Abstract
Ewing’s sarcoma (EWS) is a bone cancer arising predominantly in young children. EWSR1 (Ewing Sarcoma breakpoint region 1/EWS RNA binding protein 1) gene is ubiquitously expressed in most cell types, indicating it has diverse roles in various cellular processes and organ development. Recently, several studies have shown that missense mutations of EWSR1 genes are known to be associated with central nervous system disorders such as amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Otherwise, EWSR1 plays epigenetic roles in gene expression, RNA processing, and cellular signal transduction. Interestingly, EWSR1 controls micro RNA (miRNA) levels via Drosha, leading to autophagy dysfunction and impaired dermal development. Ewsr1 deficiency also leads to premature senescence of blood cells and gamete cells with a high rate of apoptosis due to the abnormal meiosis. Despite these roles of EWSR1 in various cellular functions, the exact mechanisms are not yet understood. In this context, the current review overviews a large body of evidence and discusses on what EWSR1 genetic mutations are associated with brain diseases and on how EWSR1 modulates cellular function via the epigenetic pathway. This will provide a better understanding of bona fide roles of EWSR1 in aging and its association with brain disorders.
Keywords: EWSR1, genetic mutation, epigenetic function, miRNA, brain disorders, autophagy
Introduction
Ewing’s sarcoma (EWS) is a bone cancer arising mostly in children. In 1921, EWS was originally described as “diffuse endothelioma of bone” by Dr. James R. Ewing, an American pathologist [1]. In 1992, Ewing sarcoma breakpoint region 1 (EWSR1)/EWS (herein termed EWSR1) gene was identified at chromosomal breakpoint t(11;22)(q24;q12) region from EWS and neuroectodermal tumors as a translocation-generated fusion gene between EWSR1 and FLI1 (Friend leukemia integration 1) [2]. The EWSR1 gene encodes a RNA/DNA binding protein and involves in various cellular processes. EWSR1 is well-known as a multifunctional protein, which regulates transcription and RNA splicing, indicating that it is involved in diverse cellular processes. N-terminal domains of EWSR1 interacts with the basal transcription factor TFIID and RNA Polymerase II. Moreover, EWSR1 protein is able to modulate gene transcription via interaction with CREB-binding protein (CBP), suggesting that EWSR1 plays a role in basal transcription process [3,4]. Previous studies discovered that EWSR1 regulates the transcriptional activity of HNF4, OCT4 and BRN3A [5, 6, 7]. In addition, EWS also serves a role in posttranscriptional mRNA splicing by cooperating with multiple splicing factors [8]. As such, growing body of evidence indicates that EWSR1 concerts various cellular pathways by itself or via multiple interactions with other molecules in a gene-context dependent manner.
EWSR1 belongs to a TET (also known as FET) family of proteins, which includes Fused in Sarcoma/Translocated in Liposarcoma (FUS/TLS, herein referred as FUS) and TATA-box binding protein Associated Factor 15 (TAF15). EWSR1, FUS and TAF15 are related in both structure and function. TET members are DNA/RNA-binding proteins (RBPs) that mainly contain an N-terminal serine-tyrosine-glycine-glutamine (SYGQ)-rich domain that acts as a transcriptional activation domain, a central RNA recognition domain (RRM) and a C-terminal zinc finger domain involved in RNA and DNA binding, respectively, and multiple Arg-Gly-Gly (RRG)-rich regions (RRGs) in the Cterminal that affect RNA binding [9,10,11,12]. The amino acid sequences of these proteins share high homology (~70%), and are evolutionarily conserved from fish to human [10]. TET proteins are expressed in most cell and tissue types, and predominantly reside in the nucleus [13]. In addition, they also have functional similarities. TET members contribute diverse roles in physiological cellular functions, and are involved in the regulation of RNA metabolism. Importantly, recent studies have shown that mutations of TET family genes are closely linked to certain neurodegenerative disorders such as amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD) (or termed as frontotemporal lobar degeneration [FTLD]), and essential tremor.
Epigenetic mechanisms unveil ambiguous and mysterious features and phenotypes derived from gene activity that cannot be explained by conventional wisdom of genetics beyond DNA mutations. In this paradigm, epigenetic changes include reversible modifications in DNA (methylation) and post-translational modifications in histone protein (acetylation, methylation, phosphorylation, and etc.) that eventually affect gene expression. Involvement of noncoding RNAs at post-transcriptional regulation of mRNA level is also categorized as a component of epigenetic pathway. It has been shown that epigenetic modifications and signatures contribute to the pathogenesis of many human disorders. Despite the strong association between EWSR1 mutations in cancers and neurodegenerative disorders, its epigenetic roles are not well known. Accordingly, if EWSR1 functions through the epigenetic pathway, it can provide novel insights to identify unseen biological markers and therapeutic targets to treat EWSR1-related disorders. Taken together, this review paper outlines and discusses the genetic and epigenetic roles of EWSR1 in cellular function, aging, and brain disorders.
1. Genetic aspects: EWSR1 and TET family gene mutations are linked to neurodegenerative disorders
Mutations in TET family genes encoding FUS, EWSR1, and TAF15 have recently been demonstrated to be associated with several neurodegenerative disorders including ALS, FTD/FTLD, and essential tremor that causes involuntary and rhythmic shaking (Figure 1) [14,15,16,17]. ALS is a neurodegenerative disease that is typically described by the degeneration of lower and upper motor neurons, which leads to a progressive and fatal muscle paralysis (amyotrophic) [18,19,20]. FTD is caused by the degeneration of neurons in the frontal and temporal lobes, and clinically characterized by progressive disorders of behaviors and personality, as well as language skills. Notably, FTD is ranked as the second most common dementia, just after Alzheimer’s disease [21]. The findings of TET mutations in ALS and FTD pathology have made a shift in understanding the pathological mechanisms, from RBP aggregation to problem of RNA metabolism [22]. Here, we focus on mutations of EWSR1 and other members of TET family underlying ALS and FTD pathogenesis.
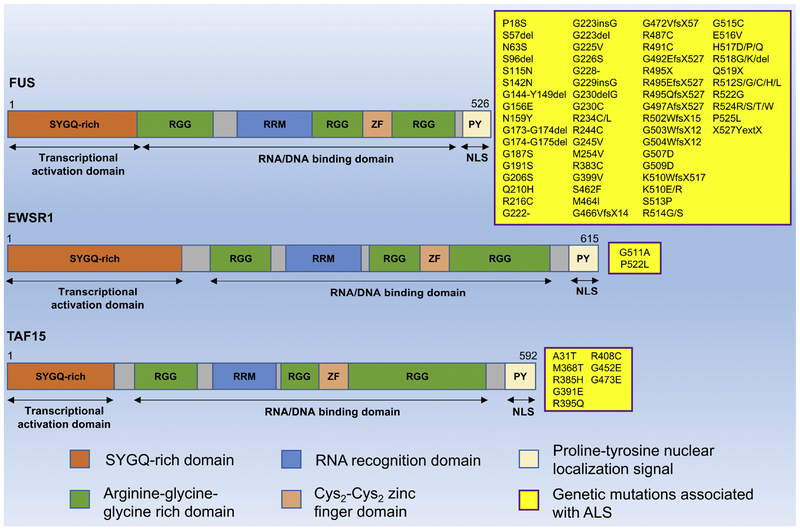
Figure 1: Scheme representing the domain structure and ALS-associated mutations of TET family (FUS, EWSR1, and TAF15).
