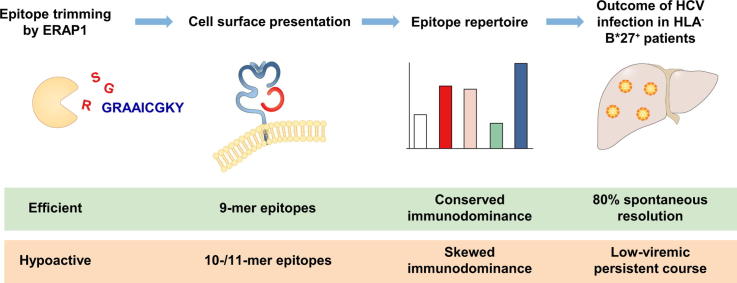
Graphical abstract
Keywords: Epitope repertoire, ERAP1, Hepatitis C virus, HLA-B*27, T cells
Highlights
-
•
ERAP1 polymorphisms are strongly linked with HLA class I-associated autoinflammatory disorders.
-
•
We identified 2 hypoactive ERAP1 allotypes in an HLA-B*27:05+ individual with acute HCV infection.
-
•
These ERAP1 allotypes modified the HCV-specific CD8+ T cell epitope repertoire in vivo, leading to altered immunodominance patterns.
-
•
Altered immunodominance patterns potentially contributed to the failure of antiviral immunity.
Abstract
Background & Aims
Endoplasmic reticulum aminopeptidase 1 (ERAP1) polymorphisms are linked with human leukocyte antigen (HLA) class I-associated autoinflammatory disorders, including ankylosing spondylitis and Behçet’s disease. Disease-associated ERAP1 allotypes exhibit distinct functional properties, but it remains unclear how differential peptide trimming in vivo affects the repertoire of epitopes presented to CD8+ T cells. The aim of this study was to determine the impact of ERAP1 allotypes on the virus-specific CD8+ T cell epitope repertoire in an HLA-B*27:05+ individual with acute hepatitis C virus (HCV) infection.
Methods
We performed genetic and functional analyses of ERAP1 allotypes and characterized the HCV-specific CD8+ T cell repertoire at the level of fine epitope specificity and HLA class I restriction, in a patient who had acquired an HCV genotype 1a infection through a needle-stick injury.
Results
Two hypoactive allotypic variants of ERAP1 were identified in an individual with acute HCV infection. The associated repertoire of virus-derived epitopes recognized by CD8+ T cells was uncommon in a couple of respects. Firstly, reactivity was directed away from classically immunodominant epitopes, preferentially targeting either novel or subdominant epitopes. Secondly, reactivity was biased towards longer epitopes (10–11-mers). Despite the patient exhibiting favorable prognostic indicators, these atypical immune responses failed to clear the virus and the patient developed persistent low-level infection with HCV.
Conclusions
ERAP1 allotypes modify the virus-specific CD8+ T cell epitope repertoire in vivo, leading to altered immunodominance patterns that may contribute to the failure of antiviral immunity after infection with HCV.
Lay summary
Endoplasmic reticulum aminopeptidase 1 (ERAP1) plays a key role in antigen presentation. Genetic variants of ERAP1 (leading to distinct allotypes) are linked with specific autoinflammatory disorders, such as ankylosing spondylitis and Behçet’s disease. We found that ERAP1 allotypes modified the repertoire of virus-specific CD8+ T cell epitopes in a patient with hepatitis C virus, leading to an altered pattern of immunodominance that may have contributed to the failure of antiviral immunity in this patient.
Introduction
Endoplasmic reticulum aminopeptidase 1 (ERAP1) trims peptides to an optimal length (usually 8 or 9 amino acids) for presentation in the context of human leukocyte antigen (HLA) class I molecules.[1], [2] Genome-wide association studies have identified single nucleotide polymorphisms (SNPs) in ERAP1 as important risk factors in several HLA class I-associated autoinflammatory disorders,3 including ankylosing spondylitis,4 especially in conjunction with HLA-B*27,5 and Behçet’s disease, which is strongly linked with HLA-B*51.6 In addition, SNPs in ERAP1 can combine to encode discrete allotypes with composite functional properties that further increase the risk of disease.7 Biochemical analyses have revealed differential peptide trimming among ERAP1 allotypes, with hypoactive forms typically generating longer fragments (10–12-mers), and hyperactive forms typically generating shorter fragments (7–8-mers).[8], [9], [10] However, the impact of specific ERAP1 allotypes on the naturally presented repertoire of peptide epitopes is currently unclear.11
Hepatitis C virus (HCV) has infected approximately 71 million people worldwide. Persistent infection ensues in 50–80% of cases, with a high risk of liver disease, which may progress to cirrhosis, liver failure, and hepatocellular carcinoma.12 Virus-specific CD8+ T cells are thought to play a key role in immune protection against HCV.13 This consensus is predicated on associative studies that have linked viral clearance with potent and broadly directed CD8+ T cell responses and the presence of certain HLA class I allotypes.14 In particular, up to 80% of HLA-B*27+ individuals clear HCV spontaneously, contrasting with only 20–50% of individuals in the general population.15 We previously attributed this protective effect to the immunodominant HLA-B*27-restricted CD8+ T cell epitope NS5B2841-2849 (ARMILMTHF), which is targeted in almost all HLA-B*27+ individuals during acute infection with HCV.[16], [17]
In this study, we performed a comprehensive analysis of ERAP1 allotypes and virus-specific CD8+ T cell responses in an HLA-B*27:05+ individual with acute HCV infection. Despite a symptomatic presentation, expression of HLA-B*27, and a favorable interleukin 28B (IL28B)/interferon lambda 4 (IFNL4) genotype, all of which predict viral clearance, this individual progressed to chronic infection, with low-level viremia (mostly <100 IU/ml) throughout a follow-up period of 20 months before the initiation of antiviral therapy. In-depth genetic, biochemical, and immunological analyses revealed the presence of ERAP1 allotypes with hypoactive trimming functions, which shaped the epitope repertoire to elicit atypical HCV-specific CD8+ T cell responses.
Patients and methods
Samples
A donor (MM) with acute HCV genotype 1a infection acquired via needle-stick injury was recruited after providing written informed consent in line with federal guidelines and the Declaration of Helsinki. Ethical approval for this study was obtained from the Ethik-Kommission der Albert-Ludwigs-Universität Freiburg (#524/14). Venous blood samples (50 ml per draw) were collected in ethylene diamine tetraacetic acid (EDTA)-anticoagulated tubes at longitudinal time points. Peripheral blood mononuclear cells (PBMCs) were isolated using lymphocyte separation medium density gradients (PAA Laboratories, Austria) and resuspended in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% fetal calf serum, 1% streptomycin/penicillin, and 1.5% HEPES buffer 1 M (complete medium; all additives from Thermo Fisher Scientific, Germany).
