Abstract
Smooth muscle cells (SMCs) are a critical component of blood vessel walls that provide structural support, regulate vascular tone, and allow for vascular remodeling. These cells also exhibit a remarkable plasticity that contributes to vascular growth and repair but also to cardiovascular pathologies, including atherosclerosis, intimal hyperplasia and restenosis, aneurysm, and transplant vasculopathy. Mouse models have been an important tool for the study of SMC functions. The development of smooth muscle-expressing Cre-driver lines has allowed for exciting discoveries, including recent advances revealing the diversity of phenotypes derived from mature SMC transdifferentiation in vivo using inducible CreERT2 lines. We review SMC-targeting Cre lines driven by the Myh11, Tagln, and Acta2 promoters, including important technical considerations associated with these models. Limitations that can complicate study of the vasculature include expression in visceral SMCs leading to confounding phenotypes, and expression in multiple non-smooth muscle cell types, such as Acta2-Cre expression in myofibroblasts. Notably, the frequently employed Tagln/SM22α-Cre driver expresses in embryonic heart but can also confer expression in non-muscular cells including perivascular adipocytes and their precursors, myeloid cells, and platelets, with important implications for interpretation of cardiovascular phenotypes. With new Cre-driver lines under development and the increasing use of fate mapping methods, we are entering an exciting new era in SMC research.
Introduction
Smooth muscle cells
Smooth muscle cells (SMCs) make up the muscular layer of arteries and veins and are essential in regulation of vascular tone and blood pressure. Under physiological conditions, SMCs are differentiated and quiescent, but in response to injury, SMCs de-differentiate, characterized by proliferation, migration, and loss of contractile protein expression. While the remarkable phenotypic plasticity of SMCs in the vessel wall is important for wound healing and growth, SMCs also play a role in pathophysiology. SMC de-differentiation is central to vascular pathologies such as atherosclerosis, restenosis, and aneurysms 1, 2. Phenotypically modulated SMCs also contribute to the formation of fibrous cap, neointima, and foam cells in atherosclerotic plaques 2. SMCs are integral to the vascular reactivity and remodeling changes that contribute to the pathophysiology of hypertension and its complications 3. Thus, it is critical to elucidate the mechanisms by which SMCs are regulated.
Differentiated SMCs are characterized by a large repertoire of genes encoding a variety of contractile/cytoskeletal proteins, including α-smooth muscle actin (ACTA2), smooth muscle myosin heavy chain (MYH11; formerly known as SM-MHC), calponin (CNN1), transgelin (TAGLN; formerly known as SM22α), h-caldesmon (CALD1) and smoothelin (SMTN) 4. These genes have been important markers in the study of SMC, and the regulatory regions of several of these genes have been used to drive expression of Cre recombinase. We will discuss the pros and cons of each in this review.
The Cre/lox system
The Cre/lox site-specific recombination system allows generation of cell-type specific and conditional deletion of a specific gene. Cre, a 38 kDa bacteriophage enzyme, has been shown to catalyze recombination between two 34 bp lox sites5. The Cre protein creates a DNA loop and then either excises or inverts the looped segment depending on the orientation of the loxP sites 6, 7. Initially, the method was developed to function efficiently in yeast to cause recombination on chromosomes and was subsequently adapted for use in mammalian cells and mouse models6, 8. The Cre/lox site-specific recombination system is now well established for generation of conditional cell type-specific deletion or overexpression of specific genes, allowing for sophisticated mouse models in which to study cardiovascular disease. DNA regions flanked by loxP sites are often referred to as “floxed”. This Cre/lox system has been elegantly summarized in the prior article in this series on endothelial-specific Cre drivers 9. An important variation in this technology is the fusion of Cre with a mutant estrogen receptor (Cre-ERT2) to allow tamoxifen-inducible cell type-specific gene expression or deletion. Feil et al. constructed two mutants of the estrogen receptor that either contain G400V/M543A/L544A or G400V/L539A/L540A in their ligand binding domain. These two mutants are termed as T1 and T2 respectively. The T2 mutant version of ER is strongly activated by tamoxifen and has been widely used in mouse genetic studies 10. Tamoxifen treatment induces translocation of Cre-ERT2 from the cytosol to the nucleus where it can induce DNA recombination between 34 bp loxP sites 11.
As reviewed and highlighted in this review, a strategy that tends to yield faithful Cre expression involves inserting Cre recombinase into the gene of interest in a bacterial artificial chromosome to express in mice. The limitations involve random site of integration and variable number of gene copies. Song et al. highlight the use of gene targeting by homologous recombination in embryonic stem cells or by using CRSPR/Cas9 methods to ensure a single copy of a Cre recombinase gene is expressed from an endogenous gene locus 7. There are also now different types of recombinases and several variants of lox sites that can be used together to create more controlled recombination events. Song et al. elegantly describe breeding strategies to avoid unwanted recombination and methods to overcome such issues if they arise 7. Proper breeding strategies, genotyping protocols, and control experiments should help investigators in Cre-dependent gene manipulations and interpret their data appropriately.
There are additional general important considerations when using Cre/lox approaches. Several studies have highlighted the potential for differential sensitivities to Cre mediated recombination in distinct targeted floxed alleles12. Liu et al investigated the recombination correlation between several floxed alleles induced by Cre-expressing mouse lines and determined that factors including potential methylation, distance between loxP sites, sequences flanking the loxP sites and the level of Cre activity per cell can contribute to differences in Cre-mediated recombination 12. Accordingly, investigators should carefully document the expression pattern of new Cre-driver mice with a reporter (lacZ, mTmG, YFP, etc). Similarly, the extent of recombination and degree of deletion should be evaluated using qPCR or Southern blotting strategies. For inducible Cre lines, optimization of the timing and dosing of tamoxifen may be required. Additional important “best practices” include maintaining the Cre mice in a hemizygous manner (for transgenes) and as heterozygous (for Cre-knockin) to avoid problems such as mutagenesis or loss of endogenous gene function. Refreshing breeders periodically can also help minimize genetic drift, and periodic re-evaluation of Cre patterns and deletion efficiency should be performed. Validating findings with multiple SMC Cre drivers, as was successfully employed by Herring et al (see below in Myh11-Cre section), is an additional rigorous approach13.
In reviewing the different approaches to generate Cre driver lines, it is apparent that fidelity in replicating endogenous gene expression patterns improves with larger promoter fragments, with enhanced recapitulation generally obtained from constructs generated from bacterial artificial chromosome (BAC) segments or with knock-in of the Cre recombinase into the endogenous locus. While these Cre-based strategies have opened new frontiers in SMC research, targeting Cre expression specifically to SMCs has proven challenging as many of these smooth muscle markers are expressed, at least transiently, in other cell types, including fibroblasts and myofibroblasts (detailed in following sections). Furthermore, expression of target genes in visceral smooth muscle can produce gastrointestinal phenotypes that complicate or preclude analysis of vascular phenotypes (discussed in the concluding section). Herein, we review smooth muscle-targeting Cre driver lines and highlight the advantages and caveats of each. We additionally summarize some of the recent paradigm changing discoveries made possible by this technology.