TET members have similar domain structure, mainly contain an N-terminal serine-tyrosine-glycine-glutamine (SYGQ)-rich domain that acts as a transcriptional activating domain, a RNA recognition domain (RRM) and a zinc finger domain involved in RNA and DNA binding, respectively, multiple Arg-Gly-Gly (RRG)-rich regions (RRGs) in C-terminal that affect RNA binding, and prolie-tyrosine (PY) region functioning as nuclear localization signal (NLS). Yellow boxes present many kinds of missense-mutations that are found in FUS, EWSR1, and TAF15 gene, respectively.
1–1. FUS
FUS mutations are identified as a common genetic cause for ALS, accounting for around 4% of familial ALS (fALS) cases, while not frequently seen in sporadic ALS (sALS) [23,24,25,26,27,28]. Over 50 mutations in the FUS gene have been identified in ALS, and about two-thirds of mutations are missense changes clustering in exons 13–15, encoding the last RRG-rich domain and a PY nuclear localization signal (Figure 1). The majority of the C-terminal mutant FUS proteins show cytoplasmic redistribution due to the defects of transportin-mediated nuclear import, resulting in a loss of nuclear function and an aggregation of cytoplasmic FUS proteins. The remaining mutations are located in exons 3, 5, and 6, encoding SYGQ-rich domain and the first RGG domain [16,25,26,27]. It has been documented that mutations in FUS account for less than 1% of sALS [28], whereas 35 different pathogenic FUS mutations have been reported in fALS in only 2 years [27,29,30,31]. The FUS-immunoreactive cytoplasmic inclusions were observed in postmortem analysis of fALS cases [26,30]. In ALS patients, a hallmark feature of FUS-associated neuropathies is the aggregated cytoplasmic FUS proteins. To analyze pathological alterations within ALS related to FUS mutations, Higelin et al. used the models of human induced pluripotent stem cells (hiPSCs) and hiPSC derived motor neurons, and observed the FUS mislocalization in motor neurons that express mutant FUS [32], which is correlated with the findings in ALS patients. Nonetheless, it remains unsolved how the mutations in FUS cause the development of ALS. FTD neuropathology in most cases is identified by abnormal cellular aggregations of tau protein (FTD-tau) or transactive response DNA-binding protein with molecular weight of 43kDa (TDP-43) (FTD-TDP). However, FUS mutations were also reported in FTD pathology, and even new subgroup of FTD has been categorized based on FUS pathology with the presence of abnormal cytoplasmic FUS-positive inclusions [33,34].
1–2. EWSR1
In contrast to FUS, a few EWSR1 mutations have been found in sALS patients. Couthouis et al. (2012) performed complete sequencing of the last four exons encoding the last RGG- and PY-NLS domains in the C-terminus, which are equivalent to regions in FUS and TDP43 contributing to ALS pathology. Two missense mutations, G511A and P552L, were identified in 2 sALS cases out of 817 samples and none in 1082 control individuals, suggesting their potential cause for ALS pathogenicity (Figure 1). G511A and P552L are located in the last RGG domain of EWSR1. Intriguingly, both ALS-linked mutant EWSR1 proteins result in a formation of cytoplasmic EWSR1 inclusion in primary motor neurons cultured from mouse spinal cord and rat embryos, whereas wild-type (WT) EWSR1 primarily reside in the nucleus [35]. It has been well known that main pathological hallmarks of TETassociated pathology in ALS include the mislocalization of TET proteins to the cytoplasm. Interestingly, ALS-associated mutant EWSR1 proteins show increased proclivity to form aggregated proteins than WT EWSR1, which suggests the possibility of these mutations causing accelerated aggregation in the affected motor neurons. Furthermore, overexpression of mutant EWSR1 in Drosophila causes neurodegeneration in the nervous system. Both in vitro and in vivo data support that mutant EWSR1 is able to confer neurodegeneration in similar manner to FUS, TAF15 and TDP43 [35]. On the other hand, Ticozzi et al. screened the coding regions of EWSR1 in fALS patients and healthy individuals; however, no coding variants were found in EWSR1 [13]. Therefore, whether mutations in EWSR1 contribute significantly to the pathogenesis of fALS or not awaits further investigation. To date, there is very little data on EWSR1 mutations in FTD pathology. EWSR1 cytoplasmic inclusions were observed in FTD cases with FUS pathology, suggesting its potential involvement in pathogenic mechanisms of FTD [28]. However, it remains to be further investigated to determine whether EWSR1 represents as a cause of FTD pathogenesis.
1–3. TAF15
TAF15 gene mutations are observed in both sALS and fALS patients (Figure 1) [13,36,37]. Using a yeast functional screening, missense variants in TAF15 gene (G391E, R408C and G473E) were found in sALS cases, while absent in a large number of healthy individuals. The ALS-associated mutant TAF15 proteins also accelerate TAF15 aggregation in primary spinal cord neurons. Notably, mutant TAF15 proteins confer neurodegeneration in Drosophila with more severe phenotypes than WT TAF15. In sALS cases, mutant TAF15 was mislocalized to the cytoplasm of motor neurons in the spinal cord [37]. Mutations of TAF15 (A31T and R395Q) were also discovered in fALS cases that belong to three unrelated pedigrees, while not in over 1100 healthy cases [13]. These studies suggest that further investigations are needed to identify whether TAF15 genetic mutations are the cause of ALS pathogenesis. TAF15 has been implied as a potential candidate for understanding FTD pathology. A recent study has shown that TAF15 is found in cytoplasmic aggregations in the neurons of FTD-FUS patients, suggesting that TAF15 is likely involved in the pathological processes of FTD-FUS [36].
2. Ewsr1 knock out (KO) mouse model reveals bona fide functions of EWSR1
In order to determine the physiological roles of WT (wild type) EWSR1, Dr. Lee’s group established a conventional Ewsr1 KO mouse [38]. Since then, the Ewsr1 KO mouse model has successfully been utilized to decipher many important in vivo roles of EWSR1 (Figure 1).
2–1. EWSR1 deficiency shows defects in multiple organs and cell types
Ewsr1-deficient mice are born according to the Mendelian ratio [38]. However, all of Ewsr1 homozygous KO (−/−) mice are born smaller than their littermates and a few surviving Ewsr1 KO mice in 129SvEv/Black Swiss mixed background (10% survival rate) remain smaller in the body size and weight in adulthood. Congenic Ewsr1 KO (−/−) mice in C57BL6/J or 129SvEv background exhibit complete postnatal lethality within 24h of birth, but the cause of death remains unresolved.
Surviving129SvEv/Black Swiss background Ewsr1 KO (−/−) mice show severe lymphopenia and a cell-autonomous pre-B cell development defect compared to littermate control mice [38]. The flow cytometric analysis indicates that the number of pre-B cells is significantly reduced compared to WT mouse, while the number of pro (Progenitor) B cells appeared to be normal. Ewsr1 KO (−/−) mice display reduced cellularity of thymi and spleens.
All Ewsr1 KO (−/−) mice are sterile and have relatively small testis and ovary compared to littermate WT mice [38]. Further analysis showed impaired development of spermatogonium to spermatid and complete lack of mature sperms in 12-week-old Ewsr1 KO (−/−) mice. Furthermore, Ewsr1 KO (−/−) spermatocytes showed a reduced meiotic recombination and XY asynapsis, resulting in massive apoptosis of spermatocytes. In 12-week-old female Ewsr1 KO (−/−) mice, most ovaries were devoid of maturing ovarian follicles, and no corpora lutea was present.
