Abstract
A key step in achieving accurate detection of somatic mutations is comparison of sequencing data from a tumor sample to its matched germline control. Sensitivity to detect somatic variants is greatly reduced when the matched normal sample is contaminated with tumor cells. To overcome this limitation, we developed deTiN, a method that first estimates the tumor-in-normal contamination (TiN) level, and then, in contaminated cases, improves sensitivity by reclassifying initially discarded variants as somatic.
Somatic mutation detection requires distinguishing between somatic and germline (inherited) variants. Comparing between tumor and patient-matched control (normal) DNA sequencing data enables the removal of patient-specific inherited variants and locus-specific (e.g., alignment) artifacts affecting both samples. This variant detection paradigm provides sensitive and specific somatic mutation calls with low false-positive rates (<0.5 mut/Mb)1, but it relies on obtaining sequencing data from matched normal healthy tissue free of contaminating tumor cells1–3. Procuring pure normal tissue, however, can be challenging4–7. Tumor-in-normal (TiN; tumor-sample DNA found in the normal sample (Methods Eq. 1)) contamination arises from cancer (or pre-cancer) cell invasion into healthy compartments and is reported in leukemias6,8,9, breast, bladder, and gastric cancers10–12, among others. TiN contamination may cause methods to reject true somatic variants based on the presence of tumor-derived reads supporting the mutation in the matched normal tissue, decreasing sensitivity for mutation detection and leading to potential misinterpretation of patient sequencing data (Supplementary Figure 1a). To overcome these challenges, we developed deTiN, a method that estimates TiN and salvages many somatic mutations otherwise filtered out as germline or artifactual variants.
DeTiN models a normal sample as a mixture of normal with an unknown fraction of contaminating tumor cells. We estimate TiN, defined as the relative tumor DNA fraction in normal and tumor samples (Methods), using two independent types of tumor-specific events: (i) somatic single nucleotide variants (SSNVs) and (ii) genomic regions of allelic imbalance (deletions, amplifications, copy-neutral loss-of-heterozygosity) extracted from allele-specific somatic copy number alterations (aSCNAs) (Supplementary Figure 1b, Methods). DeTiN calculates posterior distributions over TiN values based on each of the two somatic event types separately, and then combines them to identify the maximum a posteriori (MAP) value (and confidence interval, Methods). The estimated TiN is used to recover previously rejected SSNVs or indels (Methods), deTiN probabilistically compares two scenarios for each candidate variant: that the alternate allele count in the normal represents either (i) an underlying germline variant, or (ii) a somatic variant coming from tumor DNA mixed in the normal according to the estimated TiN value (Supplementary Figure 1b, Methods).
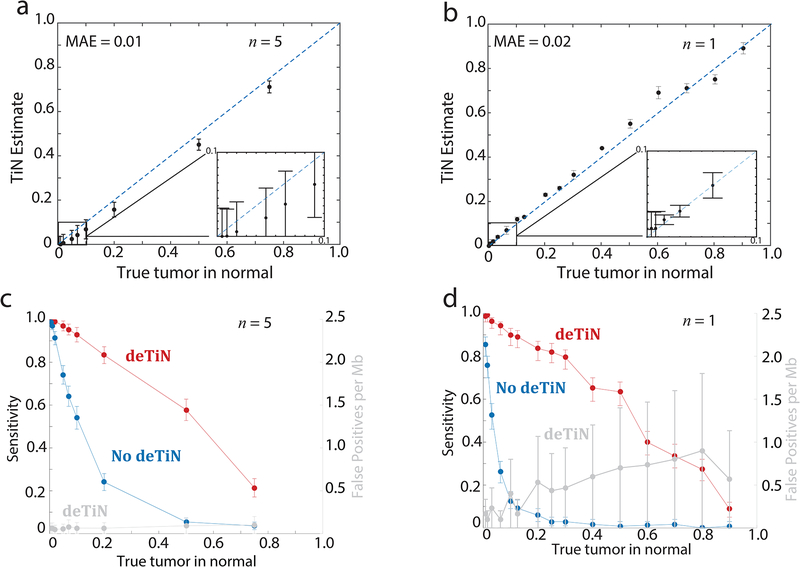
We performed in silico and in vitro simulation experiments to measure deTiN’s accuracy in estimating TiN and its ability to recover SSNVs. Somatic mutations in pairs of tumors and artificially contaminated normal samples were first called using MuTect1 (Methods), and then processed by deTiN. Comparing estimated against known simulated values, deTiN estimated TiN contamination with a mean absolute error of 0.01 (in silico) and 0.02 (in vitro) over the range of simulated TiN values (Figure 1a–b, Supplementary Table 1, Supplementary Table 2).
