Abstract
Aging is associated with a decline in heart function across the tissue, cellular, and molecular levels. The risk of cardiovascular disease grows significantly over time, and as developed countries continue to see an increase in lifespan, the cost of cardiovascular healthcare for the elderly will undoubtedly rise. The molecular basis for cardiac function deterioration with age is multifaceted and not entirely clear, and there is a limit to what investigations can be performed on human subjects or mammalian models. Drosophila melanogaster has emerged as a useful model organism for studying aging in a short timeframe, benefitting from a suite of molecular and genetic tools and displaying highly conserved traits of cardiac senescence. Here, we discuss recent advances in our understanding of cardiac aging and how the fruit fly has aided in these developments.
Keywords: cardiac aging, fruit fly, proteostasis, obesity, epigenetics
1. Introduction
Aging can be defined as the time-dependent, progressive loss of physiological integrity, leading to impaired organ, tissue, and cellular function and increased susceptibility to death [1]. It is the main risk factor for a number of debilitating and life-threatening disorders, including cardiovascular disease (CVD) [2]. As life expectancy continues to increase around the world, so does the financial burden of healthcare for the elderly and CVD in particular. Globally, it is expected that the cost to treat CVD will double to triple by 2030 [3–5]. Over time, the heart exhibits characteristic gene expression and morphological changes that are associated with declining performance, which ultimately increases mortality. Age-related changes include impaired systolic and diastolic function, extracellular matrix remodeling, and elevated propensity for arrhythmia [6–8]. As technology has advanced drastically in the field of genomics, transcriptomics, and proteomics, connections have been increasingly identified between the physiological variations observed in aging hearts and the molecular processes involved. Cellular aging, in general, is accompanied by an accumulation of damaged proteins and increased proteotoxicity, dysfunction of mitochondria, increased genomic instability, reduced autophagic flux, increased activation of NF-κB, and telomere attrition [9]. These changes are particularly detrimental to the function of post-mitotic cells like cardiomyocytes, which, when damaged beyond repair, cannot be readily replaced.
Drosophila melanogaster has proven to be a valuable animal model for studying the aging heart. Flies age in a matter of weeks (five to seven for a typical cardiac aging study) [10–15] and are amenable to extensive genetic manipulation [16, 17]. Genes that are differentially expressed over time can be readily manipulated temporally and spatially in large, isogenous populations of offspring [17–19]. The adult fruit fly heart comprises approximately 80 mature cardiomyocytes that are aligned along two opposing bilateral rows to form a linear cardiac tube (Figure 1) [20]. Coordination of contractility is governed by two pacemakers believed to exist at posterior and anterior regions of the heart. Depending on which pacemaker is dominant, the circulation can flow toward the head or posterior of the fly [21, 22]. The cardiomyocytes consist of conserved, minimally-redundant components including those involved in the cytoskeleton, calcium handling, protein homeostasis (i.e. proteostasis), metabolism, and chromatin structure [10, 12, 13, 15, 23–25]. Overall, the relative simplicity of the fruit fly heart renders it an attractive model for rapid cardiac senescence investigations.
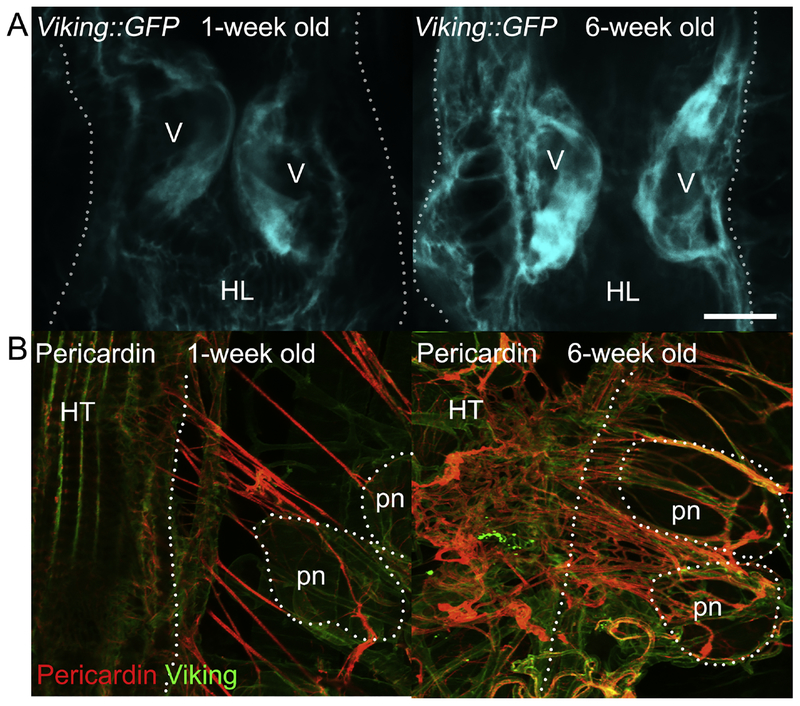
Figure 1: The adult Drosophila heart tube and associated structures.
(A) The abdominally located adult fruit fly heart tube (HT) contains roughly 80 cardiomyocytes, arranged along two opposing bilateral rows. These aligned cells form a central luminal space and are flanked by adjacent pericardial nephrocytes (pns) that filter the hemolymph (insect’s blood) and alary muscles (ams), which tether the heart to the dorsal cuticle [20, 256–258]. The alary muscles span from epidermal attachments to the heart and contact the heart tube indirectly by an interface formed from extracellular matrix components, such as the collagens Pericardin and Viking. They serve as a flexible heart suspension system to maintain the cardiac tube in an anatomically correct position. (B) Heart tubes showing cardiac collagen-IV (Viking) and Pericardin (a collagen IV-like protein) deposition. Hemolymph enters the heart through the ostia (os), which are inflow openings formed by specialized cardiomyocytes. The adult Drosophila heart has three intracardiac valves (v), which subdivide the organ into distinct chambers, close the luminal space during systole, and permit unidirectional flow. Pericardial nephrocytes are analogous to mammalian reticuloendothelial cells and, being situated close to ostia, are ideally placed to filter the passing hemolymph entering the heart tube. Scale bar = 100μm. (C) Illustration depicting the aforementioned anatomical structures.
With the emergence and advancement of techniques for assessing whole genomes, transcriptomes, and proteomes under differing conditions and with age, considerable data have been collected on cardiomyocytes from invertebrate and vertebrate models. Here, we discuss how Drosophila has been utilized to investigate the physiological, genetic, and epigenetic bases for cardiac aging. Drosophila and mammalian hearts and cardiomyocytes share many of the same traits of cardiac senescence, including systolic and diastolic dysfunction, increased arrhythmias, a decline in proteostasis, and decreased metabolic fitness [11, 12, 24, 26–28]. Additionally, the aging fruit fly has been successfully used as a model of obesity and its exacerbation of normal cardiac aging as well as to investigate the effects of exercise on heart health over time [29–31].
2. Conserved age-related myocardial changes in Drosophila
Cardiac aging is characterized by several histological, physiological, and biochemical changes. Aged myocardium from rodents and humans demonstrates structural remodeling, which includes left ventricular (LV) hypertrophy due to increased cardiomyocyte size [7, 32–39]. Concomitant with hypertrophy are alterations in LV shape and dimensions [8, 40–42]. For example, using cardiac magnetic resonance imaging, age-associated changes in LV structure and function were assessed in a multi-ethnic cohort of ~5000 individuals free of cardiovascular disease [42]. The authors reported a marked age-related increase in LV mass/volume ratio. This was ~25% higher in the old vs. young age group and was driven by a proportionately greater magnitude of age-associated decline in LV end diastolic volume compared to that of LV mass. Moreover, the significant age-dependent decrease in LV end diastolic volume exceeded the decrease in LV end systolic volume, which resulted in reduced stroke volume. While the cardiac tube of Drosophila has been reported to hypertrophy in response to genetic manipulation [43], altered wall thickness or mass with age has not been reported. However, several studies have demonstrated changes in shape and dimensions of fruit fly hearts over time [15, 23, 44], akin to those in humans described above [42]. Wild-type Drosophila heart tubes display a progressive decrease in both diastolic and systolic diameters with age [15, 23, 24]. The decline in diastolic dimensions is greater than that for systolic dimensions, which highlights deterioration in contractile performance, as manifest by significantly reduced fractional shortening [15, 23, 24].
Healthy aging is accompanied by additional variations in cardiac contraction. The mean shortening velocity during systole has been reported to decrease in mammals [8, 45–47]. The slowing of myocardial contraction with advanced age, including that observed during lightly loaded isotonic contractions using heart muscle from senescent vs. younger adults rats, may be in part due to the switching from α- to β-myosin heavy chain gene expression [8, 45, 46, 48, 49]. The cardiac tube of wild-type Drosophila lines likewise exhibits depressed shortening velocities in five- relative to one-week-old flies under basal and/or loaded conditions [13]. While there is currently no evidence of a switch in myosin heavy chain isoforms that have distinct hydrolytic and mechanical activities, aging fly hearts do show changes in the expression of several myofilamentous genes, including tropomyosin, actins, α-actinin, troponin-C, and tropomodulin [15, 24], which may contribute to reduced contractile speeds.
In conjunction with decreased mean shortening velocity, vertebrate cardiac aging is characterized by a prolongation of systolic contraction time [8, 50–52]. The Drosophila heart similarly displays increasingly longer systolic intervals with advancing age [23, 44]. The mechanistic basis of these changes likely involves variations in, and modifications to, several conserved cardiomyocyte calcium handling components across species. In mammals, various studies have documented coordinated changes in protein function and/or gene expression with aging that prolong the calcium transient that drives myocardial contraction [8, 52, 53]. Upon membrane depolarization, calcium enters the cardiomyocyte through L-type calcium channels, prompting the release of additional calcium from the sarcoplasmic reticulum (SR), the cardiomyocyte’s intracellular storage compartment. Although L-type calcium channel density is not apparently affected by age, its function seems to decline, as illustrated by a reduction in the calcium transient amplitude and slower channel inactivation [8, 41, 54, 55]. Moreover, the rate of calcium reuptake into the SR decreases in senescent myocardium [48, 53, 56, 57]. An age-associated reduction in the transcription of SERCA2, the gene that encodes the SR calcium pump, accounts in part for decreased SR pump site density and the impaired sequestration of intracellular calcium [48, 55, 58]. However, expression of the Na+/Ca2+ exchanger, which extrudes intracellular calcium from the cell, increases between adulthood and senescence, and enhanced antiporter-mediated flux may partially compensate for impaired calcium reuptake into the SR [48].
The calcium cycling properties of the Drosophila cardiac tube are also negatively impacted by age. Old flies display altered intracellular cardiomyocyte calcium dynamics, with a prolonged transient decay, which promotes the extended periods of systole observed in senescent animals [23, 44, 59]. Recently, whole-cell patch clamp recordings of calcium currents across the membrane of isolated Drosophila cardiomyocytes confirmed the cells possess a conserved compendium of L- and T-type calcium channels [60]. As in mammals, L-type (A1D in Drosophila) channels serve as the main conduits for sarcolemmal calcium flux in fly cardiomyocytes. These channels share regulatory properties, such as calcium dependent inactivation, a critical negative feedback process that modulates the rate of channel inactivation, with their vertebrate counterparts. While not studied directly, these properties may analogously deteriorate with age, slow inactivation, and extend contraction. Furthermore, microarray data reveal that the expression of genes involved with calcium handling, including both L- and T-type channels, the ryanodine receptor, SERCA, and the Na+/Ca2+ exchanger, is significantly reduced over time [15, 24], which could further engender altered calcium transients in aged fly cardiomyocytes. Importantly, cardiac arrhythmias are a byproduct of aberrant calcium handling, and their incidence increases in both elderly humans and flies [8, 23, 44, 61].
Diminished adrenergic signaling and autonomic modulation of cardiac function is an important factor in mammalian age-associated cardiovascular change [8, 48, 62]. For example, the ability of β-adrenergic receptor stimulation to increase contractility declines over time. This results from a failure of β-adrenergic receptor stimulation to augment the intracellular calcium transient to the same extent in cardiomyocytes from senescent hearts relative to those from younger hearts [48, 63]. The observed age-related reduction in β-adrenergic receptor modulation of cardiac contraction is attributable, at least in part, to insufficient enhancement of the activity of L-type calcium channels [48, 63]. The root cause of these changes is not completely understood, but several lines of evidence indicate reduced myocardial β-adrenergic receptor density, their functional decline, and deficits in the β-adrenergic signaling cascade with advanced age [48, 64, 65]. Interestingly, Drosophila express components homologous to those of the vertebrate β-adrenergic pathway, including adrenergic-like octopamine receptors (OctαRs, and OctβRs), adenylyl cyclase (rutabaga), phosphodiesterase (dunce), and both regulatory and catalytic subunits of protein kinase A (PKA), and their cardiac L-type calcium channels exhibit PKA-mediated current enhancement [60]. The expression of all aforementioned genes uniformly declines with age in Drosophila cardiomyocytes [15, 24, 66], suggesting old fly hearts may also exhibit blunted responses to adrenergic stimulation.
A profound, well-characterized, and frequently-cited hallmark of cardiac aging is impaired diastolic function. In addition to decreased LV end diastolic volumes in humans and reduced cardiac tube diastolic diameters in flies, several indices of age-related diastolic dysfunction are shared among the species. Older vertebrate hearts fill more slowly than younger hearts and exhibit increased relaxation times [8, 67]. The velocity at which the Drosophila heart reestablishes diastolic volumes is significantly lower for five- vs. one-week-old flies, at baseline and/or against elevated afterloads, consistent with impaired relaxation kinetics [13]. These changes are plausibly associated with the disruption of calcium homeostasis and cycling observed in aged myocardium and, potentially, with altered passive recoil of elastic elements compressed during systole [68].