Myh11-Cre
Of the aforementioned smooth muscle markers, MYH11 is widely considered the most specific for the smooth muscle lineage and the most definitive marker of SMC differentiation. Joe Miano, Eric Olson and colleagues were the first to report in situ hybridization analysis of Myh11 transcript expression and concluded that Myh11 was highly specific to SMCs in embryonic and adult mice 14. Subsequently, a transgene consisting of the 4.2 kb of the 5’ flanking region of the rat Myh11 gene, 88 bp of the first exon (which is untranslated), an additional 11.5 kb of the first intron followed by the lacZ reporter gene was used to generate transgenic mice 15. These Myh11-lacZ mice displayed specific transgene expression in vascular and non-vascular SMCs; however, there was wide variability in lacZ expression both within and between specific vessels 15. Taking advantage of the Cre-lox system, the Owens and Kotlikoff labs generated mouse lines that expressed Cre recombinase driven by a fragment of the Myh11 promoter to induce recombination in SMCs 16, 17. Unfortunately, subsequent studies suggested that these Cre lines also induce recombination in the germ line 18, 19.
More recently, a transgenic mouse carrying a Myh11-driven inducible Cre was generated through a distinct cloning strategy by the Offermanns group 20. A fusion protein of the Cre recombinase with the modified estrogen receptor binding domain (CreERT2) was cloned into the initial coding ATG of the mouse Myh11 gene carried by a BAC 20. Tamoxifen treatment then promotes Cre-mediated recombination and subsequent expression or excision of the target gene 11. In their initial characterization of Myh11-CreERT2 mice also carrying the ROSA26R-lacZ reporter 21, Wirth et al. reported inducible Cre-mediated beta-galactosidase (β-Gal) activity exclusively in SMCs in the tissues examined, including in blood vessels in multiple organs and in stomach, colon, and bladder visceral SMCs 20. The use of the large genomic segment in the BAC likely confers this expression pattern replicating that of endogenous Myh11 with higher fidelity. It should be noted, however, that expression from this construct has not yet been thoroughly characterized during embryogenesis.
The Myh11-CreERT2 mice generated by the Offermanns lab are available from Jackson Laboratories (B6.FVB-Tg(Myh11-cre/ERT2)1Soff/J) and have proven to be a windfall for diverse studies evaluating the role of gene products in SMCs and the fate of SMCs during development and disease22. This Cre driver was used in two studies that determined that the majority of neointimal cells in models of vascular injury are derived from mature medial SMCs. These rigorous findings provided consensus in the field on the question of neointimal origin as the results were validated by two different labs using two distinct reporters (LacZ13 vs mTmG)23 as well as two different injury models (femoral wire denudation23 and carotid ligation13). Furthermore, Herring et al additionally confirmed the finding with the Acta2-CreERT213(see later section for description of this Cre line).
Work from several laboratories has employed lineage tracing with the Myh11-CreERT2 line to uncover paradigm-shifting new roles for SMC in atherosclerosis. Gary Owens and colleagues demonstrated that tamoxifen induction of Apoe(−/−), Myh11-CreERT2, ROSA26R(YFP/YFP) mice results in efficient marking of medial SMCs in large arteries. Notably, their lineage tracing approach revealed that the contributions of SMCs to atherosclerotic plaques had been underestimated, as >80% of SMC-derived cells in plaques could not be identified by immunostaining for SMC contractile protein markers after high fat diet feeding 24. In plaques, mature YFP+ SMC were found to have lost their classical SMC markers and express markers of other cell types, including macrophages or stem cells 24. More recent work from the Greif lab found that pre-existing SMCs are recruited into atherosclerotic plaques, first coating the cap of the plaque and subsequently invading the core 25. Importantly, these investigations provide evidence that SMCs change fate during atherogenesis. Further mechanistic experiments, which include introducing a floxed allele for the Kruppel-like factor (klf) 4 into Myh11-CreERT2 lineage tracing mice, demonstrate that KLF4 plays a key role in pathological SMC transitions in atherosclerosis 24 as well as in cancer 26. Fate mapping studies using the Acta-CreERT2 and Klf4flox/flox mice also identified a key role for KLF4 in SMC transitions in pulmonary hypertension (see Acta2-Cre section below)27.
Although Myh11 transcript and the Myh11-CreERT2 transgene were originally reported to be specific to SMCs, data indicating expression in non-SMC populations are beginning to come to light in distinct contexts. For instance, the Greif lab recently showed that tamoxifen induction of Myh11-CreERT2, ROSA26R(mTmG/mTmG) mice also marked ACTA2− cells in the alveoli of the adult lung 28. In addition, hypoxia-induced alveolar ACTA2+ myofibroblasts express MYH11; indeed, in mice exposed to hypoxia (FiO2 10%) for 21 days, ~85% of alveolar ACTA2+ myofibroblasts express MYH11 28. In a recent study from another group, the authors suggest that in the adult mouse lung, MYH11 is expressed in SMCs as well as at least a subset of pericytes and that these pericytes are YFP+ in tamoxifen-induced Myh11-CreERT2, ROSA26R(YFP/YFP) mice 26. In the lungs of these mice, 10% of YFP+ cells were found to be negative for MYH11. While these may be derived from dedifferentiated pericytes or SMC, their origin is not definitively known. This study found that KLF4 upregulation also mediates phenotypic switching in YFP+ perivascular cells, which contribute to a pro-metastatic microenvironment 26.
An important consideration is that the Myh11-CreERT2 transgene initially incorporated into the Y chromosome which has limited investigations with this transgene to male mice. This limitation is profound as SMCs are key players in cardiovascular development and disease, and cardiovascular disease is the leading cause of death in women. Notably, the laboratory of Zhihua Jiang recently reported that through breeding they have obtained a colony of Myh11-CreERT2 mice – presumably by translocation to the X chromosome - in which the transgene is inherited by both male and female mice 29–31. Importantly, this colony may markedly enhance the study of SMCs in the development and disease of female mice.
Tagln-Cre
Transgelin (TAGLN), formerly known as SM22α, is an actin binding protein that regulates SMC contraction and serves as a marker of the differentiated phenotype. Accordingly, Tagln/SM22α-Cre drivers have been commonly used to study smooth muscle-specific expression. While these were originally referred to and published as “SM22α-Cre”, we will refer to them in this review using the current official nomenclature “Tagln-Cre”. These Cre drivers have been widely employed despite the well-known limitation that Tagln is transiently expressed in cardiac myocytes during embryonic development. By mid-gestation, Tagln gene expression becomes restricted to visceral and vascular SMCs. Olson and colleagues initially determined that a 455 bp fragment of the mouse SM22α promoter was sufficient to drive lacZ reporter activity in SMCs in vivo but did not completely recapitulate endogenous expression patterns 32. The first Tagln-Cre lines were made by the Feil group, using a transgenic or knock-in approach. The knock-in Tagln-Cre-ERT2 line was superior in recombination to the transgenic line, but efficiency was very low in vascular SMC compared to visceral SMC 33. Another early attempt created a transgenic Tagln-Cre with a 1.4 kb fragment of the Tagln promoter 34. A ROSA26R-LacZ reporter indicated embryonic expression in heart, aorta, and umbilical vessels, but also in the head and tail regions of the embryo at E10.5. Conditional deletion of Srf with this Tagln-Cre proved to be embryonic lethal, identifying a key role for Srf in heart and vascular development 34. The Liaw lab also took two approaches incorporating a 1.4 kb fragment of the Tagln promoter to make a transgenic Tagln-Cre and a Tet-on inducible Tagln-rtTA-Cre. When crossed to five reporter lines, these mice showed very high variability in recombination 35.