Figure 1. Results from in silico and in vitro validation of deTiN.
(a) TiN estimates at different in silico simulated TiN levels. (b) deTiN estimates at different in vitro mixed TiN levels. MAE = mean absolute error. (c, d) Sensitivity to detect mutations with deTiN (red) and without deTiN (blue) at (c) different in silico simulated TiN levels and (d) in vitro mixed TiN levels. (a, c) deTiN results from n=5 in silico independent simulation experiments. Dots represent weighted average and error bars represent standard errors. (b, d) Results from n=1 sequencing experiment. Error bars depict 95% confidence intervals on TiN estimates. (a, b) Dotted blue lines indicate y=x.
We quantified the impact of TiN contamination on SSNV detection sensitivity. MuTect1, VarScan3, and Strelka2 lost sensitivity to detect SSNVs at TiN>0.02 (Figure 1c–d [MuTect], Supplementary Table 1, Supplementary Table 2, Supplementary Figure 2 [Strelka and VarScan], Supplementary Results). TiN mostly affects mutations with high allele fraction in the tumor (AF) since they are more likely to be observed in the contaminated normal and cause the mutation caller to reject the somatic mutation. Indeed, mutations with AF>0.3 exhibited lower sensitivity than those with AF<0.3 (Mann-Whitney one tailed p=0.004 in silico TiN=0.2) (Supplementary Fig. 3a–b). Applying deTiN’s mutation recovery step improved detection sensitivity across all TiN values (Figure 1c–d). At very high TiN (>0.75), where germline SNPs were indistinguishable from somatic events, SSNV recovery was less effective. DeTiN-recovered mutations did not substantially increase false-positive rates (Figure 1c–d; Supplementary Figure 3c–f; Supplementary Results) and, as expected, were enriched with high AF events (Supplementary Figure 3a–b). High AF SSNVs are more likely clonal mutations, thus representing many initiating drivers and clinically important oncogenic events. We characterized deTiN’s performance using simulated data over a range of tumor sample purities, sequencing depths, and mutation rates (Supplementary Figure 4; Supplementary Results).
Thus far, we assumed that all the tumor cells that contaminated the normal sample share the same somatic events (e.g., SSNVs and aSCNAs) with the tumor cells in the tumor sample. However, this assumption may be invalid if: (i) the tumor cells in a tumor-adjacent normal tissue sample (a common source of “normal” tissue) contain tumor subclones that differ from the dominant clone in the tumor sample, or (ii) normal-appearing cells are the descendants of a premalignant precursor and share a subset of clonal events with the neighboring tumor cells5,11,13. Thus, multiple TiN values may be required to describe the contaminating clones in a single normal sample.. The tumor and normal cell lines selected for the in vitro experiments provided a model to test this phenomenon. At each simulated TiN fraction, deTiN identified two distinct TiN levels: (i) the intended mixing fraction and (ii) a fraction corresponding to a shared precursor subclone (Supplementary Figure 5). Presence of the parental clone did not interfere with TiN estimation.