Peptides, antibodies, and multimers
Overlapping peptides (mostly 15 amino acids each, overlapping by 11 amino acids) spanning the entire polyprotein of HCV strain H77 (genotype 1a) were obtained from BEI Resources (Manassas, VA, USA) via the National Institute of Allergy and Infectious Diseases (Bethesda, MD, USA). Additional peptides were synthesized with a free amine NH2 terminus and a free acid COOH terminus using standard Fmoc chemistry (Genaxxon Bioscience, Germany). The viability dye 7-aminoactinomycin D (7-AAD) and the directly conjugated monoclonal antibodies (mAbs) anti-CD8-PE (clone RPA-T8), anti-CD8-APC (clone SK1), anti-CD8-BV421 (clone RPA-T8), and anti-IFN-γ-FITC (clone 25723.11) were obtained from BD Biosciences (Germany). Multimeric complexes of HLA-B*27:05/NS5B2936-2944 (GRAAICGKY) and HLA-B*27:05/NS5B2936-2946 (GRAAICGKYLF) were generated as described previously.18
CD8 selection and polyclonal expansion of PBMCs
Procedures were carried out as described previously.19 More details may be found in the supplementary information.
Peptide-specific T cell lines
PBMCs were activated with peptides as described previously.17 More details may be found in the supplementary information.
Enzyme-linked immunospot assay
An IFN-γ enzyme-linked immunospot (ELISpot) kit (Diaclone, France) was used to quantify peptide-specific T cells as described previously.17 Peptides were distributed across 92 pools in a matrix format (n = 10 peptides per pool). Each peptide was present in 2 different pools, allowing identification of specific responses to individual peptides. Tests with peptide pools were performed in duplicate. In each assay, negative control wells (n = 4) contained medium alone, and positive control wells (n = 4) contained medium supplemented with phytohemagglutinin or phorbol 12-myristate 13-acetate (PMA) and ionomycin (Sigma-Aldrich, Germany). Responses were considered positive if the mean number of spots per test well was at least 3 times as high as the mean number of spots per negative control well. Positive responses were confirmed by intracellular IFN-γ staining with single peptides.
Intracellular IFN-γ staining
Procedures were carried out as described previously.20 More details may be found in the supplementary information.
HLA/peptide multimer staining
Procedures were carried out as described previously.21 More details may be found in the supplementary information.
Isolation and cloning of ERAP1 allotypes
ERAP1 was amplified from donor complementary DNA as described previously.[7], [10] The amplicon was cloned into pcDNA3.1™ (Invitrogen, UK) and sequenced to identify both chromosomal allotypes of ERAP1. Sequences from donor MM were compared with the prototype (PT) ERAP1 using DNADynamo alignment software (Blue Tractor Software, UK).
DNA constructs
The pcDNA3.1 minigenes (ES)-SHL8 and (ES)-X-SHL8 were engineered to encode an endoplasmic reticulum (ER) targeting signal sequence.10 The pcDNA3.1-(ES)-SRG-SHL8 construct was generated via the incorporation of an additional nucleotide sequence encoding serine, arginine, and glycine (SRG) in pcDNA3.1-(ES)-SHL8, engineered using the primers 5′-CTTGCGGCAGTCTGCAGCGCGAGCCGTGGCAGCATCATCAACTTCGAGCAC-3′ and 5′-GTGCTCGAAGTTGATGATGCTGCCACGGCTCGCGCTGCAGACTGCCGCAAG-3′. Three HCV epitope-specific constructs were generated via the insertion of oligonucleotides into the EcoRI/XbaI sites of pcDNA3.1, encoding an ER translocation signal sequence followed by the amino acid sequences GRAAICGKY, SRGGRAAICGKY, or GRAAICGKYLF. Plasmid constructs encoding PT ERAP1, E320A non-functional ERAP1, and HLA-B*27:05 were described previously.7 All constructs were sequence-verified using DNADynamo alignment software (Blue Tractor Software, UK).
Cell lines and transfections
ERAP1 knock-out 293T (293T E1KO) cells were transfected using the FuGENE 6 Transfection Reagent (Promega, UK).7 For assessment of ERAP1 trimming activity, 293T E1KO cells were transfected with 1 µg total DNA, consisting of 0.5 µg PT ERAP1, 0.5 µg E320A non-functional ERAP1, or 0.25 µg of each MM allotype-encoded ERAP1, together with 0.25 µg H2-Kb and 0.25 µg SHL8 or the extended precursor SRG-SHL8. For assessment of ERAP1 amino acid specificity, 293T E1KO cells were transfected with 0.1 µg total DNA, consisting of 0.05 µg MM allotype-encoded ERAP1, 0.025 µg H2-Kb, and 0.025 µg X-SHL8. All transfected cells were incubated for 24 h at 37 °C. Presentation of trimmed SHL8 and activation of the LacZ-inducible B3Z T cell hybridoma were assessed by measurement of intracellular LacZ activity using the substrate chlorophenol red-β-d-galactopyranoside (Roche, UK).10
Activation of donor MM T cells
The 293T E1KO cells were transfected with 1 µg total DNA, consisting of 0.5 µg ERAP1, 0.25 µg HLA-B*27:05, and 0.25 µg GRAAICGKY, SRGGRAAICGKY, or GRAAICGKYLF. Transfected cells were incubated for 24 h, harvested, and cultured for a further 24 h with donor MM T cells at an effector-to-target ration of 3:1. Activation of donor MM T cells was determined by IFN-γ staining as described.
Statistical analysis
Differences among multiple groups were evaluated for significance using a 1-way ANOVA with Dunnett’s post hoc test implemented in GraphPad Prism (www.graphpad.com).
For further details regarding the materials used, please refer to the CTAT table and supplementary information.
Results
Persistent low-level viremia after infection with HCV despite multiple predictors of spontaneous viral clearance
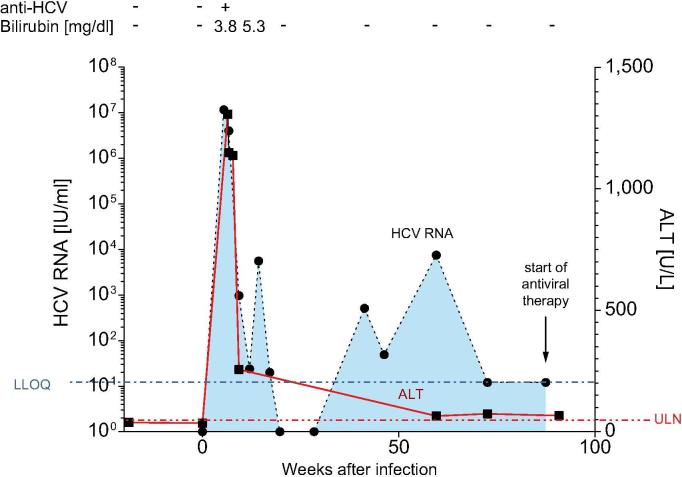
Donor MM experienced an accidental needle-stick injury from an HCV genotype 1a-infected patient and developed symptomatic HCV infection over a period of 20 months (Fig. 1). Seven weeks after the needle-stick injury, alanine aminotransferase peaked at 1,307 U/L, viremia peaked at 1.15 × 107 IU/ml, HCV-specific antibodies became detectable in the serum, and the donor suffered from fatigue, nausea, and moderate jaundice (bilirubin peaked at 5.3 mg/dl). Further virological analyses confirmed infection with HCV genotype 1a. Genetic analyses revealed that donor MM had the favorable IFNL4 (previously named IL28B) genotype CC (SNP rs12979860) and expressed the HLA class I molecules A*01:01, A*26:01, B*08:01, and B*27:05. Three positive predictors of spontaneous viral clearance were therefore present in this individual, namely symptomatic infection,22 IFNL4 genotype CC,23 and HLA-B*27.15 However, donor MM continued to display moderately elevated liver enzymes and remained positive for HCV RNA, albeit below the lower level of quantification (LLOQ, 12 IU/ml). In addition, donor MM suffered from oscillating complaints, including subfebrile temperatures and flu-like symptoms. After 20 months, treatment was initiated with direct-acting antivirals (DAAs), leading to normalization of liver enzymes, clearance of the virus (negative HCV RNA 12 and 24 weeks after discontinuation of DAAs) and resolution of symptoms.
