ABSTRACT
Genomic interactions can occur in addition to those within chromosome territories and can be organized around nuclear bodies. Several studies revealed how the nucleolus anchors higher order chromatin structures of specific chromosome regions displaying heterochromatic features. In this review, we comment on advances in this emerging field, with a particular focus on a recent study published by Quinodoz et al., that developed a new method to characterize simultaneous genomic interactions in the same cell. Highlighting studies conducted in animal and plant cells, we then discuss the establishment of inactive chromatin at nucleolus organizer region (NOR)-bearing chromosomes.
KEYWORDS: Chromatin, genome organization, nucleolus, rRNA genes
Introduction
In 1928, the cytogeneticist Heitz described two major chromatin states that differ according to their condensation level in the nucleus [1,2]. He named the condensed chromatin regions heterochromatin, while more loosely packed chromatin regions were named euchromatin. The identification of genome-wide chromatin–chromatin interactions maps confirmed this bipartite organization. Today, euchromatin and heterochromatin are generally referred to as A/B compartments [3]. This type of organization is relevant to both plant and animal cells [4]. However, this bipartite organization can be subdivided into several states according to their epigenetic signatures such as, but is not restrictive to, DNA methylation, histone modifications, histone variant distribution or nucleosome positioning [5–7]. Notably, these different states strongly participate in gene transcriptional regulation by modifying the access, or the attractiveness, of any loci to the transcriptional machineries.
At the nucleus scale, cytological analyses already revealed the existence of chromosome territories that can be distributed randomly, as in Arabidopsis thaliana or non-randomly, as in human cells, within interphase nuclei [8–11]. In addition, recent technical advances deeply increased our knowledge of how the genome is organized within the nucleus. For example, the development of the chromosome conformation capture (3C) approach to a genome-wide scale revealed in various organisms and cell-types how inter and intra-chromosomal DNA contacts shape this 3D organization [12]. These studies revealed the existence of topologically associating domains or (TADs) that are self-interacting genomic regions, separated by boundaries, and confined either in A or B compartments, depending on their chromatin signatures. In plants, although the presence of TADs is not obvious in Arabidopsis thaliana, they were described in other plant species with larger genomes [4,13].
Recent studies pushed forward our understanding of the 3D genome organization, by taking into consideration the subnuclear compartments such as the nucleolus and the nuclear speckles. In this review, we aim to combine data of recent studies that demonstrate an important role of ribosomal RNA genes distribution and expression in global genome organization.
The nucleolus, a platform for genome organization
Ribosomal RNA (rRNA) genes are organized as tandem repeats in specific genomic regions named nucleolus organizer regions (NORs) by Barbara McClintock [14]. The nucleolus is the site of ribosome biogenesis, starting with rRNA transcription, and is present in most eukaryote cells. Its size is dependent on the ribosome biogenesis activity, but can represent a large volume of the cell, especially in actively dividing cells [15]. In human cells, several nucleoli are usually present, while in A. thaliana, usually one round nucleolus is present. The nucleolus is a direct consequence of ribosome biogenesis. However, this large subnuclear compartment is also implicated in other cellular processes, especially in the maturation or sequestration of several ribonucleoproteins complexes such as the spliceosome, the proteasome, the RNA telomerase and others implicated in RNA degradation [15–20].
In 2010, two studies conducted in human cells demonstrated the direct role of the nucleolus in genome organization [21,22]. DNA co-purified with nucleoli from two types of human cells were analyzed, demonstrating that all chromosomes contain, at least, one region associating with the nucleolus. These genomic regions were named NADs for nucleolus-associated chromatin domains. In parallel, a network composed by nuclear lamina also anchors genomic regions with repressive epigenetic features at the nuclear periphery [23]. These regions are named LADs for lamina-associated domains. NADs were subsequently identified in A. thaliana cells [24]. In both plant and animal cells, excluding rRNA genes, NADs are composed of around 4% of the genome and display repressive chromatin features [24]. Interestingly, genes are also present in NADs and tend to be lowly expressed. Majority of NADs are presumed to be located at the nucleolus periphery, but cytological analyses demonstrated that the DNA inside the nucleolus is not only composed of rRNA genes [24]. Since RNA polymerase II is absent from inside the nucleolus, it has been proposed that the nucleolus would have a negative impact on gene expression [25,26].
