Abstract
KLF5 is an important regulator of cell proliferation, differentiation, and apoptosis in mammals. Little is known about the function of KLF5 in the regulation of chicken. Hence, qPCR was used to detect the expression of KLF5 in different tissues of chicken. And chicken skeletal muscle satellite cells (SMSCs) were transfected KLF5-specific small interfering RNA (siRNA) to assay SMSCs’ proliferation, differentiation, and apoptosis. The results showed that KLF5 expressed higher in skeletal muscle than in the other tissues of chicken. Knockdown of KLF5 significantly inhibited the differentiation and increased apoptosis of chicken SMSCs, but it had no significant effect on proliferation of SMSCs. These results indicate that KLF5 plays an essential role during myogenesis, which will affect muscle repair and muscle regeneration, and may ameliorate muscle aging or sarcopenia.
Keywords: KLF5, Chicken satellite cells, Proliferation, Differentiation, Apoptosis
Introduction
Skeletal muscle not only meets the physiological need of animals, but also relates to muscle aging and a variety of common diseases, such as cancer, heart failure, and kidney failure. The number of muscle fibers remains constant basically after animals birth (Stickland et al. 2004); repaired muscle fibers rely on skeletal muscle satellite cells (SMSCs. SMSCs are pluripotent stem cells with differentiation, when adult myofibers are damaged or necrotic, SMSCs will be activated, and then proliferated, differentiated, and fused to form new myofibers to repair damaged one (Buckingham et al. 2014). SMSCs can also differentiate into nerve cells and adipocytes, which can increase the fat content and provide nutrition for new muscles, and, thus, improve the effect of repair in injured muscle. Myogenesis of satellite cells is partially regulated by an orchestration of several major factors, the extracellular matrix (Shin et al. 2012; Velleman 1999), growth factors (Allen et al. 2010; Florini et al. 1991), myogenic regulatory transcription factors (Cornelison et al. 2000; Meadows et al. 2008), and apoptosis (Song et al. 2013).
KLF5 (Krüppel-like factor 5) is a member of the Krüppel-like factor subfamily of zinc-finger transcription proteins; it is a basic transcription factor which regulates genes transcription by binding to GC boxes of gene promoters. Like other KLFs, the KLF5 has three tandem Cys2His2 zinc-finger motifs located at the extreme C-terminus of the protein; the zinc-finger motifs play functions in protein–protein interactions that modulate DNA-binding specificity (Kaczynski et al. 2003). KLF5 was first identified from a human placenta library using a homology screening strategy (Sogawa et al. 1993). It mediates the signaling functions in cell proliferation, cell cycle, apoptosis, migration, differentiation, and stemness by regulating gene expression in response to environmental stimuli. In NIH3T3 cells, ectopic expression of KLF5 significantly increased the rate of cell proliferation (Sun et al. 2001), KLF5 is upregulated in oncogenic H-Ras-transformed NIH3T3 cells, knockdown of KLF5 expression leads to a decreased proliferation rate and a significant reduction in colony formation (Nandan et al. 2004). In adipocytes, KLF5 is induced at an early stage of differentiation by C/EBPβ/δ, and followed binding to the promoter of PPARγ gene, which activates PPARγ gene expression to promote adipocyte differentiation (Oishi et al. 2005). It has also been reported that KLF5 has an anti-apoptosis function. Liu et al. suggest that KLF5 can promote breast epithelial cell survival by increasing the MKP-1 protein levels. KLF5 can also make smooth muscle cell resistant to apoptosis, in vascular lesions; KLF5 confers apoptotic resistance through interacting with PARP-1, which is a nuclear enzyme important in DNA repair and apoptosis (Suzuki et al. 2007).
In summary, there are lots of evidences to show that KLF5 was essential for the proliferation, differentiation, and apoptosis in different cell types. However, these studies have only been carried out in mammals, no relevant studies have been reported in poultry. In this study, we examined the role of KLF5 in proliferation, differentiation, and apoptosis of chicken SMSCs by determining the effects of KLF5 knockdown by KLF5-specific small interfering RNA (siRNA).
Materials and methods
Isolation, culture, and transfection of chicken SMSCs
Twenty male Ross-308 chickens with 5 days old were sacrificed to isolate SMSCs. Primary chicken SMSCs were isolated as described by Bai et al. (2012) Chicken SMSCs were initially cultured in growth medium composed of 1% blue streptomycin mixture, 10% gestational horse serum, 10% fetal bovine serum (FBS), and Dulbecco’s Modified Eagle Medium (DMEM) of 0.5% chicken embryo extract at 37 °C under 5% CO2 with saturating humidity. Differentiation was induced by replacing the original growth medium with DMEM containing 2% of gestational horse serum, 5% of FBS, and 1% of blue streptomycin mixture.
In this study, three siRNAs for KLF5 were designed and synthesized (synthesized and provided by GenePharma, China) based on the mRNA sequence of Gallus gallus KLF5 gene (Gene ID: 418818) (Table 1). SMSCs were plated in 6-well plates and grown to approximately 70–80% confluence, and cells were transfected with KLF5 siRNAs and negative siRNA. Cell transfection was performed using the reagent protocol Lipofectamine 3000 (Invitrogen, USA). The Lipofectamine 3000 and siRNA were diluted with optim-MEM culture medium. The diluted siRNA and TransEasy were mixed uniformly and placed at room temperature for 15 min. The composite was added to the cell culture plate and mixed in the culture plate. Knocking down efficiency was estimated by quantitative qPCR of KLF5 mRNA.
Table 1.
