell subsets (SLECs and MPECs) and memory responses were not investigated, so it is difficult to compare their findings to work in our model in which an “early-acting” CD11c-cre leads to the absence of the CD11b+P1 DCs as well as IRF4 deficiency in CD103+ DCs. Because the major difference between the two lines of CD11c-cre-Irf4f/f mice is the absence of CD11b+P1 DCs, we hypothesized that these DCs promote CD8+ memory T cells. To determine if either the CD11b+P1 or CD103+ DCs preferentially promote MPECs, we transferred sorted WT lung DC subsets into KO mice at the time of infection. Importantly, transfer of WT CD11b+P1 DCs, but not CD103+ DCs, into KO mice led to increased numbers of MPECs on day 10 p.i. TREG numbers in the lung have waned at this time point, so it was not possible to determine if the CD11b+P1 or CD103+ DCs promote TREG differentiation in these experiments. These data support our hypothesis that the absence of the CD11b+P1 DCs is the primary driver of the CD8+ T memory defect in CD11c-cre-Irf4f/f mice. This finding also is consistent with the above mentioned study showing that CD103+ DCs are not required for complete TRM differentiation30.
Our data suggest that CD11b+P1 DCs regulate both the lung TRM and TEM pools by promoting expression of Foxo1, Bcl6 and Eomes in MPECs, thus ensuring their complete differentiation and/or maintenance. Notably, FOXO1 opposes CD8+ T cell effector programming while instructing early memory T cell fate, and Foxo1-deficient mice contain fewer memory T cells and more CD8+ T cell effectors38,39,43, comparable to our results with KO mice. Continuous FOXO1 activity also is required to maintain the memory state of CD8+ T cells43,44, suggesting that the reduced Foxo1 levels in memory T cells in KO mice may be insufficient to maintain the memory state. Levels of FOXO1 in differentiating memory CD8+ T cells are regulated by the balance of IL-10 and IL-12 in the local environment. IL-10R signaling acts via PI3K-AKT to block the phosphorylation of FOXO1, thus inhibiting its degradation and allowing nuclear translocation and transcription of downstream genes such as Bcl645. In contrast, IL-12R signaling acting via AKT promotes the phosphorylation and degradation of FOXO1, thus increasing T-BET expression38. Thus, our data suggest that the deficit of IL-10 and excess of IL-12 in the absence of the IRF4-dependent CD11b+P1 DCs leads to reduced FOXO1 expression in differentiating memory T cells.
Our finding is consistent with a report showing that the absence of IL-10 leads to decreased Foxo1 in maturing memory CD8+ T cells in LCMV infection and that TREGs are a significant source of IL-10 promoting MPEC differentiation34. IAV infection also results in a significant population of antigen-specific TREGs36, and we observed a marked deficit of FOXP3+ TREGs in lungs of KO mice on day 8 p.i. This suggests that IL-10-producing IRF4-dependent DCs first promote TREG differentiation, and the TREGs in turn produce the IL-10 needed for MPEC differentiation. Alternately or in parallel, DCs producing IL-10 or displaying other cytokines such as TGFβ or IL-15 may directly promote MPEC differentiation and/or maintenance of memory T cell populations33. Our in vitro assays showed that both CD103+ and CD11b+P1 DCs promoted differentiation of naïve CD4+ T cells into FOXP3+ TREGs. Our DC transfer experiments did not permit assessment of TREGs because their numbers in the lung have waned by day 10 p.i., the optimal time point for MPEC assessment. KO CD103+ DCs were less capable of inducing TREG differentiation, raising the possibility that Irf4 deficiency alters the ability of CD103+ DCs to produce mediators such as active TGFp that promotes Foxp3 expression during TREG generation46. Preliminary comparisons of WT and KO CD103+ DCs did not reveal significant differences in expression of genes in pathways important for TREG generation, including Tgfb1, Tgfb2, Tgfb3, Lrrc32, Itgb8, Ahr, Aldhla2, Entpdl, Ebi3, or 1127 [IH, manuscript in preparation]. Thus, the absence of CD11b+P1 DCs producing IL-10 and a defect in CD103+ DCs may contribute to the paucity of TREGs in the KO lung.
