Abstract
Cellular RNAs are naturally decorated with a variety of chemical modifications. The structural diversity of the modified nucleosides provides regulatory potential to sort groups of RNAs for organized metabolism and functions, thus affecting gene expression. Recent years have witnessed a burst of interest in and understanding of RNA modification biology, thanks to the emerging transcriptome-wide sequencing methods for mapping modified sites, highly-sensitive mass spectrometry for precise modification detection and quantification, and extensive characterization of the modification “effectors”, including enzymes (“writers” and “erasers”) that alter the modification level and binding proteins (“readers”) that recognize the chemical marks. However, challenges remain due to the vast heterogeneity in expression abundance of different RNA species, further complicated by divergent cell-type-specific and tissue-specific expression and localization of the effectors as well as modifications. In this review, we highlight recent progress in understanding the function of N6-methyladenosine (m6A), the most abundant internal mark on eukaryotic messenger RNA (mRNA), in light of the specific biological contexts of m6A effectors. We emphasize the importance of context for RNA modification regulation and function.
Keywords: RNA modifications, Epitranscriptome, N6-methyladenosine (m6A), Gene expression regulation, METTL3/14, YTHDF proteins, FTO, Context-dependent functions
eTOC Blurb
RNA N6-methyladenosine (m6A) has emerged as a multifaceted controller for gene expression regulation, mediated through its effector proteins—writers, readers, and erasers. Shi et al. review recent advances in the mechanistic understandings of m6A effectors in various biological systems and cellular responses, emphasizing cellular and molecular contexts as important determinants of RNA modification functions.
More than 150 distinct chemical marks on cellular RNA have been identified to date (Boccaletto et al., 2018) since the discovery the first structurally modified nucleoside, pseudo-uridine, in the 1950s (Cohn and Volkin, 1951). However, it was only until several years ago that board interest in RNA modification biology resurged, prompted by recognition of the prevalence and functional significance of internal messenger RNA (mRNA) modifications, most prominently N6-methyladenosine (m6A, Figure 1). These insights were sparked by the identification of enzymes capable of reversing m6A (Jia et al., 2011; Zheng et al., 2013) and through the development of detection methods with improved sensitivity and high-throughput sequencing approaches to map modified sites (Dominissini et al., 2012; Meyer et al., 2012).
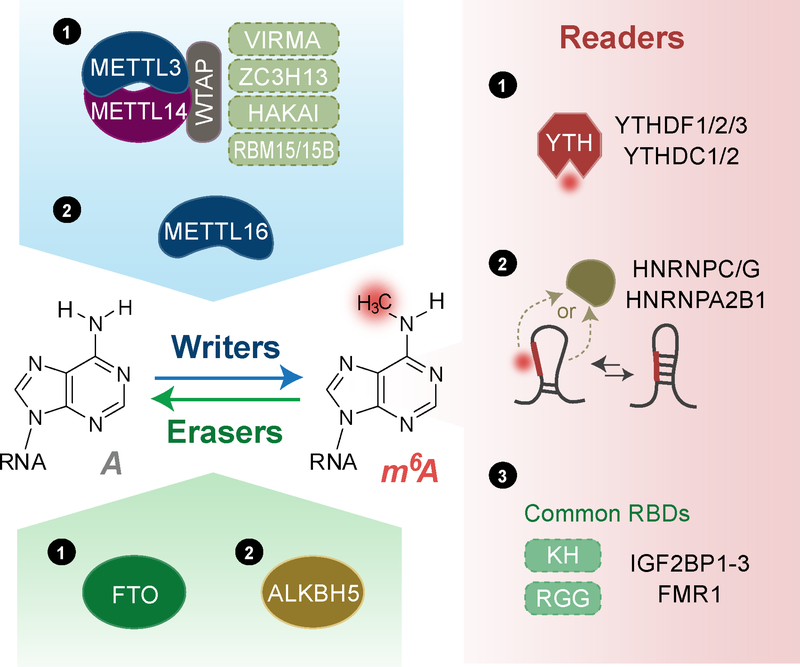
Figure 1. m6A effectors: writers, erasers, and readers.
Writers: Majority of m6A methylation on mRNA is installed by a writer complex ① composed of core subunits METTL3 and METTL14 and additional adaptors proteins including WTAP, VIRMA, ZC3H13, HAKAI, and RBM15/15B in a sequence context of RRACH (R = A or G; H = A, C, or U). The other known writer METTL16 ② installs m6A in a sequence context of UAC(m6A)GAGAA on top of a hairpin structure in transcript MAT2A. Erasers: two erasers have been characterized for m6A methylation on mRNA, including FTO and ALKBH5. Readers: Three classes of reader proteins utilize different mechanisms to prefer binding m6A-containing RNAs. ① YTH-domain containing proteins (YTHDF1–3, YTHDC1–2) use a well-characterized YTH domain to direct recognize m6A methylation. ② A local structure disrupted by the presence of m6A could favor RNA-binding events of HNRNPC/G and HNRNPA2B1. ③ RNA binding proteins including IGF2BP1–3 and FMR1 prefer m6A-containing RNAs through their tandem common RNA binding domains (RBDs) via a mechanism yet to be characterized.
m6A has been the best-characterized mRNA modification so far. It was first discovered in 1974 as the major form of internal methylation on mammalian mRNA (Desrosiers et al., 1974; Perry and Kelley, 1974) and early work showed that it occurs in a sequence context as (G/A)(m6A)C (Schibler et al., 1977; Wei and Moss, 1977). More recent transcriptome-wide m6A site mapping has given greater details on its localization and prominence, revealing its prevalence in thousands of transcripts and its unique and conserved distribution preferentially centered around stop codons and enriched at 3’ untranslated regions (3’UTRs) in the transcriptomes of human and mice (Dominissini et al., 2012; Meyer et al., 2012). Building on the understanding of its widespread prevalence on mRNA, recent work has uncovered that m6A plays an important role in gene expression regulation (Roundtree et al., 2017a), animal development (Frye et al., 2018), and human diseases (Hsu et al., 2017a).
The effectors in m6A pathways include “writers” and “erasers” that respectively install and remove the methylation, and “readers” that recognize it (Figure 1). Characterization of these effector proteins in assorted biological systems have underscored multifaceted and tunable features of their functions, emphasizing local contexts as important determinants of their biological roles. Why does context matter in m6A epitranscriptomics? On one hand, an m6A effector itself may exhibit different expression levels, post-translational modifications (PTMs), and cellular localization, depending on cell types and/or in response to environmental stimuli. For instance, in most cell lines the m6A demethylase FTO is largely nucleus localized and mediates ~5–10% of total mRNA m6A demethylation; however, in certain leukemia cells, FTO is also highly abundant in cell cytoplasm and can mediate up to ~40% m6A demethylation of total mRNA. On the other hand, the intrinsic heterogeneity of RNA molecules further complicates the local contexts facing m6A effectors and challenges interpretation of experimental data: (1) diversity in RNA species, including mRNA, tRNA, rRNA, and other abundant non-coding/regulatory RNA; (2) a wide range of cellular abundances for RNA species including individual mRNA sequences—1 to 50 copies for most protein-coding genes, as estimated from single-cell RNA-seq with spike-in quantification (Marinov et al., 2014); (3) complex RNA secondary structures (stem, loop, and bulge, etc); (4) varying compaction and therefore accessibility of individual RNA sequences depending on RNA cellular localization and translation state (Adivarahan et al., 2018); as well as other variable factors. Furthermore, cis elements in RNAs and trans elements including different RNA binding proteins (RBPs) could regulate the interactions between m6A effectors and RNA substrates. Therefore, the occurrence and biological outcome of m6A methylation at a given site on an mRNA could be highly context-dependent, providing a unique potential to tune gene expression in different biological processes.
