Abstract
Nitric oxide (NO) stimulates soluble guanylyl cyclase (sGC) activity, leading to elevated intracellular cyclic guanosine 3′,5′-monophosphate (cGMP) and subsequent vascular smooth muscle relaxation. It is known that downregulation of sGC expression attenuates vascular dilation and contributes to the pathogenesis of cardiovascular disease. However, it is not well understood how sGC transcription is regulated. Here, we demonstrate that pharmacological inhibition of Forkhead box subclass O (FoxO) transcription factors using the small-molecule inhibitor AS1842856 significantly blunts sGC α and β mRNA expression by more than 90%. These effects are concentration-dependent and concomitant with greater than 90% reduced expression of the known FoxO transcriptional targets, glucose-6-phosphatase and growth arrest and DNA damage protein 45 α (Gadd45α). Similarly, sGC α and sGC β protein expression showed a concentration-dependent downregulation. Consistent with the loss of sGC α and β mRNA and protein expression, pretreatment of vascular smooth muscle cells with the FoxO inhibitor decreased sGC activity measured by cGMP production following stimulation with an NO donor. To determine if FoxO inhibition resulted in a functional impairment in vascular relaxation, we cultured mouse thoracic aortas with the FoxO inhibitor and conducted ex vivo two-pin myography studies. Results showed that aortas have significantly blunted sodium nitroprusside-induced (NO-dependent) vasorelaxation and a 42% decrease in sGC expression after 48-hour FoxO inhibitor treatment. Taken together, these data are the first to identify that FoxO transcription factor activity is necessary for sGC expression and NO-dependent relaxation.
Introduction
Arterial blood vessel dilation is mediated, in part, through production of the endogenous endothelial vasodilator molecule, nitric oxide (NO). NO signals by binding to ferrous heme iron in soluble guanylyl cyclase (sGC) in vascular smooth muscle cells (VSMC) to catalyze the intracellular conversion of guanosine 5′-triphosphate to the second messenger molecule cyclic guanosine 3′,5′-monophosphate (cGMP) (Arnold et al., 1977). Increased cGMP, in turn, activates protein kinase G, leading to vascular smooth muscle relaxation needed to govern tissue perfusion and blood pressure (Kuo and Greengard, 1970). Indeed, several prior studies conducted in spontaneous hypertensive rats showed sGC mRNA and protein expression to be downregulated and associated with impaired NO-dependent vasodilation (Ruetten et al., 1999; Klöss et al., 2000). Moreover, smooth muscle cell–specific inducible knockout of sGC β in mice results in hypertension, suggesting sGC expression in smooth muscle cells is critical for modulating vascular tone (Groneberg et al., 2010). Genome-wide association studies of single nucleotide polymorphisms also identified genetic variants within the sGC genes GUCY1A3 and GUCY1B3 as being associated with a high risk for cardiovascular disease (Ehret et al., 2011; Wobst et al., 2015; Shendre et al., 2017). Of translational importance, small-molecule sGC modulators have been developed for clinical use to restore disease-associated loss of sGC activity and cGMP production to varying degrees of success (Yoshina et al., 1978; Stasch et al., 2001; Frey et al., 2008; Meurer et al., 2009; Mittendorf et al., 2009). For example, riociguat (BAY 63-2521), a heme-dependent sGC stimulator, has been recently approved for treatment of pulmonary arterial hypertension and chronic thromboembolic pulmonary hypertension (Ghofrani et al., 2013). Additionally, cinaciguat (BAY 58-2667), an NO-independent sGC activator, increases cGMP when sGC heme is oxidized or resides in the heme-deficient state (Stasch et al., 2006), but further studies were halted with BAY 58-2667 after it showed risk for hypotension in a phase IIb clinical trial for treatment of acute heart failure syndrome (Gheorghiade et al., 2012). Importantly, the clinical efficacy of these sGC modulators is dependent on adequate sGC protein expression.
While the NO-sGC-cGMP pathway continues to be extensively studied, there are a limited number of studies that have identified the mechanisms regulating sGC transcription. Prior work has shown interacting partners with sGC transcripts—namely, human antigen R, which stabilizes the sGC mRNA (Klöss et al., 2004, 2005), and miR-34c-5p and ARE/poly(U)-binding/degradation factor 1 (AUF1), which destabilize and promote the degradation of sGC transcripts (Pende et al., 1996; Xu et al., 2012). Furthermore, CCAAT-binding factor deletion significantly blunts production of sGC mRNA expression in a neuroblastoma cell line (Sharina et al., 2003). However, beyond these studies, our knowledge of the transcription factor(s) responsible for the constitutive expression of sGC, particularly within VSMCs, remains elusive.
The Forkhead box subclass O (FoxO) family of transcription factors—namely, FoxO1, FoxO3, and FoxO4—are the predominant isoforms in VSMCs (Salih and Brunet, 2008). Recent studies have identified the importance of this family of transcription factors in the vasculature, including their role in the promotion of angiogenesis (Furuyama et al., 2004; Potente et al., 2005), inhibition of endothelial NO production (Potente et al., 2005; Xia et al., 2013), prevention of pulmonary hypertension (Savai et al., 2014), and maintenance of pluripotency in a variety of different cell types (Liu et al., 2005; Zhang et al., 2011; Wang et al., 2013). Additionally, an important observation in the field has established that aging decreases the expression and capacity for FoxO signaling (Ogg et al., 1997; Hsu et al., 2003; Giannakou et al., 2004; Hwangbo et al., 2004; Salih and Brunet, 2008) and has also been shown to decrease the expression of sGC within the vasculature (Kloss et al., 2000; Goubareva et al., 2007; Stice et al., 2009).
However, a link connecting the sGC-cGMP and FoxO pathways has yet to be described. Therefore, with the use of in silico prediction analysis and a previously characterized FoxO inhibitor compound, AS1842856 [International Union of Pure and Applied Chemistry name: 5-amino-7-(cyclohexylamino)-1-ethyl-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid] (Nagashima et al., 2010), we sought to determine if the FoxO transcription factor family regulates sGC transcription. Herein, we provide evidence that inhibition of FoxO transcription factors results in significant downregulation of sGC transcript and protein, decreased cGMP production, and impaired NO-induced vasodilation, implicating FoxO transcription factors as a major regulator of sGC transcription.
Materials and Methods
Cell Culture and Drug Treatment.
Rat aortic smooth muscle cells (RASMCs; Lonza) were cultured as previously described (Rahaman et al., 2017). In brief, RASMCs were cultured at 37°C in Lonza Smooth Muscle Growth Medium-2 with SmGm-2 SingleQuot kit (Lonza) supplementation. Cells were passaged using Gibco Trypsin/EDTA up to passage 12. For treatment, FoxO inhibitor AS1842856 (Cayman) was dissolved in DMSO at 10 mM stock concentration and diluted in DMSO for further experiments. Control and treatment DMSO concentration was 0.1% of the total volume for treatment periods.