2–2. EWSR1 and adipogenesis
Park et al. discovered that EWSR1 is indispensable for brown fat lineage determination [39]. Brown adipose tissues (BATs) from Ewsr1 KO embryos are developmentally arrested. A mechanistic study shows that Ewsr1 deficiency halts brown preadipocyte differentiation due to loss of Bmp7, a critical brown adipogenic factor during BAT development. Upon adipogenic stimulation, EWSR1 interacts with Y-box binding protein 1 (YBX1) to activate Bmp7 transcription. Accordingly, loss of Ewsr1 or Ybx1 function results in the reduction of Bmp7 expression and brown adipogenesis. Considering the facts that EWSR1 is a transcriptional co-factor in the brown fat adipocyte fate determination and BAT is potentially applicable for reducing obesity, the EWSR1-YBX1-BMP7 pathway may be a new therapeutic target to control obesity.
2–3. EWSR1 and mitochondrial function
Interestingly, EWSR1 participates in mitochondria function and cellular energy homeostasis by modulating the stability of PGC-1α (Peroxisome proliferator-activated receptor γ Coactivator) protein, a key regulator of mitochondria biogenesis, [40]. Park et al. identified that Ewsr1 deficiency leads to a rapid degradation of PGC-1α by increasing ubiquitination and proteolysis of PGC-1α via proteasome pathway [40]. The reduction of PGC-1α, in turn, markedly reduces mitochondria abundance and activity in brown adipocytes and skeletal muscles of Ewsr1 KO mice. Rescue of EWSR1 activity in Ewsr1-deficient cells complements PGC-1α activity and restore mitochondrial abundance. How is PGC-1α protein vulnerable to ubiquitination and degradation under Ewsr1 deficiency condition? FBXW7 (F-box/WD40 domain protein 7), an E3 ubiquitin ligase, is involved in PGC-1α ubiquitination in Ewsr1-deficient cells. Knockdown of Fbxw7 in Ewsr1deficient cells restores PGC-1α protein level and mitochondria abundance. The decrease of mitochondrial biogenesis coincides with the significant reduction of respiration and fatty acid βoxidation genes in the liver of Ewsr1 KO mice.
2–4. The global effect of EWSR1 deficiency on cellular senescence and aging
Ewsr1-null fibroblasts exhibit cellular senescence while Ewsr1-deficient mice show an aging-like phenotype [38]. The immunocytochemistry analysis using senescence-associated β-galactosidase staining verified that the cellular senescence is significantly elevated in Ewsr1 homozygous (−/−) mouse embryonic fibroblasts (MEFs) compared to Ewsr1 WT (+/+) MEFs. In addition, Li et al. further determined that gamma-irradiation (7 Gy) exacerbates the death of Ewsr1 mutant mice compared to Ewsr1 WT mice. Upon irradiation, Ewsr1 mutant mice lived up to 60 days while littermate control mice (Ewsr1+/+ and Ewsr1+/–) survived beyond 120 days [38].
How is EWSR1 deficiency linked to cellular aging? It is well known that cellular senescence is triggered by the attrition of telomere due to the shortening of the telomere length beyond a critical threshold [41,42]. However, mouse cells possess long telomeres. Accordingly, it seems unlikely that the early onset of senescence in Ewsr1 mutant cells is due to telomere attrition. The other molecular pathways via p53 and RB may be involved in the cellular senescence of Ewsr1 mutant cells. However, no significant changes in p53, p21, and p19ARF levels were found in Ewsr1–/– MEFs. On the other hand, the phosphorylated Rb (pRb) level was markedly reduced in Ewsr1+/+ MEFs. Considering that the reduced pRb levels are more likely to result in proliferation rather than cellular senescence, the cellular senescence of Ewsr1+/+ MEFs might be independent of the RB pathway. Interestingly, after more than 10 passages, an elevated level of p16INK4A, a marker of cellular senescence, was observed in Ewsr1–/– MEFs compared to Ewsr1+/+ MEFs [43].
EWSR1 deficiency also contributes to hematopoietic stem cell senescence [44]. Cho et al. reported that EWSR1 is essential for stem cell quiescence by observing that Ewsr1 KO mice show the early onset of senescence in hematopoietic stem progenitor cells [44]. Ewsr1-deficient hematopoietic stem cells exhibit a phenotypic change such as an increase in β-galactosidase activity and a molecular change such as a robust increase of p16INK4a compared to Ewsr1 WT mice. These findings indicate that EWSR1 plays a significant role in maintaining the hematopoietic stem cell lineage and its deficiency may trigger aging processes in a cell type-specific manner. At the moment, it is not known whether free radicals are up or down regulated under EWSR1 deficiency condition. In addition, parameters of free radical injury are not determined in EWSR1 deficiency cells and animal models. On the other hand, it is curious about whether modulation of free radicals can ameliorate or rescue EWSR1 deficiency-associated cellular dysfunction and aging. To address these questions, the future study remains to be investigated.
2–5. EWSR1 deficiency leads to neuronal atrophy and abnormal motor function
Recently, we have reported that Ewsr1 KO mice show neuroanatomical changes followed by motor dysfunction as one of behavioral phenotypes (Figure 2) [45]. Overall, the neuronal sizes in the cortex, striatum and hippocampus were significantly reduced in homozygous Ewsr1 KO (−/−) mice than that of Ewsr1 WT mice. Furthermore, it was found that both tyrosine hydroxylase (Th) mRNA and TH protein, a dopamine synthesizing enzyme, are significantly reduced in Ewsr1 KO mice compared to Ewsr1 WT mice. The immunoreactivity of TH and Protein Phosphatase 1 Regulatory Inhibitor Subunit 1B (PPP1R1B)/dopamine- and cAMP-regulated neuronal phosphoprotein (DARPP-32) was markedly decreased in the striatum and substantia nigra of Ewsr1 KO (−/−) mice. PPP1R1B/DARPP-32 is an important molecule that mediates signal transduction in the medium spiny neurons of the striatum [46]. This finding indicates that Ewsr1 deficiency downregulates TH level and, in turn, reduces PPP1R1B/DARPP-32 phosphorylation. Concurrent with the molecular changes of TH and PPP1R1B/DARPP-32 level, Ewsr1 KO (−/−) mice exhibit a significant increase of forelimb and hindlimb clasping movements compared to Ewsr1 WT mice. Taken togehter, these results suggest that Ewsr1 deficiency deregulates dopaminergic signaling pathways by reducing TH and PPP1R1B/DARPP-32 activity and subsequently leads to motor dysfunction. However, future studies are necessary to elucidate a precise mechanism on how EWSR1 regulates the motor function and other behaviors using multiple molecular and behavioral analyses.
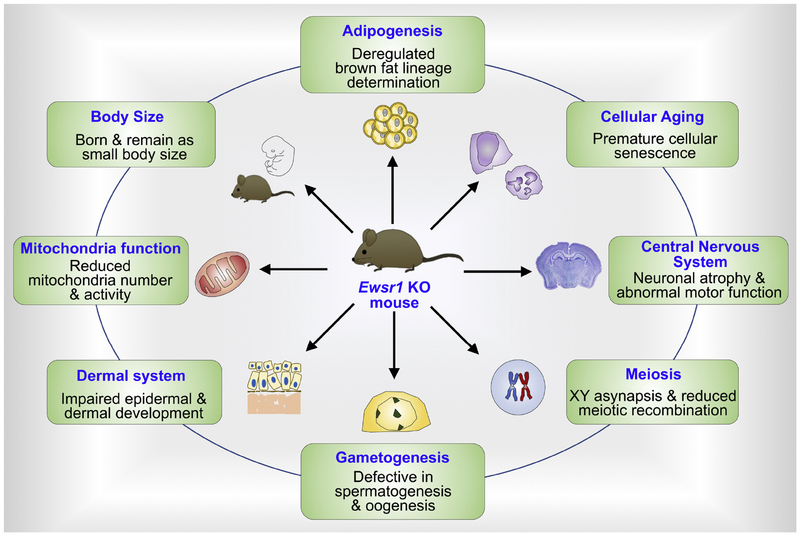
Figure 2. EWSR1 is a multifunctional protein and its deficiency impairs tissue-specific functions.