In this review, we summarize recent progresses in mechanistic studies of m6A effectors in the light of specific cellular and molecular contexts such as cell types, external stimuli, subcellular localization of effectors, and locations of m6A sites on an mRNA. It is important to take into consideration these context complexities to refine and advance our understanding of RNA modification functions.
Writers for m6A Methylation
m6A is installed on mRNA co-transcriptionally by a complex composed of multiple subunits (Figure 1) with a stable core complex formed between methyltransferase-like 3 (METTL3) and methyltransferase-like 14 (METTL14) (Bokar et al., 1997; Liu et al., 2013)— the former as the catalytic subunit and the latter as an essential component to facilitate RNA binding (Wang et al., 2016a; Wang et al., 2016b). Further studies characterized a handful of additional subunits and revealed how they contribute to the activity and specificity of the writer complex. Wilms tumor 1-associating protein (WTAP) binds to METTL3/14 and is required for optimal substrate recruitment and METTL3/14 localization (Ping et al., 2014; Zhong et al., 2008); Vir like m6A methyltransferase associated (VIRMA) is critical for deposition of m6A specifically to the 3’UTR (Yue et al., 2018); Zinc finger CCCH-type containing 13 (ZC3H13) facilitates nuclear localization of the writer complex (Wen et al., 2018); and RNA binding motif protein 15/15B (RBM15/15B) is reported to bind U-riched regions and may facilitate methylation of certain RNAs (Patil et al., 2016). In fruit flies, Zc3h13/Flacc is shown to stabilize the interaction between Wtap/Fl(2)d and Rbm15/Nito (Knuckles et al., 2018).
Multifaceted METTL3: Cellular Localization, Post-translational Modification, and Functions
The cellular distribution of METTL3 varies among cell lines, and there are cases where cellular stress induces its redistribution (Knuckles et al., 2017; Xiang et al., 2017). When interacting with WTAP in the form of a stable dimer with METTL14, METTL3 localizes to the nuclear speckle in HeLa cells (Bokar et al., 1997; Ping et al., 2014). Functional nuclear localization signals (NLS) have been identified in both METTL3 and WTAP, of which key residue mutations abolished preferential nuclear localization of ectopic METTL3 and WTAP in HeLa cells (Scholler et al., 2018). A fraction of METTL3 protein is also detected in the cytoplasm in multiple human cancer cell lines at various percentages, including HeLa cells (Chen et al., 2015; Choe et al., 2018; Lin et al., 2016), breast cancer cells (MDA-MB-231 cells) (Alarcon et al., 2015b), and acute myeloid leukemia cells (MOLM13 cells) (Barbieri et al., 2017). In a rare instance, in mouse cortical neurons METTL14 rather than METTL3 localizes to both cytoplasm and nuclei (Merkurjev et al., 2018). It is not yet clear why METTL3 localization varies. The protein abundance ratios between METTL3 and METTL14 as well as other adaptor subunits of the writer complex could vary among cell lines, which could affect localization of METTL3. It is also possible that post-translational modifications (PTMs) alter interactions between METTL3 and its partner proteins, leading to cytoplasmic presence.
m6A-containing genes are enriched in important cellular processes, and a subset of m6A sites appear dynamic in response to stimuli and stress (Dominissini et al., 2012; Meyer et al., 2012). The transcript-specificity of m6A methylation could be due to writer recruitment at desired chromatin loci, likely through transcription factors (TFs) and/or epigenetic marks (Figure 2A). After heat shock, METTL3 was observed to localize to heat-shock genes in the chromatin, and m6A installation on those heat-shock transcripts was suggested to ensure their timely clearance after the stress (Knuckles et al., 2017). Upon DNA UV damage METTL3/14 localizes within 2 min to UV-induced damage sites, co-occurring with increased m6A intensity (Xiang et al., 2017). In the TGFβ signaling in human pluripotent stem cells, activated transcription factors SMAD family member 2/3 (SMAD2/3) interact with METTL3/14-WTAP, facilitating co-transcriptional m6A installation on selective transcripts (Bertero et al., 2018). A recent study in acute myeloid leukemia cells showed that a fraction of METTL3 associates with the promoter regions of ~80 active genes specified by a transcription factor CCAAT enhancer binding protein zeta (CEBPZ) independent of METTL14 (Barbieri et al., 2017). Their results suggest that specialized “adaptor” proteins might exist in different cells to target the writer to distinct sets of genes in the chromatin, resulting in transcript-specific m6A methylation. Moreover, a gene-body enriched histone modification Histone H3 trimethylation at lysine 36 (H3K36me3) is suggested to recruit the writer complex through interaction with METTL14 in HepG2 cells, favoring m6A installation on mRNA coding sequences (CDS) and 3’UTRs (Huang et al., 2019).
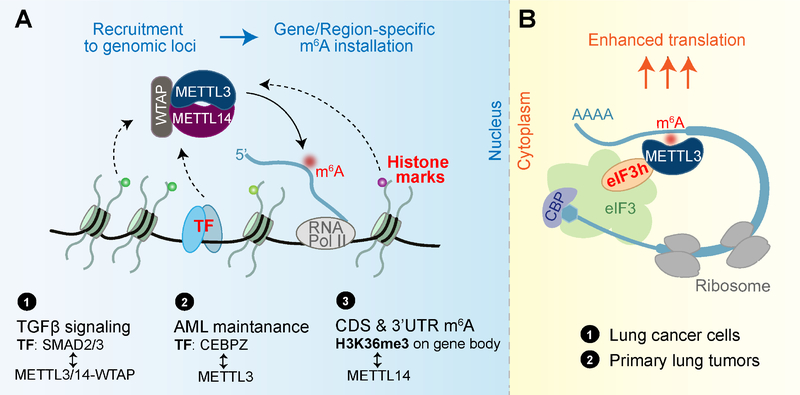
Figure 2. A model of cellular-localization dependent functions of m6A writer proteins.
(A) m6A writer complex installs m6A co-transcriptionally in the nucleus. The recruitment of the writer complex to specific genomic loci by transcription factors (TFs) or histone marks may contribute to the gene- or region-specificity in m6A installation. Examples include ① TF SMAD2/3 interacts with m6A writer complex in response to TGFβ signaling; ② In acute myeloid leukemia (AML) cells, METTL3 is recruited to TSS (transcription start sites) regions with dependence on TF CEBPZ, which subsequently mediates methylation of transcripts important for cancer maintenance; ③ Gene-body enriched histone mark H3K36me3 recruits the m6A writer complex by interacting with METTL14, a process contributing to preferential m6A installation at CDS and 3’UTR of nascent transcripts.
(B) In the cytoplasm, METTL3 itself recognizes 3’UTR m6A sites on mRNA and promotes protein translation from the transcript by facilitating translation loop formation through interaction with eIF3h.
In contrast to these nuclear roles, cytoplasmic-localized METTL3 is suggested not to function as an m6A writer, but instead as a potential m6A reader (Figure 2B). Independent of its catalytic activity, METTL3 in the cytoplasm in lung cancer cells promotes translation of a reporter mRNA when tethered to its 3’UTR (Lin et al., 2016). Further studies showed that this translation promotion effect depends on interaction between METTL3 and eIF3h, which are coordinately overexpressed in many types of cancers (Choe et al., 2018).
METTL3 could be modulated through PTMs or those of its interacting proteins, which could affect protein stability, localization, writer complex formation, and writer catalytic activity. Human METTL14 has been reported to be phosphorylated at residue Serine399 which lies on the protein-protein interface with METTL3, suggesting regulatory functions (Wang et al., 2016b). SUMOylation sites have also been detected on four lysine residues of human METTL3 and were shown to reduce activity of METTL3/14 in in vitro methylation assays (Du et al., 2018). This repressive effect was mediated by a currently unknown mechanism but not through affecting METTL3’s stability, localization, nor interaction with METTL14/WTAP. Further investigations on protein partners of METTL3 in either the nucleus or the cytosol is likely to give more clues to deciphering PTM-mediated regulation of METTL3’s functions, especially in a biological system where phosphorylation or SUMOylation pathways are misregulated.