Design of siRNA target on chicken KLF5 CDS
| siRNA | Sense strands (5′ → 3′) | Anti-sense strands (5′ → 3′) |
|---|---|---|
| KLF5 siRNA 1 | GAAGUACAGAAGAGACAGUTT | ACUGUCUCUUCUGUACUUCTT |
| KLF5 siRNA 2 | UUCACAACCCGAAUUUACCTT | GGUAAAUUCGGGUUGUGAATT |
| KLF5 siRNA 3 | GUAACCCAGAUUUGGAGAATT | UUCUCCAAAUCUGGGUUACTT |
| Negative siRNA | UUCUCCGAACGUGUCACGUTT | ACGUGACACGUUCGGAGAATT |
All experimental procedures involving animals were approved by the Animal Care and Use Committee of College of Animal Science and Technology, Sichuan Agricultural University (No. S20163637), and were carried out in accordance with the National Research Council’s Guide for the Care and Use of Laboratory Animals.
Quantitative qPCR
Total RNA from tissue or cells was isolated by Total RNA Isolation Kit reagent (ROREGENE, China). RNA quality and concentration were evaluated by an Agilent 2100 Bioanalyzer (Agilent Technologies, USA). qPCR primers (Table 2) were designed by Primer Premier 5 software. A 15 μL reaction containing 6.5 μL of SYBR premix Ex Taq TM (TaKaRa Biotechnology, China), 1.5 μL of cDNA, 0.3 μL of forward primer, 0.3 μL of reverse primer, and 6.4 μL of RNasefree H2O (Tiangen, China) was used for qPCR. The reaction was carried out with the following amplification conditions: 95 °C for 30 s, followed by 40 cycles of 95 °C for 10 s, primer-specific annealing temperature for 10 s, and 72 °C for 10 s (Yu et al. 2018). Each experiment was biologically replicated three times. The qPCR data were normalized relative to the expression of β-actin (endogenous control) and the 2−∆∆Ct method was applied to quantify mRNA expression levels.
Table 2.
Primers for qPCR
| Gene | Primer sequences (5′ → 3′) | Annealing temperature (°C) | Product (bp) |
|---|---|---|---|
| KLF5 | F: CGCGCTCGGATGAATTAACG | 60 | 164 |
| R: GATACAGAACAGCCTCGGCA | |||
| PCNA | F: AACACTCAGAGCAGAAGAC | 54 | 225 |
| R: GCACAGGAGATGACAACA | |||
| CCND1 | F: CTCCTATCAATGCCTCACA | 54 | 165 |
| R: TCTGCTTCGTCCTCTACA | |||
| MyoD | F: GCCGCCGATGACTTCTATGA | 60 | 66 |
| R: CAGGTCCTCGAAGAAGTGCAT | |||
| MYHC | F: GAAGGAGACCTCAACGAGATGG | 60 | 138 |
| R: ATTCAGGTGTCCCAAGTCATCC | |||
| Caspase-3 | F: CGGACTGTCATCTCGTTCA | 57 | 186 |
| R: TGGCTTAGCAACACACAAAC | |||
| Survivin | F: GCCTATGCTGAAATGCTGCC | 60 | 246 |
| R: CGCGGAGTGCTTTTTGTGTT | |||
| β-actin | F: GTCCACCGCAAATGCTTCTAA | 59 | 78 |
| R: TGCGCATTTATGGGTTTTGTT |
Cell proliferation assay
SMSCs viability was analyzed by cell counting kit-8 (CCK-8) (Bestbio Biotechnology, China). SMSCs were seeded at a density of 5 × 103 cells/well in a 96-well plate; after transfection negative control siRNA or KLF5 siRNA, cells were incubated with 10 μl CCK-8 for additional 3 h at 37 °C; the absorbance at 450 nm was measured using a Microplate Reader (Thermo, USA). The samples from each treatment at each point had ten replicates.
Western blot analysis
Total cellular proteins from cells were isolated by lysing cells in ice-cold radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, and 1 mM EDTA). Protein samples from individual experiments were pooled for western blotting analysis. The following primary antibodies were used: anti-MYHC (Santa Cruz Biotechnology, Santa Cruz, CA, USA; 1:500 dilution), anti-MyoG (Abcam, San Francisco, CA, USA; 1:500 dilution), and anti-β-actin (N21; Santa Cruz Biotechnology, Santa Cruz, CA, USA; 1:1000 dilution). The membranes were incubated with antibodies at 4 °C overnight and then washed in washing buffer [10 mM Tris-HCI, pH 7.5, 100 mM NaCI, and 0.1% (v/v) Tween 20]. Next, the membranes were treated with horseradish peroxidase-conjugated IgG antibody (Santa Cruz Biotechnology; 1:2000 dilution) for 2 h at room temperature, making use of chemiluminescence as a result. Each experiment was biologically replicated three times. Densitometric analysis of the bands, relative to β-actin, was performed using ImageJ software (National Health Institute, Bethesda, MD, USA). The relative expression of protein was analyzed with Quantity One software.
Cell apoptosis assay
Cells cultured in 6-well plates and the control group was incubated with 0.5 μM staurosporine (STS) at 37 °C with 5% CO2 for 12 h. Apoptosis was measured using an eBioscience™ Annexin V-FITC Apop Kit apoptosis detection kit (Invitrogen, AU). Briefly, cells cultured in 6-well plates were trypsinized, washed, and stained with Annexin V-FITC under darkness for 10 min, and then stained with PI under darkness for 5 min at room temperature, analyzed by flow cytometry (Backman; USA).
Statistical analyses
Comparisons between two groups were analyzed using two-tailed student’s t test. Differences among more than two groups were analyzed using one-way ANOVA followed by Tukey–Kramer post hoc tests. All these analyses were performed using the JMP Pro software (SAS, NC, USA). Values of p < 0.05 were considered statistically significant. All data are shown as mean ± SEM.