Ewsr1 knockout mice exhibit smaller body size, deregulated brown fat lineage determination, premature cellular senescence, neuronal atrophy and abnormal motor function, XY asynapsis and reduced meiotic recombination, defective in spermato- and oogenesis, impaired epidermal and dermal development, and reduced mitochondria number and activity.
3. EWSR1 deficiency alters microRNA processing via Drosha
As a part of epigenetic components, microRNAs (miRNAs) play significant roles on regulating gene expression. miRNAs are transcribed by RNA polymerase II from the genome. miRNA is a short noncoding RNA consisting of about 22 nucleotides and plays a role as a post-transcriptional regulator in gene expression. miRNAs bind to complementary target sequence of messenger RNA (mRNA), and degrade target mRNAs or inhibit their translation into proteins. Long primary transcript miRNAs (called pri-miRNAs) are processed in the nucleus by DROSHA, a member of the ribonuclease III family (RNase III), and converted into precursor miRNAs (pre-miRNAs) [47]. In turn, the pre-miRNA is being exported to the cytoplasm via XPO5/EXPORTIN-5 and is further processed by DICER1, a double-stranded RNA-specific endoribonuclease [48]. Although many studies have characterized the role of miRNAs under normal and disease conditions, no studies have shown whether EWSR1 is involved in the regulation of miRNA levels.
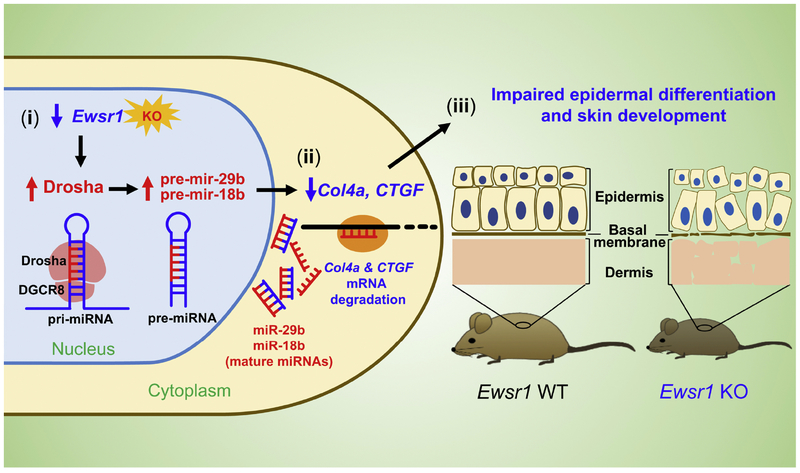
Our group found for the first time that EWSR1 indirectly regulates the expression of microRNAs (miRNAs) via an induction of DROSHA (Figure 3) [43]. Ewsr1 deficiency elevates expression of Drosha and, in turn, increases miR-29b and miR-18b levels. Interestingly, both miR29b and miR-18b directly target collagen IV alpha 1 (Col4a1) and connective tissue growth factor (CTGF) mRNAs and reduce their mRNA levels by negatively regulating the post-transcriptional pathway in Ewsr1 KO mouse MEFs. Consequently, the increased expression of Drosha, miR-29b, and miR-18b and the reduction of Col4a1 and CTGF lead to impaired epidermal and dermal development, resulting the abnormal skin development and aging in the Ewsr1 KO mice. In contrast, loss of Drosha function restores Col4a1 and CTGF protein levels by normalizing miR-29b, and miR18b expression in Ewsr1 KO mouse MEFs. Collectively, our previous study indicates that EWSR1 presents the epigenetic effector function in the post-transcriptional regulation of Col4a1 and CTGF via the Drosha-miRNA-dependent pathway. This evidence proves a novel epigenetic role of EWSR1 in miRNA biogenesis and dermal morphogenesis.
Figure 3. EWSR1 deficiency influences epithelial cell senescence through an epigenetic modulation of miRNA processing via Drosha.
(i) EWSR1 deficiency increases Drosha expression and certain miRNAs (miR-29b and miR-18b) in the nucleus of epithelial cells. (ii) In turn, miR-29b and miR-18b degrades Col4a1 and CTGF mRNAs in the cytoplasm of epithelial cells. (iii) Consequently, reduction of Col4a1 and CTGF leads to impaired epithelial cell senescence and dermal development.
4. EWSR1 regulates autophagy via an epigenetic modulation of UVRAG
Autophagy is a well-known intracellular self-digestive process that disassembles dysfunctional macro-molecular components to maintain cellular homeostasis [49,50]. In response to stress, autophagy often plays a key role by removing damaged organelles and recycling nutrients and energy within the cell [51,52,53]. Moreover, it has been reported that excessive activation or inactivation of autophagy is associated with various diseases, including neurodegenerative disorders and cancer [51,52,54,55].
miRNAs are known to regulate the autophagy-related genes and their activities. Also, miRNAs modulate autophagy at different stages such as autophagic induction, vesicle nucleation, vesicle elongation and completion, by targeting autophagy complexes via different miRNAs [56,57,58]. Recently, although there has been plenty of evidence that miRNAs modulate autophagy, their target genes and precise roles in the autophagy pathways have not been fully defined yet. In this context, our group previously investigated whether EWSR1 plays a role in the autophagy pathway or not using Ewsr1 null (−/−) MEFs. Two novel findings were identified as follows: First, Ewsr1 deficiency up regulates microprocessor complexes and miR125a and miR351. Interestingly, EWSR1 indirectly regulates UVRAG expression at the post-transcriptional level via miR125a and miR351 [59]. Second, UVRAG dysfunction subsequently leads to an aberrant deregulation of autophagy pathway. Decreased expression levels of Uvrag mRNA and protein are correlated with the altered autophagy pathway in Ewsr1 KO mice.