Methylation Specificity Driven by Both Sequence and Structure
While the METTL3/14 complex preferentially installs m6A methylation in a sequence motif RRACH (R = A or G; H = A, C, or U) (Liu et al., 2013), another m6A writer, methyltransferase-like 16 (METTL16) installs m6A in a different sequence and structure context (Pendleton et al., 2017). Two validated substrates of METTL16 are U6 small nuclear RNA (snRNA) and a hairpin (hp1) in the 3’UTR of human methionine adenosyltransferase 2A (MAT2A) mRNA that encodes the S-adenosylmethionine (SAM) synthetase (Pendleton et al., 2017). Truncation/mutation tests, in vitro methylation selection assays, and a crystal structure of METTL16 with its RNA substrates together suggest that METTL16 prefers a UAC(m6A)GAGAA sequence in the bulge of a stem-loop structured RNA (Doxtader et al., 2018; Mendel et al., 2018; Pendleton et al., 2017). An important functional implication for the METTL16-mediated MAT2A methylation is to establish a negative feedback loop for SAM homeostasis. One working model proposed was that when SAM is limiting, prolonged interaction between METTL16 and MAT2A hp1 promotes proper intron splicing in HEK293 cells (Pendleton et al., 2017). A different study in HeLa cells suggested that an increased stability of MAT2A mRNA under SAM-limiting conditions also contributes to its elevated abundance.
More recently, a new m6A writer ZCCHC4 was identified to mediate methylation of A4220 on 28S rRNA within an AAC motif and also interact with a subset of mRNAs (Ma et al., 2019).
m6A Readers
Characterization of m6A readers have provided valuable insights into the molecular mechanisms of the m6A-mediated post-transcriptional gene regulation. Methylated probe pull-down and quantitative protein mass spectrometry assays have identified multiple RBPs that favor m6A probes (Dominissini et al., 2012; Edupuganti et al., 2017) through divergent binding modes (Figure 1). Diverse regulatory or functional machineries can be recruited to m6A-containing mRNA through m6A readers, and therefore impact the fate of the target mRNA.
An Expanding List of m6A Reader Proteins Adds Complexity
One class of direct and robust m6A readers are proteins containing the YT521-B homology (YTH) domain, including YTH domain family 1–3 (YTHDF1–3) and YTH domain containing 1–2 (YTHDC1–2) in humans (Figure 1). Cytoplasmic YTHDF2 promotes degradation of its target transcripts partially through recruiting the CCR4-NOT deadenylase complex (Du et al., 2016; Wang et al., 2014). By contrast, the other two cytoplasmic m6A readers, YTHDF1 and YTHDF3, are suggested to promote translation of target transcripts in HeLa cells by recruiting translation initiation factors (Shi et al., 2017; Wang et al., 2015). Nuclear reader YTHDC1 has been suggested to play multiple roles including regulating mRNA splicing by preferably recruiting a certain splicing factor (Xiao et al., 2016), expediting mRNA export (Roundtree et al., 2017b), and accelerating decay of certain transcripts (Shima et al., 2017). YTHDC2 mediates both mRNA stability and translation and regulates spermatogenesis (Hsu et al., 2017b), while its additional RNA binding domains and helicase domains render detailed mechanistic studies enigmatic.
A different group of m6A readers utilize common RNA binding domains (RBDs) such as K homology (KH) domains, RNA recognition motif (RRM) domains, and arginine/glycine-rich (RGG) domains, to preferentially bind m6A-containing RNAs; in some of these cases RNA structure may play a role (Figure 1). The presence of m6A can remodel local RNA structure and consequently modulates RNA-protein interactions around or nearby, termed an “m6A-switch” (Liu et al., 2015). Several heterogeneous nuclear ribonucleoproteins (HNRNPs) fall into this category, including HNRNPC, HNRNPG, and HNRNPA2B1, which regulate alternative splicing or processing of target transcripts (Alarcon et al., 2015a; Liu et al., 2015; Liu et al., 2017; Wu et al., 2018). Fragile X mental retardation 1 (FMR1) contains three KH domains and one RGG domain, and has been shown to prefer m6A-containing RNA, impacting both RNA translation and RNA stability likely through interplay with YTHDF1 and YTHDF2 (Edupuganti et al., 2017; Zhang et al., 2018). An additional class of readers, insulin-like growth factor 2 mRNA-binding proteins 1–3 (IGF2BP1–3), was reported to stabilize target mRNA in an m6A-dependent manner (Huang et al., 2018), in contrast to the role of YTHDF2 in decreasing mRNA stability. Mutation and truncation assays identified domains KH3–4 of the IGF2BP proteins critical for in vitro binding to an m6A-modified RNA probe. A recent study added proline rich coiled-coil 2A (Prrc2a) to the list of m6A readers (Wu et al., 2019), showing that recombinant Prrc2a prefers to bind a methylated probe and Prrc2a stabilizes a critical m6A-modified transcript required for myelination. The exact domains of Prrc2a responsible for m6A recognition remain to be explored.
We currently do not know how readers achieve selectivity towards certain m6A sites or certain m6A-modified transcripts. One likely scenario is that readers may be localized to different regions of mRNA through interacting with other RBPs that recognize distinct features of the RNA. IGF2BP1–3 and YTHDF2, which regulate mRNA stability in opposite directions, were shown to bind distinct sites on mRNA: the former favors 3’UTR in HEK293T cells examined while the latter shows more binding to CDS (Huang et al., 2018). RNA recognition modes of RBPs can depend on multiple variables including binding site sequences, flanking sequences, and RNA secondary structures (Dominguez et al., 2018). Therefore, RBPs that interact with reader proteins might convey specificity towards certain m6A sites or m6A-modified transcripts. Secondly, the density and sequence contexts of m6A sites are likely to matter. Densely populated m6A regions could be more frequently occupied by m6A readers. In the case of FMR1, it binds consensus RNA sequences of GGA and ACU similar to the m6A-containing sequence (Ascano et al., 2012; Edupuganti et al., 2017). Perhaps the protein has evolved to further recognize the methyl group in these particular sequence contexts in order to add an additional regulation layer. Thirdly, reader proteins might enrich at specific cellular compartments, and therefore preferentially interact with local RNA species. For instance, reader proteins YTHDF1–3, FMR1, and HNRNPA2B1, were identified in mammalian stress granule cores (Jain et al., 2016). YTHDF1–3, HNRNPK, and IGF2BP2–3 were found enriched in the cell protrusion compared to the cell body in breast cancer cells (Mardakheh et al., 2015). The methylation could promote translation or affect stability of a certain group of transcripts depending on which reader is present or dominates under the specific cellular contexts (Figure 3).
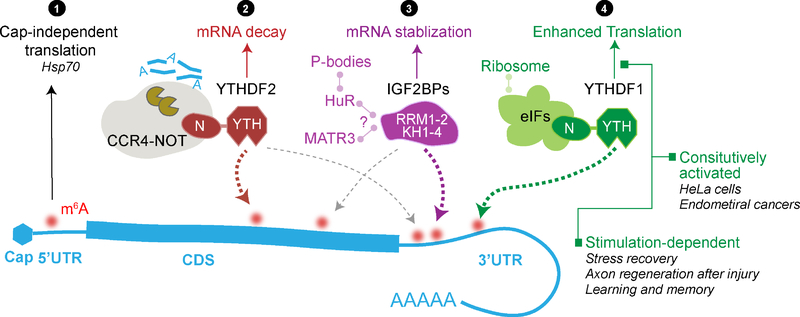
Figure 3. Region-, reader-, and stimulation-dependent roles of m6A methylation on mRNA.