Mammalian hearts normally experience changes in their material properties, including myocardial stiffening, with age [8, 69–73]. Similarly, fly hearts display changes in passive mechanical properties over time [13, 15, 26, 74]. Using an atomic force microscopy‐ based approach, five‐ week-old wild-type hearts were found to be significantly stiffer than one‐ week-old hearts [12, 13, 15, 74]. The molecular basis of altered heart wall and cardiomyocyte compliance likely involves several factors. These include aberrant diastolic calcium handling, myofilament dysfunction, and extracellular matrix modifications and remodeling [26, 68, 75–78].
3. Extracellular matrix and matricellular proteins in cardiac aging
Extracellular matrix and matricellular proteins (for brevity in this review, collectively termed ECM) are a large family of evolutionarily conserved proteins contributing to the structure and function of multicellular organisms [79]. The matrix proteins help form a structural “scaffold” that supports cell-cell and cell-matrix interactions, whereas the matricellular proteins contribute to the formation of the scaffold but do not integrate into it. ECM regulation in the heart is linked with cardiac output, and disruption of or experimental modification to ECM deposition causes cardiomyopathy in both mammalian and Drosophila models [80].
Abnormal or pathological accumulation of ECM proteins (fibrosis), particularly collagens, occurs in the aging human heart and is associated with increased mortality [81, 82]. Additionally, markers of fibrosis can be predictive of mortality in aged populations [83] and are generally associated with cardiac dysfunction [84]. Importantly, the mechanistic underpinnings of ECM turnover are of intense interest due to the clinical significance of fibrosis and intractability of its treatment.
Most experimental research requires whole organisms to establish the pathophysiology of aging on organs and systems. Although mammalian models recapitulate the fibrosis-like features of aging human hearts [85, 86], such models are costly, and long-term studies of aging are not widely reported. Thus, simpler systems to study ECM deposition in age-dependent cardiac dysfunction are beneficial. Although Drosophila may not provide the complexity of ECM components seen in humans and mammalian models, the fly provides enormous advantages in terms of genetic tractability, brevity of lifespan, and resource usage.
With reference to collagens, Drosophila development is dependent on expression of the type-IV collagen α2 and α1 chains encoded by viking and Cg25C [87]. Alternatively, cardiac development is reliant on Multiplexin (COL18A1 in humans [88]) and Pericardin, a type-IV-like collagen that tethers the Drosophila heart to the underlying cuticle and to supportive alary muscles (Figure 1, 2), which run perpendicular to the cardiac tube [89, 90]. Although Pericardin was initially described as a matrix-forming type-IV-like collagen, it adopts prominent fiber-like structures, acting as a “tendinous bridge” between the heart and alary muscles [90, 91].
Figure 2. Changes to cardiac collagen deposition in aging fly hearts.
Under normal laboratory conditions, the fly’s lifespan is approximately 8–10 weeks. Prior to the onset of mortality, in the general population at around six weeks, a significant increase in the amount of cardiac collagen-IV (Viking) and Pericardin (a collagen IV-like protein, restricted to the heart) can be seen around the heart. With advanced age, there is an increased accumulation of collagen. Confocal micrographs demonstrate age-associated changes to (A) Viking::GFP levels on the luminal side of the valve cells and (B) the Pericardin network. Scale bar = 20μm.
Both Pericardin and Viking show age-dependent accumulation in the Drosophila heart (Figure 2), a phenotype that accompanies severe cardiomyopathy, suggesting that cardiac fibrosis may develop from evolutionarily conserved mechanisms [91][92]. In mouse models of aging, ECM composition around the heart changes considerably and correlates with cardiac dysfunction [86]. Collagen deposition is mediated by direct interaction with an evolutionarily conserved matricellular protein called Secreted Protein Acidic and rich in Cysteine (SPARC). SPARC expression is required for normal heart development in the Drosophila model [93], increases in the aging mouse heart [85], but decreases significantly in the aging Drosophila heart [15]. In mammals, changes to SPARC expression are dependent on tissue type (for example, it decreases in tendons [94]), indicating that such variations are sensitive to the local signaling environment. Upstream signals coordinating collagen deposition and turnover in Drosophila remain to be elucidated.
In mice, collagen accumulation in the aging heart can be ameliorated by reducing SPARC expression [85, 86]. Interestingly, reduced dosage of Drosophila SPARC also attenuates age-dependent changes in cardiac function in flies [91]. Although overexpression of SPARC leads to dramatic deposition of Pericardin around the heart tube and causes cardiomyopathy, its role in age-dependent changes in cardiac function may be independent of gross collagen accumulation and related to a non-structural role of the protein in cell signaling [91]. Recent data from the mouse model, where cardiac fibrosis accompanies inflammation, suggests that SPARC mediates activation of pro-inflammatory macrophage pathways [95], which may explain the profibrotic shift in ECM turnover as animals age. The possibility that such phenotypes are evolutionarily conserved has not yet been examined in flies. Notably, there are few, if any, studies regarding age-dependent changes to the ECM of other organ systems in Drosophila.
4. Loss of proteostasis in cardiac aging
Proper cellular homeostasis depends upon quality control mechanisms to preserve the stability, functionality, and turnover of the proteome [1, 96, 97]. If a protein fails to fold into its native conformation, is damaged beyond repair, or is no longer needed, the cell must degrade it and, if possible, recycle its constituents. Protein synthesis, folding/refolding, and degradation are carried out in the overall process of proteostasis. Proteostasis involves the interplay of a multi-branched coordinated network, which includes the ubiquitin-proteasome system (UPS), autophagy, and chaperone-mediated protein refolding [98–102]. The pathways of this network are able to respond to deficiencies in the others to help ensure proteins are maintained in a soluble, nonaggregated, nontoxic state [98, 99, 102–106]. Maintaining this delicate balance of protein quality control (PQC) is especially critical in post-mitotic cells, including cardiomyocytes and neurons, since their limited regenerative capacity means they rely upon proper proteostasis for delaying age-related functional decline [107–109]. When PQC is not maintained in such cells, irreversible proteotoxicity and cell death can ensue [106, 108].
Reduced proteostasis is a highly conserved hallmark of non-pathological cellular aging [1, 110, 111] that has been well-documented in vertebrate cardiomyocytes and human patients [112–115]. For example, a Finnish study observed that a quarter of the hearts of patients at advanced age (85 years or older) contained amyloid plaques and protein aggregates similar to those found in the brains of Alzheimer’s disease patients [116], indicating problems potentially with all branches of the proteostasis network. The time-dependent, progressive accumulation of cellular damage and misfolded protein aggregates in heart cells has additionally been linked to several cardiovascular disorders, including desmin-related cardiomyopathy, dilated cardiomyopathy, hypertrophic cardiomyopathy, ischemic heart disease, and heart failure [97, 108, 110, 117–121].
While all branches of the proteostasis network are affected by age, a temporal decline in autophagy is believed to be a major contributor to time-dependent changes in cardiac function [122–124]. Autophagy plays an integral role in removing dysfunctional organelles and protein aggregates by lysosomal degradation [1, 108–110]. It is activated in response to many cues, including low nutrient availability and transcription factors, such as longevity-associated FoxO. Indirect enhancement of autophagy has been shown to alleviate age-associated hypertrophy, fibrosis, and apoptosis in studies using vertebrate myocytes [125, 126], and thus interventions to increase the level of autophagy have been proposed to prevent or slow the progression of aging in the heart [124]. However, evidence in which autophagy is directly and exclusively augmented in cardiomyocytes to produce the averred beneficial effects is lacking. Additionally, while maintaining basal levels of autophagy in the heart is vital for cell survival, insufficient autophagy results in the accumulation of toxic proteins, while excessive autophagy is maladaptive and induces disproportionate catabolism and cell death [127]. Therefore, more research is needed, and enthusiasm for enhanced autophagy as a therapeutic modality should be tempered [128–133].
The rapidly aging Drosophila model has been used to examine the functional impact of enhanced and repressed activity of the proteostasis network in diverse tissues. In both Drosophila cardiac and skeletal muscle an accumulation of misfolded and ubiquitinated proteins is observed over time accompanying age-associated decline in function [15, 134]. In skeletal muscle, ubiquitinated protein accumulation was attributed mainly to decreased cellular autophagy [134]. In the fly heart, however, age-associated functional decline correlated with decreased transcript levels of dozens of genes associated with UPS and to a lesser extent chaperone-mediated protein refolding, but not autophagy [15]. Only one autophagy-associated gene, Atg8b, was significantly downregulated with age according to cardiac-restricted microarray analysis [15]. It is unclear if UPS-associated genes were similarly downregulated in skeletal muscle with age. In both studies, dfoxo, the Drosophila ortholog of FoxO, was overexpressed either exclusively in the heart or in all muscle. In the heart, modest overexpression of dfoxo comprehensively ameliorated all hallmarks of cardiac aging described above [15], although increased lifespan was not observed [135]. Compared to aged controls, dfoxo-overexpressing hearts displayed significantly decreased arrhythmias, stiffness, pacing-induced heart failure, and diastolic interval, and increased cardiac output, myocardial relengthening rate, and heart rate [10, 15, 135, 136]. Furthermore, improved function was accompanied by a marked reduction of ubiquitinated cardiac proteins as well as a significant reversal of transcriptional activity of genes associated with each step along the UPS pathway [15]. In skeletal muscle, autophagy-associated transcripts reportedly increased systemically upon dfoxo overexpression, in addition to increased lifespan [134]. However, it was also determined that the dfoxo-overexpressing flies consumed less compared to controls [134]. Consequently, it is plausible that the lower nutrient intake potentially contributed to the systemic increase in autophagy rather than exclusively the increase in skeletal muscle dfoxo expression. Nonetheless, dFOXO may play different but related roles in cardiac vs. skeletal muscle over time. More evidence correlating non-pathological cardiac aging and loss of UPS function is needed, as it remains vastly understudied among invertebrate and vertebrate models [109]. It should be noted that increasing the dosage of dfoxo in the heart beyond modest overexpression, or reducing its expression, had toxic effects on cardiac function and organismal development, but the direct cause remains unknown [15]. Similarly, excessive dfoxo overexpression in skeletal muscle did not afford systemic health benefits but caused premature organismal death, possibly due to excessive catabolism and cytotoxicity [15]. Thus, there appears to be an optimal stoichiometric range that undermines the benefits of manipulating FoxO expression [127]. Because cardiac proteostasis appears to be maintained over time when dfoxo is expressed in optimal doses, and these changes seemingly correlate with improved UPS activity in particular, UPS-associated components may be attractive targets for therapeutic intervention to improve heart function in the elderly. Regardless, downstream effectors of dFOXO transcriptional control must be studied in more depth to determine a mechanism by which to reap the benefits observed in response to mild overexpression and bypass the negative outcomes of deleterious levels of dFOXO in both cardiac and skeletal muscle.
Substantiating the role of proteostasis in the aging heart remains a challenging task between species and even among individuals of the same species with similar genetic backgrounds [24]. As described above, microarray analysis of roughly 30 pooled adult Drosophila hearts highlighted an important role of the UPS in cardiac aging and dFOXO-mediated functional improvement [15]. However, a nanofluidic RNAseq evaluation of single Drosophila cardiac tubes indicated expression variation with age and no definitive role of UPS-associated genes in every fly [24, 66]. Additional investigation into transcriptomes and proteomes of aged murine and simian LVs revealed that a number of components of the proteasome were actually upregulated rather than downregulated [13, 24], a seemingly paradoxical result. While initially surprising, as there is a consensus that cellular aging is generally accompanied by decreased proteostasis [97], in many cases, decreased proteasome activity is not necessarily met by a decrease in proteasome components [118, 120]. The presence of inactive or defective proteasomal proteins may contribute to or result from the age-associated loss of proteostasis. Alternatively, it is possible that the UPS is upregulated to counteract failing and/or downregulation of other branches of the proteostasis network [97, 124]. Evidence suggests that autophagy and UPS each compensates for the other in cardiac pathologies [137], an interplay that may likewise accompany cardiac aging. Genes encoding proteins involved in chaperone-mediated refolding were also observed to exhibit disparate aging expression patterns among species [13, 15, 24]. Despite this, the mammalian heat shock protein HSPB8 (Hsp27 in Drosophila) exhibited conserved downregulation in cardiomyocytes over time among several species tested [13, 15, 114, 138]. Further examination of molecular cardiac aging data across the animal kingdom is needed to confirm these findings and to test the potential universally cardioprotective properties of conserved targets.
5. The decline of metabolic cardiac fitness with age
It is well established that obesity and its associated metabolic disturbances are major risk factors for CVD [139]. Furthermore, the risk of obesity as well as CVD increases with age [7, 140, 141]. In addition to changes in PQC, an important hallmark of the aging process is the progressive dysfunction of white adipose tissue and the related metabolic alterations that lead to multi-organ damage [142]. In humans as well as in flies, fat progressively accumulates in nonadipose tissues [143, 144]. This is a key factor in a vicious cycle that accelerates aging and the onset of age-related diseases, such as type 2 diabetes, cancer, and CVD [145].
The heart has a continuously high energetic demand. Therefore, cardiomyocytes are extremely mitochondria-rich in order to generate the ATP required for contraction, calcium handling, and cellular homeostasis in general. Heart tissue is thought to be especially sensitive to dietary changes, such as increased consumption of sugar and fat, since it is heavily dependent upon fatty acids for ATP production [146]. Excessive body fat accumulation causes maladaptive changes in the heart. Over time, in humans and vertebrate animal models, obesity may result in cardiomyocyte growth, interstitial fat infiltration, and triglyceride accumulation in cardiomyocytes and the contractile elements [30, 147–150]. These changes contribute to LV mass accrual, hypertrophy, altered chamber dimensions, and eventually dysfunction reminiscent of the age-associated myocardial disturbances described above [151–153].