Quantitative Reverse-Transcription Polymerase Chain Reaction.
RASMCs were cultured in six-well dishes until confluent and lysed in TRIzol reagent (Thermo Fisher). RNA was isolated from lysates according to the RNA purification protocol from the Direct-zol RNA miniprep plus kit (Zymo). Isolated RNA was reverse transcribed to cDNA using SuperScript III First Strand Synthesis System (Thermo Fisher). Quantitative polymerase chain reaction (PCR) was performed using PowerUp SyBr Green Master Mix (Thermo Fisher); 1 µM target primer (Supplemental Table 1) was mixed; and 40 PCR cycles with 95°C melting temperature, 58°C annealing temperature, and 72°C extension temperature were performed in a QuantStudio 5 Real-Time PCR System (Thermo Fisher).
Western Blot.
RASMCs were lysed in ice-cold 1× cell lysis buffer (Cell Signaling) containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, and 1 µg/ml leupeptin supplemented with additional protease and phosphatase inhibitors (Sigma-Aldrich). Protein lysate concentrations were quantified using a standard bicinchoninic acid kit (Thermo Fisher). Laemmli buffer was added for a final concentration containing 31.5 mM Tris-HCl (pH 6.8), 10% glycerol, 1% SDS, and 0.005% Bromophenol Blue sodium salt. Lysates were boiled and subjected to electrophoresis on 4%–12% BisTris polyacrylamide gels (Life Technologies). Proteins were transferred to nitrocellulose membrane and blocked for 1 hour at room temperature with 1% bovine serum albumin (BSA) in PBS. Membranes were rocked overnight with primary antibodies (Supplemental Table 2) diluted in 1% BSA in 0.1% Tween-20 in PBS (PBST) at 4°C. Membranes were washed and incubated with secondary antibodies from LI-COR (Supplemental Table 2) diluted in 1% BSA in PBST for 1 hour at room temperature followed by washing with PBST. Visualization and analyses were completed utilizing a LI-COR Odyssey Imager and Image Studio Software.
cGMP ELISA.
Confluent RASMCs were cultured in 12-well dishes and pretreated with 10 µM sildenafil (Sigma-Aldrich) for 45 minutes and then stimulated with the NO donor, diethylammonium (Z)-1-(N,N-diethylamino)diazen-1-ium-1,2-diolate (DEA NONOate; Cayman), for 15 minutes. Baseline measurements were performed after 45-minute treatment with 10 μM sildenafil. Cell samples were lysed in 125 µl of ice-cold 1× cell lysis buffer (Cell Signaling) supplemented with protease and phosphatase inhibitors (Sigma-Aldrich). cGMP production was determined via enzyme-linked immunosorbent assay (ELISA; Cell Signaling). Ten microliters of sample (approximately 5–10 µg of protein) was added to each well and diluted with additional lysis buffer, and exact protein concentration of each sample was quantified using a standard BCA protein assay kit (Thermo Fisher). ELISAs were performed hereafter according to the manufacturer’s protocol.
Immunofluorescence Staining.
Twelve-week-old C57BL/6J mice (Jackson Laboratories) were sacrificed via CO2 asphyxiation. Thoracic aortas were excised and placed in 4% paraformaldehyde solution for 24 hours, followed by 24 hours in 30% sucrose in PBS. Tissues were then frozen in optimum cutting temperature compound fixative (Tissue-Tek) via liquid nitrogen and cryosectioned at 8-µM thickness with three sections/slide using an FSE Cryostat Microtome slicer (Thermo Scientific). Slides were permeabilized with −20°C acetone for 10 minutes, air dried for 10 minutes, washed thrice for 5 minutes each in PBS, and then blocked with PBS + 0.25% Triton X-100 + 10% horse serum + 1% fish skin gelatin (blocking buffer) for 1 hour. Slides were incubated with rabbit anti-sGC β primary antibody and anti-ACTA2 AlexaFluor 488 conjugated primary antibody (Supplemental Table 2) diluted in blocking buffer (40-µl/section) and placed overnight at 4°C in a darkened humidity chamber. Slides were then washed twice for 5 minutes in 1x PBST, incubated with donkey anti-rabbit AlexaFluor 594 antibody for 1 hour at room temperature in a darkened humidity chamber (Supplemental Table 2), then washed twice for 5 minutes in 1x PBST. Coverslips were then mounted onto microscope slides using Duolink mounting medium containing 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI). Aortas were imaged using an Olympus FluoView 1000 confocal microscope. Fluorescence semiquantitation was calculated by quantifying the fluorescent signal from each respective channel relative to the area of aortic tissue imaged via ImageJ software (National Institutes of Health).
Treatment of Aortic Rings and Myography.
The following treatment method was performed as previously described (Rahaman et al., 2015). In brief, isolated murine thoracic aortas were isolated from mice and incubated with 10 µM FoxO inhibitor for 48 hours in one part Lonza Smooth Muscle Growth Medium-2 supplemented with SmGM-2 SingleQuot kit to nine parts unsupplemented Lonza Smooth Muscle Growth Medium-2. Following treatment, aortas were cut into 2-mm rings before being placed on a two-pin myograph (Danish Myo Technology). Aortic rings were then incubated in a physiologic salt solution (PSS) for 30 minutes of rest, after which 500-mg tension was applied to the vessels. Vessel viability was tested using potassium physiologic salt solution for 15 minutes, followed by a triplicate of PSS washes and a 60-minute resting period. Cumulative concentration responses to phenylephrine (10−9–10−5) and sodium nitroprusside (SNP; 10−9–10−4) determined vessel contractility and relaxation responses, respectively. Finally, relaxation percentage was determined by normalizing cumulative SNP relaxation to maximal contraction at 10−5 M phenylephrine and maximal dilation at 10−4 M SNP in Ca2+-free PSS.
Statistics.
TRANSFAC analysis software was used to predict FoxO binding sites on sGC promoter DNA utilizing settings to include the minimum number of false positives (Fig. 1). For drug-response and time-course cell culture experiments, Student’s t test was used to determine significance in Figs. 5, E–G and 6A and Supplemental Fig. 1 using GraphPad Prism version 7.03 software, and a one-way analysis of variance (ANOVA) was used to determine significance in Figs. 2, B, C, E, and F, 3, and 4, A, B, D, and E and Supplemental Fig. 2, A–F using GraphPad Prism version 7.03 software. A two-way ANOVA test for Fig. 6B was performed for each data point to determine significance using GraphPad Prism version 7.03 software. Calculated P values represent a significant difference from the control group, error bars represent the S.D., and symbols for confidence are represented by the following: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Fig. 1.