Dysregulation of one or more pathways may explain the increased numbers of NP-specific CD8+ and CD4+ T cells capable of producing IFNγ in the mLN after resolution of infection in KO mice. TCR and IL-12R signaling determine levels of T-BET and the terminal differentiation of IL-7Rlo SLECs31, and elevated IL-12 in KO mice was associated with increased numbers of T-BET+ T cells on day 5 p.i., perhaps indicative of increased effector differentiation. However, numbers of antigen-specific T-BET+ cells and SLECs were equivalent in lungs of WT and KO mice at later time points, suggesting that KO mice may instead have a defect in effector contraction or egress of antigen-specific T cells from the mLN. Consistent with this, NP-specific CD8+ T cells in the mLN of KO mice contained reduced Foxo1 and Klf2 RNA on day 10 p.i. FOXO1 regulates Klf2, which promotes expression of S1 Pi and CD62L that are important for naïve T cell recirculation, and Klf2 deficiency results in retention of naïve T cells in LNs40. Thus, antigen-specific CD4+ and CD8+ T cells may accumulate in the mLN of KO mice due to a reduced ability to egress from the LN.
The increased numbers of antigen-specific CD8+ and CD4+ T cells by d.12 p.i. in KO mice was associated with reduced numbers of CD8+ but not CD4+ memory cells, suggesting that signals from CD11b+P1 DCs are less important for specification of the CD4+ memory T cell program. CD4+ memory T cell formation and lung homing during IAV infection requires viral antigen and costimulatory molecule recognition at later stages of infection during the peak of effector generation47, 48. At days 6-8 p.i., iDCs and CD11b+P2 DCs predominate in the lung and their antigen presentation, costimulation and cytokine production may promote the formation and retention of CD4+ TRM cells. The absence of a defect in CD4+ memory T cells in KO mice may reflect our finding that Ly6C+ iDCs and CD11b+P2 DCs are present in comparable numbers in lungs of WT and KO mice (IH, manuscript in preparation). These CD11b+ subsets accumulating later in infection in mLN also may support our finding that CD8+ TCM cells do not differ in WT and KO mice, consistent with the observation that prolonged antigen presentation promotes CD8+ T cell memory programming during IAV infection19,49. Similarly, iDCs were required for the persistence but not differentiation of circulating and lung resident CD8+ memory T cells in respiratory poxvirus infection50.
Upon secondary infection with a heterosubtypic virus bearing conserved NP epitopes, KO mice mounted a weaker antiviral CD8+ T cell response, characterized by reduced numbers of NP-specific CD8+ T cells capable of making IFNγ, TNFα or IL-2, and reduced levels of IFNγ and TNFα in responding T cells. This is consistent with quantitative and qualitative deficits in the TRM and TEM compartments, but the response is not entirely abolished perhaps due to an intact TCM pool. Recent work shows that DCs are not absolutely required for the initial response of TRMs during secondary infection, although they likely contribute to the strength of the recall response51–53. Our data show early defects in the CD8+ T memory cell program during the primary infection of KO mice, but we cannot rule out a role for IRF4-dependent DCs in the maintenance or reactivation of TRM populations in the lung.
In contrast to nonlymphoid tissues such as skin and the female reproductive tract, numbers of IAV-specific TRM in the lower respiratory tract wane over time, leading to loss of heterosubtypic immunity24,27. Recent work showed that the gradual loss of TRMs is due to an imbalance of their apoptosis and replacement through conversion of circulating TEMs26· Since IRF4-dependent CD11b+P1 DCs regulate the differentiation of the TRM and TEM pools, vaccination strategies that target this subset of lung-resident DCs during primary and booster immunizations may maximize the retention of respiratory tract CD8+ TRM cells.