Among the important aspects pointed out by Nemeth et al. [22], is the potential redundancy between NADs and LADs. Additional studies confirmed this observation and demonstrated that a portion of LADs, identified as facultative LADs, can reshuffle stochastically either at the nuclear or the nucleolar periphery after mitosis [27,28]. Conversely, LADs remaining at the nuclear periphery in the daughter cells are now identified as constitutive LADs [29]. In plant cells, no homolog of nuclear lamina proteins have been identified so far, but putative homologs like nuclear matrix constituent protein (NMCP), nuclear envelope-associated proteins (NEAPs) or CROWDED NUCLEI (CRWN) proteins, all localized at the nuclear periphery, are suspected to play a similar role [30–33]. Chromatin domains associating with proteins located at the nuclear periphery have not been identified so far, but Chang Liu’s laboratory was able to identify the genomic regions associated with the nuclear periphery using a derived chromatin immunoprecipitation (ChIP) protocol. The nucleoporin NUP1, which is one of the subunits of the nuclear pore complex, was used as a bait [34]. As for NADs, chromosome regions identified in this study (hereafter referred to as NUP1-enriched domains) are enriched in repressive chromatin signatures, transposable elements and contain genes that tend to be lowly expressed [34]. Since at least a portion of NADs overlap with LADs in human cells, we analyzed the potential connection between NADs and NUP1-enriched domains in A. thaliana (Figure 1). At the chromosome scale, NADs correspond to genomic regions flanking telomeres, as well as the short arm of chromosome 4, where active rRNA genes are located [35,36]. In contrast, NUP1-enriched domains mainly correspond to peri-centromeric regions, and large portions of chromosome arms (Figure 1(a)) [24,34]. The partial overlap observed on the short arm of chromosome 4 could potentially link the nucleolus and the nuclear periphery. Interestingly, the heterochromatic knob, corresponding to the conspicuous gap located between the 3,5 and 4,5 MB region, only associate with the nucleolus.
Figure 1.
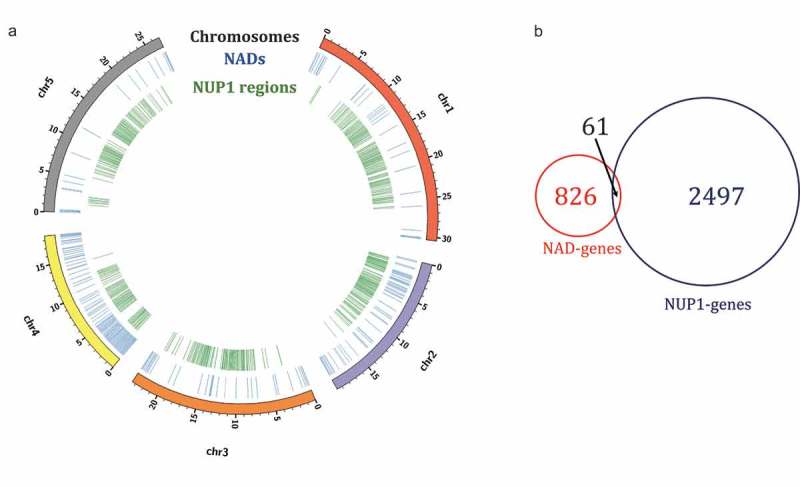
Overlap between NADs and NUP1-regions in Arabidopsis thaliana. (a) Genome-wide view of NAD-genes and the regions enriched at the nuclear periphery (NUP1-enriched regions) in the A. thaliana genome produced by Circos [49]. Considering the concentric circles from the outside inward, the outermost (and first) circle indicates the five chromosomes of A. thaliana. The next two circles are showing the location of NADs-genes (blue) and NUP-enriched regions (green) throughout the genome. (b) Venn diagram of NAD-genes and NUP1-enriched regions corresponding to protein-coding genes in TAIR10. Data from [24,34] were used to generate this figure.
Therefore, the overlap between NADs and NUP1-enriched domains appears minor, and genes usually either associate with the nuclear periphery or with the nucleolar periphery (Figure 1(b)).