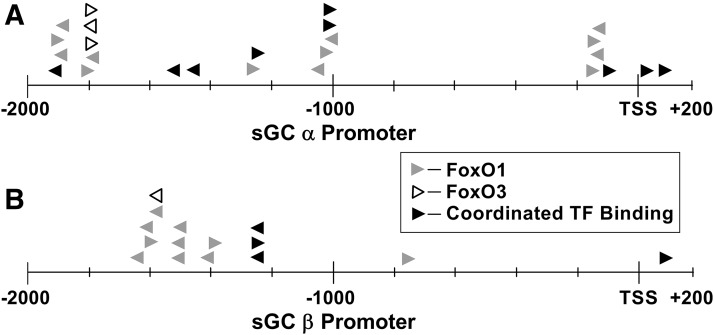
In silico analysis of human sGC promoter. Transcription factor (TF) binding site analysis of human sGC α (A) and sGC β (B) promoter sequence showed predicted FoxO transcription factor binding sites for 2200-bp promoter fragments flanking the sGC transcription start sites. Numbers indicate distance from transcription start site (TSS). Arrows facing right indicate binding sites on the positive (+) DNA strand; arrows facing left indicate binding sites on the negative (−) DNA strand.
Fig. 5.
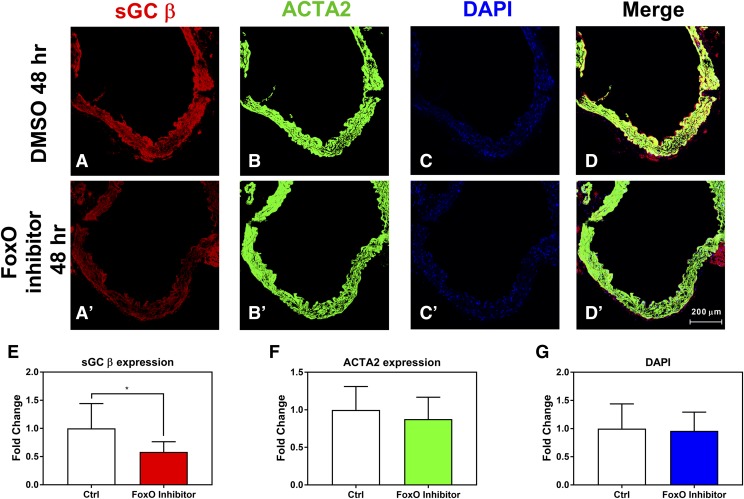
sGC expression in ex vivo murine aortas treated with FoxO inhibitor was decreased. Representative staining for ex vivo murine aortas treated with 10 μM FoxO inhibitor for 48 hours showing sGC β protein (A and A′), ACTA2 (B and B′), DAPI (C and C′), and merged channels (D and D′). Quantification of immunostaining for sGC β protein (E), ACTA2 protein (F), or DAPI staining (G). n = 3 animals. Student’s unpaired t test was used for determination of significance. *P < 0.05. Error bars represent S.D.
Fig. 6.
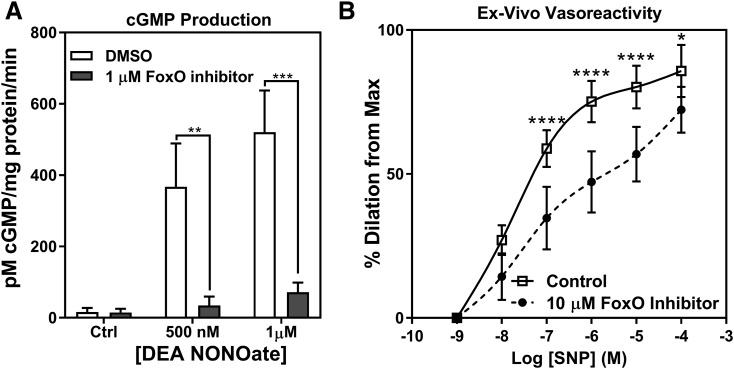
NO-dependent signaling in RASMCs and murine aortas is blunted after treatment with AS1842856. (A) cGMP produced by cultured RASMCs treated with AS1842856 and stimulated with NO donor DEA NONOate (n = 4). Student’s unpaired t test was used for determination of significance. (B) Ex vivo murine aortic vessels treated with 10 μM FoxO inhibitor or DMSO for 48 hours and dilated using the NO donor SNP (n = 5). Two-way ANOVA was used to determine significance. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Error bars represent S.D.
Fig. 2.
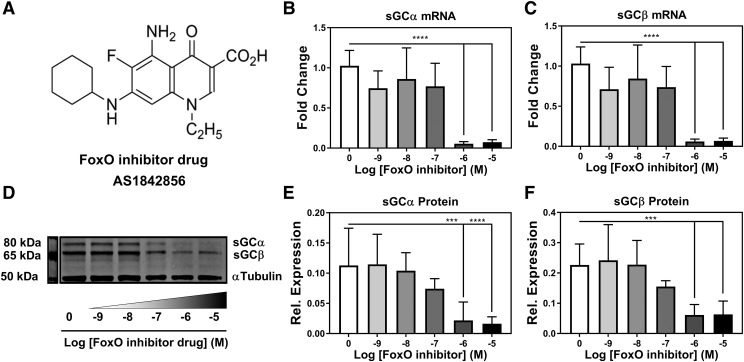
Treatment of RASMCs with FoxO inhibitor drug AS1842856 shows concentration-dependent decrease in sGC mRNA and protein expression. AS1842856 FoxO inhibitor drug structure (A) and effect on sGC α mRNA expression (B) or sGC β mRNA expression (C) following 48-hour drug treatment. (D) Western blot and quantification of 48-hour treatment with FoxO inhibitor on sGC α protein expression (E) and sGC β protein expression (F). n = 3 for all samples. One-way ANOVA test was used for determination of significance. ***P < 0.001; ****P < 0.0001. Error bars represent S.D.
Fig. 3.