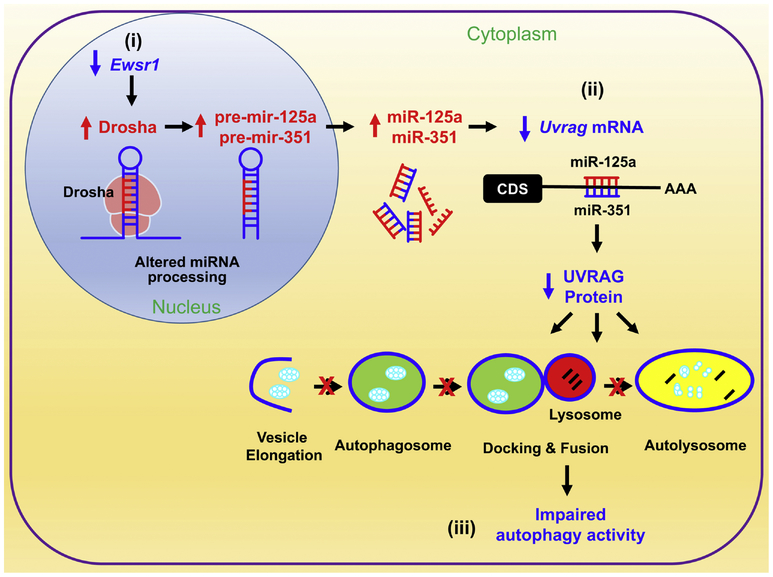
UVRAG is a mammalian ortholog of yeast Vps38 and a promoter of autophagy [51,60,61]. It forms distinct complexes with BECN1 (mammalian ortholog of yeast Vps30/Atg6) and the class III phosphatidylinositol 3-kinase (whose catalytic subunit [PIK3C3] is the mammalian ortholog of yeast Vps34) and contributes to both autophagosome formation and maturation [60,62]. UVRAG suppresses cancer cell growth by promoting autophagy, its deficiency leads to decrease of autophagy and uncontrolled cell proliferation [60]. Based on the previous finding that DROSHA level is elevated by Ewsr1 deficiency, it is hypothesized that DROSHA-miRNA dependent pathway may be involved in UVRAG expression [43]. Our group found that Uvrag mRNA is inversely correlated with elevated DROSHA levels in the cytoplasm of Ewsr1 null (−/−) MEFs [59]. In addition, miRNA microarray analysis verified that mir125a and miR351 are significantly increased in Ewsr1−/− MEFs. Indeed, miR125a and miR351 directly target and degrade Uvrag mRNA. Moreover, Ewsr1 KO mice show that the levels of miR125a and miR351 are significantly increased, whereas the levels of UVRAG and LC3-II (autophagy marker) are significantly reduced compared with littermate control mice. Together, the previous study suggests that EWSR1 indirectly regulates autophagy via an epigenetic modulation of UVRAG level (Figure 4). Thus, EWSR1-mediated regulation of UVRAG and autophagy may be a potential therapeutic target for restoration of the cellular function.
Figure 4. EWSR1 plays an epigenetic role in the regulation of autophagy via a posttranscriptional modulation of UVRAG level.
(i) EWSR1 deficiency increases the abnormal processing of pri-miRNAs to pre-mir125a and pre-mir351 by Drosha in the nucleus compartment. (ii) Next, elevated mature miRNA products (miR125a and miR351) degrade Uvrag mRNA in the cytoplasmic compartment. (iii) As a result, impaired UVRAG-dependent autolysosomal pathway leads to autophagy and cellular dysfunction. This figure is adopted from Autophagy (2015) 11 (5):796–811.
5. The role of EWSR1 in regulating stem cells
The EWSR1 gene plays a crucial role in the regulation of stem cells and the development of a number of different tumors. During early development, Oct-4 (octamer-binding transcription factor 4), also known as POU5F1 (POU domain, class 5, transcription factor 1), that encodes a key regulator of stem cell pluripotency, is expressed to maintain the totipotent status of embryonic stem and germ cells. EWSR1 is fused to POU5F1 in hidradenoma of the skin and mucoepidermoid carcinoma of salivary glands [63]. POU5F1-mediated transactivation is stimulated by EWSR1 protein in mouse and human embryonic stem cells [6]. In 85–90% of cases, Ewing’s sarcoma (ES) is characterized by the expression of the EWSR1-FLI1 chimeric protein resulting from the chromosomal translocation, which links the transcription regulating domain of EWSR1 to the ETS DNA-binding domain of FLI1 [64]. This EWSR1-FLI-1 fusion oncoprotein is responsible for the transcriptional deregulation of target genes, such as the CD99 membrane receptor. Expression of CD99 contributes to the ES oncogenesis by modulating the growth and differentiation of tumor cells [65]. Also, EWSR1 is chimerically fused to DDIT3 (DNA Damage Inducible Transcript 3) by the myxoid liposarcoma-specific chromosomal translocation. Suzuki K et al. (2012) investigated the molecular mechanisms underlying EWSR1-DDIT3 fusion protein-mediated phenotypic selection of putative target multipotent mesenchymal cells during myxoid liposarcoma development [66]. A better understanding of this mechanism is pivotal to elucidate the direct lineage reprogramming process in oncogenic sarcoma transformation mediated by EWSR1-fusion proteins [66].
6. Therapeutic approaches to target EWSR1
EWSR1, EWSR1-fusion protein, EWSR1-interacting molecules, and its downstream pathways can be ideal therapeutic targets to treat ES or EWSR1-related disorders. Interestingly, transcriptional activation of protein kinase PKC-ß (PRKCB) is directly regulated by the chimeric EWSR1-FLI1 protein in EWS. PRKCB loss induces apoptosis in vitro and prevents tumor growth in vivo. PRKCB possesses an enzymatic activity that can be directly targeted by small compounds. Accordingly, in the perspective of therapeutic strategy, blocking PRKCB activity in EWS is a new promising approach [67]. Poly (ADP-ribose) polymerase-1 (PARP) protein plays a role in the regulation of the cell cycle, apoptosis, and etc. It has shown that EWSR1-ETS fusion protein could be sensitive to PARP inhibitors such as Olaparib, Veliparib, and Iniparib [68]. Trabectedin is an antitumoral agent that modulates EWSR1-FLI1 transcriptional functions, causing DNA damage. The combination of a PARP inhibitor and Trabectedin highly inhibits proliferation and induces apoptosis in EWS cells [68]. Sanker et al. described that the Nucleosome Remodeling Deacetylase (NuRD) complex directly binds to EWS-FLI1 oncoprotein and regulates transcriptional activity of EWS-FLI1 target genes [68]. It has been widely known that the EWS-FLI1 plays a driver of proliferation and transformation in ES. Daniel et al. investigated that the combination effects of the histone deacetylases inhibitor suberoylanilide hydroxamic acid (SAHA) and Lysine-specific demethylase1 inhibitor (HCI-2509) on different biological functions in ES. The combination of SAHA and HCI-2509 inhibits the essential driver of this sarcoma and tumor growth and is proposed as a novel treatment strategy for ES patients [69].
Otherwise, ES cells express high level of histone lysine specific demethylase 1 (LSD1) expression. In this context, it is proposed that LSD1 inhibition may block the function of EWS-ETS proteins [70]. Furthermore, checkpoint kinase 1 (CHK1), a modulator of cell survival under the condition of impaired DNA replication, is a candidate of therapeutic targets in ES [71]. Smallmolecule CHK1 inhibitor combined with gemcitabine shows elevated toxicity both in vitro and in vivo models of ES.
Conclusion
EWSR1 participates in various functions which are crucial for the regulation of tissue development and cellular homeostasis. We overviewed that i) genetic mutations of EWSR1 are associated with neurodegeneration, ii) EWSR1 deficiency leads to epigenetic alteration such as miRNA processing, and iii) EWSR1, as if “Jack of all trades, master of none”, plays diverse molecular functions and its deficiency affects many cellular functions including autophagy and mitochondrial activity. Loss of EWSR1 function also contributes to the hypersensitivity of ionizing radiation and premature cellular senescence and aging. Even though several groups have studied in depth about EWSR1, there are still many areas that have not yet been explored. For example, conditional KO or knock-in EWSR1 animal models are necessary to determine the EWSR1’s multifunctional or undescribed roles in an organ-specific or a cell type-specific manner. For example, to study desmoplastic small round cell tumor characterized by EWSR1-WT1 translocation, Vanoli et al. (2017) developed a strategy using the combination of CRISPR-Cas9 genome editing and homology-directed repair to select human mesenchymal stem cells containing the EWSR1-WT1 translocation with fusion transcript expression under the control of the EWSR1 promoter and conditionally using Cre recombinase. A similar strategy was recently applied to generate conditionally inducible EWSR1-WT1 and EWSR1-FLI1 fusion genes in a human cell line (HEK293). This approach provides multiple advantages and expected to be a model for studying the tumors driven by chromosomal translocations [72,73]. Another group also developed a mouse model harboring conditional expression of EWS-FLI1 fusion transcripts under the control of Prx1-Cre, which is expressed in the primitive mesenchymal cells in the limb bud of embryo. The EWS-FLI1;Prx1-Cre mice showed developmental defects of the limbs without tumors. Thus, it was clearly demonstrated that EWS-FLI1 is not able to initiate sarcoma formation by itself. Nonetheless, conditional deletion of p53 in EWS-FLI1; p53flox/flox;Prx1-Cre triple transgenic mice produced a poorly differentiated sarcoma. These data imply that sarcomagenesis can be induced via the cooperation of EWS-FLI1 and inactivation of the p53 tumor suppressor pathway [74].