Methods
Mice
Mice (purchased from The Jackson Laboratory) bearing a conditional allele of Irf4 (B6.129S1-Irf4<tm1Rdf>/J)54 were bred to mice bearing Cre recombinase driven by the CD11c promoter (B6.Cg-Tg(Itgax-cre)1-1Reiz/J)55 and then interbred to yield CD11c-cre-Irf4f/for CD11c-cre-Irf4+/+ mice. Upon deletion of the Irf4 allele, an Egfp minigene is placed in frame such that Irf4−/− cells are GFP+. Mice used as “wild-type” were either Cre+ but bearing two WT alleles of Irf4, or Cre− and bearing either WT or conditional Irf4 alleles. The Cre activity may randomly act in CD11c− cells in the line of CD11 c-Cre mice, leading to mice that have the conditional allele deleted in many tissues6. Therefore, we screened for global deletion of Irf4 using PCR primers (forward: 5′-CAGGATGTTGCCGTCCTCCTTG-3′ and reverse: 5′-CCTGCAGCCAATAAGCTTATAAC-3′), and excluded those mice from this study. Littermate mice of each genotype and both sexes were analyzed at 10-12 weeks of age. OT-II mice bearing a transgenic TCR specific for I-Ab/OVA323-339 (B6.Cg-Tg(TcraTcrb)425Cbn/J) were purchased from The Jackson Laboratory. The Oklahoma Medical Research Foundation Institutional Animal Care and Use Committee approved the studies.
Influenza virus infection
Viruses were grown in embryonated chicken eggs, and viral end point titer (egg infectious units, EIU) was determined using allantoic fluid in a hemagglutination assay in the lab of Dr. Gillian Air (Oklahoma University Health Sciences Center). HA genes were sequenced to confirm identity of virus stocks. Mice were infected intranasally with 30 μl PBS containing 5×102 EIU of mouse adapted A/Puerto Rico/8/1934 (PR8, H1N1) virus. For sequential infections with heterosubtypic viruses, mice were initially infected with A/X31 (H3N2) virus (3×105 EIU), which carries the HA and NA genes of A/Hong Kong/1/1968 (H3N2) on the PR8 background, and then secondarily infected with 2×104 EIU of PR8 on day 42 post-infection. Alternately, mice initially infected with PR8 were infected on day 42 post-infection with 3×105 EIU of X31 virus. Mice were weighed each day to track morbidity during the infection.
Isolation of cells from tissues
Lungs were perfused with PBS + 1mM EDTA before digestion for 45-60 min with collagenase type A (1 mg/ml) or Liberase (0.1 mg/ml) and DNase I (0.1 mg/ml) (all from Roche) in PBS + 0.5% BSA pH 7.4. Mediastinal lymph nodes (mLN) were mechanically disrupted into single cell suspension. Lung and LN cells were filtered (40μM), washed with RPMI+10% FCS or HBSS without Ca2+ and Mg2+, respectively, and red cells lysed using RBC lysis buffer (BD Biociences). To distinguish populations of T cells present in lung tissue or blood, a fluorochrome-labeled anti-CD45 mAb was injected into the intra-orbital vein shortly before mice were euthanized56.
Ex vivo T cell stimulation
3-4×106 mLN cells were incubated with the H-2Db-binding NP366-374 peptide [2μg/ml] (MBLI) and the I-Ab-binding NP311-325 peptide [2μg/ml] (Bio-Synthesis Inc) and brefeldin A [5μg/ml] (BD Biosciences) in RPMI+5%FCS for 5 hours. Surface and intracellular staining identified IFNγ, TNFα or IL-2 producing CD4+ and CD8+ T cells.