Results
Expression of KLF5 gene in different tissues of chickens
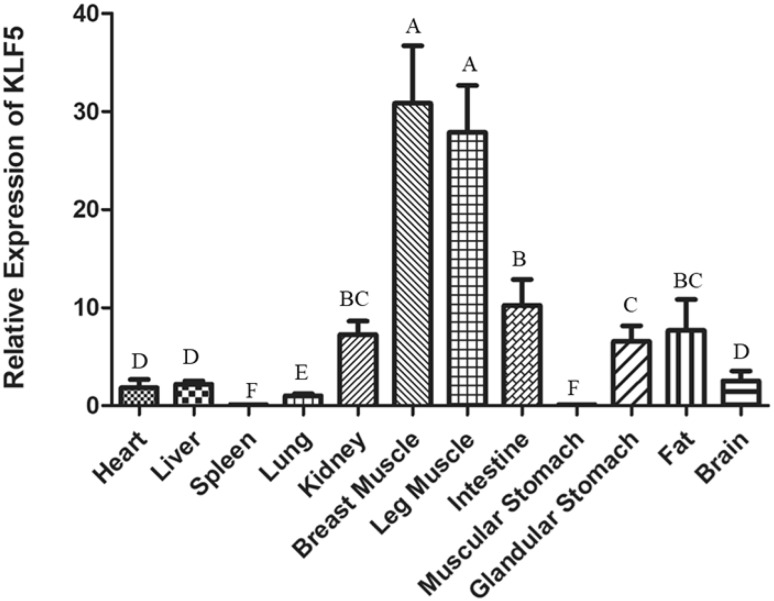
According to qPCR, the expression of KLF5 mRNA was mainly detected in breast muscle and leg muscle among 12 different chicken tissues or organs (Fig. 1). Relative expression levels followed the intestine, kidney, fat, and glandular stomach. The heart, liver, spleen, lung, muscular stomach, and brain were extremely low.
Fig. 1.
The KLF5 mRNA express in different tissues and organs of chicken. The mRNA expression level was measured by qPCR and results are averaged from three independent replicates at 4 days old. The expression level was normalized to β-actin and measured with 2(−ΔΔCt) value. Data are expressed as the mean ± SEM. n = 3
Confirmation of interference efficiency of KLF5 gene
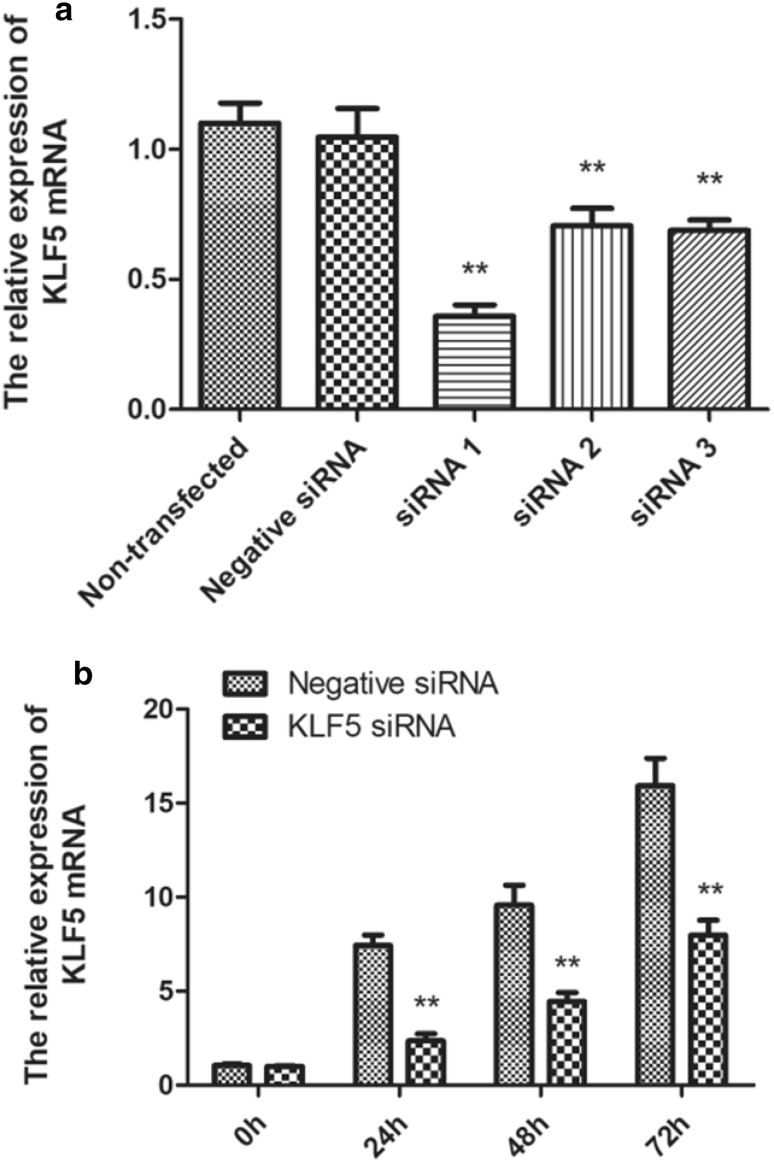
To determine the potential role of KLF5 in chicken SMSCs proliferation, differentiation, and apoptosis, three siRNA molecules were transfected in chicken SMSCs. Cells were transfected with three siRNAs for 24 h and the interference efficiency was quantitatively detected by qPCR, and the interference efficiency was 65.8, 32.6, and 34.2% respectively (Fig. 2a). We chose siRNA1 to subsequent research because of its highest interference efficiency. Based on the results, the expression levels of KLF5 showed gradually increasing at 0 h, 24 h, 48 h, and 72 h (Fig. 2b).
Fig. 2.