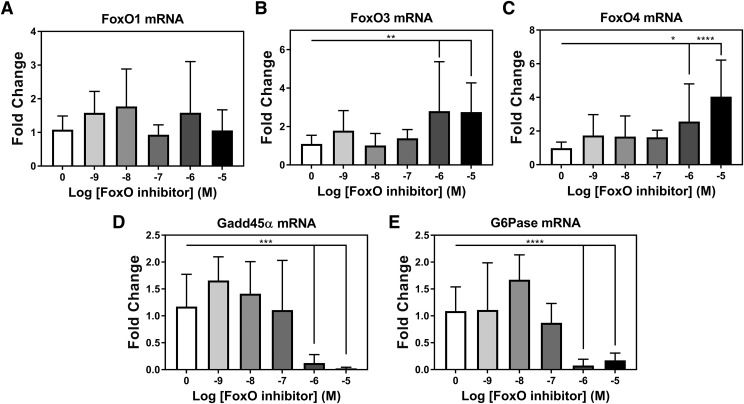
Treatment of RASMCs with FoxO inhibitor drug AS1842856 shows concentration-dependent decrease in canonical FoxO targets with compensatory increase in FoxO3 and FoxO4 mRNA expression. Quantitative reverse-transcription PCR from RASMCs treated with FoxO inhibitor. Response for 48-hour FoxO inhibitor treatment measuring FoxO1 (A), FoxO3 (B), FoxO4 (C), Gadd45α (D), or glucose-6-phosphatase (G6Pase) (E) mRNA expression. n = 3 for all samples. One-way ANOVA test was used for determination of significance. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Error bars represent S.D.
Fig. 4.
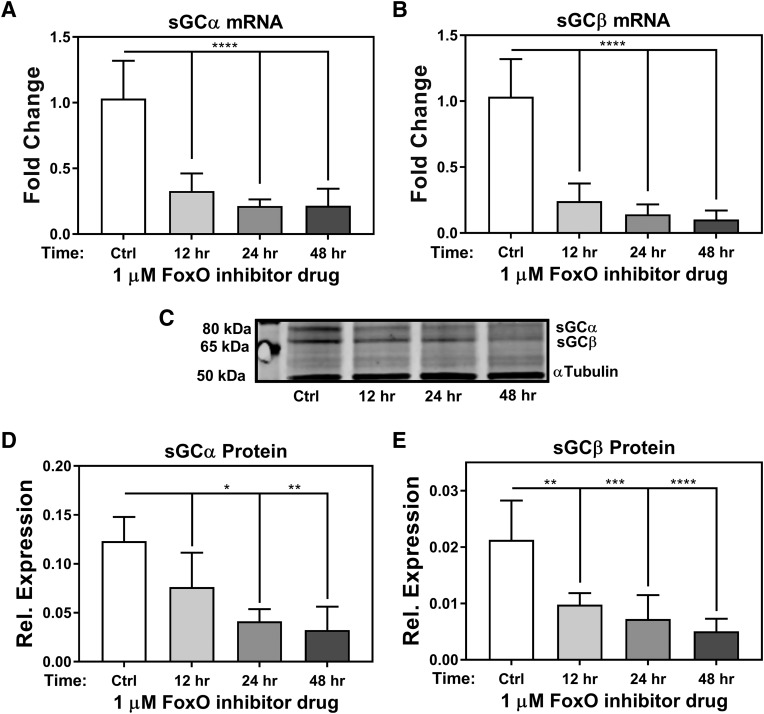
Treatment of RASMCs with FoxO inhibitor drug AS1842856 shows decrease in sGC mRNA, and protein expression occurs rapidly. Quantitative reverse-transcription PCR of 1 μM FoxO inhibitor treatment of 12, 24, and 48 hours on sGC α mRNA expression (A) or sGC β mRNA expression (B). (C) Western blot and quantification of 1 μM FoxO inhibitor treatment of 12, 24, and 48 hours on sGC α protein expression (D) and sGC β protein expression (E). n = 3 for all samples. One-way ANOVA test was used for determination of significance. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Error bars represent S.D.
Results
We began our study by analyzing the promoters for both sGC α and β subunits for potential transcription factor binding sites. TRANSFAC analysis software (Fogel et al., 2005) was used to evaluate the human sGC promoter using the GRCh38/hg38 reference genome (Miga et al., 2014), and 158 and 91 predicted binding sites were identified on the DNA ±10 kbp flanking the sGC α and sGC β transcription start sites, respectively. We discovered an abundance of FoxO family transcription factor binding sites clustered around 47 binding regions for sGC α and 36 binding regions for sGC β. Binding domains for FoxO family proteins in coordination with at least one other transcription factor were clustered around 55 regions for sGCα (Supplemental Table 3) and 10 regions for sGC β (Supplemental Table 4). A fragment of each gene’s promoter, 2000 bp upstream and 200 bp downstream of the transcription start site (TSS), was analyzed and showed 25 FoxO binding sites on the sGC α promoter (Fig. 1A) and 15 binding sites on the sGC β promoter (Fig. 1B). FoxO binding sites on sGC α were clustered around four locations—namely, 1900, 1800, 1000, and 100 bp upstream of the TSS. Likewise, the 2200-bp sGC β promoter fragment contained three clusters of predicted binding sites 1600, 1500, and 1200 bp upstream of the TSS.
The enrichment of the FoxO transcription factors based upon in silico analyses led us to investigate whether our predictive promoter analysis could be validated. We tested this by treating RASMCs with a FoxO inhibitor drug, AS1842856, which has been shown to inhibit all three FoxO isoforms expressed in VSMCs: FoxO1, FoxO3, and FoxO4 (Nagashima et al., 2010). In 48-hour AS1842856 drug (Fig. 2A) treatment experiments in RASMCs, we observed a stoichiometric loss of sGC α (Fig. 2B) and sGC β (Fig. 2C) mRNA expression of 90%–95% at concentrations of 1 µM or greater. Over the same treatment period, a loss of 80% in the protein expression of sGC α (Fig. 2, D and E) and a 74% decrease in sGC β (Fig. 2, D and F) were observed at concentrations of 1 µM or greater. Increasing concentrations of AS1842856 were accompanied by no change in FoxO1 mRNA expression (Fig. 3A), while an increase in FoxO3 (Fig. 3B) and FoxO4 (Fig. 3C) mRNA expression was observed. Classic FoxO transcriptional targets, such as growth arrest and DNA damage inducible α (Gadd45α) and glucose-6-phosphatase expression, exhibited the same degree of decreased gene expression (greater than 90%) at concentrations of 1 µM or greater (Fig. 3, D and E). These drug experiments showed that inhibition of FoxO transcriptional activity was commensurate with a loss of sGC expression, suggesting that the FoxO family plays a regulatory role in sGC gene expression. Additionally, measurement of the gene expression of cyclic nucleotide–generating enzymes adenylate cyclase (ADCY) and particulate guanylyl cyclase (pGC) showed that FoxO inhibition did not have a significant dampening effect on their expression. In fact, FoxO inhibition caused increased gene expression of ADCY1 and pGC1 with no significant changes in gene expression of ADCY3 or pGC2 (Supplemental Fig. 1).