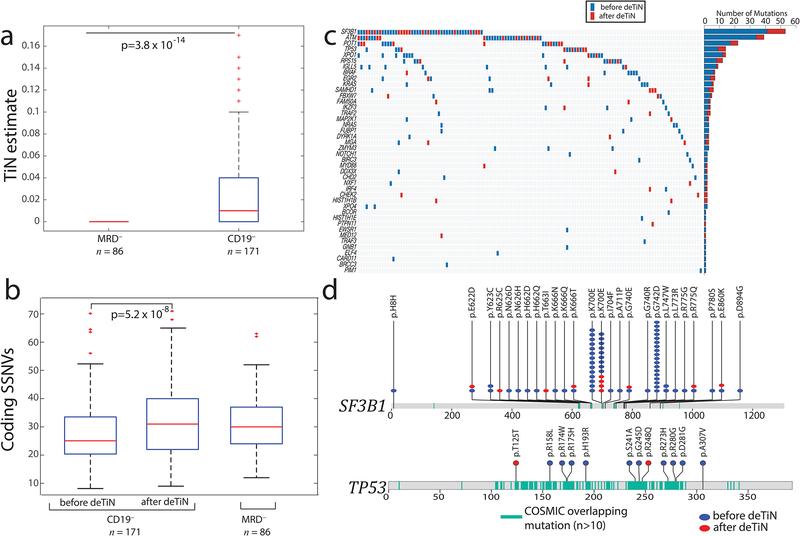
We applied deTiN to a whole-exome sequencing data cohort generated from 257 tumor-normal paired samples from chronic lymphocytic leukemia (CLL) patients9. Leukemic DNA was extracted from CD19+ selected cells; matched germline DNA was derived from either the negative fraction (‘sorted CD19– cells’) or matched post-treatment samples without molecularly detectable disease (MRD–, Figure 2a). DeTiN identified higher TiN contamination in sorted CD19– cells than MRD– samples (Figure 2a; Mann-Whitney p<0.001). In one case, the CD19–, but not the saliva-derived, normal sample was contaminated (Supplementary Figure 6, Supplementary Results). Consistent with the simulation results, mutation calling without deTiN on 171 tumors with CD19– normals resulted in a markedly lower mutation rate (Mann-Whitney p<0.001). Following deTiN application, CD19– and MRD– mutation rates became similar (p=0.56, Figure 2b, Supplementary Table 3). The fraction of candidate mutations at dbSNP sites was not statistically different between tumor samples paired with CD19– or MRD– normals, suggesting that the putative false-positive SNV rate did not increase (p=0.27; Supplementary Table 3). DeTiN recovered mutations in known CLL drivers (Figure 2c)9 at previously reported hotspots, supporting their functional oncogenic role (Figure 2d)14.
Figure 2. Application of deTiN to chronic lymphocytic leukemia (CLL) sequencing data.
(a) TiN estimates for CD19– selected (normal) blood compared with whole blood from minimal residual disease negative (MRD–) patients. Box plot: median TiN value (red line), box represents Q1 and Q3 quartiles, whiskers represent the most extreme data points that are not outliers. Outliers are denoted with red crosses and represent data points out side the range [Q1 - 1.5 IQR, Q3 + 1.5 IQR] where IQR is the interquartile range. P value is calculated using two-tailed Mann–Whitney test (n=257 independent patient samples). (b) Mutation rate in samples pre- and post-application of deTiN stratified by normal sample type. Box plot and P value as in panel a. (c) Heat map and bar plot illustrating recovery of SSNVs in the CLL cohort. Samples are in columns, genes in rows. Blue boxes indicate variants detected prior to deTiN (“without deTiN”); red boxes indicate additional variants recovered by deTiN (“with deTiN”). (d) Stick plots showing mutation data in SF3B1 and TP53. Amino acid positions of recurrent COSMIC mutations are highlighted in teal. Blue circles indicate variants detected prior to deTiN; red circles indicate variants recovered by deTiN.