Influence of RNA genes in nucleolus-associated chromatin domains and inactive hubs
The Hi-C approach allows the detection of direct or indirect chromatin interactions in a cell population [12]. However, Hi-C is not compatible to detect simultaneous chromatin interactions and cell-to-cell variability. Single-cell Hi-C has recently been developed to counteract this problem, but sequencing depth remains an issue, leading to very low genome coverage [37]. In a recent study, Quinodoz and colleagues enable the detection of simultaneous interactions via the development of a new technique named SPRITE for split-pool recognition of interactions by tag extension [1]. SPRITE allows the detection of multiple chromatin interactions mediated by an individual complex within a nucleus, revealing the context of the interactions. Briefly, unique complex, from which all RNA, DNA and protein components are covalently crosslinked, are sorted in individual wells. Therefore, all DNA reads from one individual complex are identified as a SPRITE cluster [1]. In other words, a chromatin loop can be associated with its TAD and its compartment. Using the SPRITE technique, Quinodoz et al., linked some A/B compartments with nuclear bodies. They identified active hubs, corresponding to A compartments, in association with nuclear speckles. In parallel, inactive hubs, corresponding to B compartments, were shown to associate with the nucleolus [1]. This corroborates previous observations made in both animal and plant cells [25,38], regarding the important role of the nucleolus in organizing inactive chromatin regions at the nucleolar periphery.
Importantly, inactive hubs are present on rRNA-genes bearing chromosomes [1]. In other words, inactive hubs correspond to genomic regions flanking rRNA genes. This raises the question of the role of rRNA gene expression and location on chromosomes in defining the identity of inactive hubs at the nucleolar periphery. In A. thaliana, NADs are mainly composed of genomic regions corresponding to the NOR-bearing chromosomes 2 and 4, as well as genomic regions from the subtelomeric regions [24]. In wild-type leaves, only NOR4-derived rRNA genes associate with the nucleolus and are actively transcribed, while NOR2-derived rRNA genes are silent and excluded from the nucleolus [35,36,39–42]. Interestingly, NOR2-derived rRNA genes expression provokes a complete reorganization of the short arm of chromosome 2 that associates with the nucleolus [24]. Collectively, these data suggest that rRNA genes expression or silencing impact the global heterochromatin nuclear distribution. To test the potential presence of inactive hubs in A. thaliana, we analyzed the global gene expression of nucleolus-associated genomic regions. As in mammalian cells, the short arms of NOR-bearing chromosomes tend to contain low-expressed genes compared to the entire genome (Figure 2). However, we do not see such correlation for genes of the subtelomeric regions that also associate with the nucleolus in a wild-type (Col-0) A. thaliana line [43].
Figure 2.
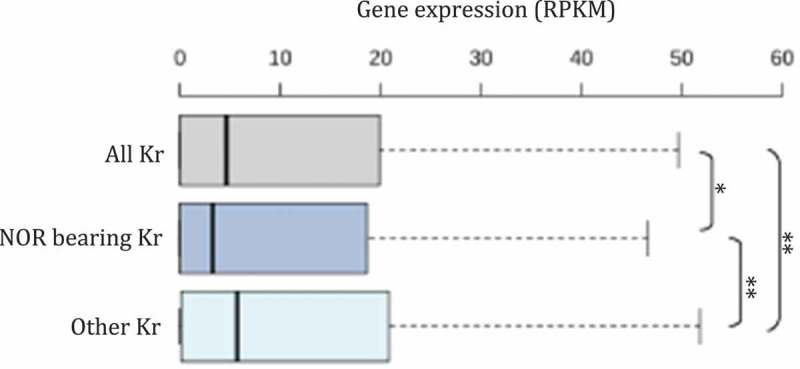
Gene expression of nucleolus-associated genomic regions in A. thaliana. Box plot representing the global expression of genes located in the short arm of NOR-bearing chromosomes 2 and 4 (NOR-bearing Kr) compare to other chromosomes 1, 3 and 5 (other Kr 1, 3 & 5) or all five chromosomes (all Kr) in A. thaliana. A p value < 0.05 *; p value < 0.01: ** (Wilcoxon rank sum test).
Conclusions and perspectives
Overall, recent studies revealed the importance of nuclear bodies in the distribution of the epigenome. In these cases, nuclear body specific proteins play crucial roles in these regulations. For example, the nucleolar protein Nucleolin was demonstrated to regulate (i) telomere nucleolar clustering in A. thaliana [24], (ii) the two-cell embryo master regulator gene DUX in human embryonic stem cells [44] or (iii) centromere nucleolar clustering in drosophila [45]. A missing aspect is an overlap between inactive hubs identified using the SPRITE method and NADs identified in human cells [1,19,20]. We indeed expect NADs and inactive hubs to overlap. To further demonstrate how rRNA genes distribution in the genome defines NADs or inactive hubs, it will be important to identify these regions in cells with the variable NOR chromosomal location, or in mutant showing reduction or increase numbers of rRNA genes. Previous observations in yeast and in drosophila demonstrated that reducing the number of rRNA genes alter heterochromatin organization and genome stability [46–48]. In the light of these new data, we expect to observe changes in global genome organization in cells with a reduced number of rRNA genes.