In addition, studies using multiple behavioral and molecular analyses are required to better understand the mechanisms by which EWSR1 regulates brain and motor neuron functions. It is with great anticipation that future studies will further scrutinize and unravel new functions of EWSR1, which could be utilized to fuel new research areas in cancer, neurodegeneration and perhaps open a new field of research.
Highlights.
EWSR1 (Ewing Sarcoma breakpoint region 1/EWS RNA binding protein 1) has diverse roles in various cellular processes such as gene expression and RNA processing, and organ development.
Missense mutations of EWSR1 genes are associated with amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD).
EWSR1 deficiency also contributes to hematopoietic stem cell senescence
EWSR1 participates in mitochondria function and cellular energy homeostasis by modulating the stability of PGC-1α (Peroxisome proliferator-activated receptor γ Coactivator) protein
EWSR1 deficiency deregulates dopaminergic signaling pathways by reducing TH and PPP1R1B/DARPP-32 activity and leads to motor dysfunction.
Acknowledgements
This study was supported by NIH Grant (R01 AG054156 to H.R.) (R01 NS109537 to J.L.) (Tulane Startup and Pilot Fund to S.B.L). This study was also supported by the National Research Foundation (NRF) Grant (NRF-2016M3C7A1904233 and NRF-2016M3C7A1905119 (S.C)), the National Research Council of Science & Technology (NST) Grant (CRC-15–04-KIST) from the South Korea Government, and Grant (2E26870) from Korea Institute of Science and Technology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cripe TP, Ewing sarcoma: an eponym window to history, Sarcoma 2011 (2011) 457532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delattre O, Zucman J, Plouqastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G, Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours, Nature 359 (1992) 162–165. [DOI] [PubMed] [Google Scholar]
- 3.Bertolotti A, Melot T, Acker J, Vigneron M, Delattre O, Tora L, EWS, but not EWS-FLI-1, is associated with both TFIID and RNA polymerase II: interactions between two members of the TET family, EWS and hTAFII68, and subunits of TFIID and RNA polymerase II complexes, Mol. Cell. Biol 18 (1998) 1489–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rossow KL, Janknecht R, The Ewing’s sarcoma gene product functions as a transcriptional activator, Cancer Res. 61 (2001) 2690–2695. [PubMed] [Google Scholar]
- 5.Araya N, Hirota K, Shimamoto Y, Miyagishi M, Yoshida E, Ishida J, Kaneko S, Kaneko M, Nakajima T, Fukamizu A, Cooperative interaction of EWS with CREB-binding protein selectively activates hepatocyte nuclear factor 4-mediated transcription, J. Biol. Chem 278 (2003) 5427–5432. [DOI] [PubMed] [Google Scholar]
- 6.Lee J, Rhee BK, Bae GY, Han YM, Kim J, Stimulation of Oct-4 activity by Ewing’s sarcoma protein, Stem Cells 23 (2005) 738–751. [DOI] [PubMed] [Google Scholar]
- 7.Kovar H, Dr. Jekyll and Mr. Hyde: the two faces of the FUS/EWS/TAF15 protein family, Sarcoma 2011 (2011) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erkizan HV, Uversky VN, Toretsky JA, Oncogenic partnerships: EWS-FLI1 protein interactions initiate key pathways of Ewing’s sarcoma, Clin. Cancer Res. 16 (2010) 4077–4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morohoshi F, Arai K, Tanigami EI, Ohki M, Cloning and mapping of a human RBP56 gene encoding a putative RNA binding protein similar to FUS/TLS and EWS proteins, Genomics 38 (1996) 51–57. [DOI] [PubMed] [Google Scholar]
- 10.Morohoshi F, Ootsuka Y, Arai K, Ichikawa H, Mitani S, Munakata N, Ohki M, Genomic structure of the human RBP56/hTAFII68 and FUS/TLS gene, Gene 221 (1998) 191–198. [DOI] [PubMed] [Google Scholar]
- 11.Tan Y, Manley JL, The TET family of proteins: functions and roles in disease, J. Mol. Cell Biol 1 (2009) 82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paronetto MP, Ewing sarcoma protein: a key player in human cancer, Int. J. Cell Biol 2013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andresson MK, Stahlberg A, Arvidsson Y, Olofsson A, Semb H, Stenman G, Nilsson O, Aman P, The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell typespecific expression patterns and involvement in cell spreading and stress response, BMC Cell Biol. 9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ticozzi N, Vance C, Leclerc AL, Keagle P, Glass JD, McKenna-Yasek D, Sapp PC, Silani V, Bosco DA, Shaw CE, Brown RH Jr., Landers JE, Mutational analysis reveals the FUS homolog TAF15 as a candidate gene for familial amyotrophic lateral sclerosis, Am. J. Med. Genet. B Neuropsychiatr. Genet 156B (2011) 285–290. [DOI] [PubMed] [Google Scholar]
- 15.Mackenzie R, Neumann M, FET proteins in frontotemporal dementia and amyotrophic lateral sclerosis, Brain Res. 1462 (2012) 40–43. [DOI] [PubMed] [Google Scholar]
- 16.Merner ND, Girard SL, Catoire H, Bourassa CV, Belzil VV, Riviere JB, Hince P, Levert A Dionne-Laporte A, Spiegelman D, Noreau A, Diab S, Szuto A, Fournier H, Raelson J, Belouchi M, Panisset M, Cossette P, Dupre N, Bernard G, Chouinard S, Dion PA, Rouleau GA, Exome sequencing identifies FUS mutations as a cause of essential tremor, Am. J. Hum. Genet 91 (2012) 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Svetoni F, Frisone P, Paronetto MP, Role of FET proteins in neurodegenerative disorders, RNA Biol. 13 (2016) 1089–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caroline E, Wim R, The genetic basis of amyotrophic lateral sclerosis: recent breakthroughs, Adv. Genomics Genet 5 (2015) 327–345. [Google Scholar]
- 19.Andersen PM, Al-Chalabi A, Clinical genetics of amyotrophic lateral sclerosis: what do we really know?, Nat. Rev. Neurol 7 (2011) 603–615. [DOI] [PubMed] [Google Scholar]
- 20.Pratt J, Getzoff ED, Perry JJ, Amyotrophic lateral sclerosis: update and new developments, Degener. Neurol. Neuromuscul. Dis 2012 (2012) 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seltman RE, Matthews BR, Frontotemporal lobar degeneration: epidemiology, pathology, diagnosis and management, CNS Drugs 26 (2012) 841–870. [DOI] [PubMed] [Google Scholar]
- 22.Boeynaems S, Bogaert E, Van Damme P, Van Den Bosch L, Inside out: the role of nucleocytoplasmic transport in ALS and FTLD, Acta Neuropathol, 132 (2016) 159–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kapeli K, Martinez FJ, Yeo GW, Genetic mutations in RNA-binding proteins and their roles in ALS, Human Genet. 136 (2017) 1193–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng H, Gao K, Jankovic J, The role of FUS gene variants in neurodegenerative diseases, Nat. Rev. Neurol 10 (2014) 337–348. [DOI] [PubMed] [Google Scholar]