FoxO temporal activity on sGC expression was then assessed using cultured RASMCs treated with 1 µM FoxO inhibitor. We observed that sGC α (Fig. 4A; Supplemental Fig. 2A) and sGC β (Fig. 4B; Supplemental Fig. 2B) mRNA expression significantly decreased after initial inhibitor administration and continued to decline with increased drug exposure time. Moreover, suppression of sGC mRNA was observed throughout the treatment period, reaching 80%–90% reduction by 48 hours (Fig. 4, A and B). As with the drug-response experiment, the loss of sGC α and β protein expression over time (Fig. 4, C–E) mirrored the observations in mRNA. sGC protein expression was marginally affected during the first few hours after FoxO inhibition, diminished by 32% loss of sGC α and 40% loss of sGC β within 6 hours (Supplemental Fig. 2, D–F), while significant losses in gene expression of sGC α (Supplemental Fig. 2A), sGC β (Supplemental Fig. 2B), and glucose-6-phosphatase (Supplemental Fig. 2C) were observed within 3 hours of drug treatment. This trend was also observed in the longer time-course experiments, wherein it reached maximal loss of 78% sGC α and 76% sGC β by the end of the 48-hour treatment period (Fig. 4, D and E). Combined, these experiments provide evidence that the loss of sGC expression in cultured RASMCs via inhibition of FoxO transcriptional activity occurs rapidly and remains impaired throughout a 48-hour treatment period.
We then assessed the impact of FoxO inhibition on sGC protein expression within isolated blood vessels. Mouse thoracic aortas were isolated and treated with 10 µM FoxO inhibitor drug for 48 hours and then immunostained for sGC β, smooth muscle α actin (ACTA2), and nuclei (DAPI). A higher concentration compared with culture experiments was necessary for loss of sGC expression, as studies have shown that sGC protein is highly stable in vivo (Groneberg et al., 2010). We chose to treat ex vivo tissue to circumvent the extensive mechanisms to stabilize sGC protein in vivo. FoxO inhibitor treatment decreased sGC β protein expression by 48% (Fig. 5, A, A′, and E) but had no significant effect on the expression of ACTA2 (Fig. 5, B, B′, and F) or on the density of nuclei staining (DAPI) within these isolated blood vessels (Fig. 5, C, C′, and G). Taken together, these data show that inhibition of FoxO activity in ex vivo tissue significantly lowers sGC expression, consistent with our previous studies in RASMCs (Fig. 5, D and D′).
Next, the extent to which FoxO inhibition impacts sGC signaling function was examined by measuring RASMC cGMP production and isolated aorta vessel relaxation. Cultured RASMC experiments showed that after 1 µM FoxO inhibitor treatment of 48-hours, cGMP production was reduced 85%–90% after stimulation with 0.5–1 µM of NO donor DEA NONOate (Fig. 6A). Similarly, ex vivo vessel myography studies on isolated murine aortas treated with 10 µM FoxO inhibitor for 48 hours showed impaired vasodilation in response to concentrations of the NO donor SNP greater than 100 nM (Fig. 6B). Collectively, these data suggest that the loss of sGC expression following FoxO inhibition results in a corresponding loss of NO-dependent, sGC-mediated cGMP production and vasoreactivity.
Discussion
sGC is crucial for NO-dependent relaxation to maintain cardiovascular health. To date, few studies have investigated the transcriptional regulation of sGC, and none have identified the transcription factors responsible for the constitutive expression of sGC within the vasculature. Prior work showed the CCAAT-binding factor regulates sGC gene expression within a neuroblastoma cell line (Sharina et al., 2003). However, no such regulation of sGC was observed in VSMCs, which prompted us to search for other transcription factors capable of initiating sGC transcription. One candidate family, the FoxO family of transcription factors, is known to regulate the development of new blood vessels and has been implicated in the prevention of pulmonary arterial hypertension (Furuyama et al., 2004; Savai et al., 2014; Deng et al., 2015). Our predictive analysis of the human GUCY1A3 and GUCY1B3 promoters showed that a plethora of potential FoxO binding sites are found along the upstream genetic regions of both sGC subunits. These findings inspired further exploration of the impact of FoxO proteins on the expression and downstream function of sGC.
In this study, we inhibited FoxO transcriptional activity using a pan-FoxO inhibitor (AS1842856) and observed significant decreases in the gene expression of sGC and well established targets of FoxO proteins. The decrease in sGC mRNA transcription demonstrated a stoichiometric decrease in both sGC α and sGC β gene expression. This inhibition rapidly attenuated sGC gene expression and signified that the decrease in expression of one gene matched the expression of the other. Proportionate sGC α and sGC β protein expression is necessary for optimal enzymatic activity of the obligate heterodimeric protein. Therefore, a symmetrical decrease in both sGC α and sGC β mRNA and protein suggests dual α and β regulation by FoxO transcriptional activity. Alternatively, it is possible that mechanisms which destabilize or stabilize sGC mRNA have the capability to alter sGC expression. For example, increases in the mRNA destabilizer AUF1 or loss of an sGC mRNA chaperone, human antigen R, could contribute to the observed loss of sGC mRNA (Pende et al., 1996; Klöss et al., 2004, 2005), but exploration of these hypotheses would require further investigation.
In a similar manner to the observations in sGC mRNA expression, FoxO inhibitor treatment decreased sGC α and β protein expression in cultured RASMCs commensurate with the swift concentration-dependent loss of sGC mRNA expression. Moreover, FoxO inhibition in mouse aortas caused a loss of sGC β protein expression and vascular reactivity. This likely suggests that the loss of protein expression and downstream activity occurs as a direct result of the loss of sGC mRNA and not through post-translational regulatory mechanisms. Signaling molecules identified to have effects on sGC protein expression, such as transforming growth factor β in the developing lung, have decreased expression of sGC α1 protein; however, no change was observed on the expression of sGC mRNA in response to hypoxia-induced transforming growth factor β expression (Bachiller et al., 2010). Furthermore, the chaperone-dependent E3-ligase protein, C terminus of heat shock cognate 70-interacting protein (CHIP), which is responsible for degradation of sGC protein, also targets the FoxO proteins for proteasomal degradation (Xia et al., 2007; Li et al., 2009). Previous studies also indicated that the half-life of sGC β in cultured cells due to CHIP breakdown is roughly 7 hours (Xia et al., 2007), which was consistent with the observed rate of sGC protein loss after our FoxO inhibition experiments and acted as the reasoning for longer drug treatment to observe a loss of protein expression and functional responses in the vasculature. As a result, FoxO inhibitor treatment alone on CHIP activity would presumably affect both sGC and the FoxO proteins, while mRNA expression would not be directly affected. In response to the effects on sGC mRNA expression, the observed loss of sGC protein expression was due to the lack of sGC transcript production. Additionally, FoxO inhibition caused a 90% reduction of cGMP production by sGC, which presumably led to the observed increase in gene expression of pGC1 as a compensatory mechanism. Despite this increase in pGC1 gene expression, the predominant method of cGMP production in smooth muscle is derived from sGC (Groneberg et al., 2010); thus, it is unclear whether this compensatory increase would rescue lost function due to impaired NO-dependent cGMP signaling. This effect on sGC protein expression in both culture and ex vivo tissue manifests in a loss of cGMP production and subsequent blunting of NO-dependent vasodilation.