Multiple layers of contexts could substantially affect how m6A methylation regulates the fate of modified mRNA. Region-dependent regulation: 5’UTR m6A is linked to cap-independent translation, especially during stress response when cap-dependent translation is repressed (①). Reader-dependent regulation: YTHDF2 and IGF2BPs can affect mRNA stability in opposite directions. YTHDF2 recruits CCR4-NOT complex via its N-terminal domain to promote mRNA decay (②) while IGF2BPs stabilizes mRNA likely through co-factors HuR and MATR3 (③). They may recognize distinct m6A sites and exhibit differential binding density in CDS and 3’UTR. Stimulation-dependent regulation exemplified by YTHDF1 (④): YTHDF1 preferentially binds 3’UTR m6A and promotes translation through interaction with translation initiation factors. While it constitutively promotes translation in cancer cells including HeLa cells and endometrial cancer cells, in post-mitotic cells the translation promotion effect only significantly manifests when induced by stimulations such as recovery/repair processes or learning signals.
Readers Acting in Response to Stimuli
It has been observed that predominately cytoplasmic YTHDF proteins could redistribute to the nuclei of cells under certain stimuli including heat shock stress and viral infection. YTHDF2 was reported to be greatly upregulated at both transcript and protein levels within hours after heat shock and the majority of YTHDF2 translocated to the nuclei (Zhou et al., 2015). The authors proposed that the nuclear YTHDF2 would compete with the demethylase fat mass and obesity-associated protein (FTO, discussed below) to prevent demethylation of 5’UTR-m6A in heat shock response genes, thereby enhancing their cap-independent translation in the cytosol. In vitro demethylation activity of FTO was inhibited by the presence of YTHDF2, supporting the competition hypothesis. In a similar scenario where monkey kidney cells (Vero cells) were infected with enterovirus type 71, YTHDF1 and YTHDF2 were upregulated and distributed into both cytosol and the nucleus 12 and 24 hours post infection (Hao et al., 2019). Although its molecular basis and outcome remain to be elucidated, this reader translocation event emphasizes the dynamic nature and versatility in m6A-mediated regulation pathways.
In post-mitotic cells, the translation-promoting effect of YTHDF1 becomes especially apparent when active translation of functional proteins is desired (Figure 3). In the dorsal root ganglion (DRG) model of injury-induced axon regeneration, regeneration-associated genes are heavily m6A methylated, and YTHDF1 is required for robust global de novo protein synthesis during this recovery process; in comparison, the basal translation enhancing effect of YTHDF1 before the injury is minor (Weng et al., 2018). This observation resembles the initial study on YTHDF1 in HeLa cells, showing that reporter transcripts tethered by YTHDF1 demonstrated a faster recovery from the translation arrest after removal of sodium arsenite stress (Wang et al., 2015). More recently, we studied in detail how the m6A- and YTHDF1-mediated translation regulation contributes to learning and memory process (Shi et al., 2018), in which the formation of long-term memory relies on production of new proteins in synapses. In the mouse model, YTHDF1 deficiency impaired hippocampus-dependent neuronal functions such as spatial learning and memory as well as contextual fear memory, which could be rescued by re-introducing YTHDF1 protein specifically in the hippocampus. At the molecular level, transcripts that are m6A-modified and also bound by YTHDF1 enrich genes functioning in synaptic transmission and long-term potentiation. Interestingly, nascent protein labelling and luciferase reporter assays revealed a stimulus-dependent feature for YTHDF1: cultured neurons stimulated by potassium chloride depolarization showed YTHDF1-dependent translation enhancement in 2 to 4 hours but not before the stimulation. These studies showed that while the YTHDF1-dependent upregulation of translation could be constitutively activated in certain cells such as cancer cells (Liu et al., 2018; Wang et al., 2015), in other cells this process could be stimulation-induced. How stimuli trigger YTHDF1 to promote translation remains to be studied.
Functions of Readers: Redundant or Convoluted?
YTHDF1–3 are similar in their domain structures: they all contain an N-terminal low-complexity sequence and a C-terminal conserved YTH domain. There have been arguments that YTHDF1–3 target almost identical m6A sites in HEK cells, supporting a model that YTHDF1–3 are redundant in function in mammals (Meyer and Jaffrey, 2017). However, in the case of endometrial cancer cells, YTHDF1 and YTHDF2 clearly target unique transcripts and respectively regulate mRNA translation and decay (Liu et al., 2018). Transcripts in the AKT pathway are highly m6A methylated. YTHDF1 upregulates protein production from the phosphatase coding mRNA PHLPP2 while YTHDF2 downregulates the transcript level of kinase coding mRNAs PRR5, PRR5L, and mTOR. Therefore, YTHDF1 and YTHDF2 collectively inhibit the downstream AKT phosphorylation, preventing the AKT pathway from being hyper-activated. Moreover, an RNA-targeting platform has been developed to study the precise functions of readers on specific transcripts by fusing the catalytically inactive Cas13b to the N-terminal domain of YTHDF1 or YTHDF2, respectively (Rauch et al., 2018). In this context, YTHDF2-dCas13b delivery resulted in roughly an equal decrease in mRNA and protein level, while YTHDF1-dCas13b enhanced translation with minor mRNA destabilization effect, supporting different functions of the two proteins. Taken together, the notion that YTHDF1–3 function redundantly is oversimplified.
There have been multiple examples showing a crosstalk or competition between m6A reader proteins (Edupuganti et al., 2017; Wu et al., 2019; Zhang et al., 2018) and even between the eraser FTO and readers during certain cellular responses (Zhou et al., 2015). These proteins form a network of physical or functional interactions. Appreciating their context-dependent regulation will be important for deciphering the roles of m6A in future studies.
m6A Erasers
m6A methylation can be reversed via active demethylation by m6A demethylases FTO or AlkB homolog 5 (ALKBH5), thereby the methylation-dependent processes can be reversed and controlled.
The RNA Demethylation by FTO is Context Dependent
FTO, the first RNA demethylase identified, was reported to remove the methyl group of N6-methyladenosine (m6A) in mRNA both in vitro and inside cells (Fu et al., 2013; Jia et al., 2011). FTO has also been reported to demethylate N6, 2-O-dimethyladenosine (m6Am), a modification that has an identical chemical structure in the base moiety to m6A and is found on the second base adjacent to the 5’ cap (cap-m6Am) in a portion of mRNAs (Adams and Cory, 1975; Wei et al., 1975), both in vitro (Fu, 2012) and inside cells (Mauer et al., 2017) (Figure 4). We have comprehensively characterized substrate spectrum of FTO by carefully validating FTO binding targets from cross-linking immunoprecipitation followed by high-throughput sequencing (CLIP-Seq) results, confirming that FTO possesses effective demethylation activity towards m1A in specific tRNAs, m6Am in some snRNAs, and internal m6A and cap-m6Am in mRNA (Wei et al., 2018).
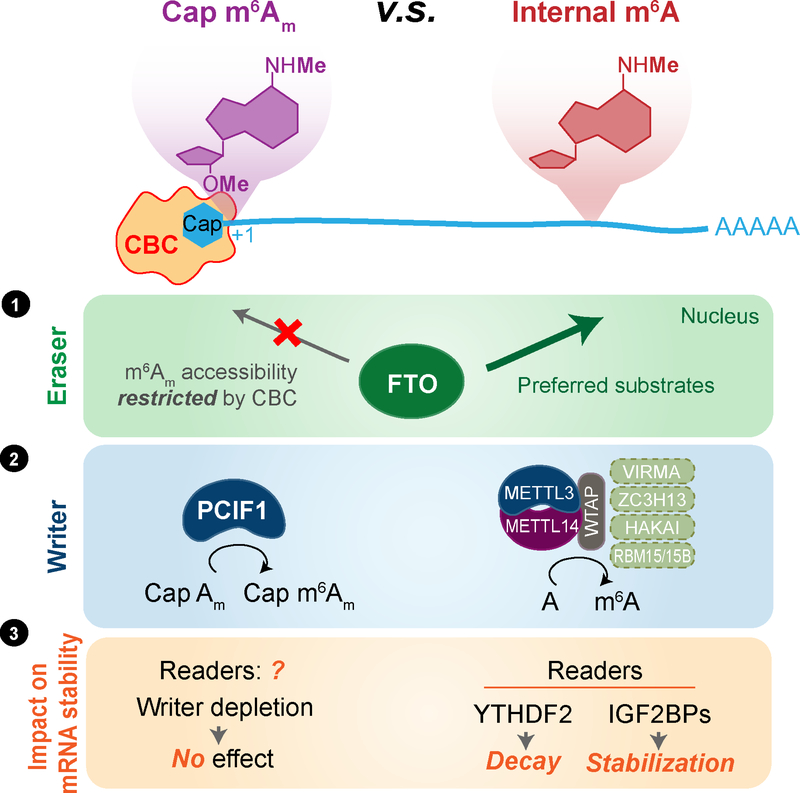
Figure 4. Distinct effectors and functions of cap N6, 2-O-dimethyladenosine (m6Am) versus internal m6A in mRNA.