- 25.van Blitterswijk M, Wang ET, Friedman BA, Keagle PJ, Lowe P, Leclerc AL, van den Berg LH, Housman DE, Veldink JH, Landers JE, Characterization of FUS mutations in amyotrophic lateral sclerosis using RNA-seq, PloS One 8 (2013) e60788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rademakers R, Stewart H, Dejesus-Hernandez M, Krieger C, Graff-Radford N, Fabros M, Briemberg H, Cashman N, Eisen A, Mackenzie IR, FUS gene mutations in familial and sporadic amyotrophic lateral sclerosis, Muscle Nerve 42 (2010) 170–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mackenzie R, Rademakers R, Neumann M, TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia, Lancet Neurol. 9 (2010) 995–1007. [DOI] [PubMed] [Google Scholar]
- 28.Lai SL, Abramzon Y, Schymick JC, Stephan DA, Dunckley T, Dillman A, Cookson M, Calvo, Battistini S, Giannini F, Caponnetto C, Mancardi GL, Spataro R, Monsurro MR, Tedeschi G, Marinou K, Sabatelli M, Conte A, Mandrioli J, Sola P, Salvi F, Bartolomei I, Lombardo F, ITALSGEN Consortium, Mora G, Restagno G, Chio A, Traynor BJ, FUS mutations in sporadic amyotrophic lateral sclerosis, Neurobiol. Aging 32 (2011) 550.e1–550.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chio, Restagno G, Brunetti M, Ossola L, Calvo A, Mora G, Sabatelli M, Monsurro MR, Battistini S, Mandrioli J, Salvi F, Spataro R, Schymick J, Traynor BJ, La Bella V, ITALSGEN Consortium, Two Itanlian kindreds with familial amyotrophic lateral sclerosis due to FUS mutation, Neurobiol. Aging 30 (2009) 1272–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corrado L, Del Bo R, Castellotti B, Ratti A, Cereda C, Penco S, Soraru G, Carlomagno Y, Ghezzi S, Pensato V, Colombrita C, Gagliardi S, Cozzi L, Orsetti V, Mancuso M, Siciliano G, Mazzini L, Comi GP, Gellera C, Ceroni M, DAlfonso S, Silani V, Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis, Science 47 (2010) 190–194. [DOI] [PubMed] [Google Scholar]
- 31.Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, Ganesalingam J, Williams KL, Tripathi V, Al-Saraj S, AlChalabi A, Leigh PN, Blair IP, Nicholson G, de Belleroche J, Gallo JM, Miller CC, Shaw CE, Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6, Science 323 (2009) 1208–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Higelin J, Demestre M, Putz S, Delling JP, Jacob C, Lutz A, Bausinger J, Huber A, Klingenstein M, Barbi G, Speit G, Huebers A, Weishaupt JH, Hermann A, Liebau S, Ludolph AC, Boeckers TM, FUS mislocalization and vulnerability to DNA damage in ALS patients derived hiPSCs and aging motorneurons, Front. Cell Neurosci 10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dormann D, Rodde R, Edbauer D, Bentmann E, Fischer I, Hruscha A, Than ME, Mackenzie IR, Capell A, Schmid B, Neumann M, Haass C, ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import, EMBO J. 29 (2010) 2841–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neumann M, Rademakers R, Roeber S, Baker M, Kretzschmar HA, Mackanzie IR, A new subtype of frontotemporal lobar degeneration with FUS pathology, Brain 132 (2009) 2922–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Couthouis J, Hart MP, Erion R, King OD, Diaz Z, Nakaya T, Ibrahim F, Kim HJ, Mojsilovic-Petrovic K, Panossian S, Kim CE, Frackelton EC, Solski JA, Williams KL, Clay-Falcone D, Elman L, McCluskey L, Greene R, Hakonarson H, Kalb RG, Lee VM, Trojanowski JQ, Nicholson GA, Blair IP, Van Deerlin VM, Mourelatos Z, Shorter J, Gitler AD, Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis, Hum. Mol. Genet 21 (2012) 2899–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neumann M, Bentmann E, Dormann D, Jawaid A, DeJesus-Hernandez M, Ansorge O, Roeber S, Kretzchmar HA, Munoz DG, Kusaka H, Yokota O, Ang L, Bilbao J, Rademakers R, Haass C, Mackenzie IR, FET proteins TAF15 and EWS are selective markers that distinguish FTLD with FUS pathology from amyotrophic lateral sclerosis with FUS mutations, Brain 134 (2011) 2595–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Couthouis J, Hart MP, Shorter J, DeJesus-Hernandez M, Erion R, Oristano R, Liu AX, Ramos D, Jethava N, Hosangadi D, Epstein J, Chiang A, Diaz Z, Nakaya T, Kim HJ, A Solski J, Williams KL, Mojsilovic-Petrovic J, Ingre C, Boylan K, Graff-Radford NR, Dickson DW, Clay-Falcone D, Elman L, McCluskey L, Greene R, Lee VM, Trojanowski JQ, Ludolph A, Robberecht W, Andersen PM, Nicholson GA, Blair IP, King OD, Bonini NM, Van Deerlin V, Rademakers R, Mourelatos Z, Gitler AD, A yeast functional screen predicts new candidate ALS disease genes, Proc. Natl. Acad. Sci 108 (2011) 20881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Watford W, Li C, Parmelee A, Bryant MA, Deng C, O’Shea J, Lee SB, Ewing sarcoma gene EWS is essential for meiosis and B lymphocyte development, J. Clin. Invest 117 (2007) 1314–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park JH, Kang HJ, Kang SI, Lee JE, Hur J, Ge K, Mueller E, Li H, Lee BC, Lee SB, A multifunctional protein, EWS, is essential for early brown fat lineage determination, Dev. Cell 26 (2013) 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park JH, Kang HJ, Lee YK, Kang H, Kim J, Chung JH, Chang JS, McPherron AC, Lee SB, Inactivation of EWS reduces PGC-1a protein stability and mitochondrial homeostasis, Proc. Natl. Acad. Sci 112 (2015) 6074–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campisi J, Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors, Cell 120 (2005) 513–522. [DOI] [PubMed] [Google Scholar]
- 42.Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW, DNA repair, genome stability, and aging, Cell 120 (2005) 497–512. [DOI] [PubMed] [Google Scholar]
- 43.Kim KY, Hwang YJ, Jung MK, Choe J, Kim Y, Kim S, Lee CJ, Ahn H, Lee J, Kowall NW, Kim YK. Kim JI, Lee SB, Ryu H, A multifunctional protein EWS regulates the expression of Drosha and microRNAs, Cell Death Differ. 21 (2014) 136–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho J, Shen H, Yu H, Li H, Cheng T, Lee SB, Lee BC, Ewing sarcoma gene Ews regulates hematopoietic stem cell senescence, Blood 117 (2011) 1156–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoon Y, Park H, Kim S, Nguyen PT, Hyeon SJ, Chung S, Im H, Lee J, Lee SB, Ryu H, Genetic ablation of EWS RNA binding protein (EWSR1) leads to neuroanaltomical changes and motor dysfunction in mice, Exp. Neurobiol 27 (2018) 103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Svenningsson P, Nishi A, Fisone G, Girault JA, Nairn AC, Greengard P, DARPP-32: An integrator of neurotransmission. Annu. Rev. Pharmacol. Toxicol 44 (2004) 269–296. [DOI] [PubMed] [Google Scholar]