We hypothesize that multiple FoxO proteins play a role in the physiologic transcription of sGC mRNA due to partial redundancy of the transcription factors as well as a nonselective effect of the inhibitor drug on all FoxO family members. While we cannot rule out off-target effects of the FoxO inhibitor, our data thus far do not suggest an off-target effect is responsible for the regulation of sGC that we observed. Based upon our pharmacological data and that of Nagashima et al. (2010) when characterizing this FoxO inhibitor drug, disparate effects on transcriptional activity of each of the FoxO proteins are observed after treatment. Pharmacodynamic studies indicate that FoxO1 is 70% inhibited, FoxO4 is 20% inhibited, and FoxO3 is 3% inhibited at a treatment concentration of 100 nM (Nagashima et al., 2010). A recent study also used several in silico modeling methods to predict a −6.3-kcal/mol binding energy of AS1842856 to FoxO1 and identified 10 amino acids in the transactivation domain which constitute drug-protein hydrogen bonding and hydrophobic interactions (Damayanti et al., 2016). These studies suggest that AS1842856 is specific for the transactivation domain of FoxO1, which governs the activity of the transcription factor after binding the target DNA sequences. Additionally, the lower affinity for AS1842856 to elicit FoxO3 and FoxO4 inhibition likely explains the necessity for higher drug concentration to induce the loss of sGC transcription in both cultured cells and isolated tissue. Our experiments also showed that FoxO3 and FoxO4 mRNA expression are elevated in response to drug treatment. These differential effects of the inhibitor drug on each of the isoforms suggest a complicated mechanism with multiple contributing factors in the compensatory expression of FoxO3 and FoxO4. Promoter analyses also showed that the FoxO transcription factors share affinity for the same conserved “TTGTTTAC” DNA motifs, which is supported by previous research (Brent et al., 2008; Obsil and Obsilova, 2008; Casper et al., 2014). This detail may indicate that loss of one transcription factor alone may not be sufficient to achieve the observed knock down of sGC expression in this study. Further investigation is required to elucidate the contributions of each FoxO protein in the transcription of sGC.
Many therapies available today, which include nitrovasodilator compounds that increase the bioavailability of NO (Arnold et al., 1977), and drugs that target sGC directly (Yoshina et al., 1978; Evgenov et al., 2007; Gheorghiade et al., 2012; Ghofrani et al., 2013) both promote second messenger cGMP signaling to improve cardiovascular health. Deficits in sGC transcription can eliminate the protein target of current therapeutic drugs, rendering many of them ineffective. Discovery of FoxO transcription factors as key regulators of the NO-sGC-cGMP signaling pathway within VSMCs also presents a potential contraindication that should be monitored when considering FoxO inhibitor drugs, such as AS1842856. Anomalies in blood pressure and other cardiovascular characteristics will be important biomarkers in future drug development studies involving the FoxO proteins. Taken together, the identification of the key transcription factors responsible for production of sGC mRNA is a vital component to understanding how cardiovascular homeostasis is regulated.
In summary, our study is the first to identify a family of transcription factors—namely, the FoxO family—capable of regulating sGC expression in vascular smooth muscle. This reveals a pivotal new role for the FoxO transcription factors in modulating vascular tone, and our next studies will investigate the specific role of each FoxO transcription factor in the regulation of sGC transcription. The discovery of the FoxO family as transcriptional regulators for sGC not only provides an alternative therapeutic approach for blood pressure control but also reveals a potentially novel mechanism that may impact sGC-related cardiovascular diseases.
Acknowledgments
We acknowledge the Center for Biological Imaging at the University of Pittsburgh for its support and confocal microscope usage. We acknowledge the contributions of Subramaniam Sanker, Nolan Carew, Heidi Schmidt, Rohan Shah, and Jacob Jerome for their aid in critical analysis of the manuscript.
Abbreviations
- ADCY
adenylate cyclase
- BSA
bovine serum albumin
- cGMP
cyclic guanosine 3′,5′-monophosphate
- CHIP
C terminus of heat shock cognate 70-interacting protein
- DAPI
4′,6-diamidino-2-phenylindole, dihydrochloride
- FoxO
Forkhead box subclass O
- NO
nitric oxide
- PBST
0.1% Tween-20 in PBS
- PCR
polymerase chain reaction
- pGC
particulate guanylyl cyclase
- PSS
physiologic salt solution
- RASMC
rat aortic smooth muscle cell
- sGC
soluble guanylyl cyclase
- SNP
sodium nitroprusside
- TSS
transcription start site
- VSMC
vascular smooth muscle cell
Authorship Contributions
Participated in research design: Galley, Durgin, Miller, Yuan, Straub.
Conducted experiments: Galley, Miller, Hahn.
Contributed new reagents or analytic tools: Galley, Miller, Hahn.
Performed data analysis: Galley, Miller, Hahn.
Wrote or contributed to the writing of the manuscript: Galley, Durgin, Yuan, Wood, Straub.
Footnotes
Financial support for this work was provided by the National Institutes of Health (NIH) [grants R01 HL 133864 and R01 HL 128304], American Heart Association (AHA) Grant-in-Aid [16GRNT27250146] (A.C.S.), Louis J. Ignarro Cardiovascular Fellowship [5T32GM008425] (J.C.G.), NIH T32 Division of Geriatrics Aging Institute Fellowship [AG021885] (J.C.G.), NIH T32 Post-Doctoral Fellowship Award [DK007052] (B.G.D.), and AHA Post-Doctoral Fellowship Award [19POST34410028] (S.Y.). This work was also supported by the Institute for Transfusion Medicine and the Hemophilia Center of Western Pennsylvania (A.C.S.).