An additional transgenic line was generated employing a longer 2.8 kb fragment of the mouse Tagln promoter to drive Cre expression. The Cre mRNA was detected at high levels in aorta, intestine, and uterus similar to endogenous Tagln. When crossed with ROSA-26R-lacZ reporter mice, efficient recombination was demonstrated in hepatic and pulmonary arteries, while non-muscle cells in these organs did not express β-Gal 36. This line (now congenic) is available from the Jackson Laboratories (B6.Cg-Tg(Tagln-cre)1Her/J) 36 and has been used by many investigators.
The Parmacek group used a similar strategy to construct Tagln-Cre using –2775-bp to +39-bp of the Tagln promoter which was similarly expressed in SMC during development and in adult tissues 37. The expected expression was found in the vascular wall of cranial vessels, the bronchial arch arteries, umbilical artery as well as the developing heart at E9.5, and in adult vascular and visceral smooth muscle. While arterial SMC were labeled with nearly 100% efficiency, as expression was also noted in the adult ventricular myocardium, this driver line did not faithfully recapitulate Tagln expression in the adult.37
Chen and colleagues employed a distinct approach, knocking the Cre recombinase coding sequence into the endogenous Tagln locus in frame just prior to the Tagln initiation site (Tagln-CreKI) 38. In this approach two targeted clones were injected into C57BL/6 blastocysts to produce chimeras, which were then further crossed with C57BL/6 female mice to generate heterozygous Tagln-CreKI mice. A homozygous knockin of Cre may cause inadvertent deletion of endogenous gene. When crossed to ROSA26R LacZ or eGFP reporter mice, expression from this Cre faithfully recapitulated expression of Tagln in adult tissues, including arteries, veins, bladder and gastrointestinal tract, with no expression in skeletal muscle. Strikingly, however, this Tagln-CreKI did not drive expression in embryonic SMC or cardiac myocytes at E9-E16.5, but could be detected in neonates after birth. While this does not mimic endogenous expression in this regard, this line could potentially circumvent adverse effects of gene expression changes driven by the Tagln-Cre during development. This line has also been available through Jackson Laboratories (B6.129S6-Taglntm2(cre)Yec/J) 38 and is widely published.
Non-smooth muscle expression of Tagln
Some early studies that predated development of Tagln-Cre lines detected Tagln expression in non-SMC or cardiac myocyte tissues. These include TAGLN expression in cultured human fetal lung myofibroblasts 39, rabbit bladder serosal myofibroblasts 40, and porcine adventitial myofibroblasts following vascular injury 41. Subsequently, other studies have revealed that Tagln-Cre lines direct expression in other cell types. One study by Shen and colleagues used both Tagln-Cre lines available at Jackson Laboratories and bred them individually to a strain to transgenically express a dominant negative mastermind-like-GFP fusion protein (DNMAML-GFP) 42. This allowed use of GFP signal to trace cells labeled by each Cre, with the caveat of potentially confounding effects of overexpression of a factor that inhibits canonical Notch-mediated transcription on the pattern of GFP expression. While GFP was detected in the expected SMC-rich tissues including aorta, bladder, stomach, and uterus, very high expression was also noted in the immune cell-rich spleen. Subsequent FACS and labeling approaches revealed that the Tg(Tagln-cre)1Her/J line expressed in a portion of circulating neutrophils and monocytes. Genotyping for a recombined allele also revealed Cre activity in peripheral blood and peritoneal macrophages of the Chen lab Tgln2-CreKI line42. While this study is not definitive due to the introduction of DNMAML, it raised the provocative possibility that Tagln-Cre lines could drive expression in cells other than smooth muscle and cardiac myocytes. Work from other labs, including our own, discussed below, supports this notion.
Importantly, megakaryocytes, which give rise to platelets, also express TAGLN 43. Diane Krause’s lab noted that in mice with a tdTomato to GFP Cre-reporter (mTmG)44 (Jax Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J), the Tagln-Cre transgene (Tg(Tagln-cre)1Her/J) causes from 30–60% of platelets to become GFP+. In Figure 1, the tdTomato and GFP fluorescence of peripheral blood platelets is shown for WT mice, mTmG mice without Cre recombinase, and mTmG mice crossed with Tagln-Cre. Note that in the mouse with the Tagln-Cre, over half of the platelets in the peripheral blood are Tomato− and GFP+, indicative of Cre-mediated recombination in megakaryocytes. Thus, in mice with a gene of interest deleted using the Tagln-Cre, loss of gene expression in platelets could impact the phenotypes observed. This is particularly notable as this Cre driver is frequently used to study vascular SMC responses in models such as wire denudation to induce intimal hyperplasia, a process heavily influenced by platelets. Given the caveats of Tagln-Cre-driven expression in myeloid cells and platelets, bone marrow transplantation experiments would be an important approach to determine the extent to which observed phenotypes are attributable to SMC versus hematopoietic cells. Additionally, Varberg and colleagues recently demonstrated fetal Tagln expression in the vascular progenitor endothelial colony-forming cells (ECFCs)45. This study revealed that upregulation of Tagln in these ECFCs in response to gestational diabetes led to abnormalities in vasculogenesis, suggesting another potential factor to consider when using Tagln-Cre constructs in developmental studies.
Figure 1. Flow cytometric analysis reveals Tagln-Cre mediated switch from tdTomato to GFP in a majority of platelets.
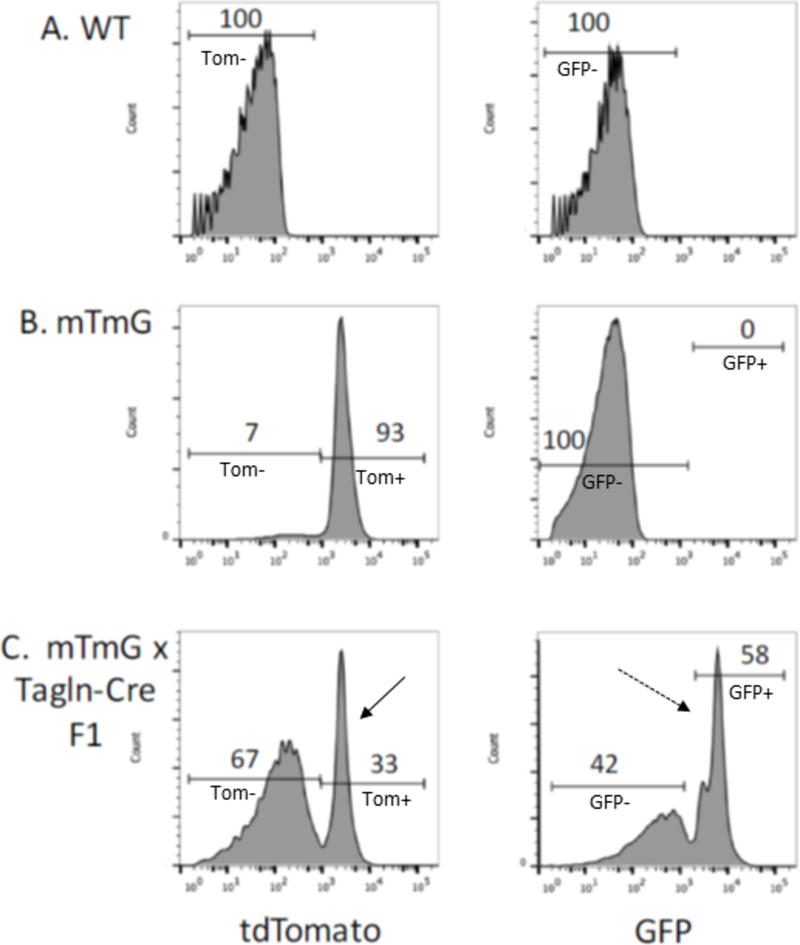
FACS analysis of platelets isolated from platelets from WT (A), mTmG (B) and Tagln-Cre x mTmG F1 (C) mice for tdTomato red (left) and GFP (right). The cutoff for positive signal is based on analysis of WT (A) nonfluorescent platelets. In 8 week old Tagln-Cre x mTmG mice (C), 33% of platelets are positive for tdTomato expression (solid arrow) while 58% are positive for GFP expression (dashed arrow) after Cre-mediated recombination.