While FTO exhibits catalytic demethylation activity towards both cap m6Am and internal m6A in mRNA, in the cell nucleus, m6A is the main substrate of FTO since cap m6Am is most likely masked by cap-binding complex (CBC) in mRNA (①). The methyl group on the N6 position of m6Am is installed by a cap-specific methyltransferase PCIF1 while internal m6A is installed by METTL3-METTL14 in a multi-component writer complex (②). The role of m6A on mRNA stability through the m6A readers have been established. However, recent characterization of the m6Am writer PCIF1 showed that depletion of the writer had minimum effect on the abundance/stability of the cap-m6Am-modified mRNA in cell lines tested(③), refusing the role of the FTO-mediated mRNA cap m6Am demethylation in affecting transcript stability.
A recent crystal structure of human FTO bound to 6mA-modified ssDNA revealed the molecular basis of its function towards internal m6A as well as cap m6Am (Zhang et al., 2019). The superimposition of FTO with NOP2/Sun RNA methyltransferase family member 6 (NSUN6), a known tRNA m5C methyltransferase (Haag et al., 2015), revealed similarities in their overall structures, supporting that FTO can also acts as a tRNA demethylase on stem-loop substrates (Wei et al., 2018).
The spatial distribution of FTO also modulates its accessibility towards physiologically relevant substrate(s). FTO was originally reported to be a nuclear protein containing an NLS in the N-terminus (Sanchez-Pulido and Andrade-Navarro, 2007) and partially colocalizes with nuclear speckles (Jia et al., 2011), but it was later shown to localize in both the nucleus and the cytoplasm in certain cell lines (Aas et al., 2017; Gulati et al., 2014). Cellular localization of FTO varies among several mammalian cell lines. In certain AML cell lines, a large portion of FTO proteins localize in cell cytoplasm (Wei et al., 2018); the spatial regulation of FTO results in the distinct substrate preference in the nucleus versus the cytoplasm. FTO mediates mRNA m6A and cap m6Am demethylation in cytoplasm but mostly mRNA m6A demethylation in cell nucleus (Wei et al., 2018), likely because the cap moiety is bound by cap-binding proteins and not accessible for demethylation. While it is true that 5–10% of mRNA m6A are subjected to the FTO-mediated demethylation in common cell lines such as HeLa and HEK (Jia et al., 2011; Wei et al., 2018), perhaps because nuclear mRNA accounts for a small portion of total cellular mRNA and they may not be adequately accessed by FTO, in some AML cells FTO is highly elevated, and mostly localizes in the cytoplasm. In these cells up to ~40% of all mRNA m6A are subjected to demethylation by FTO (Li et al., 2017; Su et al., 2018; Wei et al., 2018). The highly expressed FTO and its prevalent m6A demethylation play an oncogenic role in these AML cells; inhibition of FTO by an oncometabolite 2-hydroxyglutarate (2HG) suppresses leukemia progression (Li et al., 2017; Su et al., 2018). This effect is again context dependent, as FTO high AML cells are more sensitive to 2HG inhibition.
Consistent with this substrate preference, the transcriptome-wide distribution pattern of FTO binding sites identified in FTO CLIP-Seq also varies among cell lines (Yixing Li et al., 2019). CLIP-Seq data analysis showed that GAC- and/or GGAC-containing motifs are enriched in FTO binding sites in certain cells. Especially in HeLa cells where FTO localizes mostly inside nucleus, the binding to GAC-containing and RRACH motifs is enhanced by FTO overexpression.
What is the molecular basis for differential cellular localization of FTO? A recent study suggested that localization and expression of FTO can be regulated by its post-translational ubiquitination on Lys-216 (Zhu et al., 2018). Knocking in the ubiquitin-deficient K216R mutation reduces the nuclear expression of FTO and prohibits the nuclear translocation of FTO in response to amino acid starvation. It is also likely that FTO is recruited to different subcellular fractions by its interacting partners. It will be interesting to comprehensively investigate how the posttranslational modifications and the protein partners of FTO contribute to the spatial regulation of FTO.
m6A versus m6Am Demethylation by FTO
Cap m6Am, residing at the +1 ribose from the 5’cap in mRNA, was discovered around almost the same time as internal m6A (Adams and Cory, 1975; Wei et al., 1975). Unlike m6A, which is estimated to be 0.1%−0.4% of that of adenosines (that is, present at ~3 sites per mRNA), m6Am mainly occurs next to the cap with the second nucleoside 2’-O-methylated (m7GpppNm). This m7GpppNm contains less than 30% m6Am (Wei et al., 1976), as measured by a labeling method. Thus, the level of m6Am is at or below one tenth of that of m6A in mRNA, with the ratio m6Am/m6A ratios varying between 1/10 to 1/15 in HeLa, HEK293T, 3T3-L1, MEL624 and brain tissues, as quantified in several recent studies (Engel et al., 2018; Sendinc et al., 2018; Sun et al., 2019; Wei et al., 2018) using ultra-high-performance liquid chromatography-tandem mass spectrometry (UHPLC-QqQ-MS/MS). This ratio further decreases to ½0 to 1/30 in certain acute myeloid leukemia (AML) cells (Su et al., 2018; Wei et al., 2018).
Because of the much higher abundance of m6A than m6Am in mRNA, FTO mediates predominant demethylation of m6A versus m6Am inside cells despite m6Am being a preferred substrate in vitro (Wei et al., 2018). Both can be demethylated by cytoplasmic FTO. Since almost all mRNA caps are bound by cap-binding proteins inside the cell nucleus (Marcotrigiano et al., 1997), the lack of m6Am demethylation in most cell nucleus may reflect restricted cap-m6Am accessibility by FTO (Figure 4). In addition, recent studies (Bartosovic et al., 2017; Louloupi et al., 2018) indicated the considerable intronic binding of FTO, suggesting additional roles of FTO on pre-mRNAs or chromosome associated RNAs.
So far, all functionally relevant reports of FTO have been consistent with internal m6A but not cap m6Am in mRNA being the relevant substrate, including but not limited to facilitating the 3T3-L1 cell differentiation (Zhao et al., 2014); responding to heat shock (Zhou et al., 2015) and UV damage (Xiang et al., 2017); regulating the hepatitis C virus infection (Gokhale et al., 2016); promoting the growth and self-renewal of glioblastoma stem cells (Cui et al., 2017), and playing key oncogenic roles in leukemogenesis (Li et al., 2017; Su et al., 2018), with numerous other examples in recent years (Engel et al., 2018; Yan et al., 2018; Yu et al., 2018; Zhou et al., 2018).