Flow cytometry
For surface staining, cells were incubated with mAbs on ice for 15 min after 5 min of anti-CD16/32 treatment in FACS buffer (PBS, 5% newborn calf serum, 0.1% NaN3). Kits for intranuclear staining (ThermoFisher Scientific) and intracellular cytokine staining (BD Biosciences) were used according to manufacturer instructions. T cells were identified with fluorochrome-linked mAbs specific for CD45.2-APCCy7 (104), CD3-PECy7 (145-2C11), CD4-BV786 (GK1.5), CD8a-APCfire750 and -APC (53-6.7), IL7Rα-BIO (A7R34), KLRG1-BV421(2F1/KLRG1), CD44-BV711 (1M7), T-BET-PEDazzle (4B10), CD25-PerCPCy5.5 (PC61), CD103-BV711 (2E7), FOXP3-PE (MF-14), CD62L-PerCPCy5.5 (MEL-14), CD69-BV605 (H1.2F3), IFNγ-PE (XMG1.2), IL-2-PEDazzle (JES6-5H4), TNFα-BV711 (MP6-XT22). DC subsets were identified with mAbs specific for CD11c-PECy7 (N418), CD103-BV711 (M290), CD11b-BV786 (M1/70), SIGLECF-APCCy7 (E20-2240), MHCII-AF700 (M5/114.15.2), SIRPα-PEDazzle (P8), CD24-PerCpCy5.5 (M1/69), Ly6C-BV605 (HK 1.4), CD88-APC (20/70), CD26-PE (H194-11), IRF4-PE (3E4). mAbs were purchased from BD Biosciences, Biolegend, Tonbo Biosciences, and ThermoFisher Scientific. Tetramers of H-2Db-NP366-374-PE were purchased from MBL International. Tetramers of I-Ab-NP311-325-APC and negative control I-Ab-PVSKMRMATPLLMQA-APC were obtained from the NIH Tetramer Core Facility. Live/dead cell discrimination was done with a fixable zombie aqua dye (Biolegend). Samples were acquired on an LSRII instrument containing 4 lasers (BD Biosciences) and analyzed using FlowJo software (Treestar).
Cell sorting
CD11c+ cells were enriched from collagenase/DNAse-digested lungs on days 0 or 3 p.i. using a CD11c positive selection magnetic bead kit (StemCell Technologies). To sort lung DC subsets on day 5 p.i., DCs within a lung digest were enriched by gradient separation with OptiPrep (Axis-Shield) according to the manufacturer’s protocol. DC subsets were identified and sorted after staining with mAbs to DC surface markers listed above as in Fig. S2b. In KO mice, Irf4-deficient DCs were sorted as GFP+, since not all Ly6C+ iDCs were GFP+. Tet+CD8+ MPECs (GFP-) in lung and CD8+ T cells in the mLN were sorted on day 10 p.i. after enrichment on a 40% Percoll gradient and staining with mAbs specific for CD3-PECy7 (145-2C11), CD8a-APC or APCfire750 (53-6.7), IL7Rα-BIO (A7R34), KLRG1-BV421 (2F1/KLRG1), Streptavidin BV605 and H-2Db- NP366-374-PE tetramer as in Fig. 3. T cell and DC subsets were sorted directly into ARCTURUS PicoPure RNA extraction buffer (ThermoFisher Scientific) using a BD FACS Aria II instrument.
Adoptive transfer of DCs
CD11b+ P1 and CD103+ DC subsets were sorted from lungs of naïve WT mice (pools of 15-18 mice) as described above. Each sort yielded enough DCs for 1-2 recipients of each DC subset. 8 × 104CD11b+ P1 or CD103+ DCs were transferred intranasally into KO mice, followed by infection with 30 μl PBS containing 5×102 EIU of mouse adapted A/Puerto Rico/8/1934 (PR8, H1N1) virus. The mice were analyzed for numbers of lung CD8+ MPECs on day 10 p.i.
T regulatory cell assays
Lung DC subsets were sorted on day 3 p.i. (as in Fig. S2b) and 1000 DCs incubated in complete RPMI+5% FCS with 100nM OVA323-339 peptide (InvivoGen) and 20,000 naïve CD4+CD25~ T cells isolated from OT-II spleens using a naïve CD4+ T cell isolation kit (Biolegend). Recombinant IL-2 (5ng/ml, Peprotech) was added after 2 days. After 6 days, numbers of CD4+CD44+CD25+FOXP3+ T cells were quantified using flow cytometry.
Isolation of RNA and real-time quantitative PCR
RNA of sorted immune cells was isolated with a PicoPure™ RNA Isolation Kit and amplified with an Ovation Pico SL WTA System V2 kit (NuGEN). Quantitative real-time RT-PCR (qPCR) of genes was performed with iTaq™ Universal SyBr® Green Supermix or iTaq™ Universal Probes Supermix (Bio-Rad) on an ABI 7900HT instrument. Expression of tested genes relative to Gapdh expression was determined using the 2−ΔCt method. The primers are listed in supplementary Table 1.