Fig. 1.
Clinical course of HCV infection in donor MM. The needle-stick injury was set as day 0. HCV, hepatitis C virus; LLOQ, lower level of quantification (12 IU/ml); ULN, upper limit of normal (50 U/L).
Lack of viral clearance associated with altered HCV-specific CD8+ T cell responses
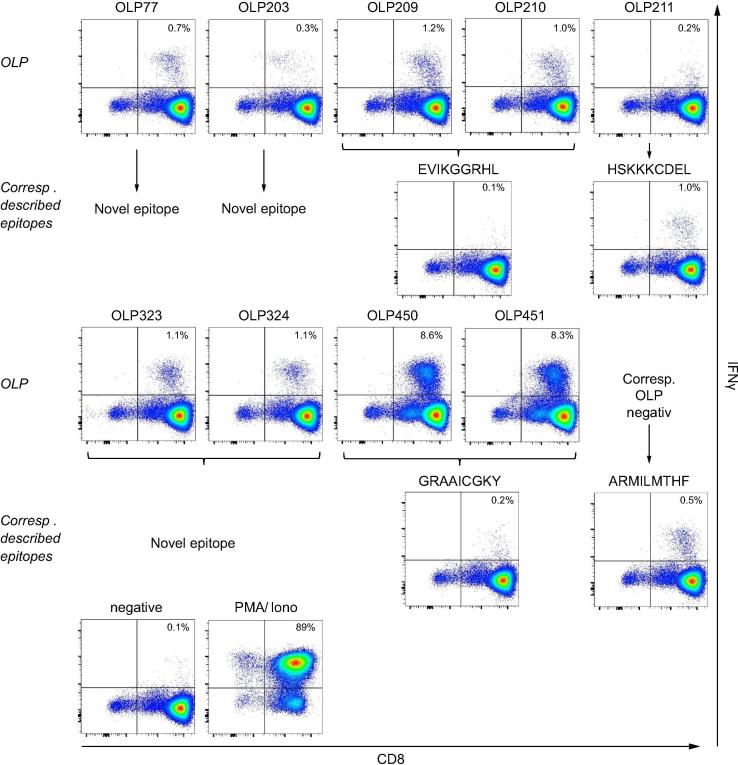
To determine the immunological correlates of persistent infection in donor MM, we performed a comprehensive analysis of HCV-specific CD8+ T cell responses, which are thought to play a key role in viral clearance.13 We used 460 overlapping peptides (OLPs) spanning the complete HCV genotype 1a polyprotein and performed IFN-γ ELIspot assays in a matrix format.[24], [25] Positive responses were confirmed individually by intracellular staining for IFN-γ. Using this approach, we identified 6 HCV-specific CD8+ T cell responses in donor MM (OLP-77, OLP-203, OLP-209/210, OLP-211, OLP-323/324, and OLP-450/451; Fig. 2 and Table 1).
Fig. 2.
Comprehensive analysis of HCV-specific CD8+ T cell responses in donor MM. OLPs spanning the complete HCV proteome (n = 460) were tested in IFN-γ ELISpot assays using a matrix format. Individual responses were confirmed via intracellular IFN-γ staining. Previously described HCV-derived epitopes restricted by HLA-A*01:01, HLA-A*26:01, HLA-B*08:01, or HLA-B*27:05 were tested individually. Only positive responses are displayed. Medium alone served as the negative control; medium supplemented with PMA and ionomycin served as the positive control. HCV, hepatitis C virus; OLPs, overlapping peptides.
Table 1.
Summary of responses to OLPs and previously described HCV-specific epitopes in donor MM.
| Peptide | HCV location | Peptide aa sequence | HLA restriction |
|---|---|---|---|
| OLP77 | E2511-527 | PSPVVVGTTDRSGAPTY | A*01:01 |
| OLP203 | NS31336-1353 | AETAGARLVVLATATPPG | A*26:01 |
| OLP209 | NS31376-1393 | YGKAIPLEVIKGGRHLIF | A*26:01 |
| OLP210 | NS31383-1399 | EVIKGGRHLIFCHSKKK | |
| predescribed | NS31383-1391 | EVIKGGRHL | |
| OLP211 | NS31389-1406 | RHLIFCHSKKKCDELAKK | B*08:01 |
| predescribed | NS31395-1403 | HSKKKCDEL | |
| OLP323 | NS5A2124-2141 | ELDGVRLHRFAPPCKPLL | B*27:05 |
| OLP324 | NS5A2131-2148 | HRFAPPCKPLLREEVSFR | |
| predescribed | NS5B2841-2849 | ARMILMTHF | B*27:05 |
| OLP450 | NS5B2930-2946 | RLLSRGGRAAICGKYLF | B*27:05 |
| OLP451 | NS5B2936-2953 | GRAAICGKYLFNWAVRTK | |
| predescribed | NS5B2936-2944 | GRAAICKGY |
Optimal epitopes are indicated in bold. HCV, hepatitis C virus; OLPs, overlapping peptides.