Another notable Tagln expression pattern was uncovered when the Chen lab employed their Tagln-Cre knock-in line in order to delete PPARγ selectively in SMCs. The previously unappreciated expression of Tagln in a perivascular adipose precursor fortuitously led to very exciting and unexpected observations due to a complete lack of development of perivascular adipose tissue (PVAT) in these mice 46. PVAT was found to be highly similar to brown adipose tissue (BAT), demonstrating a thermogenic effect to regulate intravascular temperature. PVAT was also revealed as an important source of prostacyclin such that loss of PVAT led to endothelial dysfunction. Another intriguing possibility generated by this study is the existence of a common precursor that gives rise to both PVAT and vascular SMC. While “off target” gene expression driven by the Tagln-CreKI precluded a clear analysis of PPARγ function in SMC in this study, this unexpected Tagln expression pattern revealed multiple fascinating novel insights into the functions of PVAT and vascular homeostasis.
Another intriguing and serendipitous discovery was made using the Talgn-Cre 36 to delete IκB kinase β (IKKβ) in SMC. The Tagln-Cre-IKKβf/f (Tg(Tagln-cre)1Her/J ) mice were resistant to atherosclerosis in the LDLR−/− background, but were surprisingly also found to be resistant to diet-induced obesity, hepatic steatosis, and exhibited improved glucose homeostasis 47. Crossing the Talgn-Cre to Rosa26EGFP revealed that Talgn-Cre is also active in primary adipose stromal vascular cells 47. Interestingly, deficiency of IKKβ diminished the ability of these cells to differentiate, leading to accumulation of adipocyte precursor cells in adipose tissue, and revealed a mechanistic link between IKKβ and β-catenin 47.
Data from Kathleen Martin’s lab reveal bright GFP expression in the aortic media in Tagln-Cre-mTmG reporter mice (Tg(Tagln-cre)1Her/J strain), as well as in the perivascular adipocytes (Figure 2). It is now appreciated that adipose precursors reside in a perivascular niche 48. The multiple studies mentioned herein suggest or directly indicate Tagln expression in adipogenic, vascular precursors as well as mature hematopoietic cells. This is an important caveat to consider when using Tagln-Cre drivers. The functional consequences of expression of Tagln in stem cells are unknown, but suggest an interesting new avenue for further investigation.
Figure 2. Tagln-Cre-mTmG labels the aortic media and perivascular adipose tissue.
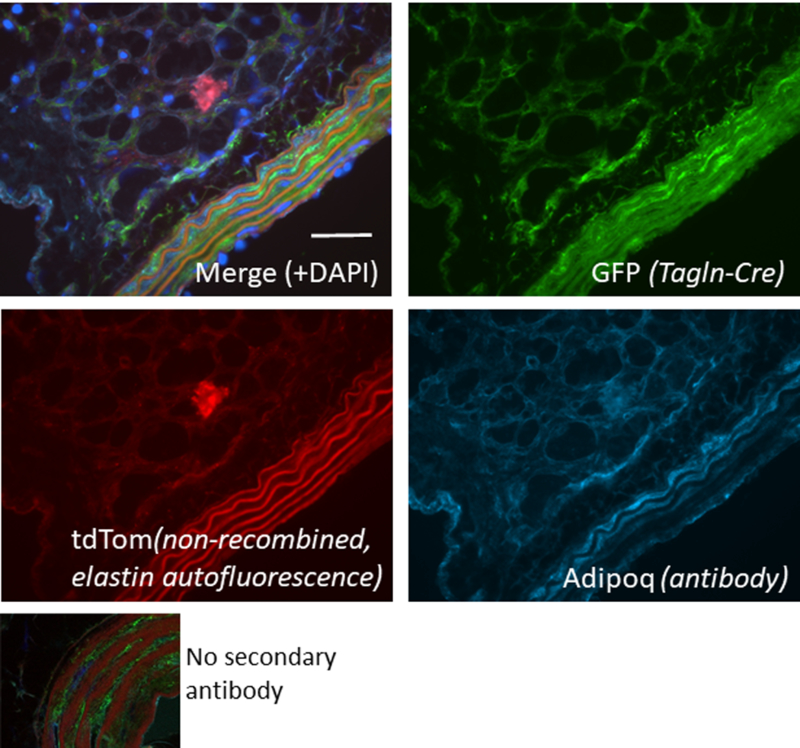
Confocal spinning disk microscopy of a section of aorta from a male Tagln-Cre-mTmG (Tg(Tagln-cre)1Her/J) mouse that was immunostained with an antibody to mouse Adiponectin (#1119 R&D Systems, 1:10 dilution) and secondary antibody (Cy5, Cyan). Merged image at top left, individual channels for GFP, tdTomato, and Cyan also shown. Scale bar = 50 mm. GFP (green) indicates cells in which Tagln-Cre is active, while tdTomato (red) indicates cells in which there is no Cre-induced recombination. DAPI (dark blue) staining indicates nuclei. A negative control with no secondary antibody is shown at bottom left. Note that adiponectin, a marker of differentiated adipocytes, strongly costains the perivascular adipocytes, as well as some medial SMCs, which we have shown to express adiponectin 65.
Acta2-Cre
Smooth muscle α-actin (ACTA2) is a key smooth muscle contractile protein whose expression increases with differentiation, but lower level expression is maintained even in “synthetic” phenotype SMC. Indeed, this marker is not a definitive marker of the smooth muscle lineage as it is expressed in many other cell types, most notably, fibroblasts and myofibroblasts. Multiple labs have generated Acta2-Cre lines. These have been primarily employed for the study of myofibroblasts and fibrosis, but under appropriate conditions, may have utility for SMC lineage tracing as described below.
The first Acta2-Cre line was generated in 1996 using a 4.7 kb fragment of the human ACTA2 gene (−891 to +3828, containing promoter region, exon 1, intron 1, and 14 bp of exon 2). Crossing to a β-Gal reporter, they reported expression only in the heart, aorta, and diencephalon in early stage embryos 49. Another early Acta2-Cre line was developed by Wu et al., using a fragment of the Acta2 gene spanning –1070 to +2582 bp including the first exon and part of first intron. When crossed to ROSA26R-LacZ, β-Gal expression was observed as expected in SMC-rich tissues including arteries, veins, and airways, and this reporter line was used to characterize mechanisms of bleomycin-induced pulmonary fibrosis 50.