Funding Statement
This work and APC PhD fellowship are supported by the ANR NucleoReg [ANR-15-CE12-0013-01] and the COST ACTION CA16212 INDEPTH.
Acknowledgments
The authors thank Gwénaëlle Détourné, Guillaume Moissiard and anonymous reviewers for critical reading of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Quinodoz SA, Ollikainen N, Tabak B, et al. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell. 2018;174:744–757.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Passarge E. Emil Heitz and the concept of heterochromatin: longitudinal chromosome differentiation was recognized fifty years ago. Am J Hum Genet. 1979;31:106–115. [PMC free article] [PubMed] [Google Scholar]
- [3].Lieberman-Aiden E, van Berkum NL, Williams L, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Dogan ES, Liu C. Three-dimensional chromatin packing and positioning of plant genomes. Nat Plants. 2018;4:521–529. [DOI] [PubMed] [Google Scholar]
- [5].Filion GJ, van Bemmel JG, Braunschweig U, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Roudier F, Ahmed I, Berard C, et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. Embo J. 2011;30:1928–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sequeira-Mendes J, Araguez I, Peiro R, et al. The functional topography of the arabidopsis genome is organized in a reduced number of linear motifs of chromatin states. Plant Cell. 2014;26:2351–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Heard E, Bickmore W. The ins and outs of gene regulation and chromosome territory organisation. Curr Opin Cell Biol. 2007;19:311–316. [DOI] [PubMed] [Google Scholar]
- [9].Pecinka A, Schubert V, Meister A, et al. Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma. 2004;113:258–269. [DOI] [PubMed] [Google Scholar]
- [10].Schubert V, Berr A, Meister A. Interphase chromatin organisation in Arabidopsis nuclei: constraints versus randomness. Chromosoma. 2012;121:369–387. [DOI] [PubMed] [Google Scholar]
- [11].Fransz P, De Jong JH, Lysak M, et al. Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc Natl Acad Sci U S A. 2002;99:14584–14589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet. 2013;14:390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dong P, Tu X, Chu P-Y, et al. 3D chromatin architecture of large plant genomes determined by local A/B compartments. Mol Plant. 2017;10:1497–1509. [DOI] [PubMed] [Google Scholar]
- [14].McClintock B. The relationship of a particular chromosomal element to the development of the nucleoli in Zea mays. Zeit Zellforsch Mik Anat. 1934;21:294–328. [Google Scholar]
- [15].Pederson T. The nucleolus. Cold Spring Harb Perspect Biol. 2011;3:a000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Boulon S, Westman BJ, Hutten S, et al. The nucleolus under stress. Mol Cell. 2010;40:216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lange H, Zuber H, Sement FM, et al. The RNA helicases AtMTR4 and HEN2 target specific subsets of nuclear transcripts for degradation by the nuclear exosome in Arabidopsis thaliana. PLoS Genet. 2014;10:e1004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pendle AF, Clark GP, Boon R, et al. Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol Biol Cell. 2005;16:260–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Montacie C, Durut N, Opsomer A, et al. Nucleolar proteome analysis and proteasomal activity assays reveal a link between nucleolus and 26S proteasome in A. thaliana. Front Plant Sci. 2017;8:1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tomlinson RL, Ziegler TD, Supakorndej T, et al. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol Biol Cell. 2006;17:955–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].van Koningsbruggen S, Gierlinski M, Schofield P, et al. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol Biol Cell. 2010;21:3735–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nemeth A, Conesa A, Santoyo-Lopez J, et al. Initial genomics of the human nucleolus. PLoS Genet. 2010;6:e1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Pombo A, Dillon N. Three-dimensional genome architecture: players and mechanisms. Nat Rev Mol Cell Biol. 2015;16:245–257. [DOI] [PubMed] [Google Scholar]