Similarly, flies fed a high fat diet (HFD, 30% coconut oil) accumulate high levels of triglycerides, become hyperglycemic, and exhibit functional and structural changes in their cardiac tubes [30, 31]. These alterations include increased heart rate, reduced fractional shortening, and increased incidence of arrhythmias and non-contractile heart regions, resembling age-related changes in function [30, 31]. Even a small, 2% increase in fat consumption during midlife, over the course of three weeks, has a significantly detrimental impact on Drosophila cardiac function and overall healthspan [154]. Restricting food consumption to only 12 daylight hours per day, compared to ad libitum-fed flies, partially protected against diet- and age-induced decline in cardiac function as evidenced by preserved fractional shortening and heart rate parameters and reduced arrhythmicity [154]. Time restricted feeding also prevented the body weight gain observed in ad libitum-fed flies over time [154]. Transcriptomic analysis of heart samples suggested that mitochondrial electron-transport chain complexes and circadian clock pathways mediate these benefits, at least to some degree [154]. These data provide evidence supporting the general idea that healthy heart aging depends on efficient metabolism and cardiac senescence can be improved by modifying diet.
A multitude of evidence suggests that metabolism is altered with age. In liver as well as in muscle tissue from old mice, glycolysis has been reported to be increased [155]. Additionally, aging murine hearts exhibit an increase in proteins involved in glycolysis and oxidative stress-response [156, 157]. Interestingly, glycolysis also increases in some animal models of heart failure [158], and augmented myocardial consumption of glucose has been reported in patients with idiopathic dilated cardiomyopathy [159]. These results are in line with a reduction in oxygen utilization and ATP synthesis in aging rat ventricles [160]. In flies, a decrease in ATP levels and in NADH/NAD+ and GSH/(GSH+GSSG) ratios was also found in muscle tissues, supporting a bioenergetic decline with age [161]. However, it is well documented that transcription of genes related to glycolysis declines with age in Drosophila heart and muscle tissues [15, 24, 161]. Furthermore, Ma et al. provided metabolomic data from fly heads supporting a reduction in glycolysis [161]. While the mechanistic underpinnings of energetic decline between certain animal models and discrete tissues might differ, it is possible that they converge and impact analogous downstream effectors. For instance, flies heterozygous for different subunits of the Polycomb Repressive Complex 2 (Pclc421/+; Su(z)12c253/+), which are long-lived, have elevated ATP and cellular redox levels [161]. Therefore, metabolomic studies on young vs. old fly hearts would be crucial to determine the conservation of mechanisms underlying the cardiac energetic imbalance that occurs with age.
The strong connection between age- and obesity-related disorders suggests that they may be controlled by similar or intersecting pathways. Accumulating evidence suggests that caloric restriction (CR) can increase longevity in yeast, worms, fruit flies, rats, and mice [162]. Conversely, humans who are overweight or obese have a higher risk of mortality [163]. This seems to be corroborated by experiments in which flies fed a HFD experienced severely shortened lifespans [144, 164]. The nutrient sensor target of Rapamycin (TOR) is believed to be a key component in mediating the CR-induced increase in lifespan [165]. TOR activation stimulates cell growth, increases lipid and protein synthesis (anabolism), and decreases autophagy (catabolism) [162]. The TOR pathway is activated by insulin, insulin-like growth factors, and amino acids and inhibited in response to stress, such as energy depletion and caloric restriction. Thus, this pathway plays an essential role in orchestrating metabolic homeostasis.
While complete depletion of TOR induced heart failure in mice [166], mild reduction may be cardioprotective [167]. For example, mTORC1 inhibition attenuated load-induced cardiac hypertrophy [168] and reduced infarct size after ischemia by restoring cardiac autophagy in obese mice [169]. These effects were proposed to be partially mediated by autophagy-induced removal of misfolded proteins and dysfunctional mitochondria. In flies, there is robust evidence indicating that HFD induces lipotoxic cardiomyopathy by activating the TOR pathway [30, 31]. For instance, hypomorphic TOR7/P mutant flies did not develop lipotoxic cardiomyopathy when fed HFD, since they have constitutively increased transcript levels of the adipose triglyceride lipase, ATGL (brummer in Drosophila), which prevented the flies from accumulating triglycerides [31]. Additionally, heart-specific inhibition of TOR activity by overexpressing the downstream effector d4EBP also prevented the deleterious effects of HFD on heart function [31]. Upon stress, cells accumulate Sestrins, a family of evolutionarily conserved antioxidant proteins, resulting in AMPK-dependent inhibition of TOR signaling. dSesn null flies displayed accumulation of triglycerides accompanying cardiac dysfunction, which could be rescued by inhibiting the TOR pathway with Rapamycin [170]. Overall, these studies suggest that partial inhibition of TOR can improve metabolic balance by reducing triglyceride accumulation and, therefore, prevent the deleterious effects of HFD on heart performance. Conversely, TOR7/P hypomorphs showed reduced physical activity levels, as assessed by a negative geotaxis assay [31], implying that each tissue may have a specific energy balance requirement. Therefore, different TOR activity levels might be required to counteract the deleterious effects of metabolic imbalance induced by obesity, stress, or aging, each posing a threat to healthy heart function.
The TOR pathway can be activated directly by amino acids and indirectly by dietary sugar or fat (see above) through insulin-IGF signaling [171]. Reduced insulin-IGF signaling is known to prolong lifespan in different animal models by regulating growth, metabolism, and stress response [172, 173]. In the fly, heart-specific reduction of signal transduction by InR, the single Drosophila insulin-like receptor, evidently improves cardiac physiology at advanced age [10, 135]. In addition, overexpression of dfoxo, a negative effector of insulin signaling, in the adipose tissue (fat body in flies) protects flies from fat accumulation and HFD-induced heart dysfunction [31]. As discussed above, modest heart-specific overexpression of dfoxo ameliorated the functional decline of the aging heart [15]. Moreover, it prevented the deleterious effects of HFD on cardiac function, though systemic fat accumulation was not avoided [31]. High-sugar diet (HSD) in flies, as in mammals, results in augmented fat content and hyperglycemia, in turn inducing insulin resistance and cardiomyopathy [146, 174–176]. Interestingly, findings in Drosophila suggest that consumption of diets high in sugar early in life leads to shortened lifespan due to maladaptive nutritional reprogramming [177]. HSD inactivates dFOXO, which seems to execute long-term transcriptional changes that affect the fitness of flies later in life [177]. Likewise, in the heart specifically, RNAi-mediated dFOXO suppression engenders a cardiac phenotype reminiscent of accelerated aging [15].
Taken together, aging and obesity seemingly share a low energy demand state, leading to pathological inhibition of FoxO and persistent TOR hyperactivation. Consequently, cells accumulate lipids, misfolded proteins, reactive oxygen species (ROS), and dysfunctional mitochondria, all of which lead to seemingly premature aging and heart function decline. The fly model, with simpler but largely conserved biochemical pathways, can aid in understanding the molecular and genetic changes induced by the metabolic imbalance that apparently stimulates accelerated cardiac aging. Thus, it can help in developing new targeted therapeutics.
6. Epigenetic modifications of the aging heart
An additional, primary hallmark of aging is the progressive accumulation of alterations to the epigenome [1]. While all cells of an organism contain the same DNA (with some exceptions, e.g. immune cells), their specific gene expression programs differ in response to internal and external cues. Such cues can trigger complex biological events including differentiation and environmental adaptation. Cellular differentiation is a highly regulated process. It is achieved in large part by altering the chromatin state, which enables proper gene expression to drive the process and thus control cellular phenotypes. Additionally, the chromatin state changes to match gene expression with fluctuating energetic demands. Thus, aging cells must face the challenge of staying healthy and remaining functionally competent by maintaining their epigenetic program and simultaneously retaining their capacity to respond to environmental fluctuations (e.g. diet or stress). This is of particular importance for long-lived cells, such as the cardiomyocytes, which are terminally differentiated very early in life [175].
Chromatin structures are dynamically controlled by epigenetic modifications. These include heritable changes in DNA methylation, histone modifications, and non-coding RNAs, all of which can regulate gene expression without changing the genetic code. While the epigenome is maintained in a state of dynamic equilibrium [180–183], evidence suggests that it is prone to “drift” over the lifespan of an organism [184]. Age-associated changes to the epigenome have been correlated with an abnormal transcriptome, which contributes to age-related pathologies including cancer, Alzheimer’s disease, dementia, and CVD [185–187]. Additionally, experimental manipulations of epigenetic factors in various animal models suggest that they can modulate healthy lifespan [184].
Hence, a dynamic and drifting epigenome likely makes it difficult for cells and tissues to achieve phenotypic stability, healthy aging, and longevity. Minor alterations to the epigenome may have major impacts on gene expression. Indeed, a significant change in transcription with age has been consistently reported at the cellular and tissue levels in different species ranging from flies to mice to humans [15, 24, 188–190]. The meta-transcriptomic analysis by Cannon et al. revealed that the pathways whose components displayed age-associated expression changes, and thus are potentially involved in cardiac aging, are conserved between fly and rodent hearts [24]. Despite conservation of the pathways, however, there was considerable variability in age-associated expression changes of particular genes among species. Interestingly, even between individual Drosophila hearts there was substantial transcriptome variability with age [24], perhaps reflecting the same phenomenon as has been reported in single cardiomyocytes from mice [191]. Thus, the accumulation of changes in gene expression over time may ultimately lead to similar cardiac aging phenotypes across species [24]. In line with these results, a recent study performed in three different tissues on 168 pairs of genetically identical, human female, monozygous twins identified a total of 137 genes with similar age-related expression changes and 42 genes with unique age-related expression changes between co-twins [192]. These data support the idea that with age, gene expression changes depend not only on genetic factors but also on environmental cues as well as stochastic variations that may happen simply by chance. These alterations could result from a combined imbalance in the activity of transcription factors, splicing factors, or epigenetic modifiers and could contribute to the characteristic individual variability observed during age-associated functional decline.
There is growing evidence that chromatin structure is altered in an age-dependent manner in different animal models and that modulation of epigenetic factors significantly impacts lifespan [193]. Indeed, deleterious age-associated epigenetic changes are well documented in many tissues. For example, an age-associated gain of methylation at promoter CpG islands, which are CG-rich regions typically located 5’ to the transcriptional start site, is thought to increase the incidence of cancer, in part by silencing tumor suppressor genes [194–196]. Conversely, age-related loss of DNA methylation at other regions of the genome may contribute to de-repression of retrotransposons, thereby causing genome instability [197]. Remarkably, the epigenetic clock, based on the methylation state of CpGs, is an accurate marker of chronological age. It is also believed to be a promising molecular biomarker of biological age, showing a strong correlation with age-associated phenotypes [187, 198, 199]. For instance, using the Horvath estimation for biological age based on DNA methylation, a 4% increase in the risk of developing CVD was observed per year of advanced biological age [200].
Epigenetic modifiers control chromatin packaging into at least two distinct states, heterochromatin or euchromatin, in part by modifying DNA methylation, by acetylation or methylation of histone tails, or by binding to the DNA or to nucleosome core particles. In flies, systemic mild overexpression of the Heterochromatin Protein 1 (HP1) led to an increase in heterochromatin levels and lifespan extension with improved muscle integrity and function [201]. HP1 helps in the maintenance of heterochromatin by interacting with nuclear lamins. Expression of nuclear lamins has been shown to decline with age in fly and human cells [202–204]. Loss of Lamin-B in the Drosophila fat body correlates with an increase in retrotransposon activation [205]. Indeed, there is a progressive de-repression of retrotransposons with age, shown by a 2-fold increase in RNA expression of previously annotated retrotransposon elements [205]. Retrotransposons are the most common type of transposable elements (TEs). Upon activation, some TEs can move to new locations in the genome, which can lead to mutations and DNA damage. Thus, the age-dependent reduction in heterochromatin documented in yeast, flies, and senescent mammalian cells could trigger activation of retrotransposons, leading to DNA instability and aging phenotypes [205–207]. Interestingly, using a position effect variegation reporter, a delay in age-related gene de-repression was found when flies underwent caloric restriction, which increases lifespan, as discussed earlier [206]. This exemplifies the plasticity of the epigenome and underscores a challenge that long-lived cardiomyocytes must overcome to maintain vital functionality over a lifetime, whether that be weeks or decades.
7. Crosstalk between epigenetics and metabolism with age
Energy balance and tight metabolic control are key determinants of organism-wide functional maintenance and healthy aging. The ability of an animal to respond to stress, such as changes in temperature, oxygen levels, and nutrient availability, gradually declines with age. Variations in the epigenome allow for the adjustment of gene expression to match environmental changes and energy requirements by regulating accessibility of the transcriptional machinery to DNA. This interplay between metabolism and epigenetics depends on the fact that histone-modifying enzymes utilize substrates, including the metabolites NAD+, ATP, S-adenosylmethionine (SAM), acetyl-CoA, and α-ketoglutarate [208–210], which are dysregulated with extreme diets and age.
In Drosophila, acute as well as maternal HFD and HSD are known to modify the expression of metabolic genes in adult offspring [211, 212]. Paternal HSD can modify the offspring’s chromatin state and gene transcription in a manner dependent upon tri-methylation of histone H3 at lysine 9 (H3K9me3) or lysine 27 (H3K27me3) [213]. In addition, HSD was found to cause lifelong changes in gene expression via inhibition of dFOXO activity [177]. Not surprisingly, null dfoxoΔ mutants and HSD-fed flies displayed overlapping changes in gene expression and specifically an enrichment in transcripts encoding epigenetic modifiers, such as Sirt-1 and −2, Histone deacetylase 1 (HDAC1), Su(z)12, and Enhancer of zeste (Ez) [177, 214].