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Arnold WP, Mittal CK, Katsuki S, Murad F. (1977) Nitric oxide activates guanylate cyclase and increases guanosine 3′:5′-cyclic monophosphate levels in various tissue preparations. Proc Natl Acad Sci USA 74:3203–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachiller PR, Nakanishi H, Roberts JD., Jr (2010) Transforming growth factor-beta modulates the expression of nitric oxide signaling enzymes in the injured developing lung and in vascular smooth muscle cells. Am J Physiol Lung Cell Mol Physiol 298:L324–L334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent MM, Anand R, Marmorstein R. (2008) Structural basis for DNA recognition by FoxO1 and its regulation by posttranslational modification. Structure 16:1407–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper SK, Schoeller SJ, Zgoba DM, Phillips AJ, Morien TJ, Chaffee GR, Sackett PC, Peterson FC, Crossgrove K, Veldkamp CT. (2014) The solution structure of the forkhead box-O DNA binding domain of Brugia malayi DAF-16a. Proteins 82:3490–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damayanti DS, Utomo DH, Kusuma C. (2016) Revealing the potency of Annona muricata leaves extract as FOXO1 inhibitor for diabetes mellitus treatment through computational study. In Silico Pharmacol 5:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Huang L, Sun Y, Heath JM, Wu H, Chen Y. (2015) Inhibition of FOXO1/3 promotes vascular calcification. Arterioscler Thromb Vasc Biol 35:175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, et al. International Consortium for Blood Pressure Genome-Wide Association Studies (2011) Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 478(7367):103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evgenov OV, Kohane DS, Bloch KD, Stasch JP, Volpato GP, Bellas E, Evgenov NV, Buys ES, Gnoth MJ, Graveline AR, et al. (2007) Inhaled agonists of soluble guanylate cyclase induce selective pulmonary vasodilation. Am J Respir Crit Care Med 176:1138–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel GB, Weekes DG, Varga G, Dow ER, Craven AM, Harlow HB, Su EW, Onyia JE, Su C. (2005) A statistical analysis of the TRANSFAC database. Biosystems 81:137–154. [DOI] [PubMed] [Google Scholar]
- Frey R, Mück W, Unger S, Artmeier-Brandt U, Weimann G, Wensing G. (2008) Pharmacokinetics, pharmacodynamics, tolerability, and safety of the soluble guanylate cyclase activator cinaciguat (BAY 58-2667) in healthy male volunteers. J Clin Pharmacol 48:1400–1410. [DOI] [PubMed] [Google Scholar]
- Furuyama T, Kitayama K, Shimoda Y, Ogawa M, Sone K, Yoshida-Araki K, Hisatsune H, Nishikawa S, Nakayama K, Nakayama K, et al. (2004) Abnormal angiogenesis in Foxo1 (Fkhr)-deficient mice. J Biol Chem 279:34741–34749. [DOI] [PubMed] [Google Scholar]
- Gheorghiade M, Greene SJ, Filippatos G, Erdmann E, Ferrari R, Levy PD, Maggioni A, Nowack C, Mebazaa A, COMPOSE Investigators and Coordinators (2012) Cinaciguat, a soluble guanylate cyclase activator: results from the randomized, controlled, phase IIb COMPOSE programme in acute heart failure syndromes. Eur J Heart Fail 14:1056–1066. [DOI] [PubMed] [Google Scholar]
- Ghofrani HA, D’Armini AM, Grimminger F, Hoeper MM, Jansa P, Kim NH, Mayer E, Simonneau G, Wilkins MR, Fritsch A, et al. CHEST-1 Study Group (2013) Riociguat for the treatment of chronic thromboembolic pulmonary hypertension. N Engl J Med 369:319–329. [DOI] [PubMed] [Google Scholar]
- Giannakou ME, Goss M, Jünger MA, Hafen E, Leevers SJ, Partridge L. (2004) Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science 305:361. [DOI] [PubMed] [Google Scholar]
- Goubareva I, Gkaliagkousi E, Shah A, Queen L, Ritter J, Ferro A. (2007) Age decreases nitric oxide synthesis and responsiveness in human platelets and increases formation of monocyte-platelet aggregates. Cardiovasc Res 75:793–802. [DOI] [PubMed] [Google Scholar]
- Groneberg D, König P, Wirth A, Offermanns S, Koesling D, Friebe A. (2010) Smooth muscle-specific deletion of nitric oxide-sensitive guanylyl cyclase is sufficient to induce hypertension in mice. Circulation 121:401–409. [DOI] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C. (2003) Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300:1142–1145. [DOI] [PubMed] [Google Scholar]
- Hwangbo DS, Gershman B, Tu MP, Palmer M, Tatar M. (2004) Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body [published correction appears in Nature (2005) 434:118]. Nature 429:562–566. [DOI] [PubMed] [Google Scholar]
- Klöss S, Bouloumié A, Mülsch A. (2000) Aging and chronic hypertension decrease expression of rat aortic soluble guanylyl cyclase. Hypertension 35:43–47. [PubMed] [Google Scholar]
- Klöss S, Rodenbach D, Bordel R, Mülsch A. (2005) Human-antigen R (HuR) expression in hypertension: downregulation of the mRNA stabilizing protein HuR in genetic hypertension. Hypertension 45:1200–1206. [DOI] [PubMed] [Google Scholar]
- Klöss S, Srivastava R, Mülsch A. (2004) Down-regulation of soluble guanylyl cyclase expression by cyclic AMP is mediated by mRNA-stabilizing protein HuR. Mol Pharmacol 65:1440–1451. [DOI] [PubMed] [Google Scholar]
- Kuo JF, Greengard P. (1970) Cyclic nucleotide-dependent protein kinases. VI. Isolation and partial purification of a protein kinase activated by guanosine 3′,5′-monophosphate. J Biol Chem 245:2493–2498. [PubMed] [Google Scholar]
- Li F, Xie P, Fan Y, Zhang H, Zheng L, Gu D, Patterson C, Li H. (2009) C terminus of Hsc70-interacting protein promotes smooth muscle cell proliferation and survival through ubiquitin-mediated degradation of FoxO1. J Biol Chem 284:20090–20098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZP, Wang Z, Yanagisawa H, Olson EN. (2005) Phenotypic modulation of smooth muscle cells through interaction of Foxo4 and myocardin. Dev Cell 9:261–270. [DOI] [PubMed] [Google Scholar]
- Meurer S, Pioch S, Pabst T, Opitz N, Schmidt PM, Beckhaus T, Wagner K, Matt S, Gegenbauer K, Geschka S, et al. (2009) Nitric oxide-independent vasodilator rescues heme-oxidized soluble guanylate cyclase from proteasomal degradation. Circ Res 105:33–41. [DOI] [PubMed] [Google Scholar]