Metzger and colleagues generated a transgenic tamoxifen-inducible Acta2-CreERT2 line using a large segment of the mouse Acta2 gene in a BAC 51. This construct was found to drive expression in both vascular and visceral smooth muscle with limited expression in cardiomyocytes 51. This line (Tg(Acta2-cre/ERT2)Pcn) and has been the most utilized of all published Acta2-Cre drivers to date with at least 23 citations. The Greif laboratory has employed this line to study changes in the muscularization of lung arterioles in response to hypoxia, a model of the pathological hypermuscularization in pulmonary hypertension (PH) 28. Acta2-CreERT2, ROSA26RmTmG/mTmG mice were used to induce GFP labeling of mature SMCs in pulmonary arterioles. By assessing these vessels over a time course following hypoxia treatment, it was determined that GFP+ vascular SMCs present in the normal adult were the source of nascent distal muscularization 28. This study also made the surprising observation that the vast majority of alveolar myofibroblasts that arise in response to hypoxia are not derived from ACTA2+ cells. Conversely, the Myh11-Cre-ERT2 line crossed to ROSA26RmTmG/mTmG revealed that some GFP labeled cells near the lung arterioles lacked SMC markers in the absence of hypoxia. Thus, in this particular organ, vascular bed, and time point, Acta2-Cre-ERT2 was a more rigorous marker for SMCs than Myh11-Cre-ERT227. Further studies using the Acta2-CreERT2 crossed to a multi-color reporter line (ROSA26RRainbow/+) extended these PH findings, revealing a clonal origin of SMCs in hypermuscularized distal arteriolar regions 27.
In addition to studies in pulmonary arteries, the Acta2-CreERT2 (Tg(Acta2-cre/ERT2)#Pcn) was used in a groundbreaking study assessing vasomotion in brain vessels. By comparing different Cre reporters and immunostaining approaches, it was determined that ACTA2 was expressed in arterioles but not in capillary-associated pericytes. In vivo live imaging revealed that SMC but not pericyte constrictions are critical in pathological responses to brain ischemia 52. The Acta2-CreERT2: ROSA26R transgenic line was also used for lineage tracing in a recent study which noted that myoepithelial cells in the proximal trachea also express ACTA2.53Another transgenic Acta2-Cre line employing 5.2kb of the mouse Acta2 gene, including 2.4kb of promoter as well as exon 1, intron 1 and part of exon 2, has been generated by the Kalluri lab 54, 55. An Acta2-RFP reporter line demonstrated RFP expression in SMCs and rare interstitial cells in the kidney 43, and the Acta2-Cre crossed to a YFP reporter was employed in a study of myofibroblasts in kidney fibrosis 44. This Acta2-Cre (B6.FVB-Tg(Acta2-cre)1Rkl/J), backcrossed to C57BL/6J) 44 and the reporter line ACTA2-RFP (B6.FVB-tg(Acta2-DsRed)1Rkl/J) 43 are available from The Jackson Laboratory.
Grcevic and colleagues produced and applied an Acta2-CreERT2 crossed to a reporter line to identify ACTA2+ mesenchymal progenitors within bone marrow and adipose tissue with osteogenic potential 56. Stappenbeck and colleagues used the BAC approach to generate an independent Acta2-CreERT2 line and crossed it to an mTmG reporter, generating strong GFP labeling of SMC in the gastrointestinal tract and demonstrating that loss of Acta2 gene expression in a mouse colonic injury model was due to SMC death as opposed to dedifferentiation 57.
Summary and Conclusions
In reviewing Cre driver lines used to study smooth muscle, we have noted that expression in non-SMCs is a common caveat which needs to be considered when designing and interpreting studies. While all of the current SMC-targeting Cre driver lines have pros and cons, we highlight the need to select the most appropriate Cre for studies, depending on the processes one is attempting to model. Target organ biology, vascular bed, visceral SMC, and spatiotemporal considerations are all key variables to consider.
The Myh11-CreERT2 is currently regarded as the most specific for SMCs, yet expression in SMC-like cells, such as pericytes, have been noted in the lung. Platelet-derived growth factor receptor-β and neuron-glial 2 are well known pericyte markers. However, these markers are also expressed in other cell types, including subsets of SMCs, thereby limiting the utility of mice carrying a Cre driven by the promoter of one of these genes. A Cre that specifically labels pericytes would be a major advancement for the field. Analysis of single cell transcriptomic studies of pericytes is yielding candidate genes, and promoters of these genes will need to be assayed as to whether they can be used to specifically drive Cre expression in pericytes58.
The Tagln-Cre lines, while very widely used, are now appreciated to be expressed in cardiac myocytes, adipocytes and their precursors in the vascular adventitia, megakaryoctyes, platelets, and myeloid cells. This complex expression pattern requires caution when evaluating phenotypes as these cell types may collectively influence cardiovascular physiology and pathophysiology through cardiometabolic and inflammatory effects. Bone marrow transplant, platelet transfusion, or complementary experiments employing myeloid- or platelet-targeting Cre drivers are potential strategies to address myeloid and platelet contributions in Tagln-Cre mice. The perivascular adipose expression is more difficult to address. Murine PVAT transplants are challenging but may be a potential alternative in some models 59. The Acta2-Cre lines strongly express in SMC,but may be confounded by myofibroblast and stem cell expression 56. Depending on the context, an Acta2-CreERT2 approach may have utility for the study of vascular 28,52 or visceral 46 SMC as described above.
A limitation to SMC-targeting Cre driver lines is the potential to label both vascular and visceral SMCs. When these tools are used to overexpress or delete genes that may cause a phenotype in non-vascular SMCs, there is potential for disruptive respiratory, genitourinary, and/or gastrointestinal complications that may preclude evaluation of the role of the gene in the vasculature. Examples include inducible deletion of Serum response factor (Srf) with the inducible Tagln-CreERT2 (Feil lab) which led to cachexia and death due to chronic intestinal pseudo-obstruction 60,61. Lethal gastrointestinal phenotypes were also observed with Myh11-CreERT2-mediated deletion of Srf 62 or Dicer 63. Promoters that distinguish between vascular and visceral, or arterial and venous SMC expression would be useful reagents for in vivo studies. In an early transgenic lacZ reporter driven by 445 bp of the Tagln promoter was found to express in embryonic vascular smooth, cardiac, and skeletal muscle, but notably did not express in venous or visceral SMC32. A subsequent study identified the converse reagent using a 370 bp fragment of the telokin gene. This telokin construct was reported to be visceral-specific in vivo, while chimeras between this telokin sequence and the Tagln sequence were found to modulate the vascular vs visceral expression patterns64. A smooth muscle Cre driver line with preferential activity for vascular SMCs is currently under development (personal communication, Joe Miano). It is possible that additional SMC-specific or -restricted enhancers may be discovered as well, especially with the advent of genome-wide epigenetic investigations in SMC.