Ez is the primary catalytic subunit of the evolutionarily-conserved Polycomb repressive complex 2 (PRC2). PRC2 mediates gene silencing by promoting H3K27me3 [215]. Upon aging, there are also profound tissue-specific changes in metabolism and energy balance analogous to those induced by HFD and HSD, as discussed earlier. In addition to an age-dependent decline in glycolysis in fly heads, and reduced ATP levels and NADH/NAD+ ratio in muscles, Ma et al. also identified a dramatic drift in the H3K27me3 repressive marks in Drosophila musculature [161] (note the brain, as opposed to the heart, preferentially uses glycolysis over fatty acid oxidation for ATP production [216, 217]). Interestingly, mutants heterozygous for PRC2 components, including Pcl, Su(z)12, Ez, and esc, showed an increase in lifespan that correlated with reduced levels of H3K27me3 and a reduction in the age-associated shift in H3K27me3 modifications in muscles [161, 218]. The authors additionally found an increase in transcripts involved in glycolysis in muscle tissues from Pcl and Su(z)12 compound heterozygous mutant flies [161]. Similarly, PRC2 heterozygous mutants had ATP levels and NADH/NAD+ ratios comparable to those of young wild-type flies, as mentioned previously [161]. These data suggest that aging leads to a genome-wide drift in H3K27me3 repressive modifications, which causes changes in transcription that result in reduced glycolysis and/or defective glucose metabolism. In aging fly hearts, an opposite shift from fatty acid oxidation to glycolysis is expected with age, but definitive studies have yet to be conducted. Since histone methyltransferases rely on the availability of the universal methyl donor SAM, it would be interesting to study the regulation of this metabolite in old flies and their hearts. Evidence currently suggests that SAM is important for heart function and healthy aging [219], possibly in part by regulating the levels of histone methylation. In fact, the Radical S-Adenosyl methionine Domain containing 1 (RSAD1) enzyme has been linked to heart development, and mutations lead to congenital heart disease [220].
NAD+ is an important co-factor for the Sirtuin group of protein deacetylases. The age-associated reduction in NADH/NAD+ ratio reported by Ma, et al. in Drosophila muscles [161] contrasts with findings in mammals (various tissues) in which NAD+ levels have been shown to decline with age, and replenishment of the NAD+ pool by supplementation with dietary precursors could drive the observed increase in healthspan [221]. However, we would like to emphasize again that tissue and species discrepancies would have to be more extensively scrutinized to determine potential fundamental differences. Indeed, overexpression of the NAD+ synthase CG9940 in flies resulted in a mild but significant extension of lifespan [222]. Consistently, NAD+ dependent Sir2 (Sirt1 in mammals), has been implicated in lifespan extension not only in flies but also in yeast and mammals in a tissue- and dose-dependent manner [223–225]. For instance, in Drosophila, modest overexpression of dSir2 increased lifespan, while excessive levels of dSir2 resulted in decreased lifespan [225]. Importantly, the extension in lifespan achieved by CR in flies has been shown to be dependent on dSir2 expression levels [226]. A loss in age-related gene silencing of a position effect variegation reporter was also observed upon global mild overexpression of dSir2 [206]. Finally, evidence suggests that replenishing the NAD+ pool by overexpressing NAD+ synthase decreases arrhythmia and fibrillations in old flies compared to age-matched controls [222]. However, these studies are not entirely conclusive, and the specific roles of NAD+ and Sir2 in cardiac aging need to be characterized in greater detail.
While there is increasing evidence showing that manipulation of the chromatin state and epigenetic modifiers can alter/extend lifespan [184], little is known about the epigenetic mechanisms that may preserve healthy cardiac aging, specifically. To make headway, its low genetic redundancy makes the fly a well-poised model for epigenetic cardiac aging studies [20, 178, 227, 228]. The histone deacetylases HDAC1 and HDAC2 regulate the expression of genes involved in cardiac morphogenesis and heart physiology in mammals [229]. The Drosophila protein reduced potassium dependency-3 (Rpd3) shows high homology with human HDAC1 and moderate homology with human HDAC2 [138]. Heterozygous rpd3 mutants (rpd3+/−) exhibited increased lifespan in flies [230] and in yeast [231]. Rpd3 works in a complex with SIN3 to repress expression of multiple glycolytic genes, genes involved in the oxidation of fatty acids into acyl-CoA in the mitochondrial matrix, and genes involved in ROS response [232]. It remains unclear if the rpd3+/− mutant fly lifespan extension is accompanied by improved cardiac aging. However, heart-restricted knockdown of rpd3 was found to extend lifespan and prevent the age-related reduction in heart rate of old flies [233]. While transcriptomic analysis of Drosophila heart samples suggested that rpd3 transcript levels are not modified with age [15], the beneficial effects of rpd3 knockdown on healthspan might result from a change in global acetylation levels. Heart-specific rpd3-knockdown flies experienced an increase in resistance against oxidative stress (20mM methyl viologen hydrate), elevated ambient temperature (37°C), and starvation compared to single transgenic controls. Whole-fly transcript levels of sod2, dfoxo, and sir2 were also found to be increased upon heart-specific reduction of rpd3 [233]. sod2 expression could potentially lead to a more efficient removal of superoxide radicals produced in the mitochondria upon induction of oxidative stress. These results are in agreement with the reported lifespan extension achieved upon ubiquitous moderate overexpression of sir2 [225] and improved cardiac aging, but not lifespan, upon modest heart-specific overexpression of dfoxo [15, 135]. Altogether, these data suggest that mild reduction of rpd3 can alter gene expression, thus improving the aging heart by enhancing the responses to stress and de-repressing expression of genes involved in energy balance such as dfoxo. Therefore, during normal aging, other epigenetic modifiers, including those potentially controlled by dFOXO [177], may have profound effects on gene programs, possibly leading to energy imbalance and cardiomyocyte malfunction.
Epigenetic modifications have been shown to be at the intersection of metabolic equilibrium and gene expression. The exact mechanisms underlying the energy imbalance and loss of chromosome homeostasis upon aging remain unknown. However, there is increasing evidence indicating that epigenetic modifications can have a significant impact on the quality of aging [234]. Because epigenetic changes are reversible, reestablishing a healthy epigenome might reduce the functional decline observed in cardiomyocytes over time. A potential mechanism by which epigenetic cardioprotection can be promoted is through pro-longevity metabolic interventions such as CR, rapamycin, and exercise [235, 236].
8. Reduction of age-related heart deterioration as result of aerobic exercise
Accumulating evidence suggests that switching to a healthier lifestyle (e.g. reduced sugar and fat consumption, increased physical activity, etc.) can reduce the risk of heart failure [237]. Many animal models and cohort analyses have shown that aerobic exercise reduces the risk of death from CVD [238, 239]. Indeed, the strongest inverse correlation between exercise and heart failure was observed in the elderly [240]. A two-year clinical trial showed that high-intensity exercise increased maximal oxygen uptake (VO2max) and reduced LV stiffness in previously sedentary, but otherwise healthy, middle-aged male participants [241]. These studies suggest that aerobic exercise not only improves cardiac function but also reverses the effects of sedentary aging on the heart to some extent. While there is compelling evidence supporting the benefits of exercise on heart function and healthy aging, the mechanisms underlying these phenomena are poorly understood.
The Drosophila model is well-suited to study the effects of exercise on heart function in a controlled manner. Because flies have an innate reaction to climb up in response to a negative geotaxis stimulus, they can easily be subjected to an exercise routine on a large scale [242]. Evidence supports the idea that regulated exercise training prevents age-related heart dysfunction in flies and has beneficial effects similar to those from aerobic exercise in humans [29, 243–245]. A pacing protocol developed for Drosophila allows for the study of cardiac performance in response to increasing the normal heart rate through external electrical field stimulation [135, 136]. This is comparable to an exercise stress test used to evaluate the fitness of the human heart [10, 136, 227]. As flies age, they normally exhibit an increase in cardiac-arrest rate after external electrical pacing, a measurement of heart failure, and a decrease in arrest recovery [136]. However, in exercise-trained elderly flies, a decrease in cardiac-arrest rate and increase in recovery compared to same-age unexercised flies was observed [29]. These findings suggest a beneficial effect of exercise on the Drosophila aging heart. A line of flies selectively bred for increased longevity demonstrated similar cardioprotection accompanied by improved mitochondrial efficiency [244]. Interestingly, such longevity-bred flies as well as three-week-old exercised flies have overlapping changes in whole-body transcript levels, including those for gustatory receptors and genes involved in carbohydrate metabolism, xenobiotic/drug metabolism, and folate biosynthesis [244]. Of note, the extension in lifespan observed upon CR was significantly reduced when flies undergoing CR were allowed to smell, but not to eat, yeast [246]. In line with these results, null mutant flies for the odorant and gustatory receptors Or83b and Gr64a, exhibited improved cardiac stress tolerance in a similar manner to ubiquitous knockdown of the G-protein coupled receptor Mthl3 [244]. The authors suggested that reducing flies’ sensory perception of food can increase exercise capacity, improve cardiac aging, and increase longevity in a manner similar to CR [244]. Indeed, experimental evidence suggests that the beneficial effects of exercise are mediated by octopaminergic activity [247].
Sirt1 (dSir2 in Drosophila) is a NAD+-dependent histone deacetylase, which functions as an energy status sensor and has been implicated in longevity, obesity, and cancer [248]. Upon increased energy demand, when NAD+ levels are highest, Sirt1 deacetylates PGC-1α, producing NADH in the process. This deacetylation activates PGC-1α and in turn induces the transcription of genes involved in gluconeogenesis, glycolysis, and fatty-acid oxidation in a tissue-specific manner [249]. Exercise is known to increase mitochondrial activity [29, 250, 251] and, more specifically, improve cardiac mitochondrial biogenesis by activating PGC-1α [252]. PGC-1xp heterozygous mutant flies exhibited cardiac dysfunction remarkably similar to that observed in HFD-fed control flies [30]. Interestingly, another mutant line for PGC-1, srl1, was reported to have delayed development and reduced lifespan [253]. Moreover, overexpression of PGC-1 in adult muscles, including the cardiac tube, protects against heart failure in response to electrical pacing [253]. This protection was increased when PGC-1-overexpressing flies were subjected to exercise training, suggesting that PGC-1 and exercise both have cardioprotective roles [253]. However, cardioprotection achieved by exercise and PGC-1 overexpression may also be mediated by a reduction in triglyceride accumulation [30, 144]. In fact, knockdown of dSir2 in the fat body led to a significant increase in total triglyceride levels and free fatty acids [254]. In line with these data, whole-fly dSir2 transcript levels were found to be reduced with age but increased upon exercise [144]. Microarray data revealed the expression of dSir2 was not changed in the Drosophila heart with age. However, expression of sirt2, which has high identity to human SIRT2 and SIRT3, was found to be significantly decreased [15]. The direct effect of dSir2 overexpression on heart function and cardiac aging remains to be determined. Overall, research in Drosophila has shown that exercise ameliorates heart function decline [29, 244, 245] and prolongs healthspan [144, 244], as similarly demonstrated in vertebrate models [255].
9. Conclusions
Drosophila melanogaster has increased our understanding of the molecular basis of cardiac aging over recent decades. During which, the tools available for investigating the gene expression patterns and physiological changes involved have advanced considerably. In humans, cardiac aging studies are frequently limited to analysis of heart performance, genomic testing, and family history records. While these are highly valuable for understanding genetic and phenotypic trends in cardiac function over time, the ability to non-invasively investigate acute transcriptomic and proteomic changes in human tissue is impossible. Cardiac senescence is a complex event, and evidence supporting the use of Drosophila melanogaster to examine various facets of it that are difficult to study in humans grows each year. It is clear from the information presented here that multiple conserved changes inside the cell and in the ECM contribute to the overall decline in heart function over time, and there is not likely a single molecular target that will reverse this deterioration (Figure 3). Thus, the molecular mechanisms behind the physiological changes of the aging heart must be studied further, and the Drosophila model with its limited redundancy of conserved pathways continues to be valuable for such studies. Moreover, the fly is ideally suited for probing epigenetic plasticity and drift since environmental cues can be tightly controlled and the genetics and epigenetics precisely manipulated. Overall, understanding the mechanisms behind known physiological and molecular hallmarks of cardiac aging is essential to improving heart health in our aging populations.
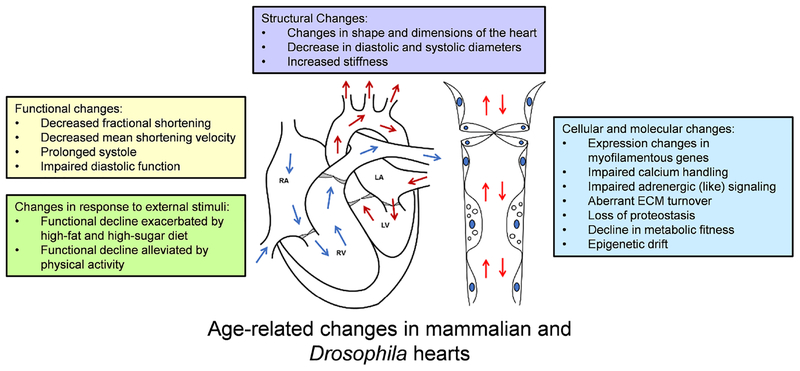
Figure 3: Hallmarks of cardiac aging are shared across species.
The changes reported above are well-conserved, highly interdependent, and contribute to a decline in overall cardiac function over time. Note that some changes may arise in response to others and should be studied in more depth. RA = right atrium, RV = right ventricle, LA = left atrium, and LV = left ventricle. Arrows indicate flow direction.
Highlights:
We review evidence supporting the use of Drosophila melanogaster as a model for studying age-associated changes to cardiac function in humans.
The review highlights important organ, tissue, cellular, and molecular similarities that exist between Drosophila and mammalian models.
There is specific reference to the technical and genetic tractability of Drosophila as a model for ageing studies, especially evolutionarily-conserved phenotypes associated with the heart’s contractility, calcium handling, extracellular matrix deposition, proteostasis, metabolism, and epigenetics.