- 47.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN, The Drosha-DGCR8 complex in primary microRNA processing, Genes Dev. 18 (2004) 3016–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bratkovic T, Glavan G, Strukelj B, Zivin M, Rogelj B, Exploiting microRNAs for cell engineering and therapy, Biotechnol. Adv 30 (2012) 753–765. [DOI] [PubMed] [Google Scholar]
- 49.Mizushima N, Levine B, Cuervo AM, Klionsky DJ, Autophagy fights disease through cellular self-digestion, Nature 451 (2008) 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glick D, Barth S, Macleod KF, Autophagy: cellular and molecular mechanisms, J. Pathol 221 (2010) 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G, Selfeating and self-killing: crosstalk between autophagy and apoptosis, Nat. Rev. Mol. Cell Biol 8 (2007) 741–752. [DOI] [PubMed] [Google Scholar]
- 52.Portt L, Norman G, Clapp C, Greenwood M, Greenwood MT, Anti-apoptosis and cell survival: a review, Biochim. Biophys. Acta 1813 (2011) 238–259. [DOI] [PubMed] [Google Scholar]
- 53.Levine B, Klionsky DJ, Development by self-digestion: molecular mechanisms and biological functions of autophagy, Dev. Cell 6 (2004) 463–477. [DOI] [PubMed] [Google Scholar]
- 54.Huang J, Klionsky DJ, Autophagy and human disease, Cell Cycle 6 (2007) 1837–1849. [DOI] [PubMed] [Google Scholar]
- 55.Kim Y, Kim YS, Kim DE, S Lee J, Song JH, Kim HG, Cho DH, Jeong SY, Jin DH, Jang SJ, Seol HS, Suh YA, Lee SJ, Kim CS, Koh JY, Hwang JJ, BIX-01294 induces autophagy-associated cell death via EHMT2/G9a dysfunction and intracellular reactive oxygen species production, Autophagy 9 (2013) 2126–2139. [DOI] [PubMed] [Google Scholar]
- 56.Fu LL, Wen X, Bao JK, Liu B, MicroRNA-modulated autophagic signaling networks in cancer, Int. J. Biochem. Cell Biol 44 (2012) 733–736. [DOI] [PubMed] [Google Scholar]
- 57.Liu B, Cheng Y, Liu Q, Bao JK, Yang JM, Autophagic pathways as new targets for cancer drug development, Acta. Pharmacol. Sin 31 (2010) 1154–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Korkmaz G, le Sage C, Tekirdag KA, Agami R, Gozuacik D, miR-376b controls starvation and mTOR inhibitionrelated autophagy by targeting ATG4C and BECN1. Autophagy 8 (2012) 165–176. [DOI] [PubMed] [Google Scholar]
- 59.Kim YH, Kim KY, Hwang YJ, Kowall NW, Lee SB, Lee J, Ryu H, Uvrag targeting by Mir125a and Mir351 modulates autophagy associated with Ewsr1 deficiency, Autophagy 11 (2015) 796–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy, Autophagy 8 (2012) 445–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU, Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG, Nat. Cell Biol 8 (2006) 688699. [DOI] [PubMed] [Google Scholar]
- 62.Funderburk SF, Wang QJ, Yue Z, The Beclin 1-VPS34 complex--at the crossroads of autophagy and beyond, Trends Cell Biol. 20 (2010) 355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Möller E, Stenman G, Mandahl N, Hamberg H, Mölne L, van den Oord JJ, Brosjö O, Mertens F, Panagopoulos I, POU5F1, encoding a key regulator of stem cell pluripotency, is fused to EWSR1 in hidradenoma of the skin and mucoepidermoid carcinoma of the salivary glands, J. Pathol 215 (2008) 78–86. [DOI] [PubMed] [Google Scholar]
- 64.Torres-Ruiz R, Martinez-Lage M, Martin MC, Garcia A, Bueno C, Castaño J, Ramirez JC, Menendez P, Cigudosa JC, Rodriguez-Perales S, Efficient Recreation of t(11;22) EWSR1-FLI1(+) in Human Stem Cells Using CRISPR/Cas9, Stem Cell Reports. 8 (2017) 1408–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rocchi A, Manara MC, Sciandra M, Zambelli D, Nardi F, Nicoletti G, Garofalo C, Meschini S, Astolfi A, Colombo MP, Lessnick SL, Picci P, Scotlandi K, CD99 inhibits neural differentiation of human Ewing sarcoma cells and thereby contributes to oncogenesis, J. Clin. Invest 120 (2010) 668–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Suzuki K, Matsui Y, Higashimoto M, Kawaguchi Y, Seki S, Motomura H, Hori T, Yahara Y, Kanamori M, Kimura T, Myxoid liposarcoma-associated EWSR1-DDIT3 selectively represses osteoblastic and chondrocytic transcription in multipotent mesenchymal cells, PLoS One. 7 (2012) 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Surdez D, Benetkiewicz M, Perrin V, Han ZY, Pierron G, Ballet S, Lamoureux F, Rédini F, Decouvelaere AV, Daudigeos-Dubus E, Geoerger B, de Pinieux G, Delattre O, Tirode F, Targeting the EWSR1-FLI1 oncogene-induced protein kinase PKC-βabolishes Ewing sarcoma growth, Cancer Res. 72 (2012) 4494–4503. [DOI] [PubMed] [Google Scholar]
- 68.Sankar S, Bell R, Stephens B, Zhuo R, Sharma S, Bearss DJ, Lessnick SL, Mechanism and relevance of EWS/FLI-mediated transcriptional repression in Ewing sarcoma, Oncogene. 32 (2013) 5089–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.García-Domínguez DJ, Hontecillas-Prieto L, Rodríguez-Núñez P, Pascual-Pasto G, Vila-Ubach M, García-Mejías R, Robles MJ, Tirado OM, Mora J, Carcaboso AM, de Álava E, The combination of epigenetic drugs SAHA and HCI-2509 synergistically inhibits EWS-FLI1 and tumor growth in Ewing sarcoma, Oncotarget. 9 (2018) 31397–31410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Theisen ER, Pishas KI, Saund RS, Lessnick SL, Therapeutic opportunities in Ewing sarcoma: EWS-FLI inhibition via LSD1 targeting, Oncotarget. 7 (2016) 17616–17630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goss KL, Koppenhafer SL, Harmoney KM, Terry WW, Gordon DJ, Inhibition of CHK1 sensitizes Ewing sarcoma cells to the ribonucleotide reductase inhibitor gemcitabine, Oncotarget. 8 (2017) 87016–87032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vanoli F, Tomishima M, Feng W, Lamribet K, Babin L, Brunet E, Jasin M, CRISPR-Cas9guided oncogenic chromosomal translocations with conditional fusion protein expression in human mesenchyme cells, Proc. Natl. Acad. Sci 114 (2017) 3696–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Spraggon L, Martelotto LG, Hmeljak J, Hitchman TD, Wang J, Slotkin EK, Fan PD, Reis-Filho JS, Ladanyi M, Generation of conditional oncogenic chromosomal translocations using CRISPT-Cas9 genomic editing and homology-directed repair, J. Pathol 242 (2017) 102–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dodd RD, Mito JK, Kirsch DG, Animal models of soft-tissue sarcoma, Dis. Model Mech 3 (2010) 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]