Some of the limitations associated with temporal patterns in Cre expression may be circumvented by using inducible constructs such as the Cre-ERT2. For example, induction in adult mice can avoid the developmental expression of Tagln-Cre in the heart. The ability to induce recombination at a specific point in time is an essential hallmark of lineage tracing studies. The permanent marking and tracing of mature, differentiated SMCs with a Cre-driven reporter allows these cells to be identified as SMCs even after downregulating their classic contractile markers beyond detection. While SMCs are well known to comprise fibrous caps, lineage tracing has identified key roles for SMCs in the initial formative processes of atherosclerotic lesions 25, as well as their previously unappreciated role in giving rise to macrophage-like and stem cell-like cells 24. This strategy also identified primed SMCs that are source of hypermuscularization in PH 27,28. Such fate mapping approaches, when rigorously applied, will continue to expand our knowledge of the physiological consequences of SMC plasticity.
Supplementary Material
Table 1. Commonly employed SMC-expressing Cre driver lines.
These lines have been widely published (Jackson Laboratories (JAX) strain name in italics). All express in vascular and visceral smooth muscle. Advantages and limitiations of each Cre line have been discussed in the main text and are summarized here.
| Mouse strain, Reference | Advantages | Limitations |
|---|---|---|
|
Myh11-CreERT2 20 Offermanns laboratory JAX: B6.FVB-Tg(Myh11-cre/ERT2)1Soff/J |
• BAC construct faithfully recapitulates endogenous expression • Inducible |
• Also marks pericytes in the lung • Y chromosomal transgene limits study to males • (Jiang lab variant line expresses in male and female 29–31) |
|
Tagln-Cre 36 Herz laboratory JAX: B6.Cg-Tg(Tagln-cre)1Her/J |
• Expresses appropriately in SMC • B6 Congenic |
• Also expresses in myeloid cells, platelets, and perivascular adipocytes and their precursors, and ECFCs • Not inducible |
|
Tagln-CreKI 38 Chen laboratory JAX: B6.129S6- Taglntm2(cre)Yec/J |
• Knock-in line expresses appropriately in adult SMC • Does not express in embryonic SMC or cardiac myocytes |
• Also expresses in perivascular adipocytes and adipose precursors • (Not determined whether also expresses in myeloid cells or platelets) • Not inducible |
|
Acta2-Cre-ERT2 51 Metzger laboratory Tg(Acta2-cre/ERT2)Pcn (NOT at JAX.) |
• BAC-derived, appropriate SMC expression • Inducible • Validated for lineage tracing in atherosclerotic plaques and pulmonary arterioles |
• Expresses in myofibroblasts, myoepithelial cells (normal expression of endogenous Acta2 beyond SMC) |
Highlights:
We summarize smooth muscle-expressing Cre lines and their advantages and caveats.
Acknowledgments
Sources of Funding: This work was supported by grants from the NIH to D.M.G. (R01HL125815, R01HL133016, R01HL142674, R21AG062202), and to K.A.M. (HL091013, HL118430, RHL119529, and R01HL142090), and to D.M.G. from the Department of Defense (W81XWH-17-PRMRP-IIRA) and March of Dimes (Gene Discovery and Translational Research Grant, #6-FY15–223). Trainees were supported by fellowships from Yale University to F.Z.S (Brown-Coxe Fellowship), and from the American Heart Association to R.C. (Postdoctoral Fellowship).
Nonstandard Abbreviations and Acronyms
- ACTA2
Smooth muscle alpha actin
- BAC
bacterial artificial chromosome
- ER
estrogen receptor
- ERT2
tamoxifen-inducible mutant estrogen receptor
- GFP
green fluorescent protein
- lacZ
encodes β-galactosidase
- MYH11
Smooth muscle myosin heavy chain
- PVAT
perivascular adipose tissue
- RFP
red fluorescent protein
- SMCs
Smooth muscle cells
- SRF
serum response factor
- TAGLN
Transgelin (also known as SM22α)
- YFP
yellow fluorescent protein
Footnotes
Disclosures: Nothing to disclose
References
- 1.Liu R, Leslie KL and Martin KA. Epigenetic regulation of smooth muscle cell plasticity. Biochimica et biophysica acta 2015;1849:448–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett MR, Sinha S and Owens GK. Vascular Smooth Muscle Cells in Atherosclerosis. Circ Res 2016;118:692–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee RM, Owens GK, Scott-Burden T, Head RJ, Mulvany MJ and Schiffrin EL. Pathophysiology of smooth muscle in hypertension. Canadian journal of physiology and pharmacology 1995;73:574–84. [DOI] [PubMed] [Google Scholar]
- 4.Alexander MR and Owens GK. Epigenetic control of smooth muscle cell differentiation and phenotypic switching in vascular development and disease. Annu Rev Physiol 2012;74:13–40. [DOI] [PubMed] [Google Scholar]
- 5.Sauer B and Henderson N. The cyclization of linear DNA in Escherichia coli by site-specific recombination. Gene 1988;70:331–41. [DOI] [PubMed] [Google Scholar]
- 6.Sauer B and Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proceedings of the National Academy of Sciences of the United States of America 1988;85:5166–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song AJ and Palmiter RD. Detecting and Avoiding Problems When Using the Cre-Iox System. Trends in Genetics 2018;34:333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lakso M, Sauer B, Mosinger B Jr., Lee EJ, Manning RW, Yu SH, Mulder KL and Westphal H. Targeted oncogene activation by site-specific recombination in transgenic mice. Proceedings of the National Academy of Sciences of the United States of America 1992;89:6232–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DE VAL SJ. NEAL A PS. Endothelial-Specific Cre Mouse Models: Is Your Cre CREdibile? Arterioscler Thromb Vasc Biol 2018. [DOI] [PMC free article] [PubMed]
- 10.Feil R, Wagner J, Metzger D and Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochemical and biophysical research communications 1997;237:752–757. [DOI] [PubMed] [Google Scholar]
- 11.Feil R, Brocard J, Mascrez B, LeMeur M, Metzger D and Chambon P. Ligand-activated site-specific recombination in mice. Proc Natl Acad Sci U S A 1996;93:10887–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, Willet SG, Bankaitis ED, Xu YW, Wright CVE and Gu GQ. Non-parallel recombination limits cre-loxP-based reporters as precise indicators of conditional genetic manipulation. Genesis 2013;51:436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herring BP, Hoggatt AM, Burlak C and Offermanns S. Previously differentiated medial vascular smooth muscle cells contribute to neointima formation following vascular injury. Vascular cell 2014;6:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miano JM, Cserjesi P, Ligon KL, Periasamy M and Olson EN. Smooth muscle myosin heavy chain exclusively marks the smooth muscle lineage during mouse embryogenesis. Circulation research 1994;75:803–12. [DOI] [PubMed] [Google Scholar]