Acknowledgements
This work was supported by NHLBI R01HL124091 (A.C.), NIA P01 AG031862 (P.A.), R01 HL54732 (R.B.), P01 AG033456 (R.B.), institutional support from Cabrini University (A.C.B.) and a British Heart Foundation Fellowship to PSH (FS_13 17_29905).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.López-Otín C, Blasco MA, Partridge L, et al. (2013) The hallmarks of aging. Cell 153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Go AS, Mozaffarian D, Roger VL, et al. (2014) Heart disease and stroke statistics−-2014 update: a report from the American Heart Association. Circulation 129:e28–e292. doi: 10.1161/01.cir.0000441139.02102.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barquera S, Pedroza-Tobías A, Medina C, et al. (2015) Global overview of the epidemiology of atherosclerotic cardiovascular disease. Arch. Med. Res 46:328–338 [DOI] [PubMed] [Google Scholar]
- 4.Heidenreich PA, Trogdon JG, Khavjou OA, et al. (2011) Forecasting the future of cardiovascular disease in the United States: A policy statement from the American Heart Association. Circulation 123:933–944. doi: 10.1161/CIR.0b013e31820a55f5 [DOI] [PubMed] [Google Scholar]
- 5.Schmid T (2015) Costs of treating cardiovascular events in Germany: a systematic literature review. Health Econ. Rev 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai D-F, Chen T, Johnson SC, et al. (2012) Cardiac aging: From molecular mechanisms to significance in human health and disease. Antioxid Redox Signal 16:1492–1526. doi: 10.1089/ars.2011.4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lakatta EG, Levy D (2003) Arterial and cardiac aging: Major shareholders in cardiovascular disease enterprises: Part I: Aging arteries: A “set up” for vascular disease. Circulation 107:139–146 [DOI] [PubMed] [Google Scholar]
- 8.Strait JB, Lakatta EG (2012) Aging-associated cardiovascular changes and their relationship to heart failure. Heart Fail. Clin 8:143–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiLoreto R, Murphy CT (2015) The cell biology of aging. Mol Biol Cell 26:4524–4531. doi: 10.1091/mbc.E14-06-1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wessells R, Fitzgerald E, Piazza N, et al. (2009) d4eBP acts downstream of both dTOR and dFoxo to modulate cardiac functional aging in Drosophila. Aging Cell 8:542–552. doi: 10.1111/j.1474-9726.2009.00504.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ocorr K, Akasaka T, Bodmer R (2007) Age-related cardiac disease model of Drosophila. Mech Ageing Dev 128:112–116. doi: 10.1016/j.mad.2006.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishimura M, Kumsta C, Kaushik G, et al. (2014) A dual role for integrin-linked kinase and β1-integrin in modulating cardiac aging. Aging Cell 13:431–440. doi: 10.1111/acel.12193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaushik G, Spenlehauer A, Sessions AO, et al. (2015) Vinculin network-mediated cytoskeletal remodeling regulates contractile function in the aging heart. Sci Transl Med 7:. doi: 10.1126/scitranslmed.aaa5843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cammarato A, Ahrens CH, Alayari NN, et al. (2011) A mighty small heart: The cardiac proteome of adult Drosophila melanogaster. PLoS One 6:e18497. doi: 10.1371/journal.pone.0018497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blice-Baum AC, Zambon AC, Kaushik G, et al. (2017) Modest overexpression of FOXO maintains cardiac proteostasis and ameliorates age-associated functional decline. Aging Cell 16:93–103. doi: 10.1111/acel.12543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu Y, Comjean A, Perrimon N, Mohr SE (2017) The Drosophila Gene Expression Tool (DGET) for expression analyses. BMC Bioinformatics 18:. doi: 10.1186/s12859-017-1509-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401–15. doi: 10.1101/lm.1331809 [DOI] [PubMed] [Google Scholar]
- 18.Brand AH, Manoukian AS, Perrimon N (1994) Ectopic expression in Drosophila. Methods Cell Biol 44:635–654. doi: 10.1016/S0091-679X(08)60936-X [DOI] [PubMed] [Google Scholar]
- 19.Pfeiffer BD, Ngo TTB, Hibbard KL, et al. (2010) Refinement of tools for targeted gene expression in Drosophila. Genetics 186:735–755. doi: 10.1534/genetics.110.119917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rotstein B, Paululat A (2016) On the morphology of the Drosophila heart. J Cardiovasc Dev Dis 3:15. doi: 10.3390/jcdd3020015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dulcis D (2005) Glutamatergic innervation of the heart initiates retrograde contractions in adult Drosophila melanogaster. J Neurosci. doi: 10.1523/JNEUROSCI.2906-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wasserthal LT (2007) Drosophila flies combine periodic heartbeat reversal with a circulation in the anterior body mediated by a newly discovered anterior pair of ostial valves and `venous’ channels. J Exp Biol. doi: 10.1242/jeb.007864 [DOI] [PubMed] [Google Scholar]
- 23.Cammarato A, Dambacher CM, Knowles AF, et al. (2008) Myosin transducer mutations differentially affect motor function, myofibril structure, and the performance of skeletal and cardiac muscles. Mol Biol Cell 19:553–62. doi: 10.1091/mbc.E07-09-0890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cannon L, Zambon AC, Cammarato A, et al. (2017) Expression patterns of cardiac aging in Drosophila. Aging Cell 16:82–92. doi: 10.1111/acel.12559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolf MJ, Amrein H, Izatt JA, et al. (2006) Drosophila as a model for the identification of genes causing adult human heart disease. Proc Natl Acad Sci U S A 103:1394–9. doi: 10.1073/pnas.0507359103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaushik G, Zambon AC, Fuhrmann A, et al. (2012) Measuring passive myocardial stiffness in Drosophila melanogaster to investigate diastolic dysfunction. J Cell Mol Med 16:1656–1662. doi: 10.1111/j.1582-4934.2011.01517.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ren J, Sowers JR, Zhang Y (2018) Metabolic stress, autophagy, and cardiovascular aging: from pathophysiology to therapeutics. Trends Endocrinol. Metab [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu X, Pang J, Chen Y, et al. (2016) Macrophage migration inhibitory factor (MIF) deficiency exacerbates aging-induced cardiac remodeling and dysfunction despite improved inflammation: Role of autophagy regulation. Sci Rep. doi: 10.1038/srep22488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piazza N, Gosangi B, Devilla S, et al. (2009) Exercise-training in young Drosophila melanogaster reduces age-related decline in mobility and cardiac performance. PLoS One 4:. doi: 10.1371/journal.pone.0005886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diop S, Bisharat-Kernizan J, Birse R, et al. (2015) PGC-1/spargel counteracts High-fat-diet-induced obesity and cardiac lipotoxicity downstream of TOR and brummer ATGL lipase. Cell Rep 10:1572–1584. doi: 10.1016/j.celrep.2015.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Birse RT, Choi J, Reardon K, et al. (2010) High-fat-diet-induced obesity and heart dysfunction are regulated by the TOR pathway in Drosophila. Cell Metab 12:533–544. doi: 10.1016/j.cmet.2010.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olivetti G, Melissari M, Capasso JM, Anversa P (1991) Cardiomyopathy of the aging human heart. Myocyte loss and reactive cellular hypertrophy. Circ Res 68:1560–1568. doi: 10.1161/01.RES.68.6.1560 [DOI] [PubMed] [Google Scholar]
- 33.Gerstenblith G, Frederiksen J, Yin FC, et al. (1977) Echocardiographic assessment of a normal adult aging population. Circulation 56:273–278. doi: 10.1161/01.CIR.56.2.273 [DOI] [PubMed] [Google Scholar]
- 34.Ganau A, Saba PS, Roman MJ, et al. (1995) Ageing induces left ventricular concentric remodelling in normotensive subjects. J Hypertens 13:1818–22. doi: 10.1097/00004872-199512010-00058 [DOI] [PubMed] [Google Scholar]
- 35.Capasso JM, Palackal T, Olivetti G, Anversa P (1990) Severe myocardial dysfunction induced by ventricular remodeling in aging rat hearts. Am J Physiol 259:H1086–96 [DOI] [PubMed] [Google Scholar]
- 36.Dai DF, Santana LF, Vermulst M, et al. (2009) Overexpression of catalase targeted to mitochondria attenuates murine cardiac aging. Circulation 119:2789–2797. doi: 10.1161/CIRCULATIONAHA.108.822403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fraticelli A, Josephson R, Danziger R, et al. (1989) Morphological and contractile characteristics of rat cardiac myocytes from maturation to senescence. Am J Physiol 257:H259–H265 [DOI] [PubMed] [Google Scholar]
- 38.Gardin JM, Siscovick D, Anton-Culver H, et al. (1995) Sex, age, and disease affect echocardiographic left ventricular mass and systolic function in the free-living elderly: The cardiovascular health study. Circulation 91:1739–1748. doi: 10.1161/01.CIR.91.6.1739 [DOI] [PubMed] [Google Scholar]
- 39.Olgar Y, Degirmenci S, Turak AT, et al. (2018) Aging related functional and structural changes in the heart and aorta: MitoTEMPO improves aged-cardiovascular performance. Exp Gerontol 110:172–181 [DOI] [PubMed] [Google Scholar]
- 40.Kitzman DW, Scholz DG, Hagen PT, et al. (1988) Age-related changes in normal human hearts during the first 10 decades of life. Part II (Maturity): A quantitative anatomic study of 765 specimens from subjects 20 to 99 years old. Mayo Clin Proc 63:137–146. doi: 10.1016/S0025-6196(12)64946-5 [DOI] [PubMed] [Google Scholar]
- 41.Steenman M, Lande G (2017) Cardiac aging and heart disease in humans. Biophys. Rev 9:131–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng S, Fernandes VRS, Bluemke DA, et al. (2009) Age-related left ventricular remodeling and associated risk for cardiovascular outcomes / Clinical perspective. Circ Cardiovasc Imaging 2:191–198. doi: 10.1161/CIRCIMAGING.108.819938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu L, Daniels J, Glaser AE, Wolf MJ (2013) Raf-mediated cardiac hypertrophy in adult Drosophila. Dis Model Mech 6:964–976. doi: 10.1242/dmm.011361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ocorr K, Reeves NL, Wessells RJ, et al. (2007) KCNQ potassium channel mutations cause cardiac arrhythmias in Drosophila that mimic the effects of aging. Proc Natl Acad Sci 104:3943–3948. doi: 10.1073/pnas.0609278104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strait JB, Lakatta EG (2012) Cardiac aging: From humans to molecules. In: Muscle; pp 639–659 [Google Scholar]
- 46.O’Neill L, Holbrook NJ, Fargnoli J, Lakatta EG (1991) Progressive changes from young adult age to senescence in mRNA for rat cardiac myosin heavy chain genes. Cardioscience 2:1–5 [PubMed] [Google Scholar]
- 47.Svanborg A (1997) Age-related changes in cardiac physiology. Can they be postponed or treated by drugs? Drugs and Aging 10:463–472 [DOI] [PubMed] [Google Scholar]
- 48.Lakatta EG, Sollott SJ (2002) Perspectives on mammalian cardiovascular aging: Humans to molecules. In: Comparative Biochemistry and Physiology - A Molecular and Integrative Physiology. pp 699–721 [DOI] [PubMed] [Google Scholar]
- 49.Buttrick P, Malhotra A, Factor S, et al. (1991) Effect of aging and hypertension on myosin biochemistry and gene expression in the rat heart. Circ Res 68:645–652. doi: 10.1161/01.RES.68.3.645 [DOI] [PubMed] [Google Scholar]
- 50.Wei JY, Spurgeon HA, Lakatta EG (1984) Excitation-contraction in rat myocardium: alterations with adult aging. Am J Physiol 246:H784–91 [DOI] [PubMed] [Google Scholar]
- 51.Lakatta EG (1987) Cardiac muscle changes in senescence. Annu Rev Physiol 49:519–531 [DOI] [PubMed] [Google Scholar]
- 52.Orchard CH, Lakatta EG (1985) Intracellular calcium transients and developed tension in rat heart muscle. A mechanism for the negative interval-strength relationship. J Gen Physiol 86:637–51. doi: 10.1085/jgp.86.5.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lakatta EG, Spurgeon HA, Janczewski AM (2014) Changes in the heart that accompany advancing age: Humans to molecules. In: Aging and Heart Failure: Mechanisms and Management. pp 319–337 [Google Scholar]
- 54.Fares E, Howlett SE (2010) Effect of age on cardiac excitation-contraction coupling. Clin. Exp. Pharmacol. Physiol 37:1–7 [DOI] [PubMed] [Google Scholar]
- 55.Feridooni HA, Dibb KM, Howlett SE (2015) How cardiomyocyte excitation, calcium release and contraction become altered with age. J. Mol. Cell. Cardiol 83:62–72 [DOI] [PubMed] [Google Scholar]
- 56.Froehlich JP, Lakatta EG, Beard E, et al. (1978) Studies of sarcoplasmic reticulum function and contraction duration in young adult and aged rat myocardium. J Mol Cell Cardiol 10:427–438. doi: 10.1016/0022-2828(78)90364-4 [DOI] [PubMed] [Google Scholar]
- 57.Babušíková E, Lehotský J, Dobrota D, et al. (2012) Age-associated changes in Ca2+-atpase and oxidative damage in sarcoplasmic reticulum of rat heart. Physiol Res 61:453–460 [DOI] [PubMed] [Google Scholar]
- 58.Lompre AM, Lambert F, Lakatta EG, Schwartz K (1991) Expression of sarcoplasmic reticulum Ca2+-ATPase and calsequestrin genes in rat heart during ontogenic development and aging. Circ Res 69:1380–1388. doi: 10.1161/01.RES.69.5.1380 [DOI] [PubMed] [Google Scholar]
- 59.Santalla M, Valverde CA, Harnichar E, et al. (2014) Aging and CaMKII alter intracellular Ca2+ transients and heart rhythm in Drosophila melanogaster. PLoS One 9:. doi: 10.1371/journal.pone.0101871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Limpitikul WB, Viswanathan MC, O’Rourke B, et al. (2018) Conservation of cardiac L-type Ca2+channels and their regulation in Drosophila: A novel genetically-pliable channelopathic model. J Mol Cell Cardiol 119:64–74. doi: 10.1016/j.yjmcc.2018.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Paternostro G, Vignola C, Bartsch DU, et al. (2001) Age-associated cardiac dysfunction in Drosophila melanogaster. Circ Res 88:1053–1058. doi: 10.1161/hh1001.090857 [DOI] [PubMed] [Google Scholar]