Histologic analyses
The medium lung lobe was inflated and fixed with 10% neutral buffered formalin (Sigma-Aldrich) and embedded in paraffin. Lung sections (5μM thickness) were stained with hematoxylin and eosin (H&E) or Periodic Acid - Schiff stain. Images were acquired using a Zeiss Axioplan Microscope, with a 20X objective. Lung pathology was evaluated for severity of peri-bronchial inflammation, and bronchial epithelial cell and goblet cell hyperplasia and epithelial shedding or necrosis. Each criterion was scored on a scale of 0 to 4 with 0=no involvement and 1-4 representing increasing severity. The results are represented as a cumulative bronchial score. Extent of inflammation in the parenchyma with hyperplasia or thickening of the alveolar wall was also assessed and the results are presented as alveolar space pathology. All slides were read by an observer blinded to experimental details.
Cytokine assays
Bronchoalveolar lavage (BAL) fluid was collected by washing the lung cavity with PBS three times using an intratracheal catheter (Tom cat 3.5 Frx5.5”, Santa Cruz Animal Health). IFNγ in the BAL fluid was measured using xMAP multiplex assays in the OMRF Serum Analyte and Biomarker core facility. ProcartaPlex kits (ThermoFisher Scientific) were used according to the manufacturer’s instructions.
Viral RNA detection
Lung cells were lysed with Trizol reagent (Ambion), and RNA was isolated with a Trizol-RNeasy® mini kit (QIAGEN) hybrid protocol. cDNA was synthesized using iScript™ gDNA Clear cDNA Synthesis kit (BIO-RAD). qPCR was performed with iTaq™ Universal Probes Supermix (BIO-RAD) using a FAM-labeled probe 5′-CTCAGTTATTCTGCTGGTGCACTTGCCA-3′, PCR forward primer 5′-GGACTGCAGCGTAGACGCTT-3′ and reverse primer 5′-CATCCTGTTGTATATGAGGCCCAT-3′ targeting the gene encoding the influenza M1 protein on an ABI 7900HT instrument57. Relative expression of the gene was determined using the ΔΔt method with normalization to Gapdh expression.
Statistical analyses
Comparison between two groups was performed using a two-tailed Mann-Whitney or student’s t test. A one-way ANOVA with a Tukey’s post-test was performed for multiple comparisons. Weight loss curves were analyzed by a multiple t test using the Bonferroni-Sidak method. The differences were considered significant with a p value <0.05. Prism 7 software was used for all analyses.
Supplementary Material
Acknowledgements
We thank Dr. Gillian Air and Shelly Gulati at OUHSC for assistance with growing and titering virus in eggs, Dr. Diana Hamilton and Jacob Bass in the OMRF Flow Cytometry Core Facility, and the staff of the Comparative Medicine Facility. This work was supported by NIH HL119501 (to S.Kovats) and by an OMRF Patricia and Don Capra Predoctoral Fellowship (to S.Kadel).
Footnotes
Conflict of Interest Statement. The authors have no financial conflict of interest with this work.