- [24].Pontvianne F, Carpentier M-C, Durut N, et al. Identification of nucleolus-associated chromatin domains reveals a role for the nucleolus in 3D organization of the A. thaliana genome. Cell Rep. 2016;16:1574–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Picart C, Pontvianne F. Plant nucleolar DNA: green light shed on the role of Nucleolin in genome organization. Nucl Austin Tex. 2017;8:11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Schubert V, Weisshart K. Abundance and distribution of RNA polymerase II in Arabidopsis interphase nuclei. J Exp Bot. 2015;66:1687–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kind J, Pagie L, de Vries SS, et al. Genome-wide maps of nuclear lamina interactions in single human cells. Cell. 2015;163:134–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kind J, Pagie L, Ortabozkoyun H, et al. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153:178–192. [DOI] [PubMed] [Google Scholar]
- [29].van Steensel B, Belmont AS. Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell. 2017;169:780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Meier I, Richards EJ, Evans DE. Cell biology of the plant nucleus. Annu Rev Plant Biol. 2017;68:139–172. [DOI] [PubMed] [Google Scholar]
- [31].Pawar V, Poulet A, Detourne G, et al. A novel family of plant nuclear envelope-associated proteins. J Exp Bot. 2016;67:5699–5710. [DOI] [PubMed] [Google Scholar]
- [32].Ciska M, Masuda K. Moreno Diaz de la Espina S. Lamin-like analogues in plants: the characterization of NMCP1 in Allium cepa. J Exp Bot. 2013;64:1553–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Dittmer TA, Stacey NJ, Sugimoto-Shirasu K, et al. LITTLE NUCLEI genes affecting nuclear morphology in Arabidopsis thaliana. Plant Cell. 2007;19:2793–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bi X, Cheng Y-J, Hu B, et al. Nonrandom domain organization of the Arabidopsis genome at the nuclear periphery. Genome Res. 2017;27:1162–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pontvianne F, Blevins T, Chandrasekhara C, et al. Subnuclear partitioning of rRNA genes between the nucleolus and nucleoplasm reflects alternative epiallelic states. Genes Dev. 2013;27:1545–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chandrasekhara C, Mohannath G, Blevins T, et al. Chromosome-specific NOR inactivation explains selective rRNA gene silencing and dosage control in Arabidopsis. Genes Dev. 2016;30:177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Nagano T, Lubling Y, Yaffe E, et al. Single-cell Hi-C for genome-wide detection of chromatin interactions that occur simultaneously in a single cell. Nat Protoc. 2015;10:1986–2003. [DOI] [PubMed] [Google Scholar]
- [38].Nemeth A, Langst G. Genome organization in and around the nucleolus. Trends Genet. 2011;27:149–156. [DOI] [PubMed] [Google Scholar]
- [39].Pontvianne F, Blevins T, Chandrasekhara C, et al. Histone methyltransferases regulating rRNA gene dose and dosage control in Arabidopsis. Genes Dev. 2012;26:945–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Pontvianne F, Abou-Ellail M, Douet J, et al. Nucleolin is required for DNA methylation state and the expression of rRNA gene variants in Arabidopsis thaliana. PLoS Genet. 2010;6:e1001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Mohannath G, Pontvianne F, Pikaard CS. Selective nucleolus organizer inactivation in Arabidopsis is a chromosome position-effect phenomenon. Proc Natl Acad Sci U S A. 2016;113:13426–13431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Earley KW, Pontvianne F, Wierzbicki AT, et al. Mechanisms of HDA6-mediated rRNA gene silencing: suppression of intergenic Pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes Dev. 2010;24:1119–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. [DOI] [PubMed] [Google Scholar]
- [44].Percharde M, Lin C-J, Yin Y, et al. A LINE1-Nucleolin partnership regulates early development and ESC identity. Cell. 2018;174:391–405.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Padeken J, Mendiburo MJ, Chlamydas S, et al. The nucleoplasmin homolog NLP mediates centromere clustering and anchoring to the nucleolus. Mol Cell. 2013;50:236–249. [DOI] [PubMed] [Google Scholar]
- [46].Ide S, Miyazaki T, Maki H, et al. Abundance of ribosomal RNA gene copies maintains genome integrity. Science. 2010;327:693–696. [DOI] [PubMed] [Google Scholar]
- [47].Paredes S, Maggert KA. Ribosomal DNA contributes to global chromatin regulation. Proc Natl Acad Sci U S A. 2009;106:17829–17834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Paredes S, Branco AT, Hartl DL, et al. Ribosomal DNA deletions modulate genome-wide gene expression: “rDNA-sensitive” genes and natural variation. PLoS Genet. 2011;7:e1001376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Krzywinski M, Schein J, Birol I, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]