- Miga KH, Newton Y, Jain M, Altemose N, Willard HF, Kent WJ. (2014) Centromere reference models for human chromosomes X and Y satellite arrays. Genome Res 24:697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittendorf J, Weigand S, Alonso-Alija C, Bischoff E, Feurer A, Gerisch M, Kern A, Knorr A, Lang D, Muenter K, et al. (2009) Discovery of riociguat (BAY 63-2521): a potent, oral stimulator of soluble guanylate cyclase for the treatment of pulmonary hypertension. ChemMedChem 4:853–865. [DOI] [PubMed] [Google Scholar]
- Nagashima T, Shigematsu N, Maruki R, Urano Y, Tanaka H, Shimaya A, Shimokawa T, Shibasaki M. (2010) Discovery of novel forkhead box O1 inhibitors for treating type 2 diabetes: improvement of fasting glycemia in diabetic db/db mice. Mol Pharmacol 78:961–970. [DOI] [PubMed] [Google Scholar]
- Obsil T, Obsilova V. (2008) Structure/function relationships underlying regulation of FOXO transcription factors. Oncogene 27:2263–2275. [DOI] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. (1997) The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389:994–999. [DOI] [PubMed] [Google Scholar]
- Pende A, Tremmel KD, DeMaria CT, Blaxall BC, Minobe WA, Sherman JA, Bisognano JD, Bristow MR, Brewer G, Port J. (1996) Regulation of the mRNA-binding protein AUF1 by activation of the beta-adrenergic receptor signal transduction pathway. J Biol Chem 271:8493–8501. [DOI] [PubMed] [Google Scholar]
- Potente M, Urbich C, Sasaki K, Hofmann WK, Heeschen C, Aicher A, Kollipara R, DePinho RA, Zeiher AM, Dimmeler S. (2005) Involvement of Foxo transcription factors in angiogenesis and postnatal neovascularization. J Clin Invest 115:2382–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahaman MM, Nguyen AT, Miller MP, Hahn SA, Sparacino-Watkins C, Jobbagy S, Carew NT, Cantu-Medellin N, Wood KC, Baty CJ, et al. (2017) Cytochrome b5 reductase 3 modulates soluble guanylate cyclase redox state and cGMP signaling. Circ Res 121:137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahaman MM, Reinders FG, Koes D, Nguyen AT, Mutchler SM, Sparacino-Watkins C, Alvarez RA, Miller MP, Cheng D, Chen BB, et al. (2015) Structure guided chemical modifications of propylthiouracil reveal novel small molecule inhibitors of cytochrome b5 reductase 3 that increase nitric oxide bioavailability. J Biol Chem 290:16861–16872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruetten H, Zabel U, Linz W, Schmidt HH. (1999) Downregulation of soluble guanylyl cyclase in young and aging spontaneously hypertensive rats. Circ Res 85:534–541. [DOI] [PubMed] [Google Scholar]
- Salih DA, Brunet A. (2008) FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr Opin Cell Biol 20:126–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savai R, Al-Tamari HM, Sedding D, Kojonazarov B, Muecke C, Teske R, Capecchi MR, Weissmann N, Grimminger F, Seeger W, et al. (2014) Pro-proliferative and inflammatory signaling converge on FoxO1 transcription factor in pulmonary hypertension. Nat Med 20:1289–1300. [DOI] [PubMed] [Google Scholar]
- Sharina IG, Martin E, Thomas A, Uray KL, Murad F. (2003) CCAAT-binding factor regulates expression of the beta1 subunit of soluble guanylyl cyclase gene in the BE2 human neuroblastoma cell line. Proc Natl Acad Sci USA 100:11523–11528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendre A, Irvin MR, Wiener H, Zhi D, Limdi NA, Overton ET, Shrestha S. (2017) Local ancestry and clinical cardiovascular events among African Americans from the atherosclerosis risk in communities study. J Am Heart Assoc 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasch JP, Becker EM, Alonso-Alija C, Apeler H, Dembowsky K, Feurer A, Gerzer R, Minuth T, Perzborn E, Pleiss U, et al. (2001) NO-independent regulatory site on soluble guanylate cyclase. Nature 410:212–215. [DOI] [PubMed] [Google Scholar]
- Stasch JP, Schmidt PM, Nedvetsky PI, Nedvetskaya TY, H S AK, Meurer S, Deile M, Taye A, Knorr A, Lapp H, et al. (2006) Targeting the heme-oxidized nitric oxide receptor for selective vasodilatation of diseased blood vessels. J Clin Invest 116:2552–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stice JP, Eiserich JP, Knowlton AA. (2009) Role of aging versus the loss of estrogens in the reduction in vascular function in female rats. Endocrinology 150:212–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tian C, Zheng JC. (2013) FoxO3a contributes to the reprogramming process and the differentiation of induced pluripotent stem cells. Stem Cells Dev 22:2954–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobst J, Rumpf PM, Dang TA, Segura-Puimedon M, Erdmann J, Schunkert H. (2015) Molecular variants of soluble guanylyl cyclase affecting cardiovascular risk. Circ J 79:463–469. [DOI] [PubMed] [Google Scholar]
- Xia N, Strand S, Schlufter F, Siuda D, Reifenberg G, Kleinert H, Förstermann U, Li H. (2013) Role of SIRT1 and FOXO factors in eNOS transcriptional activation by resveratrol. Nitric Oxide 32:29–35. [DOI] [PubMed] [Google Scholar]
- Xia T, Dimitropoulou C, Zeng J, Antonova GN, Snead C, Venema RC, Fulton D, Qian S, Patterson C, Papapetropoulos A, et al. (2007) Chaperone-dependent E3 ligase CHIP ubiquitinates and mediates proteasomal degradation of soluble guanylyl cyclase. Am J Physiol Heart Circ Physiol 293:H3080–H3087. [DOI] [PubMed] [Google Scholar]
- Xu X, Wang S, Liu J, Dou D, Liu L, Chen Z, Ye L, Liu H, He Q, Raj JU, et al. (2012) Hypoxia induces downregulation of soluble guanylyl cyclase β1 by miR-34c-5p. J Cell Sci 125:6117–6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshina S, Tanaka A, Kuo SC. (1978) Studies on heterocyclic compounds. XXXVI. Synthesis of furo[3,2-c]pyrazole derivatives. (4) Synthesis of 1,3-diphenylfuro[3,2-c]pyrazole-5-carboxaldehyde and its derivatives (author’s transl). Yakugaku Zasshi 98:272–279. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yalcin S, Lee DF, Yeh TY, Lee SM, Su J, Mungamuri SK, Rimmelé P, Kennedy M, Sellers R, et al. (2011) FOXO1 is an essential regulator of pluripotency in human embryonic stem cells. Nat Cell Biol 13:1092–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]