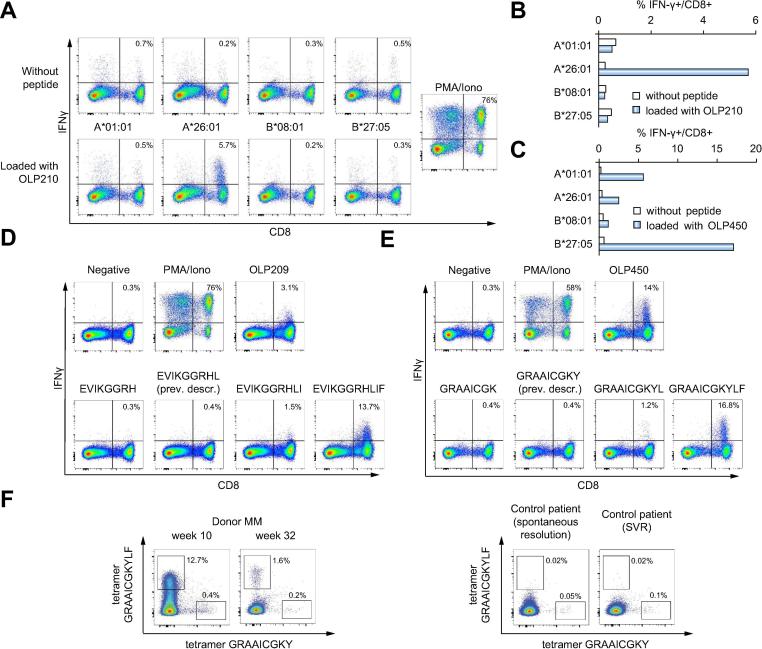
The responses elicited by OLP-209/210, OLP-211, and OLP-450/451 potentially targeted previously described epitopes restricted by HLA-A*26:01 (NS31383-1391, EVIKGGRHL),26 HLA-B*08:01 (NS31395-1403, HSKKKCDEL),[27], [28] and HLA-B*27:05 (NS5B2936-2944, GRAAICGKY).[17], [29] We therefore tested these optimal 9-mer epitopes in peptide-specific cell lines. Unexpectedly, we only observed a response to NS31395-1403 (HSKKKCDEL), corresponding to OLP-211 (Fig. 2). Responses to NS31383-1391 (EVIKGGRHL) and NS5B2936-2944 (GRAAICGKY) were either absent or very weak (Fig. 2). Using Epstein-Barr virus-immortalized B cell lines partially matched for HLA class I, we confirmed that CD8+ T cells targeting OLP-209/210 were restricted by HLA-A*26:01 (Fig. 3A and 3B), and that CD8+ T cells targeting OLP-450/451 were restricted by HLA-B*27:05 (Fig. 3C). These findings suggested that CD8+ T cells from donor MM were specific for length variants of NS31383-1391 (EVIKGGRHL) and NS5B2936-2944 (GRAAICGKY). We therefore performed in vitro analyses using peptides of different lengths. For both reactivities, the cognate epitopes were defined by 11-mer peptides (Fig. 3D and 3E; Table 2). The immunodominant response to the extended HLA-B*27:05-restricted 11-mer peptide GRAAICGKYLF was further confirmed using HLA-B*27:05/peptide multimers. In dual staining experiments, the HLA-B*27:05/GRAAICGKYLF multimer identified substantially higher frequencies of cognate CD8+ T cells among PBMCs from donor MM at week 10 and week 32 post-infection compared with the HLA-B*27:05/GRAAICGKY multimer (Fig. 3F, upper panel), whereas the HLA-B*27:05/GRAAICGKY multimer identified substantially higher frequencies of cognate CD8+ T cells among PBMCs from 2 HLA-B*27:05+ donors with resolved HCV infection compared with the HLA-B*27:05/GRAAICGKYLF multimer (Fig. 3F, lower panel).
Fig. 3.
HLA class I restriction and fine mapping of HCV-derived CD8+ T cell epitopes in donor MM. (A) Representative HLA class I restriction analysis for CD8+ T cells targeting OLP-209/210. A CD8+ T cell line specific for OLP-210 was tested for IFN-γ production after incubation with Epstein-Barr virus-immortalized B cell lines partially matched for the indicated HLA-class I alleles, either unloaded (top row) or loaded with OLP-210 (bottom row). The incorporated peptide was restricted by HLA-A*26:01. (B) Representative HLA class I restriction analysis for CD8+ T cells targeting OLP-209/210 (as in panel A). White bars: unloaded; blue bars: loaded with OLP-210. (C) Representative HLA class I restriction analysis for CD8+ T cells targeting OLP-450/451. (D, E) Fine mapping experiments for CD8+ T cell epitopes in OLP-209/210 (D) and OLP-450/451 (E). CD8+ T cell lines specific for each OLP were incubated with the indicated 8–11-mer peptides and assessed for production of IFN-γ. (F) Ex vivo staining with HLA-B*27:05/GRAAICGKY and HLA-B*27:05/GRAAICGKYLF multimers labeled with distinct fluorochromes. Left panel: donor MM (week 10 and week 32 post-infection). Right panel: control HLA-B*27:05+ donors with spontaneous resolution or treatment-induced resolution of HCV infection. HCV, hepatitis C virus; OLPs, overlapping peptides.
Table 2.
Epitopes targeted in donor MM and previously described HCV-specific CD8+ T cell epitopes restricted by the HLA class I alleles present in donor MM.
| HLA allele | Described HCV-specific epitopes (immunodominant epitopes in bold) | Epitopes targeted in donor MM (dominant responses in bold) | ||||
|---|---|---|---|---|---|---|
| A*01:01 | NS31436-1444 | ATDALMTGY | 9-mer | |||
| E2518-527 | TTDRSGAPTY | 10-mer | ||||
| A*26:01 | NS31383-1391 | EVIKGGRHL | 9-mer | NS31383-1393 | EVIKGGRHL-IF | 11-mer |
| NS31582-1590 | ENLPYLVAY | 9-mer | ||||
| NS3/4A1654-1662 | EVVTSTWVL | 9-mer | ||||
| NS5A/B2416-2424 | DVVCCSMSY | 9-mer | ||||
| NS31337-1347 | ETAGARLVVLA | 11-mer | ||||
| B*08:01 | NS31395-1403 | HSKKKCDEL | 9-mer | NS31395-1403 | HSKKKCDEL | 9-mer |
| NS31402-1410 | ELAAKLVAL | 9-mer | ||||
| NS31611-1618 | LIRLKPTL | 8-mer | ||||
| B*27:05 | P7780-788 | GRWVPGAAY | 9-mer | |||
| NS31492-1501 | GRGKPGIYRF | 10-mer | ||||
| NS5B2841-2849 | ARMILMTHF | 9-mer | NS5B2841-2849 | ARMILMTHF | 9-mer | |
| NS5B2936-2944 | GRAAICGKY | 9-mer | NS5B2936-2946 | GRAAICGKY-LF | 11-mer | |
| NS5A2131-2141 | HRFAPPCKP-LL | 11-mer | ||||
HCV, hepatitis C virus.
The responses elicited by OLP-77, OLP-203, and OLP-323/324 did not correspond with previously described HCV-specific CD8+ T cell epitopes. In silico and in vitro analyses revealed 3 novel reactivities: (i) OLP-77 – HLA-A*01:01-restricted 10-mer epitope (E2518-527, TTDRSGAPTY; Fig. S1A and S1B); (ii) OLP-203 – HLA-A*26:01-restricted 11-mer epitope (NS31337-1347, ETAGARLVVLA; Fig. S1C and S1D); and (iii) OLP-323/324 – HLA-B*27:05-restricted 11-mer epitope (NS5A2131-2141, HRFAPPCKPLL; Fig. S1E and S1F; Table 2).
Surprisingly, we did not observe HCV-specific CD8+ T cell responses to overlapping peptides incorporating the previously defined immunodominant epitopes NS31436-1444 (ATDALMTGY, restricted by HLA-A*01:01), NS5A/B2416-2424 (DVVCCSMSY, restricted by HLA-A*26:01), or NS5B2841-2849 (ARMILMTHF, restricted by HLA-B*27:05).[17], [26], [27] To confirm the absence of these epitope-specific responses, we tested the optimal 9-mer peptides in intracellular IFN-γ staining assays. A weak response was detected against ARMILMTHF (Fig. 2), but no responses were detected against ATDALMTGY or DVVCCSMSY (Fig. S2).