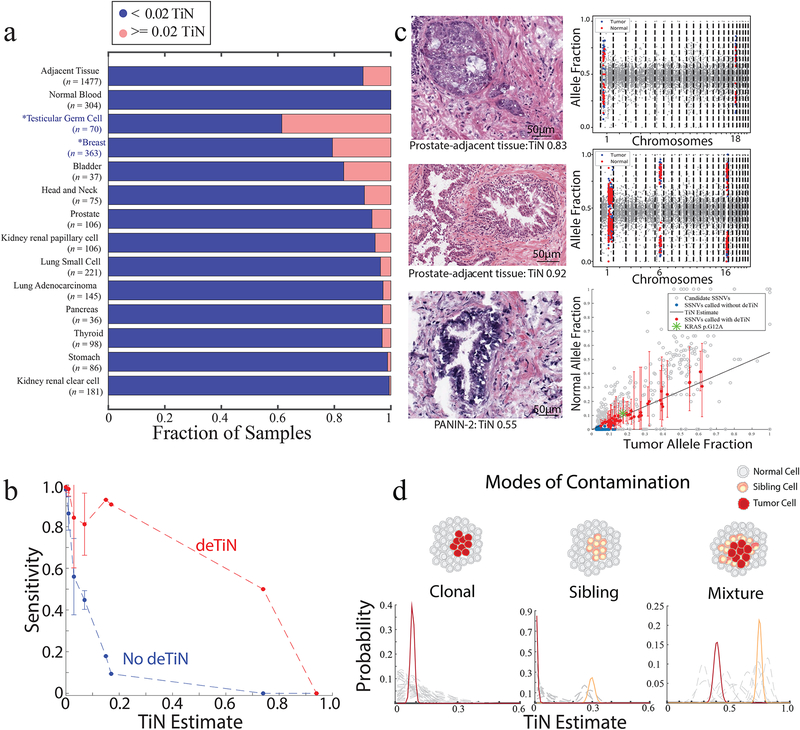
We also assessed TiN prevalence in tumor-adjacent histologically normal tissue7,15–17. Significant TiN was found in sequencing data from 161/1477 tumor and adjacent normal sample pairs (Prob[TiN>0.02]>0.95) (Supplementary Table 4). The fraction of samples containing detectable TiN varied by tumor type. Breast invasive carcinoma and testicular germ cell tumors (both non-TCGA cohorts) displayed significantly higher fraction of TiN>0.02 cases (Mann-Whitney p<0.01) and TiN levels/case (Fig. 3a, Supplementary Figure 7), perhaps due to different tissue-collection protocols than TCGA. For 304/1477 cases, a matched germline peripheral blood sample was also available and was uncontaminated. Comparing the mutation calls detected using the tissue-adjacent and blood normal samples demonstrated deTiN’s improved sensitivity (Figure 3b; Supplementary Results).
Figure 3. Application of deTiN to analysis of solid tumors with adjacent normal controls.
(a) Fraction of contaminated samples (pink; TiN≥0.02) when using different sources for normal tissue (tumor-adjacent normal tissue and peripheral blood) and, in cases with tumor-adjacent normal, stratified by tumor type. Asterisks represent non-TCGA cohorts. (b) Points show mean sensitivity for detecting mutations with deTiN (red) and without deTiN (blue). Means were derived from 256 of the 304 tumors that were matched with both a tumor-adjacent and a blood normal sample and had a sufficient number of somatic events to robustly estimate TiN (TiN = 0 [n=230]; TiN=0.01 [n=9]; TiN = 0.03 [n=9]; TiN=0.07 [n=4]; TiN=0.15 [n=1]; TiN=0.17 [n=1]; TiN=0.74 [n=1]; TiN=0.94 [n=1]). Error bars indicate standard error. (c) Histology images of selected adjacent tissue samples with evidence supporting TiN (n=1 patient sample for each image and plot). deTiN aSCNA data supporting TiN estimate is displayed for top two samples; points indicate allele-fraction of heterozygous germline SNPs, blue (tumor) and red (normal) points are used for TiN estimation, and grey points are not used by deTiN. The bottom plot displays deTiN somatic variant data supporting the TiN estimate for the bottom sample. Points indicate allele-fraction of variants in the tumor (x-axis) and normal (y-axis) samples; error bars indicate 95% beta confidence intervals. The green asterisk represents the KRAS G12V mutation, red points represent SSNVs recovered by deTiN, blue points are called before deTiN, and grey points are rejected by deTiN and MuTect as germline or artifact. Each plot displays data supporting TiN from a single tumor-normal pair corresponding to the image on the left (n = 1). (d) Illustration of three modes of contamination. Posterior distribution functions for TiN based on aSCNA data are shown clustered (red and orange) and unclustered for individual events (dashed grey). In the mixture scenario, TiN has two possible values: the lower represents events unique to the tumor cells (red) and the higher represents events shared between the tumor cells and the sibling precursor cells (orange).