The mRNA cap m6Am Methylation Does Not Affect Transcript Stability
A main evidence to assign the FTO-mediated cap m6Am demethylation as functionally relevant was the reported role of cap m6Am in protecting mRNA from DCP2-mediated decapping and microRNA-mediated mRNA degradation (Mauer et al., 2017). When FTO was knocked down, substantial changes in the levels of FTO target mRNAs were observed; these effects were assigned to cap m6Am demethylation as to show its functional effect (Mauer et al., 2017). This conclusion was not supported by a more careful analysis of the correlations between changes of expression levels of transcripts containing m6A only or m6Am only upon FTO knockdown (Wei et al., 2018). Only internal m6A-modified transcripts showed correlation with the transcript level changes induced by FTO knockdown.
The confusion was recently resolved when three groups independently identified PCIF1 as the mRNA cap m6Am methyltransferase (Akichika et al., 2019; Boulias et al., 2018; Sendinc et al., 2018). PCIF1 possess only cap m6Am but not internal methylation activity. Two research groups independently showed that the PCIF1-mediated deposition of m6Am does not alter gene expression or transcript stability (Akichika et al., 2019; Sendinc et al., 2018), which refutes the results and conclusion that the mRNA cap m6Am demethylation by FTO alters mRNA levels (Mauer et al., 2017). Thus, the transcript stabilizing effect previously observed for cap m6Am upon FTO knockdown is mostly derived from the stabilizing role of internal m6As but not cap m6Am.
Cap m6Am was also proposed to promote translation efficiency of m6Am-containing mRNA with the FTO knockdown leading to decreased translation of several target genes (Mauer et al., 2017). This result was recapitulated in one of the recent cap-m6Am methyltransferase studies (Akichika et al., 2019). However, two recent studies reported that cap m6Am has negligible effects on translation under basal conditions (Boulias et al., 2018), or even suppresses translation based on in vitro translation assays and in vivo reporter assays (Sendinc et al., 2018). The effect of cap m6Am on translation could also be context dependent and remains to be clarified.
Substrate and Roles of ALKBH5
ALKBH5 is the second RNA demethylase discovered with m6A as the only known substrate so far (Zheng et al., 2013). ALKBH5 plays an important role in mouse spermatogenesis. Its crystal structures revealed mechanism for m6A recognition and demethylation (Xu et al., 2014). The expression patterns of ALKBH5 and FTO are distinct among tissues. For instance, in mouse, while Alkbh5 is most highly expressed in testis, Fto has the highest expression in the brain. Differential expression of ALKBH5 and FTO among tissues could be one reason that these two m6A demethylases participate in different biological pathways. They also interact with different protein partners, leading to different substrate repertoire.
ALKBH5 knockdown in human cell lines can accelerate the export of target RNAs from the nucleus to the cytoplasm and this function is affected by its demethylation activity (Zheng et al., 2013). Almost all reported ALKBH5 functional studies revealed a similar molecular pathway—ALKBH5 mediates the demethylation of 3’UTR m6A in certain transcripts—including facilitating hypoxia induced HIF-dependent breast cancer stem cell phenotype (Zhang et al., 2016), regulating glioblastoma proliferation and tumorigenesis through the ALKBH5-FOXM1 pathway (Zhang et al., 2017), and modulating splicing and stability of long 3’ UTR mRNAs in male germ cells (Tang et al., 2018).
Conclusions and Future Perspectives
Recent years have witnessed a rapid advance in the understanding of the complexity of gene expression regulation at the level of RNA modifications. The regulatory roles of m6A could be modulated by multiple layers of contexts: (1) cell differentiation and developmental status. The mRNA decay function of m6A mediated by YTHDF2 is critical for cell state transitions where a transcriptome switch is typically needed (Ivanova et al., 2017; Li et al., 2018; Zhao et al., 2017). In cultured cell lines or post-mitotic cells, the role of YTHDF2 might not be as significant; (2) effects from environmental stimuli or cellular signaling. The functions of m6A may not be activated in the absence of relevant cellular cues. Thus, simply investigating cell line systems under standard growth conditions may not adequately capture the real biology of RNA methylation; (3) subcellular localization of m6A effectors. Changes in protein PTMs or the presence or absence of protein partners might account for variable distribution of writer, reader, and erasers in different biological systems. The writer METTL3 shows multifaceted functions likely depending on its cellular localization and PTMs. The eraser FTO exhibits differential substrate preferences in the cytoplasm versus in the nucleus, and can mediate substantial mRNA demethylation if the level is elevated in cell cytoplasm. Certain RNA modifications may enrich and exhibit functions in sub-cellular compartments; (4) along the same transcript, the exact locations of m6A or other modifications may matter and have different regulatory functions. The diversity in the binding modes of and crosstalk between different classes of m6A readers and their protein partners may confer specificity in the fate regulation of m6A-modified mRNA.
Moving forward, some of the key questions remaining to be addressed in the field are most likely to be context dependent. The writers and erasers likely target different groups of transcripts in different cell types and in different biological processes. The erasers and different readers may also modulate or recognize m6A at specific regions of a transcript, leading to different functional outcomes. How are the selectivity, both transcript selectivity and site selectivity, achieved? Different readers may affect distinct sets of transcripts in different cell types or tissues. How are these readers recognizing their target transcripts? How are writers, readers and erasers regulated and integrated into diverse biological signaling and regulation? RNA and RNA modifications effector proteins can exist in different cellular compartments. The correct cellular context should be an essential element of future functional investigations of RNA modifications.
Acknowledgements
C.H. thanks support from the U.S. National Institutes of Health (HG008935). C.H. is an investigator of the Howard Hughes Medical Institute. We apologize to colleagues whose work could not be cited due to space constraints.
Footnotes
Declaration of Interests
C.H. is a scientific founder of Accent Therapeutics, Inc. and a member of its scientific advisory board.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aas A, Isakson P, Bindesboll C, Alemu EA, Klungland A, and Simonsen A (2017). Nucleocytoplasmic Shuttling of FTO Does Not Affect Starvation-Induced Autophagy. PLoS One 12, e0168182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams JM, and Cory S (1975). Modified nucleosides and bizarre 5[prime]-termini in mouse myeloma mRNA. Nature 255, 28–33. [DOI] [PubMed] [Google Scholar]
- Adivarahan S, Livingston N, Nicholson B, Rahman S, Wu B, Rissland OS, and Zenklusen D (2018). Spatial Organization of Single mRNPs at Different Stages of the Gene Expression Pathway. Mol Cell 72, 727–738 e725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akichika S, Hirano S, Shichino Y, Suzuki T, Nishimasu H, Ishitani R, Sugita A, Hirose Y, Iwasaki S, Nureki O, et al. (2019). Cap-specific terminal N (6)-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science 363, eaav0080. [DOI] [PubMed] [Google Scholar]
- Alarcon CR, Goodarzi H, Lee H, Liu X, Tavazoie S, and Tavazoie SF (2015a). HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell 162, 1299–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon CR, Lee H, Goodarzi H, Halberg N, and Tavazoie SF (2015b). N6-methyladenosine marks primary microRNAs for processing. Nature 519, 482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascano M, Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, et al. (2012). FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature 492, 382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millan-Zambrano G, Robson SC, Aspris D, Migliori V, Bannister AJ, Han N, et al. (2017). Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature 552, 126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosovic M, Molares HC, Gregorova P, Hrossova D, Kudla G, and Vanacova S (2017). N6methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3’-end processing. Nucleic Acids Res 45, 11356–11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertero A, Brown S, Madrigal P, Osnato A, Ortmann D, Yiangou L, Kadiwala J, Hubner NC, de los Mozos IR, Sadée C, et al. (2018). The SMAD2/3 interactome reveals that TGFβ controls m6A mRNA methylation in pluripotency. Nature 555, 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crecy-Lagard V, Ross R, Limbach PA, Kotter A, et al. (2018). MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 46, D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokar JA, Shambaugh ME, Polayes D, Matera AG, and Rottman FM (1997). Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3, 1233–1247. [PMC free article] [PubMed] [Google Scholar]