Our detailed analysis of HCV-specific CD8+ T cell responses in donor MM therefore revealed that: (i) reactivity was directed away from typically immunodominant epitopes, preferentially targeting either novel or subdominant epitopes; and (ii) reactivity was biased towards longer epitopes (10–11-mers) (Table 2).
Altered immunodominance associated with hypoactive variants of ERAP1
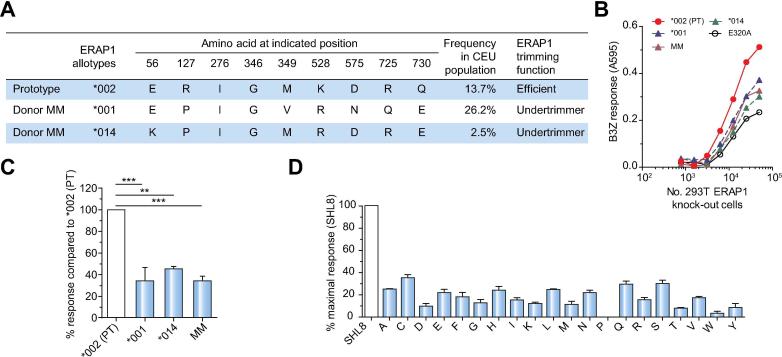
The immunoreactivity patterns identified in donor MM suggested either that the previously described optimal epitopes were not presented or that the longer targeted epitopes were more abundant on the surface of infected cells. We therefore hypothesized a determinative role for hypoactive allotypes of ERAP1, which allow surface expression of C-terminally extended peptides in the context of HLA class I.8 Molecular cloning revealed the presence of 2 ERAP1 allotypes in this donor (Fig. 4A): (i) ERAP1*001 (P127, V349, R528, N575, Q725, E730), a known hypoactive variant;7 and (ii) a previously described allotype (Hap7),30 designated here as ERAP1*014 (K56, P127, R528, E730). To determine the trimming function of these 2 ERAP1 allotypes individually and in combination, we used the well characterized SIINFEHL (SHL8) murine model system, in which an ER-targeted N-terminally extended precursor of SHL8 was transfected into 293T E1KO cells alongside each or both allotypes of ERAP1. The expression of trimmed SHL8 presented by H2-Kb on the cell surface was measured via stimulation of the SHL8-specific LacZ reporter T cell hybridoma B3Z. We further related these experiments to the poor immunogenicity of GRAAICGKY in donor MM by using an N-terminally extended version of SHL8 (SRG-SHL8) corresponding to the predicted precursor peptide generated via proteasomal cleavage of the parent protein NS5B (SRG-GRAAICGKY). As shown in Fig. 4B and 4C, both ERAP1 allotypes expressed by donor MM were hypoactive, with trimming activities between those of the negative control (a non-functional ERAP1 variant known as E320A, which carries a mutation in the active-site GAMEN motif) and the prototype (PT) control (ERAP1*002). Similar trimming efficiencies were observed with both allotypes in combination (Fig. 4B and 4C). To analyze the effect of these allotypes on trimming in the context of other HCV-derived epitopes, we quantified the generation of SHL8 from precursors with different N-terminal extensions (X-SHL8). The combined ERAP1 allotypes of donor MM again performed suboptimally in most cases (Fig. 4D), displaying marked hierarchical differences compared with the trimming activity of ERAP1*002.10 The altered immunodominance pattern in donor MM therefore correlated with the expression of 2 hypoactive variants of ERAP1.
Fig. 4.
Hypoactive trimming function of ERAP1 allotypes expressed in donor MM. (A) ERAP1 allotypes expressed in donor MM vs. the PT control allotype ERAP1*002 and their frequency in the central European (CEU) population.30 (B, C) The 3 amino acid N-terminally extended epitope precursor SRG-SIINFEHL (SRG-SHL8) was transfected into 293T E1KO cells together with the ERAP1 allotypes ERAP1*002 (PT), ERAP1*001 (expressed in donor MM), ERAP1*014 (expressed in donor MM), both donor ERAP1 allotypes (labelled MM), or E320A (a non-functional active-site mutant of ERAP1). The expression of trimmed SHL8 presented by H2-Kb on the cell surface was measured via stimulation of the SHL8-specific T cell hybridoma B3Z. (B) A representative titration curve showing activation of B3Z with increasing numbers of target cells. (C) Results normalized to ERAP1*002 activity. Bars show results pooled from 3 independent experiments ± SEM. 1-way ANOVA with Dunnett’s post hoc test was performed. ***p <0.0003, **p <0.001. (D) The ability of combined donor MM allotypes to generate the optimal SHL8 peptide from precursors with different N-terminal extensions (X-SHL8). Results are normalized to the response observed with SHL8, which does not require trimming by ERAP1. Bars show results pooled from 3 independent experiments ± SEM.
Differential generation of the GRAAICGKYLF epitope by allotypic variants of ERAP1
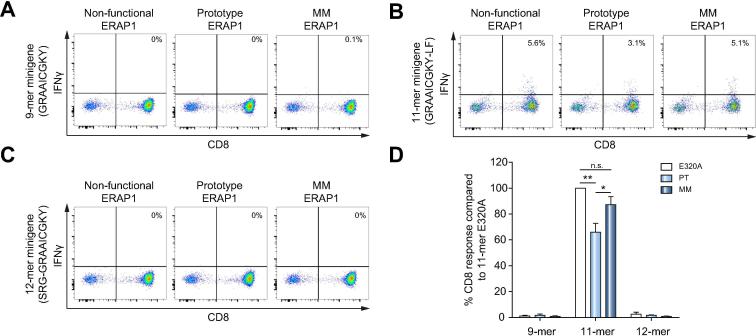
To extend these findings, we transfected 293T E1KO cells with: (i) HLA-B*27:05; (ii) minigenes expressing the 9-mer GRAAICGKY, the 11-mer GRAAICGKYLF, or an N-terminally extended 12-mer precursor of the 9-mer epitope (SRG-GRAAICGKY); and DNA encoding (iii) the non-functional ERAP1 variant (E320A), the active PT control ERAP1*002, or the donor MM allotypes ERAP1*001 and ERAP1*014. Presentation of the 11-mer epitope by HLA-B*27:05 on the surface of transfected cells was measured using a GRAAICGKYLF-specific CD8+ T cell line from donor MM. We first confirmed that CD8+ T cells from donor MM failed to recognize the 9-mer epitope, irrespective of the N-terminal sequence and the expressed ERAP1 allotype (Fig. 5A, 5C, and 5D). As expected, transfection of the 11-mer peptide minigene with inactive ERAP1 generated a strong response (Fig. 5B, left panel), whereas cells expressing the PT control ERAP1*002 elicited a significantly weaker response, consistent with overtrimming and destruction of GRAAICGKYLF (Fig. 5B, middle panel). Interestingly, cells expressing the ERAP1 allotypes from donor MM also elicited a strong response, indicating preservation of GRAAICGKYLF (Fig. 5B, right panel). The immunodominant 9-mer epitope was therefore generated from an N-terminally extended precursor, but not from a C-terminally extended precursor, in the presence of functional ERAP1. In the presence of hypoactive ERAP1, however, the N-terminally extended precursor was not trimmed to the optimal length, hence the absence of the usual 9-mer peptide, whereas the C-terminally extended 11-mer version escaped destruction to prime and activate virus-specific CD8+ T cells.