The siRNA molecules interfere with mRNA expression of KLF5 in the chicken SMSCs. a Interference efficiency among the three different KLF5-siRNA on 24 h. b Expression of KLF5 at 0, 24, 48, and 72 h after transfection of siRNA1 and negative siRNA. The mRNA expression level was measured by qPCR, normalized to β-actin and measured with 2(−ΔΔCt) value. Data are expressed as the mean ± SEM. n = 3. **p < 0.01 vs the negative siRNA groups
Effects of KLF5 gene knockdown on the proliferation of chicken SMSCs cells
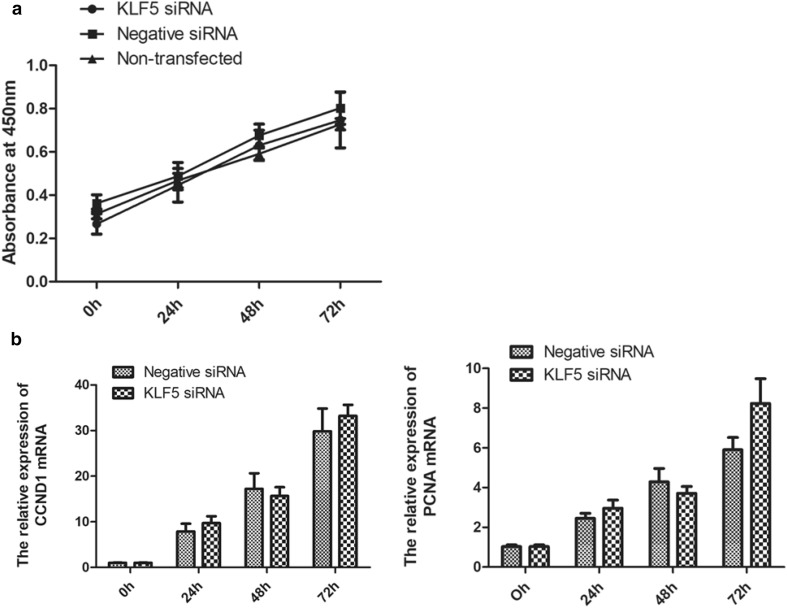
To determine the effects of KLF5 knockdown on cell proliferation, SMSCs were transfected with KLF5 siRNA and negative siRNA after SMSCs achieving 30% cell confluence for 0 h, 24 h, 48 h, and 72 h. Interference effects of KLF5 on SMSCs proliferation rate were detected by CCK-8. The number of cells displayed almost linearly growth during the 72 h period, but there was no significant difference between KLF5 siRNA group and negative siRNA group (Fig. 3a). The regulated factors of CCND1 and PCNA were closely related with cell proliferation (Chen et al. 2017; Guzińska-Ustymowicz et al. 2009), their expression levels were detected by qPCR, and the results also showed that the mRNA expression levels of CCND1, aznd PCNA were no significant difference between KLF5 siRNA group and negative siRNA group (Fig. 3b). These results showed that KLF5 had no effect on the proliferation of chicken SMSCs.
Fig. 3.