- 62.Lakatta E (1993) Cardiovascular regulatory mechanisms in advanced age. Physiol Rev 73:37–49. doi: 10.1152/physrev.1993.73.2.413 [DOI] [PubMed] [Google Scholar]
- 63.Xiao RP, Spurgeon HA, O’Connor F, Lakatta EG (1994) Age-associated changes in beta-adrenergic modulation on rat cardiac excitation-contraction coupling. J Clin Invest 94:2051–9. doi: 10.1172/JCI117559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakou ES, Parthenakis FI, Kallergis EM, et al. (2016) Healthy aging and myocardium: A complicated process with various effects in cardiac structure and physiology. Int. J. Cardiol 209:167–175 [DOI] [PubMed] [Google Scholar]
- 65.Ferrara N, Komici K, Corbi G, et al. (2014) β-adrenergic receptor responsiveness in aging heart and clinical implications. Front. Physiol 4 January [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Monnier V, Iché-Torres M, Rera M, et al. (2012) dJun and Vri/dNFIL3 are major regulators of cardiac aging in Drosophila. PLoS Genet 8:. doi: 10.1371/journal.pgen.1003081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bryg RJ, Williams GA, Labovitz AJ (1987) Effect of aging on left ventricular diastolic filling in normal subjects. Am J Cardiol 59:. doi: 10.1016/0002-9149(87)91136-2 [DOI] [PubMed] [Google Scholar]
- 68.Sharma K, Kass DA (2014) Heart failure with preserved ejection fraction: Mechanisms, clinical features, and therapies. Circ. Res 115:79–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sheydina A, Riordon DR, Boheler KR (2011) Molecular mechanisms of cardiomyocyte aging. Clin Sci 121:315–329. doi: 10.1042/CS20110115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lakatta EG, Yin FC (1982) Myocardial aging: functional alterations and related cellular mechanisms. Am J Physiol Circ Physiol 242:H927–H941. doi: 10.1152/ajpheart.1982.242.6.H927 [DOI] [PubMed] [Google Scholar]
- 71.Dai DF, Rabinovitch PS, Ungvari Z (2012) Mitochondria and cardiovascular aging. Circ. Res 110:1109–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Asif M, Egan J, Vasan S, et al. (2000) An advanced glycation endproduct cross-link breaker can reverse age-related increases in myocardial stiffness. Proc Natl Acad Sci 97:2809–2813. doi: 10.1073/pnas.040558497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arbab-Zadeh A, Dijk E, Prasad A, et al. (2004) Effect of aging and physical activity on left ventricular compliance. Circulation 110:1799–1805. doi: 10.1161/01.CIR.0000142863.71285.74 [DOI] [PubMed] [Google Scholar]
- 74.Kaushik G, Fuhrmann A, Cammarato A, Engler AJ (2011) In situ mechanical analysis of myofibrillar perturbation and aging on soft, bilayered Drosophila myocardium. Biophys J 101:2629–2637. doi: 10.1016/j.bpj.2011.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Van Der Velden J (2011) Diastolic myofilament dysfunction in the failing human heart. Pflugers Arch. Eur. J. Physiol 462:155–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ouzounian M, Lee DS (2008) Diastolic heart failure: Mechanisms and controversies. Nat. Clin. Pract. Cardiovasc. Med 5:375–386 [DOI] [PubMed] [Google Scholar]
- 77.Kass D, Bronzwaer J, Paulus W (2004) What mechanisms underlie diastolic dysfunction in heart failure? Circ Res 94:1533–1542 [DOI] [PubMed] [Google Scholar]
- 78.Borbely A, Papp Z, Edes I, Paulus WJ (2009) Molecular determinants of heart failure with normal left ventricular ejection fraction. Pharmacol Rep 61:139–145 [DOI] [PubMed] [Google Scholar]
- 79.Adams JC (2018) Matricellular proteins: Functional insights from non-mammalian animal models. In: Current Topics in Developmental Biology. pp 39–105 [DOI] [PubMed] [Google Scholar]
- 80.Hughes C, Jacobs J (2017) Dissecting the role of the extracellular matrix in heart disease: Lessons from the Drosophila genetic model. Vet Sci 4:24. doi: 10.3390/vetsci4020024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meschiari CA, Ero OK, Pan H, et al. (2017) The impact of aging on cardiac extracellular matrix. GeroScience 39:7–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu L, Guo J, Hua Y, et al. (2017) Cardiac fibrosis in the ageing heart: Contributors and mechanisms. Clin Exp Pharmacol Physiol 44:55–63. doi: 10.1111/1440-1681.12753 [DOI] [PubMed] [Google Scholar]
- 83.Larocca G, Aspelund T, Greve AM, et al. (2017) Fibrosis as measured by the biomarker, tissue inhibitor metalloproteinase-1, predicts mortality in Age Gene Environment Susceptibility-Reykjavik (AGES-Reykjavik) Study. Eur Heart J 38:3423–3430. doi: 10.1093/eurheartj/ehx510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cannata A, Merlo M, Artico J, et al. (2018) Cardiovascular aging: the unveiled enigma from bench to bedside. J Cardiovasc Med [DOI] [PubMed] [Google Scholar]
- 85.De Castro Brás LE, Toba H, Baicu CF, et al. (2014) Age and SPARC change the extracellular matrix composition of the left ventricle. Biomed Res Int 2014:. doi: 10.1155/2014/810562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bradshaw AD, Baicu CF, Rentz TJ, et al. (2010) Age-dependent alterations in fibrillar collagen content and myocardial diastolic function: role of SPARC in post-synthetic procollagen processing. Am J Physiol Heart Circ Physiol 298:H614–H622. doi: 10.1152/ajpheart.00474.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yasothornsrikul S, Davis WJ, Cramer G, et al. (1997) viking: Identification and characterization of a second type IV collagen in Drosophila. Gene 198:17–25. doi: 10.1016/S0378-1119(97)00274-6 [DOI] [PubMed] [Google Scholar]
- 88.Harpaz N, Ordan E, Ocorr K, et al. (2013) Multiplexin promotes heart but not aorta morphogenesis by polarized enhancement of Slit/Robo activity at the heart lumen. PLoS Genet 9:. doi: 10.1371/journal.pgen.1003597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chartier A, Zaffran S, Astier M, et al. (2002) Pericardin, a Drosophila type IV collagen-like protein is involved in the morphogenesis and maintenance of the heart epithelium during dorsal ectoderm closure. Development 129:3241–3253 [DOI] [PubMed] [Google Scholar]
- 90.Wilmes AC, Klinke N, Rotstein B, et al. (2018) Biosynthesis and assembly of the Collagen IV-like protein Pericardin in Drosophila melanogaster. Biol Open 7:bio030361. doi: 10.1242/bio.030361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vaughan L, Marley R, Miellet S, Hartley PS (2017) The impact of SPARC on age-related cardiac dysfunction and fibrosis in Drosophila. Exp. Gerontol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sessions AO, Kaushik G, Parker S, et al. (2017) Extracellular matrix downregulation in the Drosophila heart preserves contractile function and improves lifespan. Matrix Biol 62:15–27. doi: 10.1016/j.matbio.2016.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hartley PS, Motamedchaboki K, Bodmer R, Ocorr K (2016) SPARC-dependent cardiomyopathy in drosophila. Circ Cardiovasc Genet 9:119–129. doi: 10.1161/CIRCGENETICS.115.001254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gehwolf R, Wagner A, Lehner C, et al. (2016) Pleiotropic roles of the matricellular protein Sparc in tendon maturation and ageing. Sci Rep 6:. doi: 10.1038/srep32635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Toba H, de Castro Brás LE, Baicu CF, et al. (2015) Secreted protein acidic and rich in cysteine facilitates age-related cardiac inflammation and macrophage M1 polarization. Am J Physiol Physiol 308:C972–C982. doi: 10.1152/ajpcell.00402.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kulak NA, Geyer PE, Mann M (2017) Loss-less nano-fractionator for high sensitivity, high coverage proteomics. Mol Cell Proteomics 16:694–705. doi: 10.1074/mcp.O116.065136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Klaips CL, Jayaraj GG, Hartl FU (2018) Pathways of cellular proteostasis in aging and disease. J. Cell Biol 217:51–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Brehme M, Voisine C, Rolland T, et al. (2014) A chaperome subnetwork safeguards proteostasis in aging and neurodegenerative disease. Cell Rep 9:1135–1150. doi: 10.1016/j.celrep.2014.09.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.García-Prat L, Martínez-Vicente M, Perdiguero E, et al. (2016) Autophagy maintains stemness by preventing senescence. Nature 529:37–42. doi: 10.1038/nature16187 [DOI] [PubMed] [Google Scholar]
- 100.Li W, Bengtson MH, Ulbrich A, et al. (2008) Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS One 3:. doi: 10.1371/journal.pone.0001487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nijman SMB, Luna-Vargas MPA, Velds A, et al. (2005) A genomic and functional inventory of deubiquitinating enzymes. Cell 123:773–786 [DOI] [PubMed] [Google Scholar]
- 102.Sowa ME, Bennett EJ, Gygi SP, Harper JW (2009) Defining the human deubiquitinating enzyme interaction landscape. Cell 138:389–403. doi: 10.1016/j.cell.2009.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Varshavsky A (2012) The ubiquitin system, an immense realm. Annu Rev Biochem 81:167–176. doi: 10.1146/annurev-biochem-051910-094049 [DOI] [PubMed] [Google Scholar]
- 104.Sala AJ, Bott LC, Morimoto RI (2017) Shaping proteostasis at the cellular, tissue, and organismal level. J. Cell Biol 216:1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim YE, Hipp MS, Bracher A, et al. (2013) Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem 82:323–355. doi: 10.1146/annurevbiochem-060208-092442 [DOI] [PubMed] [Google Scholar]
- 106.Hipp MS, Park SH, Hartl UU (2014) Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 24:506–514 [DOI] [PubMed] [Google Scholar]
- 107.Van Berlo JH, Molkentin JD (2014) An emerging consensus on cardiac regeneration. Nat. Med 20:1386–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang X, Robbins J (2014) Proteasomal and lysosomal protein degradation and heart disease. J. Mol. Cell. Cardiol 71:16–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Quarles EK, Dai DF, Tocchi A, et al. (2015) Quality control systems in cardiac aging. Ageing Res Rev 23:101–115. doi: 10.1016/j.arr.2015.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lavandero S, Troncoso R, Rotherme BA, et al. (2013) Cardiovascular autophagy: Concepts, controversies, and perspectives. Autophagy 9:1455–1466 [DOI] [PubMed] [Google Scholar]
- 111.Willis M, Patterson C (2013) Proteotoxicity and cardiac dysfunction — Alzheimer’s disease of the heart? N Engl J Med 368:455–464. doi: doi: 10.1056/nejmra1106180 [DOI] [PubMed] [Google Scholar]
- 112.Andreou AM, Tavernarakis N (2010) Protein metabolism and homeostasis in aging. Preface. [PubMed] [Google Scholar]
- 113.Bulteau AL, Szweda LI, Friguet B (2002) Age-dependent declines in proteasome activity in the heart. Arch. Biochem. Biophys 397:298–304 [DOI] [PubMed] [Google Scholar]
- 114.Lin S, Wang Y, Zhang X, et al. (2016) HSP27 alleviates cardiac aging in mice via a mechanism involving antioxidation and mitophagy activation. Oxid Med Cell Longev 2016:. doi: 10.1155/2016/2586706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Powell SR (2006) The ubiquitin-proteasome system in cardiac physiology and pathology. Am J Physiol Heart Circ Physiol 291:H1–H19. doi: 10.1152/ajpheart.00062.2006 [DOI] [PubMed] [Google Scholar]
- 116.Tanskanen M, Peuralinna T, Polvikoski T, et al. (2008) Senile systemic amyloidosis affects 25% of the very aged and associates with genetic variation in alpha2-macroglobulin and tau: A population-based autopsy study. Ann Med 40:232–239. doi: 10.1080/07853890701842988 [DOI] [PubMed] [Google Scholar]
- 117.Weekes J, Morrison K, Mullen A, et al. (2003) Hyperubiquitination of proteins in dilated cardiomyopathy. Proteomics 3:208–216. doi: 10.1002/pmic.200390029 [DOI] [PubMed] [Google Scholar]
- 118.Tsukamoto O, Minamino T, Okada KI, et al. (2006) Depression of proteasome activities during the progression of cardiac dysfunction in pressure-overloaded heart of mice. Biochem Biophys Res Commun 340:1125–1133. doi: 10.1016/j.bbrc.2005.12.120 [DOI] [PubMed] [Google Scholar]
- 119.Selcen D, Ohno K, Engel AG (2004) Myofibrillar myopathy: Clinical, morphological and genetic studies in 63 patients. Brain 127:439–451. doi: 10.1093/brain/awh052 [DOI] [PubMed] [Google Scholar]
- 120.Predmore JM, Wang P, Davis F, et al. (2010) Ubiquitin proteasome dysfunction in human hypertrophic and dilated cardiomyopathies. Circulation 121:997–1004. doi: 10.1161/CIRCULATIONAHA.109.904557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Goldfarb LG, Vicart P, Goebel HH, Dalakas MC (2004) Desmin myopathy. Brain 127:723–734 [DOI] [PubMed] [Google Scholar]
- 122.Taneike M, Yamaguchi O, Nakai A, et al. (2010) Inhibition of autophagy in the heart induces age-related cardiomyopathy. Autophagy 6:600–606. doi: 10.4161/auto.6.5.11947 [DOI] [PubMed] [Google Scholar]
- 123.Huang C, Yitzhaki S, Perry CN, et al. (2010) Autophagy induced by ischemic preconditioning is essential for cardioprotection. J Cardiovasc Transl Res 3:365–373. doi: 10.1007/s12265-010-9189-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shirakabe A, Ikeda Y, Sciarretta S, et al. (2016) Aging and autophagy in the heart. Circ. Res 118:1563–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Alcendor RR, Gao S, Zhai P, et al. (2007) Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ Res 100:1512–1521. doi: 10.1161/01.RES.0000267723.65696.4a [DOI] [PubMed] [Google Scholar]
- 126.Hariharan N, Maejima Y, Nakae J, et al. (2010) Deacetylation of FoxO by Sirt1 plays an essential role in mediating starvation-induced autophagy in cardiac myocytes. Circ Res 107:1470–1482. doi: 10.1161/CIRCRESAHA.110.227371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ferdous A, Battiprolu PK, Ni YG, et al. (2010) FoxO, autophagy, and cardiac remodeling. J. Cardiovasc. Transl. Res 3:355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gurusamy N, Das DK (2009) Is autophagy a double-edged sword for the heart? Acta Physiol Hung 96:267–76. doi: 10.1556/APhysiol.96.2009.3.2 [DOI] [PubMed] [Google Scholar]