In 8 selected high-TiN cases, histological review by a pathologist, blinded to the TiN estimates, identified areas of malignant cells in 3/8 cases (prostate adenocarcinoma cells; evidence of dysplastic glands; areas of pancreatic intraepithelial neoplasia-2 [PANIN-2] [Fig. 3c]) but none in 8 uncontaminated (TiN=0) control cases. Notably, deTiN detected KRAS G12A mutations in one sample pair, and large copy number events were found in all 8 contaminated samples (Figure 3c, Supplementary Figure 8), suggesting that somatic lesions can be present in histologically non-malignant tissue and occur before full transformation18. Since the sequencing samples originated from tissue blocks and the histologically evaluated image reflects only a single slice, we cannot rule out the presence of cancer cells in the sequenced sample due to spatial heterogeneity.
Spatial heterogeneity can result in 3 TiN contamination types: (i) clonal, sharing all somatic events at a consistent ratio; (ii) one or more sibling clones (e.g., precursor cells), sharing only a subset of events; and (iii) both (i) and (ii) (Figure 3d). We identified 13 sample pairs from 6 different tumor types demonstrating sibling or mixture relationships (Supplementary Table 5). In one breast invasive carcinoma/adjacent normal pair, chr1q and chr16q amplifications were present in both samples but all other aSCNAs were absent, suggesting the amplifications occurred in a shared precursor clone (Figure 3d––sibling model, Supplementary Table 5). In a prostate adenocarcinoma-adjacent normal, most aSCNAs were consistent with TiN=0.4, but some focal deletions were present at 0.7 TiN (Fig. 3d––mixture model). Upon manual review of deTiN’s output, 2 adjacent normal samples contained arm-level aSCNAs absent in the tumor. In one particularly striking case, deTiN’s allele-specific model discerned that a chr1q amplification appearing in both breast carcinoma and its adjacent normal but on opposite alleles, demonstrating convergent evolution (Supplementary Figure 9).
In summary, deTiN is a mixture model integrating evidence from candidate somatic events and copy-number alterations to provide robust TiN estimates used to infer the somatic status of candidate variants. Our analysis quantified TiN in cases with both adjacent normal tissue and normal blood. In particular, TiN contamination may affect normal samples derived retrospectively from formalin-fixed, paraffin-embedded tumor blocks. Although no TiN was identified in 304 TCGA blood normal samples, TiN may be a factor in metastatic cases. TCGA samples, mostly obtained from untreated resected primary tumors, may have lower circulating tumor cells and DNA levels19,20. DeTiN is currently used in large-scale cancer analyses and in the ICGC/TCGA Pan-Cancer Analysis of Whole Genomes (PCAWG, https://dcc.icgc.org/pcawg) project (See Supplementary Note, Supplementary Table 6 and Supplementary Figure 10 for details relating to running deTiN). Future developments of deTiN (or similar) methods can exploit additional data sources to improve accuracy, including independent sequencing (e.g., RNA-seq), additional patient-matched biopsies, and structural variants.
Online Methods:
Overview of deTiN
DeTiN measures TiN (θ) contamination by comparing sequencing data from matched tumor and normal samples. DeTiN uses two statistical (generative mixture) models to estimate TiN. The first uses allelic somatic copy number alterations (aSCNAs) and the second utilizes somatic single nucleotide variants (SSNVs). Each model generates a posterior probability distribution for TiN. If both models are used, deTiN computes the joint posterior distribution. DeTiN reports the maximum a posteriori point estimate for TiN and a 95% confidence interval based on each model and their combination. Next, deTiN uses the TiN estimate to reclassify candidate variants detected in the tumor sample, as either somatic or germline, based on the allele counts observed in the normal at these sites. Below, we describe the inference steps in which we estimate TiN using an Expectation-Maximization (EM) procedure using SSNVs, and maximum a posteriori estimation using aSCNAs, as well as the application of these estimates for somatic variant re-classification (i.e. rescuing previously rejected somatic variants).