Effect of KLF5 knockdown on the proliferation of chicken SMSCs. a Proliferation rate of SMSCs’ cells transfected with KLF5 siRNAs, negative siRNA, or non-transfected. Cell proliferation rate was estimated by CCK-8 at 0 h, 24 h, 48 h, and 72 h after transfection. The absorbance at 450 nm on the y-axis represents the number of viable cells. The proliferation rate was not different between cells transfected with KLF5 siRNAs and negative siRNA or non-transfected. b Relative mRNA expression levels of CCND1 and PCNA at 0 h, 24 h, 48 h, and 72 h detected by qPCR. Data are expressed as the mean ± SEM. n = 3
Effects of KLF5 gene knockdown on the differentiation of chicken SMSCs cells
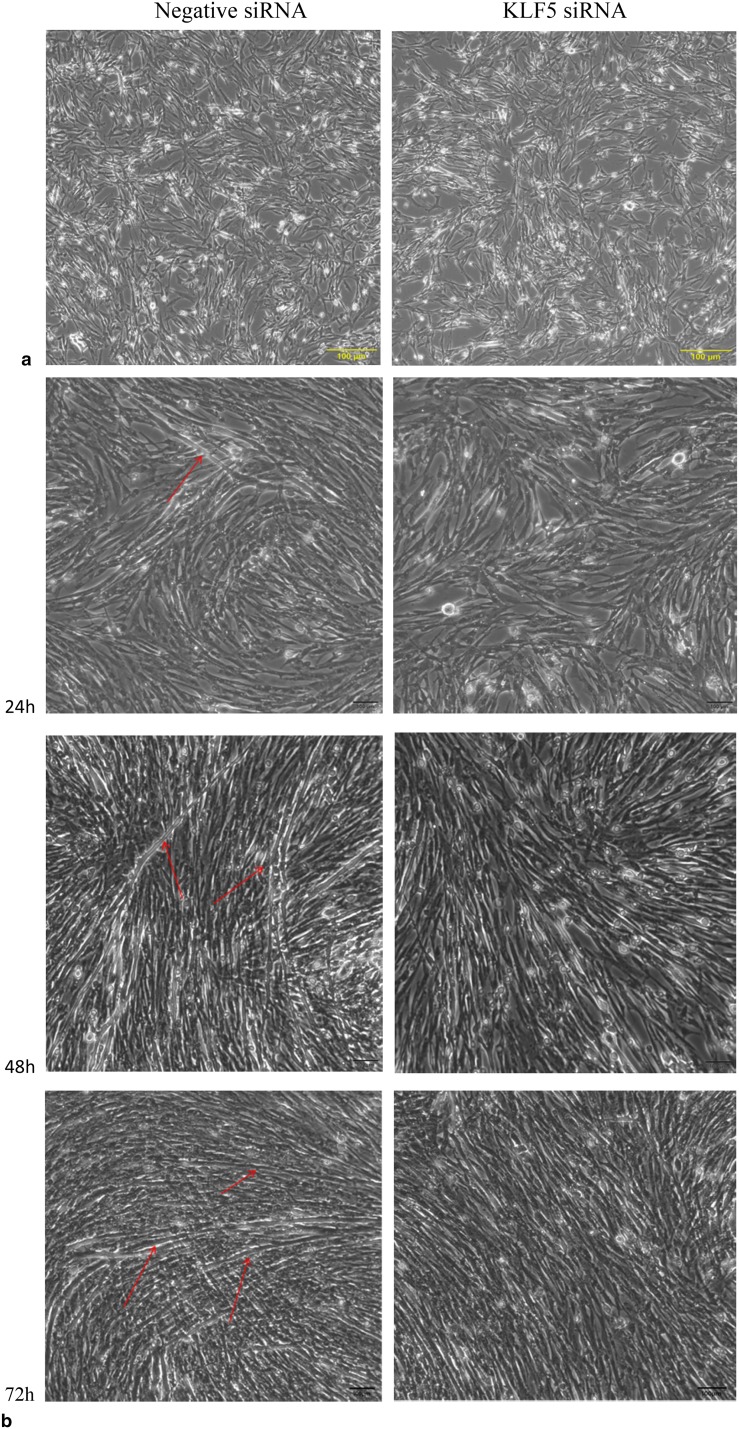
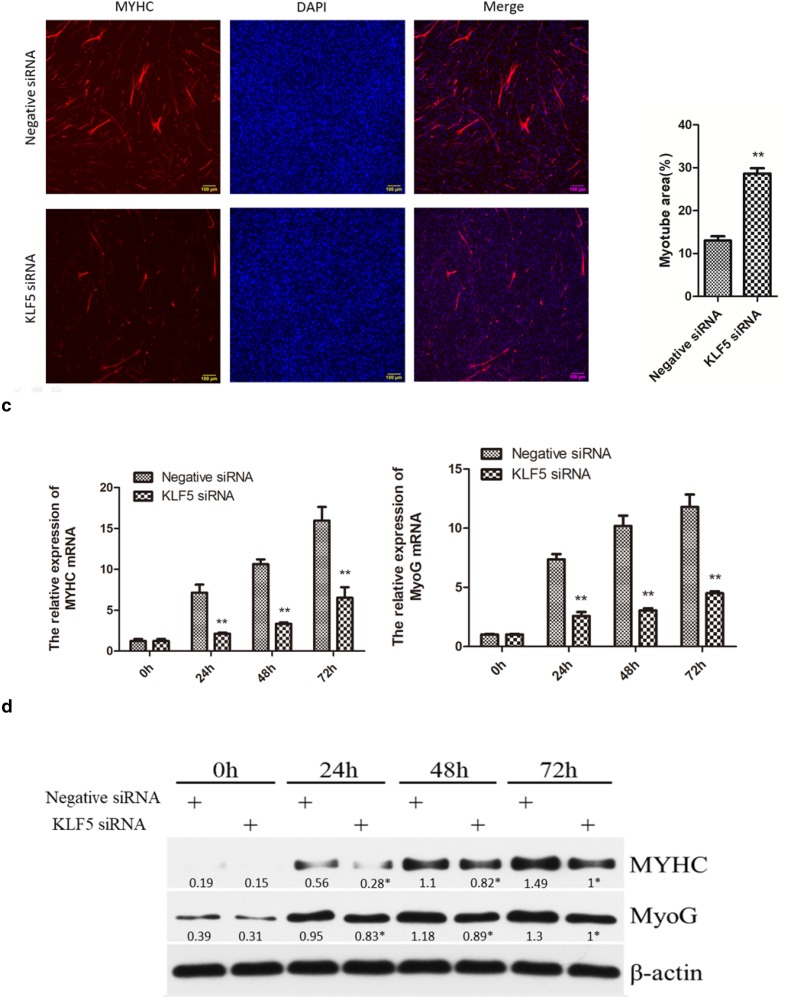
To investigate the effects of KLF5 knockdown on cell differentiation, SMSCs were transfected with negative siRNA and KLF5 siRNA, respectively, after SMSCs achieving 80% cell confluence. As shown in Fig. 4a, morphologically, in negative siRNA group, the SMSCs began to differentiate into myotubes at 24 h after transfecting negative siRNA. At 48 h, the SMSCs were well differentiated into larger myotubes, and the number of myotubes was increased dramatically at 72 h. In contrast, the cells transfected with KLF5 siRNA did not appear to undergo myotube formation. The bright-field images confirmed that differentiation in the SMSCs transfected with KLF5 siRNA was decreased significantly at 24 h, 48 h, and 72 h fusion compared with negative siRNA group. After immunofluorescence staining, we found that the knockdown of KLF5 gene inhibited the differentiation of SMSCs and significantly reduced the total areas of myotubes (Fig. 4b).
Fig. 4.