- 129.Salabei JK, Conklin DJ (2013) Cardiovascular autophagy: Crossroads of pathology, pharmacology and toxicology. Cardiovasc. Toxicol 13:220–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rifki OF, Hill JA (2012) Cardiac autophagy: Good with the bad. J. Cardiovasc. Pharmacol 60:248–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nemchenko A, Chiong M, Turer A, et al. (2011) Autophagy as a therapeutic target in cardiovascular disease. J. Mol. Cell. Cardiol 51:584–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Xie M, Morales CR, Lavandero S, Hill JA (2011) Tuning flux: Autophagy as a target of heart disease therapy. Curr Opin Cardiol 26:216–222. doi: 10.1097/HCO.0b013e328345980a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schiattarella GG, Hill JA (2016) Therapeutic targeting of autophagy in cardiovascular disease. J. Mol. Cell. Cardiol 95:86–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Demontis F, Perrimon N (2010) FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell 143:813–825. doi: 10.1016/j.cell.2010.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wessells RJ, Fitzgerald E, Cypser JR, et al. (2004) Insulin regulation of heart function in aging fruit flies. Nat Genet 36:1275–1281. doi: 10.1038/ng1476 [DOI] [PubMed] [Google Scholar]
- 136.Wessells RJ, Bodmer R (2004) Screening assays for heart function mutants in Drosophila. Biotechniques 37:58. [DOI] [PubMed] [Google Scholar]
- 137.Wang C, Wang X (2015) The interplay between autophagy and the ubiquitin-proteasome system in cardiac proteotoxicity. Biochim. Biophys. Acta - Mol. Basis Dis [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hu Y, Flockhart I, Vinayagam A, et al. (2011) An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics 12:357. doi: 10.1186/1471-2105-12-357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.GBD 2015 Obesity Collaborators, Afshin A, Forouzanfar MH, et al. (2017) Health effects of overweight and obesity in 195 countries over 25 years. N Engl J Med 377:13–27. doi: 10.1056/NEJMoa1614362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Villareal DT, Apovian CM, Kushner RF, Klein S (2005) Obesity in older adults: Technical review and position statement of the American Society for Nutrition and NAASO, the Obesity Society. Obes Res 13:1849–1863. doi: 10.1038/oby.2005.228 [DOI] [PubMed] [Google Scholar]
- 141.Canning KL, Brown RE, Jamnik VK, Kuk JL (2014) Relationship between obesity and obesity-related morbidities weakens with aging. Journals Gerontol - Ser A Biol Sci Med Sci 69:87–92. doi: 10.1093/gerona/glt026 [DOI] [PubMed] [Google Scholar]
- 142.Pérez LM, Pareja-Galeano H, Sanchis-Gomar F, et al. (2016) ‘Adipaging’: ageing and obesity share biological hallmarks related to a dysfunctional adipose tissue. J. Physiol 594:3187–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hunter GR, Gower BA, Kane BL (2010) Age related shift in visceral fat. Int J Body Compos Res 8:103–108. doi: 10.1002/nbm.3066.Non-invasive [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wen D, Zheng L, Yang F, et al. (2018) Endurance exercise prevents high-fat-diet induced locomotor impairment, cardiac dysfunction, lifespan shortening, and dSir2 expression decline in aging Drosophila. Exp Gerontol. doi: 10.1016/j.exger.2018.01.017 [DOI] [PubMed] [Google Scholar]
- 145.Jura M, Kozak LP (2016) Obesity and related consequences to ageing. Age (Omaha) 38:. doi: 10.1007/s11357-016-9884-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mellor KM, Ritchie RH, Davidoff AJ, Delbridge LMD (2010) Elevated dietary sugar and the heart: experimental models and myocardial remodeling. Can J Physiol Pharmacol 88:525–540. doi: 10.1139/Y10-005 [DOI] [PubMed] [Google Scholar]
- 147.Murdolo G, Angeli F, Reboldi G, et al. (2015) Left ventricular hypertrophy and obesity: Only a matter of fat? High Blood Press Cardiovasc Prev 22:29–41. doi: 10.1007/s40292-014-0068-x [DOI] [PubMed] [Google Scholar]
- 148.Szczepaniak LS, Dobbins RL, Metzger GJ, et al. (2003) Myocardial triglycerides and systolic function in humans: in vivo evaluation by localized proton spectroscopy and cardiac imaging. Magn Reson Med 49:417–423. doi: 10.1002/mrm.10372 [DOI] [PubMed] [Google Scholar]
- 149.Christoffersen C, Bollano E, Lindegaard MLS, et al. (2003) Cardiac lipid accumulation associated with diastolic dysfunction in obese mice. Endocrinology 144:3483–3490. doi: 10.1210/en.2003-0242 [DOI] [PubMed] [Google Scholar]
- 150.Buchanan J, Mazumder PK, Hu P, et al. (2005) Reduced cardiac efficiency and altered substrate metabolism precedes the onset of hyperglycemia and contractile dysfunction in two mouse models of insulin resistance and obesity. Endocrinology 146:5341–5349. doi: 10.1210/en.2005-0938 [DOI] [PubMed] [Google Scholar]
- 151.Ku CS, Lin SL, Wang DJ, et al. (1994) Left ventricular filling in young normotensive obese adults. Am J Cardiol 73:613–615. doi: 10.1016/0002-9149(94)90347-6 [DOI] [PubMed] [Google Scholar]
- 152.Kaltman AJ, Goldring RM (1976) Role of circulatory congestion in the cardiorespiratory failure of obesity. Am J Med 60:645–653. doi: 10.1016/0002-9343(76)90499-X [DOI] [PubMed] [Google Scholar]
- 153.Messerli FH, Nunez BD, Ventura HO, Snyder DW (1987) Overweight and sudden death: increased ventricular ectopy in cardiopathy of obesity. Arch Intern Med 147:1725–1728. doi: 10.1001/archinte.1987.00370100039008 [DOI] [PubMed] [Google Scholar]
- 154.Gill S, Le HD, Melkani GC, Panda S (2015) Time-restricted feeding attenuates age-related cardiac decline in Drosophila. Science (80-) 347:1265–1269. doi: 10.1126/science.1256682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Houtkooper RH, Argmann C, Houten SM, et al. (2011) The metabolic footprint of aging in mice. Sci Rep 1:. doi: 10.1038/srep00134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Dai DF, Karunadharma PP, Chiao YA, et al. (2014) Altered proteome turnover and remodeling by short-term caloric restriction or rapamycin rejuvenate the aging heart. Aging Cell 13:529–539. doi: 10.1111/acel.12203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Grant JE, Bradshaw AD, Schwacke JH, et al. (2009) Quantification of protein expression changes in the aging left ventricle of Rattus norvegicus. J Proteome Res. doi: 10.1021/pr900297f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Doenst T, Nguyen TD, Abel ED (2013) Cardiac metabolism in heart failure: implications beyond ATP production. Circ Res 113:709–724. doi: 10.1161/CIRCRESAHA.113.300376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dávila-Román VG, Vedala G, Herrero P, et al. (2002) Altered myocardial fatty acid and glucose metabolism in idiopathic dilated cardiomyopathy. J Am Coll Cardiol 40:271–277 [DOI] [PubMed] [Google Scholar]
- 160.Preston CC, Oberlin AS, Holmuhamedov EL, et al. (2008) Aging-induced alterations in gene transcripts and functional activity of mitochondrial oxidative phosphorylation complexes in the heart. Mech Ageing Dev. doi: 10.1016/j.mad.2008.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ma Z, Wang H, Cai Y, et al. (2018) Epigenetic drift of H3K27me3 in aging links glycolysis to healthy longevity in Drosophila. Elife 7:e35368. doi: 10.7554/eLife.35368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lushchak O, Strilbytska O, Piskovatska V, et al. (2017) The role of the TOR pathway in mediating the link between nutrition and longevity. Mech. Ageing Dev. 164:127–138 [DOI] [PubMed] [Google Scholar]
- 163.Di Angelantonio E, Bhupathiraju SN, Hu FB, et al. (2017) Body-mass index and all-cause mortality – Authors’ reply. Lancet 389:2285–2286 [DOI] [PubMed] [Google Scholar]
- 164.Woodcock KJ, Kierdorf K, Pouchelon CA, et al. (2015) Macrophage-derived upd3 cytokine causes impaired glucose homeostasis and reduced lifespan in Drosophila fed a lipid-rich diet. Immunity 42:133–144. doi: 10.1016/j.immuni.2014.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mazucanti C, Cabral-Costa J, Vasconcelos A, et al. (2015) Longevity pathways (mTOR, SIRT, Insulin/IGF-1) as key modulatory targets on aging and neurodegeneration. Curr Top Med Chem 15:2116–2138. doi: 10.2174/1568026615666150610125715 [DOI] [PubMed] [Google Scholar]
- 166.Zhang D, Contu R, Latronico MVG, et al. (2010) MTORC1 regulates cardiac function and myocyte survival through 4E-BP1 inhibition in mice. J Clin Invest 120:2805–2816. doi: 10.1172/JCI43008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sciarretta S, Volpe M, Sadoshima J (2014) mTOR Signaling in cardiac physiology and disease. Circ Res 114:549–564. doi: 10.1161/CIRCRESAHA.114.302022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Shioi T, McMullen JR, Tarnavski O, et al. (2003) Rapamycin attenuates load-induced cardiac hypertrophy in mice. Circulation 107:1664–1670. doi: 10.1161/01.CIR.0000057979.36322.88 [DOI] [PubMed] [Google Scholar]
- 169.Sciarretta S, Zhai P, Shao D, et al. (2012) Rheb is a critical regulator of autophagy during myocardial ischemia: pathophysiological implications in obesity and metabolic syndrome. Circulation 125:1134–1146. doi: 10.1161/CIRCULATIONAHA.111.078212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lee JH, Budanov AV, Park EJ, et al. (2010) Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science (80-) 327:1223–1228. doi: 10.1126/science.1182228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Saxton RA, Sabatini DM (2017) mTOR Signaling in growth, metabolism, and disease. Cell [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Altintas O, Park S, Lee SJV. (2016) The role of insulin/IGF-1 signaling in the longevity of model invertebrates, C. elegans and D. melanogaster. BMB Rep 49:81–92. doi: 10.5483/BMBRep.2016.49.2.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Shimokawa I (2017) Growth hormone and IGF-1 axis in aging and longevity In: Rattan S, Sharma R (eds) Hormones in Ageing and Longevity. Springer, pp 91–106 [Google Scholar]
- 174.Carbone S, Mauro AG, Mezzaroma E, et al. (2015) A high-sugar and high-fat diet impairs cardiac systolic and diastolic function in mice. Int J Cardiol 198:66–69. doi: 10.1016/j.ijcard.2015.06.136 [DOI] [PubMed] [Google Scholar]
- 175.Musselman LP, Fink JL, Narzinski K, et al. (2011) A high-sugar diet produces obesity and insulin resistance in wild-type Drosophila. Dis Model Mech 4:842–849. doi: 10.1242/dmm.007948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Na J, Musselman LP, Pendse J, et al. (2013) A Drosophila model of high sugar diet-induced cardiomyopathy. PLoS Genet 9:e1003175. doi: 10.1371/journal.pgen.1003175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Dobson AJ, Ezcurra M, Flanagan CE, et al. (2017) Nutritional programming of lifespan by FOXO inhibition on sugar-rich diets. Cell Rep 18:299–306. doi: 10.1016/j.celrep.2016.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bodmer R, Frasch M (2010) Heart development and regeneration, Rosetha N. Elsevier, Amsterdam [Google Scholar]
- 179.Rogina B, Helfand SL (2004) Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci 101:15998–16003. doi: 10.1073/pnas.0404184101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Elgin SCR, Reuter G (2013) Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb Perspect Biol 5:. doi: 10.1101/cshperspect.a017780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Groth A, Rocha W, Verreault A, Almouzni G (2007) Chromatin challenges during DNA replication and repair. Cell 128:721–733 [DOI] [PubMed] [Google Scholar]
- 182.Cheutin T, McNairn AJ, Jenuwein T, et al. (2003) Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science (80-) 299:721–725. doi: 10.1126/science.1078572 [DOI] [PubMed] [Google Scholar]
- 183.Berger SL (2007) The complex language of chromatin regulation during transcription. Nature 447:407–412 [DOI] [PubMed] [Google Scholar]
- 184.Booth LN, Brunet A (2016) The aging epigenome. Mol. Cell 62:728–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Nativio R, Donahue G, Berson A, et al. (2018) Dysregulation of the epigenetic landscape of normal aging in Alzheimer’s disease. Nat Neurosci 21:497–505. doi: 10.1038/s41593-018-0101-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Muka T, Koromani F, Portilla E, et al. (2016) The role of epigenetic modifications in cardiovascular disease: A systematic review. Int. J. Cardiol 212:174–183 [DOI] [PubMed] [Google Scholar]
- 187.Horvath S, Raj K (2018) DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet 19:371–384 [DOI] [PubMed] [Google Scholar]
- 188.Zahn JM, Poosala S, Owen AB, et al. (2007) AGEMAP: A gene expression database for aging in mice. PLoS Genet 3:2326–2337. doi: 10.1371/journal.pgen.0030201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Budovskaya YV, Wu K, Southworth LK, et al. (2008) An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell 134:291–303. doi: 10.1016/j.cell.2008.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Arslan-Ergul A, Adams MM (2014) Gene expression changes in aging Zebrafish (Danio rerio) brains are sexually dimorphic. BMC Neurosci 15:29. doi: 10.1186/1471-2202-15-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bahar R, Hartmann CH, Rodriguez KA, et al. (2006) Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature 441:1011–1014. doi: 10.1038/nature04844 [DOI] [PubMed] [Google Scholar]