Fig. 5.
The 11-mer GRAAICGKYLF epitope is not destroyed by ERAP1 allotypes expressed in donor MM. 293T E1KO cells were transfected with: (i) HLA-B*27:05; (ii) minigenes expressing the 9-mer GRAAICGKY (A), the 11-mer GRAAICGKYLF (B), or the N-terminally extended 12-mer precursor of the 9-mer epitope SRG-GRAAICGKY (C); and DNA encoding (iii) a non-functional ERAP1 mutant (E320A, left column), the PT allotype ERAP1*002 (middle column), or the ERAP1 allotype combination from donor MM (right column). Presentation of the 11-mer peptide by HLA-B*27:05 was assessed by measuring IFN-γ production after stimulation of a GRAAICGKYLF-specific CD8+ T cell line established from donor MM. (A–C) Representative dot plots. (D) Summary of 3 independent experiments ± SEM. 1-way ANOVA with Dunnett’s post hoc test was performed. **p <0.0039, *p <0.05, n.s., not significant.
Additional factors that may impact immunodominance patterns
Immunodominance patterns can be influenced by factors other than antigen presentation.31 Donor MM was infected with a viral strain that contained nonsynonymous mutations in 4 of the previously described epitopes restricted by HLA-A*01:01, HLA-A*26:01, HLA-B*08:01, or HLA-B*27:05 (Table S1). However, the typically immunodominant HLA-B*27:05-restricted epitope NS5B2841-2849 (ARMILMTHF) incorporated a conservative isoleucine to valine switch at position 4 (ARMVLMTHF), which did not affect immune recognition (Fig. S3), consistent with a previous report.16 In contrast, the typically immunodominant HLA-A*01:01-restricted epitope NS31436-1444 (ATDALMTGY) incorporated a tyrosine to phenylalanine switch at the C-terminus (ATDALMTGF), which has been shown previously to escape immune recognition.32 Therefore, the lack of ATDALMTGY-specific CD8+ T cells in donor MM likely reflected sequence variation in the infecting viral strain rather than SNPs in ERAP1. Competition among epitopes and cognate CD8+ T cell avidity can also affect immunodominance patterns. In donor MM, epitopes restricted by HLA-B*08:01, an HLA allele that usually restricts dominant CD8+ T cell responses in HCV infection, were only weakly targeted. The immundominant HLA-B*27:05-restricted epitope NS5B2936-2946 (GRAAICGKYLF), however, was targeted with relatively high avidity (Fig. S4), but it remains to be determined if this attribute contributed to the numerical dominance of GRAAICGKYLF-specific CD8+ T cells. It is also important to point out, however, that the virus-specific CD8+ T cell responses in donor MM decreased in strength over time, while their immunodominance pattern remained unchanged even at week 68 after infection (Fig. S5).
Discussion
ERAP1 has garnered interest as a consequence of genetic associations with several autoinflammatory disorders, such as ankylosing spondylitis, birdshot chorioretinopathy, psoriasis, and Behçet’s disease.3 Epistatic interactions between variants of ERAP1 and the corresponding disease-linked HLA alleles further implicate peptide handling in the immunopathology of these disorders.[5], [6] Much effort has therefore been dedicated to understanding the impact of ERAP1 polymorphisms on the naturally presented epitope repertoire.[8], [9], [33] However, previous studies have not addressed the physiological relevance of these effects on the immune response to exogenous antigens, for example in the context of viral infections. We identified 2 hypoactive ERAP1 allotypes in a donor with acute HCV infection. Donor MM expressed 4 distinct HLA class I allotypes (A*01:01, A*26:01, B*08:01, and B*27:05), all of which have been shown to present HCV-derived epitopes that reproducibly elicit hierarchical CD8+ T cell responses.[17], [26], [28], [32] In contrast to these classical immune profiles, CD8+ T cells from donor MM targeted either longer versions of the typically immunodominant 9-mer epitopes or novel 10-mer or 11-mer epitopes, characterized here for the first time. These results suggested that the relatively subtle effects of ERAP1 polymorphisms in vitro profoundly influenced the antigen-specific CD8+ T cell repertoire in vivo.
Presentation of the B*27:05-restricted 11-mer epitope GRAAICGKYLF was reduced only slightly in 293T E1KO cells harboring active prototype control ERAP1*002 relative to 293T E1KO cells expressing the hypoactive donor MM allotypes ERAP1*001 and ERAP1*014. Nonetheless, GRAAICGKYLF-specific CD8+ T cells vastly outnumbered GRAAICGKY-specific CD8+ T cells in donor MM. These divergent in vitro and in vivo observations suggested a degree of leakiness in the ERAP1 system, enabling the presentation of epitopes spanning different lengths, which subsequently elicited a highly focused CD8+ T cell response. Minor differences in antigen presentation were therefore sufficient to invert the archetypal immunodominance hierarchy in donor MM. In clinical terms, donor MM had no history of severe autoinflammatory disorders, but suffered from protracted viral infections during childhood and developed mild psoriasis on completion of this study. It therefore remains to be determined how the corresponding ERAP1 allotypes impact adaptive immunity beyond the context of HCV.
Despite the presence of several favorable prognostic indicators (symptomatic course of infection, IL28B/IFNL4 genotype CC, and expression of HLA-B*27), donor MM failed to clear the virus and developed persistent low-level infection with HCV. Although it is tempting to link this outcome with the hypoactive ERAP1 allotypes and the skewed repertoire of HCV-specific CD8+ T cells, there was no direct evidence for a causal association between these variables. Moreover, longitudinal sequence variations emerged in 2 of the 3 NS3-derived epitopes targeted in donor MM, consistent with mutational escape: K1386R in the HLA-A*26:01-restricted 11-mer epitope NS31383-1393 and L1403F in the HLA-B*08:01-restricted 9-mer epitope NS31395-1403. This observation suggested a potential role for viral adaptation in the outcome of infection, but equivalent data were not available for targeted NS5B-derived epitopes, in part due to the persistent low-level viremia in donor MM. However, previous studies have demonstrated an association between ERAP2 polymorphisms and resistance to HIV-1.[34], [35] In addition, the ERAP1 polymorphism I276M (rs26618 T>C) was found to be enriched in individuals with chronic HCV infection relative to healthy controls.36 The non-risk residue was present at this position in both ERAP1 allotypes expressed by donor MM. It is also notable that CD8+ T cell responses to the 11-mer epitopes HRFAPPCKPLL and GRAAICGKYLF were not detected in a cohort of 10 HLA-B*27+ patients with chronic HCV genotype 1 infection (data not shown). Rare combinations of ERAP1 allotypes may therefore shape the repertoire of CD8+ T cell epitopes more distinctly than common variants of ERAP1.7
The peptide trimming function of ERAP1 can be profoundly altered by the allotype expressed.[37], [38] These allotypic differences lead to a significantly altered peptide repertoire presented at the cell surface, which as observed for patient MM, may affect the presentation of immunodominant HCV epitopes, and thus, CD8+ T cell responses. In addition to peptide trimming, ERAP1 has been suggested to function in the shedding of the cytokine receptors IL-6R, TNFR1, and IL-1RII.39 These cytokines (IL-6, TNF, IL-1) are important in inflammatory responses to viral infection. It is not currently known how ERAP1 works to facilitate cytokine receptor shedding, as it does not possess any ectodomain cleavage activity.39 However, ERAP1 binds to the receptors and may therefore act as an important bridge to promote cytokine receptor cleavage through a third party. The cytokine receptor cleavage activity was not examined in this study, although it is tempting to speculate that allotype variants may have differential functional capacities in this pathway. This may alter levels of cytokine receptor shedding and levels of these receptors at the cell surface and in serum as previously observed for ERAP1 with reduced function, which resulted in higher levels of IL-1β, TNF and IL-6,40 modulating the inflammatory environment in infection.