Effect of KLF5 knockdown on the differentiation of chicken SMSCs. SMSCs were transfected with KLF5 siRNA or negative siRNA, and cultured in differentiation medium for the following experiments. a The myotube formation of cells transfected at 0 h, 24 h, 48 h, and 72 h was observed under the light microscope. Red arrows indicate myotube. Yellow scale bar = 100 μm. b Immunofluorescence SMSCs transfected with KLF5 siRNA or negative siRNA were induced to differentiate for 72 h, and then, cells were stained with MYHC antibody and DAPI (nuclei). Scale bars = 100 μm. c Relative mRNAs’ expression levels of MyoG and MYHC at 0 h, 24 h, 48 h, and 72 h of differentiation. d Western blots of MyoG, MYHC, and β-actin (loading control) proteins at 0 h, 24 h, 48 h, and 72 h of differentiation. Densitometric analysis was determined with ImageJ and average values from three separate experiments. *p < 0.05, ** p < 0.01 v.s. the negative siRNA groups. Data are expressed as the mean ± SEM. n = 3
To further test the differentiation effect of KLF5 on SMSCs, the mRNA and protein expression levels of MYHC and MyoG related to muscle differentiation were detected in 0 h, 24 h, 48 h, and 72 h after transfected with KLF5 siRNA and negative siRNA. Results showed that the mRNA and protein expression levels of MYHC and MyoG in SMSCs with KLF5 siRNA were significantly lower than negative siRNA groups in 24 h, 48 h, and 72 h (p < 0.01) (Fig. 4b, c). These results indicated that KLF5 had a positive relationship with the differentiation of chicken SMSCs.
Effects of KLF5 gene knockdown on the apoptosis of chicken SMSCs’ cells
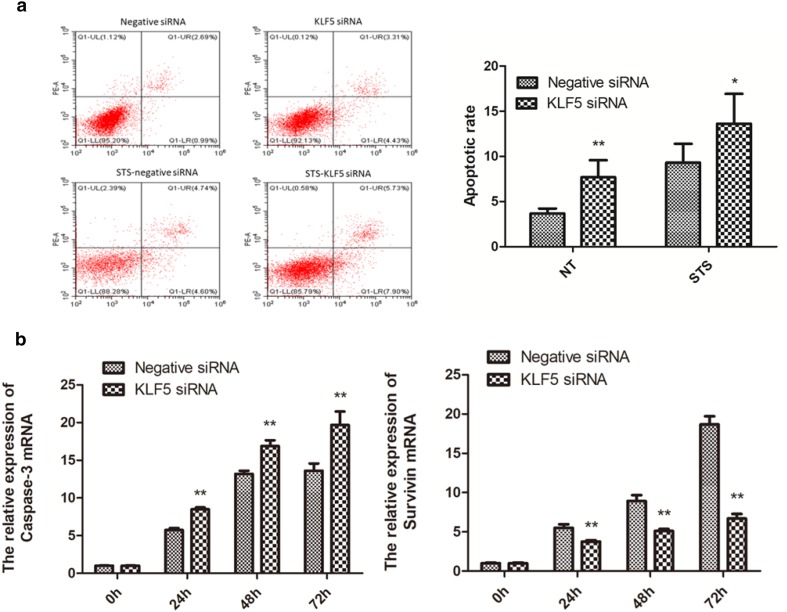
To explore the effects of KLF5 knockdown on cell apoptosis, flow cytometry was performed to detect the cell apoptosis rate after transfected with KLF5 siRNA and negative siRNA on 48 h, and the control group was transfected STS which can induce apoptosis before 12 h of detection. The results showed that the apoptotic rate of KLF5 siRNA group was significantly higher than negative siRNA group whether STS was transfected or not (Fig. 5a). Caspase-3 is widely believed to be a pro-apoptotic factor, the survivin is a survival factor making cells resistant to apoptosis, and their expression levels are closely related with the apoptosis. In the present study, the mRNA levels of caspase-3 were significantly upregulated and survivin were significantly downregulated in the KLF5 knockdown group (p < 0.01) in 24 h, 48 h, and 72 h after transfected with KLF5 siRNA (Fig. 5b). Altogether, knockdown of KLF5 could induce the apoptosis in chicken SMSCs.
Fig. 5.
Effect of KLF5 knockdown on the apoptosis of chicken SMSCs. a Apoptosis in SMSCs was measured by flow cytometry following KLF5 siRNA or negative siRNA transfection at 48 h; the control groups were treated with 0.5 μM STS for 12 h. b Relative mRNA expression levels of caspase-3 and survivin at 0 h, 24 h, 48 h, and 72 h *p < 0.05, **p < 0.01 vs the negative siRNA groups. Data are expressed as the mean ± SEM. n = 3
Discussion
The previous studies have shown that KLF5 played an essential role for the cell proliferation, differentiation, and apoptosis in mammals (Dong and Chen 2009), but its regulatory role in poultry is not clear. In this study, we determined the expression pattern of KLF5 gene in chicken. KLF5 was highly expressed in skeletal muscle compared with other tissues in chicken, which indicated that KLF5 is important in the growth, maintenance, and function of chicken skeletal muscle. Hence, we knocked down KLF5 to identify the potential regulation mechanisms of KLF5 in chicken SMSCs proliferation, differentiation, and apoptosis.
After KLF5 knockdown, the proliferation rate of SMSCs had no significant difference compared with control group (p > 0.05), and both CCND1 and PCNA related to SMSCs proliferation also had no significant difference (p > 0.05) on mRNA levels compared with the control group. Although the previous reports have shown the involvement of KLF5 in controlling cell proliferation in several cancer cell lines (Chen et al. 2010; Dong et al. 2012; Yang et al. 2005), but Hayashi et al. (2016) found that KLF5 did not affect C2C12 myoblasts cell proliferation. Combined with our present results suggests that KLF5 has no direct regulatory effect on the proliferation in SMSCs of chicken. And we speculate that KLF5 is not involved in the proliferation of muscle cells.