- 192.Viñuela A, Brown AA, Buil A, et al. (2018) Age-dependent changes in mean and variance of gene expression across tissues in a twin cohort. Hum Mol Genet 27:732–741. doi: 10.1093/hmg/ddx424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Benayoun BA, Pollina EA, Brunet A (2015) Epigenetic regulation of ageing: Linking environmental inputs to genomic stability. Nat. Rev. Mol. Cell Biol 16:593–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Rakyan VK, Down TA, Maslau S, et al. (2010) Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res 20:434–439. doi: 10.1101/gr.103101.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Teschendorff AE, Menon U, Gentry-Maharaj A, et al. (2010) Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res 20:440–446. doi: 10.1101/gr.103606.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ohm JE, McGarvey KM, Yu X, et al. (2007) A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet 39:237–242. doi: 10.1038/ng1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Fraga MF, Agrelo R, Esteller M (2007) Cross-talk between aging and cancer: The epigenetic language. In: Annals of the New York Academy of Sciences. pp 60–74 [DOI] [PubMed] [Google Scholar]
- 198.Jylhävä J, Pedersen NL, Hägg S (2017) Biological age predictors. EBioMedicine 21:29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Field A, Robertson N, Wang T, et al. (2018) No Title. Mol Cell 71:882–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lind L, Ingelsson E, Sundström J, et al. (2018) Methylation-based estimated biological age and cardiovascular disease. Eur J Clin Invest 48:e12872. doi: 10.1111/eci.12872 [DOI] [PubMed] [Google Scholar]
- 201.Larson K, Yan SJ, Tsurumi A, et al. (2012) Heterochromatin formation promotes longevity and represses ribosomal RNA synthesis. PLoS Genet 8:. doi: 10.1371/journal.pgen.1002473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Dreesen O, Chojnowski A, Ong PF, et al. (2013) Lamin B1 fluctuations have differential effects on cellular proliferation and senescence. J Cell Biol 200:605–617. doi: 10.1083/jcb.201206121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Chen H, Zheng X, Zheng Y (2014) Age-associated loss of lamin-b leads to systemic inflammation and gut hyperplasia. Cell 159:829–843. doi: 10.1016/j.cell.2014.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Shimi T, Butin-Israeli V, Adam SA, et al. (2011) The role of nuclear lamin B1 in cell proliferation and senescence. Genes Dev. doi: 10.1101/gad.179515.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Chen H, Zheng X, Xiao D, Zheng Y (2016) Age-associated de-repression of retrotransposons in the Drosophila fat body, its potential cause and consequence. Aging Cell 15:542–552. doi: 10.1111/acel.12465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Jiang N, Du G, Tobias E, et al. (2013) Dietary and genetic effects on age-related loss of gene silencing reveal epigenetic plasticity of chromatin repression during aging. Aging (Albany NY) 5:813–824. doi: 10.18632/aging.100614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.De Cecco M, Criscione SW, Peckham EJ, et al. (2013) Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell 12:247–256. doi: 10.1111/acel.12047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Orlandi I, Pellegrino Coppola D, Strippoli M, et al. (2017) Nicotinamide supplementation phenocopies SIR2 inactivation by modulating carbon metabolism and respiration during yeast chronological aging. Mech Ageing Dev 161:277–287. doi: 10.1016/j.mad.2016.06.006 [DOI] [PubMed] [Google Scholar]
- 209.Klose RJ, Zhang Y (2007) Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol 8:307–318. doi: 10.1038/nrm2143 [DOI] [PubMed] [Google Scholar]
- 210.Cai L, Sutter BM, Li B, Tu BP (2011) Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell 42:426–437. doi: 10.1016/j.molcel.2011.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Buescher JL, Musselman LP, Wilson CA, et al. (2013) Evidence for transgenerational metabolic programming in Drosophila. Dis Model Mech 6:1123–1132. doi: 10.1242/dmm.011924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Heinrichsen ET, Zhang H, Robinson JE, et al. (2014) Metabolic and transcriptional response to a high-fat diet in Drosophila melanogaster. Mol Metab 3:42–54. doi: 10.1016/j.molmet.2013.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Öst A, Lempradl A, Casas E, et al. (2014) Paternal diet defines offspring chromatin state and intergenerational obesity. Cell 159:1352–1364. doi: 10.1016/j.cell.2014.11.005 [DOI] [PubMed] [Google Scholar]
- 214.Alic N, Andrews TD, Giannakou ME, et al. (2011) Genome‐wide dFOXO targets and topology of the transcriptomic response to stress and insulin signalling. Mol Syst Biol 7:502. doi: 10.1038/msb.2011.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kuzmichev A, Nishioka K, Erdjument-Bromage H, et al. (2002) Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16:2893–905. doi: 10.1101/gad.1035902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Magistretti PJ, Allaman I (2015) A cellular perspective on brain energy metabolism and functional imaging. Neuron [DOI] [PubMed] [Google Scholar]
- 217.Grynberg A, Demaison L (1996) Fatty acid oxidation in the heart. JCardiovascPharmacol. doi: 10.1016/j.electacta.2008.02.085 [DOI] [PubMed] [Google Scholar]
- 218.Siebold AP, Banerjee R, Tie F, et al. (2010) Polycomb Repressive Complex 2 and Trithorax modulate Drosophila longevity and stress resistance. Proc Natl Acad Sci U S A 107:169–174. doi: 10.1073/pnas.0907739107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Loenen WAM (2018) Chapter 3 - S-adenosylmethionine metabolism and aging In: Moskalev A, Vaiserman AM (eds) Epigenetics of Aging and Longevity. Academic Press, Boston, pp 59–93 [Google Scholar]
- 220.Landgraf BJ, McCarthy EL, Booker SJ (2016) Radical S-adenosylmethionine enzymes in human health and disease. Annu Rev Biochem 85:485–514. doi: 10.1146/annurevbiochem-060713-035504 [DOI] [PubMed] [Google Scholar]
- 221.Bonkowski MS, Sinclair DA (2016) Slowing ageing by design: the rise of NAD+ and sirtuin-activating compounds. Nat Rev Mol Cell Biol 17:679–690. doi: 10.1038/nrm.2016.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wen D tai, Zheng L, Ni L, et al. (2016) The expression of CG9940 affects the adaptation of cardiac function, mobility, and lifespan to exercise in aging Drosophila. Exp Gerontol 83:6–14. doi: 10.1016/j.exger.2016.07.006 [DOI] [PubMed] [Google Scholar]
- 223.Kaeberlein M, McVey M, Guarente L (1999) The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev 13:2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Giblin W, Skinner ME, Lombard DB (2014) Sirtuins: Guardians of mammalian healthspan. Trends Genet. 30:271–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Whitaker R, Faulkner S, Miyokawa R, et al. (2013) Increased expression of Drosophila sir2 extends life span in a dose-dependent manner. Aging (Albany NY) 5:682–691. doi: 10.18632/aging.100599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Rogina B, Helfand SL (2004) Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A 101:15998–16003. doi: 10.1073/pnas.0404184101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Nishimura M, Ocorr K, Bodmer R, Cartry J (2011) Drosophila as a model to study cardiac aging. Exp Gerontol 46:326–330. doi: 10.1016/j.exger.2010.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Bodmer R, Venkatesh TV. (1998) Heart development in Drosophila and vertebrates: Conservation of molecular mechanisms. Dev. Genet 22:181–186 [DOI] [PubMed] [Google Scholar]
- 229.Montgomery RL, Davis CA, Potthoff MJ, et al. (2007) Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev 21:1790–1802. doi: 10.1101/gad.1563807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Rogina B, Helfand SL, Frankel S (2002) Longevity regulation by Drosophila Rpd3 deacetylase and caloric restriction. Science (80-) 298:1745. doi: 10.1126/science.1078986 [DOI] [PubMed] [Google Scholar]
- 231.Kim S, Benguria A, Lai C-Y, Jazwinski SM (1999) Modulation of life-span by histone deacetylase genes in Saccharomyces cerevisiae. Mol Biol Cell 10:3125–3136. doi: 10.1091/mbc.10.10.3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Pile L a, Spellman PT, Katzenberger RJ, Wassarman D a (2003) The SIN3 deacetylase complex represses genes encoding mitochondrial proteins: implications for the regulation of energy metabolism. J Biol Chem 278:37840–8. doi: 10.1074/jbc.M305996200 [DOI] [PubMed] [Google Scholar]
- 233.Kopp ZA, Hsieh JL, Li A, et al. (2015) Heart-specific Rpd3 downregulation enhances cardiac function and longevity. Aging (Albany NY) 7:648–663. doi: 10.18632/aging.100806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Passarino G, De Rango F, Montesanto A (2016) Human longevity: Genetics or Lifestyle? It takes two to tango. Immun. Ageing 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Denham J, Marques FZ, O’Brien BJ, Charchar FJ (2014) Exercise: Putting action into our epigenome. Sport. Med 44:189–209 [DOI] [PubMed] [Google Scholar]
- 236.Wallace RG, Twomey LC, Custaud M-A, et al. (2017) The role of epigenetics in cardiovascular health and ageing: A focus on physical activity and nutrition. Mech Ageing Dev. doi: 10.1016/j.mad.2017.11.013 [DOI] [PubMed] [Google Scholar]
- 237.Wang L, Ai D, Zhang N (2017) Exercise benefits coronary heart disease In: Exercise for Cardiovascular Disease Prevention and Treatment. Springer, Singapore, pp 3–7 [Google Scholar]
- 238.Lee DC, Pate RR, Lavie CJ, et al. (2014) Leisure-time running reduces all-cause and cardiovascular mortality risk. J Am Coll Cardiol 64:472–481. doi: 10.1016/j.jacc.2014.04.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Muller-Delp JM, Hotta K, Chen B, et al. (2017) Effects of age and exercise training on coronary microvascular smooth muscle phenotype and function. J Appl Physiol 10.1152/japplphysiol.00459.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Echouffo-Tcheugui JB, Butler J, Yancy CW, Fonarow GC (2015) Association of physical activity or fitness with incident heart failure: A systematic review and meta-analysis. Circ Hear Fail 8:. doi: 10.1161/CIRCHEARTFAILURE.115.002070 [DOI] [PubMed] [Google Scholar]
- 241.Howden EJ, Sarma S, Lawley JS, et al. (2018) Reversing the cardiac effects of sedentary aging in middle age—A randomized controlled trial: Implications for heart failure prevention. Circulation 10.1161/CIRCULATIONAHA.117.030617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Tinkerhess MJ, Ginzberg S, Piazza N, Wessells RJ (2012) Endurance training protocol and longitudinal performance assays for Drosophila melanogaster. J Vis Exp 1–5. doi: 10.3791/3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Sujkowski A, Wessells R (2018) Using Drosophila to understand biochemical and behavioral responses to exercise. Exerc Sport Sci Rev 46:112–120. doi: 10.1249/JES.0000000000000139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Sujkowski A, Bazzell B, Carpenter K, et al. (2015) Endurance exercise and selective breeding for longevity extend Drosophila healthspan by overlapping mechanisms. Aging (Albany NY) 7:535–552. doi: 10.18632/aging.100789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Zheng L, Li QF, Ni L, et al. (2017) Lifetime regular exercise affects the incident of different arrhythmias and improves organismal health in aging female Drosophila melanogaster. Biogerontology 18:97–108. doi: 10.1007/s10522-016-9665-5 [DOI] [PubMed] [Google Scholar]
- 246.Pletcher SD (2009) The modulation of lifespan by perceptual systems. Ann N Y Acad Sci 1170:693–697. doi: 10.1111/j.1749-6632.2009.04926.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Sujkowski A, Ramesh D, Brockmann A, Wessells R (2017) Octopamine drives endurance exercise adaptations in Drosophila. Cell Rep 21:1809–1823. doi: 10.1016/j.celrep.2017.10.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Chang H-C, Guarente L (2014) SIRT1 and other sirtuins in metabolism. Trends Endocrinol Metab 25:138–145. doi: 10.1016/j.tem.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Fernandez-Marcos PJ, Auwerx J (2011) Regulation of PGC-1α, a nodal regulator of mitochondrial biogenesis. In: American Journal of Clinical Nutrition [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Picard M, Gentil BJ, McManus MJ, et al. (2013) Acute exercise remodels mitochondrial membrane interactions in mouse skeletal muscle. J Appl Physiol 115:. doi: 10.1152/japplphysiol.00819.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Scalzo RL, Peltonen GL, Binns SE, et al. (2014) Greater muscle protein synthesis and mitochondrial biogenesis in males compared with females during sprint interval training. FASEB J 28:2705–2714. doi: 10.1096/fj.13-246595 [DOI] [PubMed] [Google Scholar]
- 252.Tao L, Bei Y, Zhang H, et al. (2015) Exercise for the heart: signaling pathways. Oncotarget 6:20773–20784. doi: 10.18632/oncotarget.4770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Tinkerhess MJ, Healy L, Morgan M, et al. (2012) The Drosophila PGC-1α homolog spargel modulates the physiological effects of endurance exercise. PLoS One 7:. doi: 10.1371/journal.pone.0031633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Banerjee KK, Ayyub C, Sengupta S, Kolthur-Seetharam U (2013) Fat body dSir2 regulates muscle mitochondrial physiology and energy homeostasis nonautonomously and mimics the autonomous functions of dSir2 in muscles. Mol Cell Biol 33:252–264. doi: 10.1128/MCB.00976-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Warburton DE, Nicol CW, Bredin SS (2006) Health benefits of physical activity: the evidence. Can Med Assoc J 174:801–809. doi: 10.1503/cmaj.051351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Miller A (1950) The internal anatomy and histology of the imago of Drosophila melanogaster. In: Biology of Drosophila. pp 420–531 [Google Scholar]
- 257.Rizki TM (1978) The circulatory system and associated cells and tissues. In: The Genetics and Biology of Drosophila. pp 397–452 [Google Scholar]
- 258.Lehmacher C, Abeln B, Paululat A (2012) The ultrastructure of Drosophila heart cells. Arthropod Struct Dev 41:459–474. doi: 10.1016/j.asd.2012.02.002 [DOI] [PubMed] [Google Scholar]