References
- 1.Braciale TJ, Sun J & Kim TS Regulating the adaptive immune response to respiratory virus infection. Nat Rev Immunol 12, 295–305 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moskophidis D & Kioussis D Contribution of virus-specific CD8+ cytotoxic T cells to virus clearance or pathologic manifestations of influenza virus infection in a T cell receptor transgenic mouse model. J Exp Med 188, 223–232 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krammer F et al. Influenza. Nat Rev Dis Primers 4, 3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zens KD, Chen JK & Farber DL Vaccine-generated lung tissue-resident memory T cells provide heterosubtypic protection to influenza infection. JCI Insight 1, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wakim LM, Smith J, Caminschi I, Lahoud MH & Villadangos JA Antibody-targeted vaccination to lung dendritic cells generates tissue-resident memory CD8 T cells that are highly protective against influenza virus infection. Mucosal Immunol 8, 1060–1071 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Bajaña S, Turner S, Paul J, Ainsua-Enrich E & Kovats S IRF4 and IRF8 Act in CD11c+ Cells To Regulate Terminal Differentiation of Lung Tissue Dendritic Cells. J Immunol 196, 1666–1677 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schlitzer A et al. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity 38, 970–983 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edelson BT et al. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J Exp Med 207, 823–836 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ginhoux F et al. The origin and development of nonlymphoid tissue CD103+ DCs. J Exp Med 206, 3115–3130 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haniffa M et al. Human tissues contain CD141hi cross-presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells. Immunity 37, 60–73 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakano H et al. Blood-derived inflammatory dendritic cells in lymph nodes stimulate acute T helper type 1 immune responses. Nat Immunol 10, 394–402 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aldridge JRJ et al. TNF/iNOS-producing dendritic cells are the necessary evil of lethal influenza virus infection. Proc Natl Acad Sci U S A 106, 5306–5311 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim TS & Braciale TJ Respiratory dendritic cell subsets differ in their capacity to support the induction of virus-specific cytotoxic CD8+ T cell responses. PLoS One 4, e4204 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.GeurtsvanKessel CH et al. Clearance of influenza virus from the lung depends on migratory langerin+CD11b- but not plasmacytoid dendritic cells. J Exp Med 205, 1621–1634 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ballesteros-Tato A, Leon B, Lund FE & Randall TD Temporal changes in dendritic cell subsets, cross-priming and costimulation via CD70 control CD8+ T cell responses to influenza. Nat Immunol 11, 216–224 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho AW et al. Lung CD103+ dendritic cells efficiently transport influenza virus to the lymph node and load viral antigen onto MHC class I for presentation to CD8 T cells. J Immunol 187, 6011–6021 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Helft J et al. Cross-presenting CD103+ dendritic cells are protected from influenza virus infection. J Clin Invest 122, 4037–4047 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krishnaswamy JK et al. Migratory CD11b+ conventional dendritic cells induce T follicular helper cell-dependent antibody responses. Sci Immunol 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim TS, Gorski SA, Hahn S, Murphy KM & Braciale TJ Distinct dendritic cell subsets dictate the fate decision between effector and memory CD8+ T cell differentiation by a CD24-dependent mechanism. Immunity 40, 400–413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MartÍnez-López M, Iborra S, Conde-Garrosa R & Sancho D Batf3-dependent CD103+ dendritic cells are major producers of IL-12 that drive local Th1 immunity against Leishmania major infection in mice. Eur J Immunol 45, 119–129 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams JW et al. Transcription factor IRF4 drives dendritic cells to promote Th2 differentiation. Nat Commun 4, 2990 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosato PC, Beura LK & Masopust D Tissue resident memory T cells and viral immunity. Curr Opin Virol 22, 44–50 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gebhardt T, Mueller SN, Heath WR & Carbone FR Peripheral tissue surveillance and residency by memory T cells. Trends Immunol 34, 27–32 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Wu T et al. Lung-resident memory CD8 T cells (TRM) are indispensable for optimal crossprotection against pulmonary virus infection. JLeukoc Biol 95, 215–224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McMaster SR, Wilson JJ, Wang H & Kohlmeier JE Airway-Resident Memory CD8 T Cells Provide Antigen-Specific Protection against Respiratory Virus Challenge through Rapid IFN-γ Production. J Immunol 195, 203–209 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slütter B et al. Dynamics of influenza-induced lung-resident memory T cells underlie waning heterosubtypic immunity. Sci Immunol 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shane HL, Reagin KL & Klonowski KD The Respiratory Environment Diverts the Development of Antiviral Memory CD8 T Cells. J Immunol 200, 3752–3761 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Purwar R et al. Resident memory T cells (T(RM)) are abundant in human lung: diversity, function, and antigen specificity. PLoS One 6, el6245 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar BV et al. Human Tissue-Resident Memory T Cells Are Defined by Core Transcriptional and Functional Signatures in Lymphoid and Mucosal Sites. Cell Rep 20, 2921–2934 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iborra S et al. Optimal Generation of Tissue-Resident but Not Circulating Memory T Cells during Viral Infection Requires Crosspriming by DNGR-1+ Dendritic Cells. Immunity 45, 847–860 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaech SM & Cui W Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol 12, 749–761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang JT, Wherry EJ & Goldrath AW Molecular regulation of effector and memory T cell differentiation. Nat Immunol 15, 1104–1115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackay LK & Rallies A Transcriptional Regulation of Tissue-Resident Lymphocytes. Trends Immunol 38, 94–103 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Laidlaw BJ et al. Production of IL-10 by CD4(+) regulatory T cells during the resolution of infection promotes the maturation of memory CD8+ T cells. Nat Immunol 16, 871–879 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Croom HA et al. Memory precursor phenotype of CD8+ T cells reflects early antigenic experience rather than memory numbers in a model of localized acute influenza infection. Eur J Immunol 41, 682–693 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Betts RJ et al. Influenza A virus infection results in a robust, antigen-responsive, and widely disseminated Foxp3+ regulatory T cell response. J Virol 86, 2817–2825 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akbari M et al. IRF4 in dendritic cells inhibits IL-12 production and controls Thl immune responses against Leishmania major. J Immunol 192, 2271–2279 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Rao RR, Li Q, Gubbels Bupp MR & Shrikant PA Transcription factor Foxo1 represses T-bet-mediated effector functions and promotes memory CD8(+) T cell differentiation. Immunity 36, 374–387 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hess Michelini R, Doedens AL, Goldrath AW & Hedrick SM Differentiation of CD8 memory T cells depends on Foxo1. J Exp Med 210, 1189–1200 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carlson CM et al. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature 442, 299–302 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Li MO & Flavell RA Contextual regulation of inflammation: a duet by transforming growth factor-beta and interleukin-10. Immunity 28, 468–476 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Kreijtz JH et al. Primary influenza A virus infection induces cross-protective immunity against a lethal infection with a heterosubtypic virus strain in mice. Vaccine 25, 612–620 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Tejera ΜM, Kim EH, Sullivan JA, Plisch EH & Suresh M Foxo1 controls effector-to-memory transition and maintenance of functional CD8 T cell memory. J Immunol 191, 187–199 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delpoux A et al. Continuous activity of Foxo1 is required to prevent anergy and maintain the memory state of CD8+ T cells. J Exp Med 215, 575–594 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim EH & Suresh M Role of PI3K/Akt signaling in memory CD8 T cell differentiation. Front Immunol 4, 20 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen W et al. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med 198, 1875–1886 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jelley-Gibbs DM et al. Unexpected prolonged presentation of influenza antigens promotes CD4 T cell memory generation. J Exp Med 202, 697–706 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang YH et al. Dichotomous Expression of TNF Superfamily Ligands on Antigen-Presenting Cells Controls Post-priming Anti-viral CD4+ T Cell Immunity. Immunity 47, 943–958.e9 (2017). [DOI] [PubMed] [Google Scholar]
- 49.León B, Ballesteros-Tato A, Randall TD & Lund FE Prolonged antigen presentation by immune complex-binding dendritic cells programs the proliferative capacity of memory CD8 T cells. J Exp Med 211, 1637–1655 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Desai P, Tahiliani V, Stanfield J, Abboud G & Salek-Ardakani S Inflammatory monocytes contribute to the persistence of CXCR3hi CX3CR1lo circulating and lung-resident memory CD8+ T cells following respiratory virus infection. Immunol Cell Biol 96, 370–378 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shin H, Kumamoto Y, Gopinath S & Iwasaki A CD301b+ dendritic cells stimulate tissue-resident memory CD8+ T cells to protect against genital HSV-2. Nat Commun 7, 13346 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park SL et al. Local proliferation maintains a stable pool of tissue-resident memory T cells after antiviral recall responses. Nat Immunol 19, 183–191 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Beura LK et al. Intravital mucosal imaging of CD8+ resident memory T cells shows tissue-autonomous recall responses that amplify secondary memory. Nat Immunol 19, 173–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klein U et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol 7, 773–782 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Caton ML, Smith-Raska MR & Reizis B Notch-RBP-J signaling controls the homeostasis of CD8-dendritic cells in the spleen. J Exp Med 204, 1653–1664 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anderson KG et al. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat Protoc 9, 209–222 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Crowe CR et al. Critical role of IL-17Rα in immunopathology of influenza infection. J Immunol 183, 5301–5310 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.