In conclusion, we have demonstrated that hypoactive ERAP1 allotypes can impact the repertoire of epitopes targeted by antigen-specific CD8+ T cells in a relevant human viral infection, thereby supporting the general concept that peptide handling underlies the immunopathogenesis of HLA class I-associated autoinflammatory disorders. Further studies are therefore warranted to explore similar phenomena in other clinical settings, especially where disease susceptibility has been genetically linked with functional polymorphisms in ERAP1.
Financial support
This study was funded in part by the Deutsche Forschungsgemeinschaft (SFB1160 “Immune-mediated pathology as a consequence of impaired immune reactions [IMPATH]”, projects 8 and 10 awarded to R.T. and C.N.H.) and Cancer Research UK (Programme Grant A16997 awarded to E.J.). D.A.P. is a Wellcome Trust Senior Investigator (100326/Z/12/Z).
Conflict of interest
The authors declare no conflicts of interest that pertain to this work.
Please refer to the accompanying ICMJE disclosure forms for further details.
Authors’ contributions
J.K., E.R., T.H., D.A.P., M.H., R.T., E.J., and C.N.H. developed the concept and designed the study; J.K., E.R., K.N., V.W., F.E., E.G., and A.W. performed experiments and procedures; J.K., E.R., K.N., J.T., D.A.P., M.H., R.T., E.J., and C.N.H. wrote the manuscript.
Acknowledgements
We thank all individuals who donated blood for our studies. We also thank BEI Resources and the National Institute of Allergy and Infectious Diseases for providing overlapping peptides spanning the complete polyprotein of HCV.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhep.2019.01.034.
Supplementary data
The following are the Supplementary data to this article:
References
Author names in bold designate shared co-first authorship
- 1.Alvarez-Navarro C., Lopez de Castro J.A. ERAP1 structure, function and pathogenetic role in ankylosing spondylitis and other MHC-associated diseases. Mol Immunol. 2014;57:12–21. doi: 10.1016/j.molimm.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 2.Reeves E., Elliott T., James E., Edwards C.J. ERAP1 in the pathogenesis of ankylosing spondylitis. Immunol Res. 2014;60:257–269. doi: 10.1007/s12026-014-8576-2. [DOI] [PubMed] [Google Scholar]
- 3.Lopez de Castro J.A., Alvarez-Navarro C., Brito A., Guasp P., Martin-Esteban A., Sanz-Bravo A. Molecular and pathogenic effects of endoplasmic reticulum aminopeptidases ERAP1 and ERAP2 in MHC-I-associated inflammatory disorders: towards a unifying view. Mol Immunol. 2016;77:193–204. doi: 10.1016/j.molimm.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 4.WTCC Consortium, A-A-AS Consortium. Burton P.R., Clayton D.G., Cardon L.R., Craddock N. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39:1329–1337. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans D.M., Spencer C.C., Pointon J.J., Su Z., Harvey D., Kochan G. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat Genet. 2011;43:761–767. doi: 10.1038/ng.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirino Y., Bertsias G., Ishigatsubo Y., Mizuki N., Tugal-Tutkun I., Seyahi E. Genome-wide association analysis identifies new susceptibility loci for Behcet's disease and epistasis between HLA-B*51 and ERAP1. Nat Genet. 2013;45:202–207. doi: 10.1038/ng.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reeves E., Colebatch-Bourn A., Elliott T., Edwards C.J., James E. Functionally distinct ERAP1 allotype combinations distinguish individuals with ankylosing spondylitis. Proc Natl Acad Sci U S A. 2014;111:17594–17599. doi: 10.1073/pnas.1408882111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L., Fischer R., Peng Y., Reeves E., McHugh K., Ternette N. Critical role of endoplasmic reticulum aminopeptidase 1 in determining the length and sequence of peptides bound and presented by HLA-B27. Arthritis Rheumat. 2014;66:284–294. doi: 10.1002/art.38249. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Medel N., Sanz-Bravo A., Van Nguyen D., Galocha B., Gomez-Molina P., Martin-Esteban A. Functional interaction of the ankylosing spondylitis-associated endoplasmic reticulum aminopeptidase 1 polymorphism and HLA-B27 in vivo. Mol Cell Proteomics MCP. 2012;11:1416–1429. doi: 10.1074/mcp.M112.019588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reeves E., Edwards C.J., Elliott T., James E. Naturally occurring ERAP1 haplotypes encode functionally distinct alleles with fine substrate specificity. J Immunol. 2013;191:35–43. doi: 10.4049/jimmunol.1300598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vitulano C., Tedeschi V., Paladini F., Sorrentino R., Fiorillo M.T. The interplay between HLA-B27 and ERAP1/ERAP2 aminopeptidases: from anti-viral protection to spondyloarthritis. Clin Exp Immunol. 2017;190:281–290. doi: 10.1111/cei.13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Webster D.P., Klenerman P., Dusheiko G.M. Hepatitis C. Lancet. 2015;385:1124–1135. doi: 10.1016/S0140-6736(14)62401-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin E.C., Sung P.S., Park S.H. Immune responses and immunopathology in acute and chronic viral hepatitis. Nat Rev Immunol. 2016;16:509–523. doi: 10.1038/nri.2016.69. [DOI] [PubMed] [Google Scholar]
- 14.Nitschke K., Luxenburger H., Kiraithe M.M., Thimme R., Neumann-Haefelin C. CD8+ T-cell responses in hepatitis B and C: the (HLA-) A, B, and C of hepatitis B and C. Dig Dis. 2016;34:396–409. doi: 10.1159/000444555. [DOI] [PubMed] [Google Scholar]
- 15.McKiernan S.M., Hagan R., Curry M., McDonald G.S., Kelly A., Nolan N. Distinct MHC class I and II alleles are associated with hepatitis C viral clearance, originating from a single source. Hepatology. 2004;40:108–114. doi: 10.1002/hep.20261. [DOI] [PubMed] [Google Scholar]
- 16.Dazert E., Neumann-Haefelin C., Bressanelli S., Fitzmaurice K., Kort J., Timm J. Loss of viral fitness and cross-recognition by CD8+ T cells limit HCV escape from a protective HLA-B27-restricted human immune response. J Clin Invest. 2009;119:376–386. doi: 10.1172/JCI36587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neumann-Haefelin C., McKiernan S., Ward S., Viazov S., Spangenberg H.C., Killinger T. Dominant influence of an HLA-B27 restricted CD8+ T cell response in mediating HCV clearance and evolution. Hepatology. 2006;43:563–572. doi: 10.1002/hep.21049. [DOI] [PubMed] [Google Scholar]