Knockdown KLF5 in differentiated SMSCs, myotube formation was blocked. In addition, muscle differentiation-related genes MYHC and MyoG exhibited significantly lower expression levels of mRNA and protein compared with the control group. These results were consistent with Hayashi et al. (2016), they found that myotube formation was significantly impaired in the KLF5-null C2C12 cells of mice using CRISPR-Cas 9 system compared with the control group, and expression of MYHC and MyoG was significantly reduced in KLF5-null group. To determine whether forced expression of KLF5 could rescue the compromised differentiation of KLF5-null cells, they exogenously introduced a retrovirus harboring murine KLF5, found KLF5-null cells successfully formed myotubes when stimulated to differentiate, and the expression of MYHC and MyoG was partially rescued by the forced expression of KLF5. These evidences indicate that KLF5 is essential for myogenic differentiation. KLF5 regulates skeletal muscle differentiation acting in concert with myogenic transcription factors such as MyoD and Mef2, and MyoD recruitment will greatly reduce in the absence of KLF5 (Hayashi et al. 2016). In the other hand, KLF5 is also a regulator of many others’ cell differentiation, such as embryonic stem cells (Parisi et al. 2008), smooth muscle cell (Nagai et al. 2003), and epithelial cell (Yang et al. 2007). In summary, KLF5 plays an important role in promoting differentiation of many kinds of cells including skeletal muscle cells.
Knockdown KLF5 in chicken SMSCs, the apoptotic rate was significantly higher than negative siRNA group, the same as the results with transfecting STS which can induce apoptosis, and the mRNA expression levels of survivin were significantly downregulated and caspase-3 were significantly upregulated compared with negative siRNA group. The results were similar to the previous studies. Zhu et al. (2006) found that KLF5 could induce the expression of survivin, and KLF5 binded to the promoter of survivin and interacted with p53 to abrogate p53-repressed survivin expression. KLF5 can also regulate apoptosis independent of p53, knockdown KLF5 would induce apoptosis, the apoptotic phenotype was associated with reduced bad phosphorylation, and downregulation of Pim1 survival kinase, and transfection of wild-type Pim1 was sufficient to rescue the phenotype (Zhao et al. 2008). And Xiaochen et al. (2014) found that KLF5 knockdown inhibited hypoxia-induced cell survival and promoted cell apoptosis by direct interaction with HIF-1α to actively downregulate cyclinB1 and survivin, and upregulate caspase-3. Combined with these studies, we suggest that KLF5 negatively regulates SMSCs’ apoptosis, which promoted cell survival.
In summary, this study analyzed the effects of KLF5 on proliferation, differentiation, and apoptosis in SMSCs of chicken. We found that knockdown KLF5 suppressed SMSCs differentiation and induced SMSCs apoptosis, but there was no direct effect on SMSCs proliferation, suggesting that KLF5 plays an essential role during myogenesis, which will affect muscle repair and regeneration of muscle, and may ameliorate muscle aging or sarcopenia. Further studies will be needed to investigate the application of KLF5 as a skeletal muscle repair factor in chicken muscle development. Verification of the anti-apoptotic function of KLF5 in future studies may allow the development of new therapies for poultry disease.
Acknowledgements
This research was supported by the Open Fund of Farm Animal Genetic Resources Exploration and Innovation Key Laboratory of Sichuan Province (Grant No. 2016NYZ0043 and 2017JZ0033).
Compliance with ethical standards
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
References
- Allen RE, Sheehan SM, Taylor RG, Kendall TL. Hepatocyte growth factor activates quiescent skeletal muscle satellite cells in vitro. J Cell Physiol. 2010;165:307. doi: 10.1002/jcp.1041650211. [DOI] [PubMed] [Google Scholar]
- Bai C, Hou L, Li F, He X, Zhang M, Guan W. Isolation and biological characteristics of beijing Fatty chicken skeletal muscle satellite cells Cell. Commun Adhes. 2012;19:69–77. doi: 10.3109/15419061.2012.743998. [DOI] [PubMed] [Google Scholar]
- Buckingham Margaret, Rigby PWJ. Gene regulatory networks and transcriptional mechanisms that control myogenesis developmental. Cell. 2014;28:225–238. doi: 10.1016/j.devcel.2013.12.020. [DOI] [PubMed] [Google Scholar]
- Chen C, Benjamin MS, Sun X, Otto KB, Guo P. KLF5 promotes cell proliferation and tumorigenesis through gene regulation and the TSU-Pr1 human bladder cancer cell line. Int J Cancer. 2010;118:1346–1355. doi: 10.1002/ijc.21533. [DOI] [PubMed] [Google Scholar]
- Chen DG, Zhu B, Lv SQ, ZhuH Tang J. Inhibition of EGR1 inhibits glioma proliferation by targeting CCND1 promoter. J Exp Clinl Cancer Res Cr. 2017;36:186. doi: 10.1186/s13046-017-0656-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelison DD, Olwin BB, Rudnicki MA, Wold BJ. MyoD(-/-) satellite cells in single-fiber culture are differentiation defective and MRF4 deficient. Dev Biol. 2000;224:122–137. doi: 10.1006/dbio.2000.9682. [DOI] [PubMed] [Google Scholar]