- 15.Madsen CS, Regan CP, Hungerford JE, White SL, Manabe I and Owens GK. Smooth muscle-specific expression of the smooth muscle myosin heavy chain gene in transgenic mice requires 5’-flanking and first intronic DNA sequence. Circulation research 1998;82:908–17. [DOI] [PubMed] [Google Scholar]
- 16.Regan CP, Manabe I and Owens GK. Development of a smooth muscle-targeted cre recombinase mouse reveals novel insights regarding smooth muscle myosin heavy chain promoter regulation. Circ Res 2000;87:363–9. [DOI] [PubMed] [Google Scholar]
- 17.Xin HB, Deng KY, Rishniw M, Ji G and Kotlikoff MI. Smooth muscle expression of Cre recombinase and eGFP in transgenic mice. Physiol Genomics 2002;10:211–5. [DOI] [PubMed] [Google Scholar]
- 18.Frutkin AD, Shi H, Otsuka G and Dichek DA. Targeted rearrangement of floxed alleles in smooth muscle cells in vivo. Circ Res 2007;101:e124–5. [DOI] [PubMed] [Google Scholar]
- 19.Frutkin AD, Shi H, Otsuka G, Leveen P, Karlsson S and Dichek DA. A critical developmental role for tgfbr2 in myogenic cell lineages is revealed in mice expressing SM22-Cre, not SMMHC-Cre. J Mol Cell Cardiol 2006;41:724–31. [DOI] [PubMed] [Google Scholar]
- 20.Wirth A, Benyo Z, Lukasova M, Leutgeb B, Wettschureck N, Gorbey S, Orsy P, Horvath B, Maser-Gluth C, Greiner E, Lemmer B, Schutz G, Gutkind JS and Offermanns S. G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat Med 2008;14:64–8. [DOI] [PubMed] [Google Scholar]
- 21.Soriano P Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 1999;21:70–1. [DOI] [PubMed] [Google Scholar]
- 22.Chappell J, Harman JL, Narasimhan VM, Yu HX, Foote K, Simons BD, Bennett MR and Jorgensen HF. Extensive Proliferation of a Subset of Differentiated, yet Plastic, Medial Vascular Smooth Muscle Cells Contributes to Neointimal Formation in Mouse Injury and Atherosclerosis Models. Circulation research 2016;119:1313–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nemenoff RA, Horita H, Ostriker AC, Furgeson SB, Simpson PA, VanPutten V, Crossno J, Offermanns S and Weiser-Evans MC. SDF-1alpha induction in mature smooth muscle cells by inactivation of PTEN is a critical mediator of exacerbated injury-induced neointima formation. Arteriosclerosis, thrombosis, and vascular biology 2011;31:1300–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shankman LS, Gomez D, Cherepanova OA, Salmon M, Alencar GF, Haskins RM, Swiatlowska P, Newman AA, Greene ES, Straub AC, Isakson B, Randolph GJ and Owens GK. KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nat Med 2015;21:628–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Misra A, Feng ZH, Chandran RR, Kabir I, Rotllan N, Aryal B, Sheikh AQ, Ding L, Qin LF, Fernandez-Hernando C, Tellides G and Greif DM. Integrin beta3 regulates clonality and fate of smooth muscle-derived atherosclerotic plaque cells. Nature Communications 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murgai M, Ju W, Eason M, Kline J, Beury DW, Kaczanowska S, Miettinen MM, Kruhlak M, Lei H, Shern JF, Cherepanova OA, Owens GK and Kaplan RN. KLF4-dependent perivascular cell plasticity mediates pre-metastatic niche formation and metastasis. Nat Med 2017;23:1176–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheikh AQ, Misra A, Rosas IO, Adams RH and Greif DM. Smooth muscle cell progenitors are primed to muscularize in pulmonary hypertension. Sci Transl Med 2015;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheikh AQ, Lighthouse JK and Greif DM. Recapitulation of developing artery muscularization in pulmonary hypertension. Cell Rep 2014;6:809–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao M, Yang P, Wang F, Berceli SA, Ali YH, Chan KL and Jiang Z. Smooth muscle cell-specific Tgfbr1 deficiency attenuates neointimal hyperplasia but promotes an undesired vascular phenotype for injured arteries. Physiol Rep 2016;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang P, Schmit BM, Fu C, DeSart K, Oh SP, Berceli SA and Jiang Z. Smooth muscle cell-specific Tgfbr1 deficiency promotes aortic aneurysm formation by stimulating multiple signaling events. Sci Rep 2016;6:35444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao M, Zhou J, Wang F, Ali YH, Chan KL, Zou F, Offermanns S, Jiang Z and Jiang Z. An X-linked Myh11-CreER(T2) mouse line resulting from Y to X chromosome-translocation of the Cre allele. Genesis 2017;55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L, Miano JM, Mercer B and Olson EN. Expression of the SM22alpha promoter in transgenic mice provides evidence for distinct transcriptional regulatory programs in vascular and visceral smooth muscle cells. The Journal of cell biology 1996;132:849–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuhbandner S, Brummer S, Metzger D, Chambon P, Hofmann F and Feil R. Temporally controlled somatic mutagenesis in smooth muscle. Genesis 2000;28:15–22. [DOI] [PubMed] [Google Scholar]
- 34.Miano JM, Ramanan N, Georger MA, Bentley KLD, Emerson RL, Balza RO, Qi X, Weiler H, Ginty DD and Misra RP. Restricted inactivation of serum response factor to the cardiovascular system. Proceedings of the National Academy of Sciences of the United States of America 2004;101:17132–17137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hara-Kaonga B, Gao YA, Havrda M, Harrington A, Bergquist I and Liaw L. Variable recombination efficiency in responder transgenes activated by cre recombinase in the vasculature. Transgenic Res 2006;15:101–106. [DOI] [PubMed] [Google Scholar]
- 36.Holtwick R, Gotthardt M, Skryabin B, Steinmetz M, Potthast R, Zetsche B, Hammer RE, Herz J and Kuhn M. Smooth muscle-selective deletion of guanylyl cyclase-A prevents the acute but not chronic effects of ANP on blood pressure. Proceedings of the National Academy of Sciences of the United States of America 2002;99:7142–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lepore JJ, Cheng L, Min Lu M, Mericko PA, Morrisey EE and Parmacek MS. High-efficiency somatic mutagenesis in smooth muscle cells and cardiac myocytes in SM22alpha-Cre transgenic mice. Genesis 2005;41:179–84. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, Zhong W, Cui T, Yang M, Hu X, Xu K, Xie C, Xue C, Gibbons GH, Liu C, Li L and Chen YE. Generation of an adult smooth muscle cell-targeted Cre recombinase mouse model. Arteriosclerosis, thrombosis, and vascular biology 2006;26:e23–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ehler E, Babiychuk E and Draeger A. Human foetal lung (IMR-90) cells: Myofibroblasts with smooth muscle-like contractile properties. Cell Motil Cytoskel 1996;34:288–298. [DOI] [PubMed] [Google Scholar]
- 40.Roelofs M, Faggian L, Pampinella F, Paulon T, Franch R, Chiavegato A and Sartore S. Transforming growth factor beta 1 involvement in the conversion of fibroblasts to smooth muscle cells in the rabbit bladder serosa. Histochem J 1998;30:393–404. [DOI] [PubMed] [Google Scholar]