- Boulias K, Toczydlowska-Socha D, Hawley BR, Liberman-Isakov N, Takashima K, Zaccara S, Guez T, Vasseur J-J, Debart F, Aravind L, et al. (2018). Identification of the m6Am methyltransferase PCIF1 reveals the location and functions of m6Am in the transcriptome. bioRxiv, 485862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Hao YJ, Zhang Y, Li MM, Wang M, Han W, Wu Y, Lv Y, Hao J, Wang L, et al. (2015). m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell 16, 289–301. [DOI] [PubMed] [Google Scholar]
- Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J, Du P, Kim W, Tang S, Sliz P, et al. (2018). mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature 561, 556–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn WE, and Volkin E (1951). Nucleoside-5′-Phosphates from Ribonucleic Acid. Nature 167, 483–484. [Google Scholar]
- Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun G, Lu Z, Huang Y, Yang CG, et al. (2017). m(6)A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep 18, 2622–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers R, Friderici K, and Rottman F (1974). Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A 71, 3971–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez D, Freese P, Alexis MS, Su A, Hochman M, Palden T, Bazile C, Lambert NJ, Van Nostrand EL, Pratt GA, et al. (2018). Sequence, Structure, and Context Preferences of Human RNA Binding Proteins. Mol Cell 70, 854–867 e859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. [DOI] [PubMed] [Google Scholar]
- Doxtader KA, Wang P, Scarborough AM, Seo D, Conrad NK, and Nam Y (2018). Structural Basis for Regulation of METTL16, an S-Adenosylmethionine Homeostasis Factor. Mol Cell 71, 1001–1011 e1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J, and Wu L (2016). YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4–NOT deadenylase complex. Nat Commun 7, 12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Hou G, Zhang H, Dou J, He J, Guo Y, Li L, Chen R, Wang Y, Deng R, et al. (2018). SUMOylation of the m6A-RNA methyltransferase METTL3 modulates its function. Nucleic Acids Res 46, 5195–5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edupuganti RR, Geiger S, Lindeboom RGH, Shi H, Hsu PJ, Lu Z, Wang SY, Baltissen MPA, Jansen P, Rossa M, et al. (2017). N(6)-methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nat Struct Mol Biol 24, 870–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel M, Eggert C, Kaplick PM, Eder M, Roh S, Tietze L, Namendorf C, Arloth J, Weber P, Rex-Haffner M, et al. (2018). The Role of m(6)A/m-RNA Methylation in Stress Response Regulation. Neuron 99, 389–403 e389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M, Harada BT, Behm M, and He C (2018). RNA modifications modulate gene expression during development. Science 361, 1346–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y (2012). Dynamic Regulation of Rna Modifications by AlkB Family Dioxygenases In Chemistry Department (Proquest, Order No. 3548228: The University of Chicago; ). [Google Scholar]
- Fu Y, Jia G, Pang X, Wang RN, Wang X, Li CJ, Smemo S, Dai Q, Bailey KA, Nobrega MA, et al. (2013). FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat Commun 4, 1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale NS, McIntyre ABR, McFadden MJ, Roder AE, Kennedy EM, Gandara JA, Hopcraft SE, Quicke KM, Vazquez C, Willer J, et al. (2016). N6-Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection. Cell Host Microbe 20, 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati P, Avezov E, Ma M, Antrobus R, Lehner P, O’Rahilly S, and Yeo GS (2014). Fat mass and obesity-related (FTO) shuttles between the nucleus and cytoplasm. Biosci Rep 34, e00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag S, Warda AS, Kretschmer J, Gunnigmann MA, Hobartner C, and Bohnsack MT (2015). NSUN6 is a human RNA methyltransferase that catalyzes formation of m5C72 in specific tRNAs. RNA 21, 1532–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H, Hao S, Chen H, Chen Z, Zhang Y, Wang J, Wang H, Zhang B, Qiu J, Deng F, et al. (2019). N6-methyladenosine modification and METTL3 modulate enterovirus 71 replication. Nucleic Acids Res 47, 362–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PJ, Shi H, and He C (2017a). Epitranscriptomic influences on development and disease. Genome Biol 18, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, Qi M, Lu Z, Shi H, Wang J, et al. (2017b). Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res 27, 1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al. (2018). Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol 20, 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Weng H, Zhou K, Wu T, Zhao BS, Sun M, Chen Z, Deng X, Xiao G, Auer F, et al. (2019). Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature 567, 414–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova I, Much C, Di Giacomo M, Azzi C, Morgan M, Moreira PN, Monahan J, Carrieri C, Enright AJ, and O’Carroll D (2017). The RNA m(6)A Reader YTHDF2 Is Essential for the Post-transcriptional Regulation of the Maternal Transcriptome and Oocyte Competence. Mol Cell 67, 1059–1067 e1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 7, 885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuckles P, Carl SH, Musheev M, Niehrs C, Wenger A, and Buhler M (2017). RNA fate determination through cotranscriptional adenosine methylation and microprocessor binding. Nat Struct Mol Biol 24, 561–569. [DOI] [PubMed] [Google Scholar]
- Knuckles P, Lence T, Haussmann IU, Jacob D, Kreim N, Carl SH, Masiello I, Hares T, Villasenor R, Hess D, et al. (2018). Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev 32, 415–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Zhao X, Wang W, Shi H, Pan Q, Lu Z, Perez SP, Suganthan R, He C, Bjoras M, et al. (2018). Ythdf2-mediated m(6)A mRNA clearance modulates neural development in mice. Genome Biol 19, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, Huang H, Nachtergaele S, Dong L, Hu C, et al. (2017). FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N(6)-Methyladenosine RNA Demethylase. Cancer Cell 31, 127–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Choe J, Du P, Triboulet R, and Gregory RI (2016). The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell 62, 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Eckert MA, Harada BT, Liu SM, Lu Z, Yu K, Tienda SM, Chryplewicz A, Zhu AC, Yang Y, et al. (2018). m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol 20, 1074–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. (2013). A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Cell Biol 10, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Dai Q, Zheng G, He C, Parisien M, and Pan T (2015). N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, and Pan T (2017). N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res 45, 6051–6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louloupi A, Ntini E, Conrad T, and Orom UAV (2018). Transient N-6-Methyladenosine Transcriptome Sequencing Reveals a Regulatory Role of m6A in Splicing Efficiency. Cell Rep 23, 3429–3437. [DOI] [PubMed] [Google Scholar]
- Ma H, Wang X, Cai J, Dai Q, Natchiar SK, Lv R, Chen K, Lu Z, Chen H, Shi YG, et al. (2019). N(6-)Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat Chem Biol 15, 88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano J, Gingras AC, Sonenberg N, and Burley SK (1997). Cocrystal structure of the messenger RNA 5’ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89, 951–961. [DOI] [PubMed] [Google Scholar]
- Mardakheh FK, Paul A, Kumper S, Sadok A, Paterson H, McCarthy A, Yuan Y, and Marshall CJ (2015). Global Analysis of mRNA, Translation, and Protein Localization: Local Translation Is a Key Regulator of Cell Protrusions. Dev Cell 35, 344–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinov GK, Williams BA, McCue K, Schroth GP, Gertz J, Myers RM, and Wold BJ (2014). From singlecell to cell-pool transcriptomes: stochasticity in gene expression and RNA splicing. Genome Res 24, 496–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, Linder B, Pickering BF, Vasseur JJ, Chen Q, et al. (2017). Reversible methylation of m(6)Am in the 5’ cap controls mRNA stability. Nature 541, 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendel M, Chen KM, Homolka D, Gos P, Pandey RR, McCarthy AA, and Pillai RS (2018). Methylation of Structured RNA by the m(6)A Writer METTL16 Is Essential for Mouse Embryonic Development. Mol Cell 71, 986–1000 e1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkurjev D, Hong WT, Iida K, Oomoto I, Goldie BJ, Yamaguti H, Ohara T, Kawaguchi SY, Hirano T, Martin KC, et al. (2018). Synaptic N(6)-methyladenosine (m(6)A) epitranscriptome reveals functional partitioning of localized transcripts. Nat Neurosci 21, 1004–1014. [DOI] [PubMed] [Google Scholar]