- Dong JT, Chen C. Essential role of KLF5 transcription factor in cell proliferation and differentiation and its implications for human diseases. Cell Mol Life Sci. 2009;66:2691–2706. doi: 10.1007/s00018-009-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z, Xu Z, Zhou Z, Chen C. TAZ antagonizes the WWP1-mediated KLF5 degradation and promotes breast cell proliferation and tumorigenesis. Carcinogenesis. 2012;33:59–67. doi: 10.1093/carcin/bgr242. [DOI] [PubMed] [Google Scholar]
- Florini JR, Ewton DZ, Roof SL. Insulin-like growth factor-I stimulates terminal myogenic differentiation by induction of myogenin gene expression. Mol Endocrinol. 1991;5:718. doi: 10.1210/mend-5-5-718. [DOI] [PubMed] [Google Scholar]
- Guzińska-Ustymowicz K, Pryczynicz A, Kemona A, Czyzewska J. Correlation between proliferation markers: pCNA, Ki-67, MCM-2 and antiapoptotic protein Bcl-2 in colorectal cancer. Anticancer Res. 2009;29:3049. [PubMed] [Google Scholar]
- Hayashi S, Manabe I, Suzuki Y, Relaix F, Oishi Y. Klf5 regulates muscle differentiation by directly targeting muscle-specific genes in cooperation with MyoD in mice. Elife. 2016 doi: 10.7554/elife.17462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003;4:1–8. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows E, Cho JH, Flynn JM, Klein WH. Myogenin regulates a distinct genetic program in adult muscle stem cells. Dev Biol. 2008;322:406–414. doi: 10.1016/j.ydbio.2008.07.024. [DOI] [PubMed] [Google Scholar]
- Nagai R, Shindo T, Manabe I, Suzuki T, Kurabayashi M. KLF5/BTEB2, a Krüppel-like zinc-finger type transcription factor, mediates both smooth muscle cell activation and cardiac hypertrophy. Adv Exp Med Biol. 2003;538:57. doi: 10.1007/978-1-4419-9029-7_5. [DOI] [PubMed] [Google Scholar]
- Nandan MO, Hong S, Yoon Weidong Z, Ouko LA, Sengthong C, Yang VW. Krüppel-like factor 5 mediates the transforming activity of oncogenic. H-Ras Oncogene. 2004;23:3404. doi: 10.1038/sj.onc.1207397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi Y, Manabe I, Tobe K, Tsushima K, Shindo T. Krüppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab. 2005;1:27–39. doi: 10.1016/j.cmet.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Parisi S, Passaro F, Aloia L, Manabe I, Nagai R, Pastore L, Russo T. Klf5 is involved in self-renewal of mouse embryonic stem cells. J Cell Sci. 2008;121:2629–2634. doi: 10.1242/jcs.027599. [DOI] [PubMed] [Google Scholar]
- Shin J, Mcfarland DC, Velleman SG. Heparan sulfate proteoglycans, syndecan-4 and glypican-1, differentially regulate myogenic regulatory transcription factors and paired box 7 expression during turkey satellite cell myogenesis: implications for muscle growth. Poult Sci. 2012;91:201–207. doi: 10.3382/ps.2011-01695. [DOI] [PubMed] [Google Scholar]
- Sogawa K, Imataka H, Yamasaki Y, Kusume H, Abe H, Fujiikuriyama Y. cDNA cloning and transcriptional properties of a novel GC box-binding protein, BTEB2. Nucleic Acids Res. 1993;21:1527–1532. doi: 10.1093/nar/21.7.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Mcfarland DC, Velleman SG. Role of syndecan-4 side chains in turkey satellite cell apoptosis and focal adhesion formation. Cell Biol Int. 2013;36:433–440. doi: 10.1042/CBI20110467. [DOI] [PubMed] [Google Scholar]
- Stickland NC, Bayol S, Ashton C, Rehfeldt C, Pas MFWT, Everts ME, Haagsman HP. Manipulation of muscle fibre number during prenatal development. Muscle Dev Livest Anim. 2004;2004:69–82. [Google Scholar]
- Sun R, Chen X, Yang VW. Intestinal-enriched Krüppel-like factor (Krüppel-like Factor 5) Is a positive regulator of cellular proliferation. Jbiolchem. 2001;276:6897–6900. doi: 10.1074/jbc.C000870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Nishi T, Nagino T, Suzuki T, Sasaki K, Aizawa K, Kada N. Functional interaction between the transcription factor krüppel-like factor 5 and poly(adp-ribose) polymerase-1 in cardiovascular apoptosis. J Biol Chem. 2007;282(13):9895–9901. doi: 10.1074/jbc.M608098200. [DOI] [PubMed] [Google Scholar]
- Velleman SG. The role of the extracellular matrix in skeletal muscle development. Poult Sci. 1999;79:778–784. doi: 10.1093/ps/78.5.778. [DOI] [PubMed] [Google Scholar]
- Xiaochen L, Xiansheng L, Yongjian X, Jin L, Min X, Wang N, Shixin C. KLF5 promotes hypoxia-induced survival and inhibits apoptosis in non-small cell lung cancer cells via HIF-1α. Int J Oncol. 2014;45:1507. doi: 10.3892/ijo.2014.2544. [DOI] [PubMed] [Google Scholar]
- Yang Y, Goldstein BG, Chao HH, Katz J. KLF4 and KLF5 regulate proliferation, apoptosis and invasion in esophageal cancer cells. Cancer Biol Ther. 2005;4:1216–1221. doi: 10.4161/cbt.4.11.2090. [DOI] [PubMed] [Google Scholar]
- Yang Y, Goldstein BG, Nakagawa H, Katz JP. Krüppel-like factor 5 activates MEK/ERK signaling via EGFR in primary squamous epithelial cells. Faseb J. 2007;21:543–550. doi: 10.1096/fj.06-6694com. [DOI] [PubMed] [Google Scholar]
- Yu LT, Xiao YP, Li JJ, Ran JS, Yin LQ, Liu YP, Zhang L. Molecular characterization of a novel ovodefensin gene in chickens. Gene. 2018;10:233–240. doi: 10.1016/j.gene.2018.08.029. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Hamza MS, Leong HS, Lim CB, Pan YF. Kruppel-like factor 5 modulates p53-independent apoptosis through Pim1 survival kinase in cancer cells. Oncogene. 2008;27:1. doi: 10.1038/sj.onc.1210625. [DOI] [PubMed] [Google Scholar]
- Zhu N, Gu L, Findley HW, Chen C, Dong JT, Yang L, Zhou M. KLF5 Interacts with p53 in regulating survivin expression in acute lymphoblastic leukemia. J Biol Chem. 2006;281:14711–14718. doi: 10.1074/jbc.M513810200. [DOI] [PubMed] [Google Scholar]