- 41.Faggin E, Puato M, Zardo L, Franch R, Millino C, Sarinella F, Pauletto P, Sartore S and Chiavegato A. Smooth muscle-specific SM22 protein is expressed in the adventitial cells of balloon-injured rabbit carotid artery. Arterioscl Throm Vas 1999;19:1393–1404. [DOI] [PubMed] [Google Scholar]
- 42.Shen ZX, Li C, Frieler RA, Gerasimova AS, Lee SJ, Wu J, Wang MM, Lumeng CN, Brosius FC, Duan SZ and Mortensen RM. Smooth muscle protein 22 alpha-Cre is expressed in myeloid cells in mice. Biochemical and biophysical research communications 2012;422:639–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith EC, Teixeira AM, Chen RC, Wang L, Gao Y, Hahn KL and Krause DS. Induction of megakaryocyte differentiation drives nuclear accumulation and transcriptional function of MKL1 via actin polymerization and RhoA activation. Blood 2013;121:1094–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muzumdar MD, Tasic B, Miyamichi K, Li L and Luo L. A global double-fluorescent Cre reporter mouse. Genesis 2007;45:593–605. [DOI] [PubMed] [Google Scholar]
- 45.Varberg KM, Garretson RO, Blue EK, Chu CH, Gohn CR, Tu WZ and Haneline LS. Transgelin induces dysfunction of fetal endothelial colony-forming cells from gestational diabetic pregnancies. Am J Physiol-Cell Ph 2018;315:C502–C515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang L, Villacorta L, Li R, Hamblin M, Xu W, Dou C, Zhang J, Wu J, Zeng R and Chen YE. Loss of perivascular adipose tissue on peroxisome proliferator-activated receptor-gamma deletion in smooth muscle cells impairs intravascular thermoregulation and enhances atherosclerosis. Circulation 2012;126:1067–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sui YP, Park SH, Xu JX, Monette S, Helsley RN, Han SS and Zhou CC. IKK beta links vascular inflammation to obesity and atherosclerosis. J Exp Med 2014;211:869–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang W, Zeve D, Suh JM, Bosnakovski D, Kyba M, Hammer RE, Tallquist MD and Graff JM. White fat progenitor cells reside in the adipose vasculature. Science 2008;322:583–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miwa T, Koyama T and Shirai M. Muscle specific expression of Cre recombinase under two actin promoters in transgenic mice. Genesis 2000;26:136–8. [PubMed] [Google Scholar]
- 50.Wu Z, Yang L, Cai L, Zhang M, Cheng X, Yang X and Xu J. Detection of epithelial to mesenchymal transition in airways of a bleomycin induced pulmonary fibrosis model derived from an alpha-smooth muscle actin-Cre transgenic mouse. Respir Res 2007;8:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wendling O, Bornert JM, Chambon P and Metzger D. Efficient temporally-controlled targeted mutagenesis in smooth muscle cells of the adult mouse. Genesis 2009;47:14–8. [DOI] [PubMed] [Google Scholar]
- 52.Hill RA, Tong L, Yuan P, Murikinati S, Gupta S and Grutzendler J. Regional Blood Flow in the Normal and Ischemic Brain Is Controlled by Arteriolar Smooth Muscle Cell Contractility and Not by Capillary Pericytes. Neuron 2015;87:95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lynch TJ, Anderson PJ, Rotti PG, Tyler SR, Crooke AK, Choi SH, Montoro DT, Silverman CL, Shahin W, Zhao R, Jensen-Cody CW, Adamcakova-Dodd A, Evans TIA, Xie WL, Zhang YL, Mou HM, Herring BP, Thorne PS, Rajagopal J, Yeaman C, Parekh KR and Engelhardt JF. Submucosal Gland Myoepithelial Cells Are Reserve Stem Cells That Can Regenerate Mouse Tracheal Epithelium. Cell stem cell 2018;22:653–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.LeBleu VS, Teng Y, O’Connell JT, Charytan D, Muller GA, Muller CA, Sugimoto H and Kalluri R. Identification of human epididymis protein-4 as a fibroblast-derived mediator of fibrosis. Nat Med 2013;19:227–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.LeBleu VS, Taduri G, O’Connell J, Teng Y, Cooke VG, Woda C, Sugimoto H and Kalluri R. Origin and function of myofibroblasts in kidney fibrosis. Nat Med 2013;19:1047–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grcevic D, Pejda S, Matthews BG, Repic D, Wang L, Li H, Kronenberg MS, Jiang X, Maye P, Adams DJ, Rowe DW, Aguila HL and Kalajzic I. In vivo fate mapping identifies mesenchymal progenitor cells. Stem Cells 2012;30:187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Manieri NA, Mack MR, Himmelrich MD, Worthley DL, Hanson EM, Eckmann L, Wang TC and Stappenbeck TS. Mucosally transplanted mesenchymal stem cells stimulate intestinal healing by promoting angiogenesis. The Journal of clinical investigation 2015;125:3606–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vanlandewijck M, He L, Mae MA, Andrae J, Ando K, Del Gaudio F, Nahar K, Lebouvier T, Lavina B, Gouveia L, Sun Y, Raschperger E, Rasanen M, Zarb Y, Mochizuki N, Keller A, Lendahl U and Betsholtz C. A molecular atlas of cell types and zonation in the brain vasculature. Nature 2018;554:475–480. [DOI] [PubMed] [Google Scholar]
- 59.Tian Z, Miyata K, Tazume H, Sakaguchi H, Kadomatsu T, Horio E, Takahashi O, Komohara Y, Araki K, Hirata Y, Tabata M, Takanashi S, Takeya M, Hao H, Shimabukuro M, Sata M, Kawasuji M and Oike Y. Perivascular adipose tissue-secreted angiopoietin-like protein 2 (Angptl2) accelerates neointimal hyperplasia after endovascular injury. Journal of molecular and cellular cardiology 2013;57:1–12. [DOI] [PubMed] [Google Scholar]
- 60.Mericskay M, Blanc J, Tritsch E, Moriez R, Aubert P, Neunlist M, Feil R and Li Z. Inducible mouse model of chronic intestinal pseudo-obstruction by smooth muscle-specific inactivation of the SRF gene. Gastroenterology 2007;133:1960–70. [DOI] [PubMed] [Google Scholar]
- 61.Angstenberger M, Wegener JW, Pichler BJ, Judenhofer MS, Feil S, Alberti S, Feil R and Nordheim A. Severe intestinal obstruction on induced smooth muscle-specific ablation of the transcription factor SRF in adult mice. Gastroenterology 2007;133:1948–59. [DOI] [PubMed] [Google Scholar]
- 62.Park C, Lee MY, Slivano OJ, Park PJ, Ha S, Berent RM, Fuchs R, Collins NC, Yu TJ, Syn H, Park JK, Horiguchi K, Miano JM, Sanders KM and Ro S. Loss of serum response factor induces microRNA-mediated apoptosis in intestinal smooth muscle cells. Cell Death Dis 2015;6:e2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Albinsson S, Skoura A, Yu J, DiLorenzo A, Fernandez-Hernando C, Offermanns S, Miano JM and Sessa WC. Smooth muscle miRNAs are critical for post-natal regulation of blood pressure and vascular function. PLoS One 2011;6:e18869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoggatt AM, Simon GM and Herring BP. Cell-specific regulatory modules control expression of genes in vascular and visceral smooth muscle tissues. Circulation research 2002;91:1151–9. [DOI] [PubMed] [Google Scholar]
- 65.Ding M, Carrao AC, Wagner RJ, Xie Y, Jin Y, Rzucidlo EM, Yu J, Li W, Tellides G, Hwa J, Aprahamian TR and Martin KA. Vascular smooth muscle cell-derived adiponectin: a paracrine regulator of contractile phenotype. Journal of molecular and cellular cardiology 2012;52:474–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