- Meyer KD, and Jaffrey SR (2017). Rethinking m(6)A Readers, Writers, and Erasers. Annu Rev Cell Dev Biol 33, 319–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, and Jaffrey SR (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149, 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil DP, Chen C-K, Pickering BF, Chow A, Jackson C, Guttman M, and Jaffrey SR (2016). m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, and Conrad NK (2017). The U6 snRNA m(6)A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell 169, 824–835 e814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RP, and Kelley DE (1974). Existence of Methylated Messenger-Rna in Mouse L Cells. Cell 1, 37–42. [Google Scholar]
- Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res 24, 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch S, He C, and Dickinson BC (2018). Targeted m(6)A Reader Proteins To Study Epitranscriptomic Regulation of Single RNAs. J Am Chem Soc 140, 11974–11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree IA, Evans ME, Pan T, and He C (2017a). Dynamic RNA Modifications in Gene Expression Regulation. Cell 169, 1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, Sha J, Huang X, Guerrero L, Xie P, et al. (2017b). YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife 6, e31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pulido L, and Andrade-Navarro MA (2007). The FTO (fat mass and obesity associated) gene codes for a novel member of the non-heme dioxygenase superfamily. BMC Biochem 8, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U, Kelley DE, and Perry RP (1977). Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. J Mol Biol 115, 695–714. [DOI] [PubMed] [Google Scholar]
- Scholler E, Weichmann F, Treiber T, Ringle S, Treiber N, Flatley A, Feederle R, Bruckmann A, and Meister G (2018). Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. RNA 24, 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendinc E, Valle-Garcia D, Dhall A, Chen H, Henriques T, Sheng W, Adelman K, and Shi Y (2018). PCIF1 catalyzes m6Am mRNA methylation to regulate gene expression. bioRxiv, 484931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, Liu C, and He C (2017). YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res 27, 315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Zhang X, Weng YL, Lu Z, Liu Y, Lu Z, Li J, Hao P, Zhang Y, Zhang F, et al. (2018). m(6)A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature 563, 249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima H, Matsumoto M, Ishigami Y, Ebina M, Muto A, Sato Y, Kumagai S, Ochiai K, Suzuki T, and Igarashi K (2017). S-Adenosylmethionine Synthesis Is Regulated by Selective N(6)-Adenosine Methylation and mRNA Degradation Involving METTL16 and YTHDC1. Cell Rep 21, 3354–3363. [DOI] [PubMed] [Google Scholar]
- Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, Deng X, Wang Y, Weng X, Hu C, et al. (2018). R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell 172, 90–105 e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Zhang M, Li K, Bai D, and Yi C (2019). Cap-specific, terminal N(6)-methylation by a mammalian m(6)Am methyltransferase. Cell Res 29, 80–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C, Klukovich R, Peng H, Wang Z, Yu T, Zhang Y, Zheng H, Klungland A, and Yan W (2018). ALKBH5-dependent m6A demethylation controls splicing and stability of long 3’-UTR mRNAs in male germ cells. Proc Natl Acad Sci U S A 115, E325–E333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Doxtader KA, and Nam Y (2016a). Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol Cell 63, 306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, Tang C, et al. (2016b). Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex. Nature 534, 575. [DOI] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. (2014). N6methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, and He C (2015). N(6)methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 161, 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C-M, and Moss B (1977). Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry 16, 1672–1676. [DOI] [PubMed] [Google Scholar]
- Wei CM, Gershowitz A, and Moss B (1975). Methylated nucleotides block 5’ terminus of HeLa cell messenger RNA. Cell 4, 379–386. [DOI] [PubMed] [Google Scholar]
- Wei CM, Gershowitz A, and Moss B (1976). 5’-Terminal and internal methylated nucleotide sequences in HeLa cell mRNA. Biochemistry 15, 397–401. [DOI] [PubMed] [Google Scholar]
- Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, Shi H, Cui X, Su R, Klungland A, et al. (2018). Differential m(6)A, m(6)Am, and m(1)A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol Cell 71, 973–985 e975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, Lv R, Ma H, Shen H, He C, Wang J, Jiao F, Liu H, Yang P, Tan L, et al. (2018). Zc3h13 Regulates Nuclear RNA m(6)A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol Cell 69, 1028–1038 e1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng YL, Wang X, An R, Cassin J, Vissers C, Liu Y, Liu Y, Xu T, Wang X, Wong SZH, et al. (2018). Epitranscriptomic m(6)A Regulation of Axon Regeneration in the Adult Mammalian Nervous System. Neuron 97, 313–325 e316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Su S, Patil DP, Liu H, Gan J, Jaffrey SR, and Ma J (2018). Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat Commun 9, 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Li A, Sun B, Sun JG, Zhang J, Zhang T, Chen Y, Xiao Y, Gao Y, Zhang Q, et al. (2019). A novel m(6)A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res 29, 23–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W, Xu C, Chen H, Ouyang J, Wang S, et al. (2017). RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature 543, 573–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al. (2016). Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell 61, 507–519. [DOI] [PubMed] [Google Scholar]
- Xu C, Liu K, Tempel W, Demetriades M, Aik W, Schofield CJ, and Min J (2014). Structures of human ALKBH5 demethylase reveal a unique binding mode for specific single-stranded N6-methyladenosine RNA demethylation. J Biol Chem 289, 17299–17311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F, Al-Kali A, Zhang Z, Liu J, Pang J, Zhao N, He C, Litzow MR, and Liu S (2018). A dynamic N(6)-methyladenosine methylome regulates intrinsic and acquired resistance to tyrosine kinase inhibitors. Cell Res 28, 1062–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Yixing, Wu Kejing, Quan Weili, Yu Lin, Chen Shuang, Cheng Chao, Wu Qijia, Zhao Shuhong, Zhang Yi, and Zhou L (2019). The dynamics of FTO binding and demethylation from the m6A motifs. elife under revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Chen M, Huang H, Zhu J, Song H, Zhu J, Park J, and Ji SJ (2018). Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res 46, 1412–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, Cheng T, Gao M, Shu X, Ma H, et al. (2018). VIRMA mediates preferential m(6)A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell Discov 4, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, He X, and Semenza GL (2016). Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc Natl Acad Sci U S A 113, E2047–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Kang Y, Wang M, Li Y, Xu T, Yang W, Song H, Wu H, Shu Q, and Jin P (2018). Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum Mol Genet 27, 3936–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, Chen Y, Sulman EP, Xie K, Bogler O, et al. (2017). m(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 31, 591–606 e596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wei LH, Wang Y, Xiao Y, Liu J, Zhang W, Yan N, Amu G, Tang X, Zhang L, et al. (2019). Structural insights into FTO’s catalytic mechanism for the demethylation of multiple RNA substrates. Proc Natl Acad Sci U S A 116, 2919–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao BS, Wang X, Beadell AV, Lu Z, Shi H, Kuuspalu A, Ho RK, and He C (2017). m(6)A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 542, 475–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, Hao YJ, Ping XL, Chen YS, Wang WJ, et al. (2014). FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res 24, 1403–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 49, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M, and Fray RG (2008). MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20, 1278–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, and Qian S-B (2015). Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 526, 591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wan J, Shu XE, Mao Y, Liu XM, Yuan X, Zhang X, Hess ME, Bruning JC, and Qian SB (2018). N(6)-Methyladenosine Guides mRNA Alternative Translation during Integrated Stress Response. Mol Cell 69, 636–647 e637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Yong XLH, Xia D, Widagdo J, and Anggono V (2018). Ubiquitination Regulates the Proteasomal Degradation and Nuclear Translocation of the Fat Mass and Obesity-Associated (FTO) Protein. J Mol Biol 430, 363–371. [DOI] [PubMed] [Google Scholar]