- 18.Price D.A., Brenchley J.M., Ruff L.E., Betts M.R., Hill B.J., Roederer M. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J Exp Med. 2005;202:1349–1361. doi: 10.1084/jem.20051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thimme R., Bukh J., Spangenberg H.C., Wieland S., Pemberton J., Steiger C. Viral and immunological determinants of hepatitis C virus clearance, persistence, and disease. Proc Natl Acad Sci U S A. 2002;99:15661–15668. doi: 10.1073/pnas.202608299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thimme R., Oldach D., Chang K.M., Steiger C., Ray S.C., Chisari F.V. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J Exp Med. 2001;194:1395–1406. doi: 10.1084/jem.194.10.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bengsch B., Spangenberg H.C., Kersting N., Neumann-Haefelin C., Panther E., von Weizsacker F. Analysis of CD127 and KLRG1 expression on hepatitis C virus-specific CD8+ T cells reveals the existence of different memory T-cell subsets in the peripheral blood and liver. J Virol. 2007;81:945–953. doi: 10.1128/JVI.01354-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerlach J.T., Diepolder H.M., Zachoval R., Gruener N.H., Jung M.C., Ulsenhei-mer A. Acute hepatitis C: high rate of both spontaneous and treatment-induced viral clearance. Gastroenterology. 2003;125:80–88. doi: 10.1016/s0016-5085(03)00668-1. [DOI] [PubMed] [Google Scholar]
- 23.Thomas D.L., Thio C.L., Martin M.P., Qi Y., Ge D., O'Huigin C. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lauer G.M., Barnes E., Lucas M., Timm J., Ouchi K., Kim A.Y. High resolution analysis of cellular immune responses in resolved and persistent hepatitis C virus infection. Gastroenterology. 2004;127:924–936. doi: 10.1053/j.gastro.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 25.Neumann-Haefelin C., Timm J., Spangenberg H.C., Wischniowski N., Nazarova N., Kersting N. Virological and immunological determinants of intrahepatic virus-specific CD8+ T-cell failure in chronic hepatitis C virus infection. Hepatology. 2008;47:1824–1836. doi: 10.1002/hep.22242. [DOI] [PubMed] [Google Scholar]
- 26.Neumann-Haefelin C., Killinger T., Timm J., Southwood S., McKinney D., Blum H.E. Absence of viral escape within a frequently recognized HLA-A26-restricted CD8+ T-cell epitope targeting the functionally constrained hepatitis C virus NS5A/5B cleavage site. J Gen Virol. 2007;88:1986–1991. doi: 10.1099/vir.0.82826-0. [DOI] [PubMed] [Google Scholar]
- 27.Koziel M.J., Dudley D., Afdhal N., Grakoui A., Rice C.M., Choo Q.L. HLA class I-restricted cytotoxic T lymphocytes specific for hepatitis C virus. Identification of multiple epitopes and characterization of patterns of cytokine release. J Clin Invest. 1995;96:2311–2321. doi: 10.1172/JCI118287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Timm J., Lauer G.M., Kavanagh D.G., Sheridan I., Kim A.Y., Lucas M. CD8 epitope escape and reversion in acute HCV infection. J Exp Med. 2004;200:1593–1604. doi: 10.1084/jem.20041006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neumann-Haefelin C., Oniangue-Ndza C., Kuntzen T., Schmidt J., Nitschke K., Sidney J. Human leukocyte antigen B27 selects for rare escape mutations that significantly impair hepatitis C virus replication and require compensatory mutations. Hepatology. 2011;54:1157–1166. doi: 10.1002/hep.24541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ombrello M.J., Kastner D.L., Rem-mers E.F. Endoplasmic reticulum-associated amino-peptidase 1 and rheumatic disease: genetics. Curr Opin Rheumatol. 2015;27:349–356. doi: 10.1097/BOR.0000000000000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yewdell J.W. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity. 2006;25:533–543. doi: 10.1016/j.immuni.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 32.Neumann-Haefelin C., Frick D.N., Wang J.J., Pybus O.G., Salloum S., Narula G.S. Analysis of the evolutionary forces in an immunodominant CD8 epitope in hepatitis C virus at a population level. J Virol. 2008;82:3438–3451. doi: 10.1128/JVI.01700-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanz-Bravo A., Campos J., Mazariegos M.S., Lopez de Castro J.A. Dominant role of the ERAP1 polymorphism R528K in shaping the HLA-B27 peptidome through differential processing determined by multiple peptide residues. Arthritis Rheumatol. 2015;67:692–701. doi: 10.1002/art.38980. [DOI] [PubMed] [Google Scholar]
- 34.Biasin M., Sironi M., Saulle I., de Luca M., la Rosa F., Cagliani R. Endoplasmic reticulum aminopeptidase 2 haplotypes play a role in modulating susceptibility to HIV infection. AIDS. 2013;27:1697–1706. doi: 10.1097/QAD.0b013e3283601cee. [DOI] [PubMed] [Google Scholar]
- 35.Cagliani R., Riva S., Biasin M., Fumagalli M., Pozzoli U., Lo Caputo S. Genetic diversity at endoplasmic reticulum aminopeptidases is maintained by balancing selection and is associated with natural resistance to HIV-1 infection. Hum Mol Genet. 2010;19:4705–4714. doi: 10.1093/hmg/ddq401. [DOI] [PubMed] [Google Scholar]
- 36.Liu S., Cao D., Shen Y., Li Y., Li Y., Shi L. The ERAP gene is associated with HCV chronic infection in a Chinese Han population. Hum Immunol. 2017;78:731–738. doi: 10.1016/j.humimm.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 37.Guasp P., Barnea E., Gonzalez-Escribano M.F., Jimenez-Reinoso A., Regueiro J.R., Admon A. The Behcet's disease-associated variant of the aminopeptidase ERAP1 shapes a low-affinity HLA-B*51 peptidome by differential subpeptidome processing. J Biol Chem. 2017;292:9680–9689. doi: 10.1074/jbc.M117.789180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seregin S.S., Rastall D.P., Evnouchidou I., Aylsworth C.F., Quiroga D., Kamal R.P. Endoplasmic reticulum aminopeptidase-1 alleles associated with increased risk of ankylosing spondylitis reduce HLA-B27 mediated presentation of multiple antigens. Autoimmunity. 2013;46:497–508. doi: 10.3109/08916934.2013.819855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cui X., Hawari F., Alsaaty S., Lawrence M., Combs C.A., Geng W. Identification of ARTS-1 as a novel TNFR1-binding protein that promotes TNFR1 ectodomain shedding. J Clin Invest. 2002;110:515–526. doi: 10.1172/JCI13847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aldhamen Y.A., Pepelyayeva Y., Rastall D.P., Seregin S.S., Zervoudi E., Koumantou D. Autoimmune disease-associated variants of extracellular endoplasmic reticulum aminopeptidase 1 induce altered innate immune responses by human immune cells. J Innate Immun. 2015;7:275–289. doi: 10.1159/000368899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.