Defining TiN:
DeTiN estimates the relative abundance of tumor DNA in the normal sample compared to the tumor sample.
Note that, for simplicity, we define TiN as the relative abundance of DNA to circumvent the need to estimate the purity (percent tumor cells) and ploidy (average DNA content of the tumor cells) of the tumor sample. As such, in the uncommon scenario that the normal sample has a higher fraction of tumor-derived DNA compared to the tumor sample, TiN may theoretically exceed one. In our analysis, we assume that TiN ≤ 1 and in reality it is typically ≪ 1. If the purity (α) and ploidy (τ) of the tumor cells are known (or estimated, e.g. using ABSOLUTE21) then the TiN estimate (θ) can be used to calculate the actual fraction of tumor cells in the normal sample (β) using this equation (Supplementary Figure 10):
Input Data:
The raw inputs to deTiN are: (i) pre-filtered variants (including SNVs and indels, both somatic and germline, (see Filtering of SSNVs) that are observed in the tumor sample, annotated with the corresponding read counts from both tumor and normal samples; and (ii) segmented tumor allele-specific copy number alterations (aSCNAs).
- For each variant v, we denote by fvn and fvt the underlying alternate allele fractions in the tumor (t) and normal (n), respectively. The variables follow Beta distributions, fvn and fvt, conditional on the observed read counts for the reference and alternate alleles in the tumor and normal, (rvt, rvn) and (avt, avn). The total coverage in each sample (hvn, hvt) are taken as the sum of the alternate and reference counts (ignoring the other alleles).
- The aSCNA input data for the tumor is represented as S segments representing aSCNAs (see Filtering of segments and SNPs), each with a corresponding tumor total copy ratio Rst and a set of associated heterozygous germline SNPs within the segment, (v1…vNs). Using the normal data, we first calculate the mean allele fraction (of the non-reference allele) across all heterozygous SNPs (N) to represent the balanced allele fraction (which can slightly deviate from 0.5 due to hybrid capture bias towards reference);
Model:
DeTiN compares two models: (i) no tumor-in-normal, H0 where θ = 0; and (ii) some tumor-in-normal, H1 where 0 < θ ≤ 1. The prior probability of H1, π, is set based on the estimated risk of contamination from malignant cells in the normal, which can depend on the tumor type and the type of the normal sample. For example, when using a tissue adjacent normal, we set π = 0.5, and when using a blood normal we use π = 0.05. Under model H1 we assume a uniform prior distribution for θ.
Model based on aSCNAs:
The model based on aSCNAs compares the tumor allelic imbalance with the allelic imbalance observed in the normal sample at the same genomic segment. Since aSCNAs may arise independently, we treat each segment as an independent measure of TiN. This enables the detection of multiple TiN values in one normal sample, representing different modes of contamination. Assuming we knew the segments TiN value (θs), we could calculate, for each heterozygous SNP in the segment, the expected underlying allele fraction of non-reference reads in the normal sample () (see Derivation of as a function of and θ):
The expected normal allele fraction is equal to the tumor allele imbalance (ψ(fvt)) relative to the midpoint (μsn) multiplied by TiN and the ratio of total copy ratios (Rst, Rsn). The phase of the SNP, with respect to its neighbors, (dvt) is based on the tumor data and equals 1 if it is above the mid-point and −1 otherwise. Since the true somatic allele fraction of each SNP is unknown we integrate over the distribution of possible allele fractions (f) given the observed tumor reads. To calculate the likelihood function for each segment, we calculate the joint likelihood considering all SNPs in each segment.
We perform k-means clustering on the segment TiN estimates (see Clustering of aSCNA data) and calculate the posterior distribution of TiN over all clustered segments in a chosen cluster K:
Inference using aSCNAs:
We calculate the posterior probability for each value of θ (over a grid [0, 0.01, 0.02, …, 1]) and determine θaSCNA*, the MAP estimate of θ.
Model based on SSNVs:
The model based on SSNVs compares the tumor allele fractions of candidate variants with the allele fractions in the normal sample (fvn). For each candidate SSNV, i, we assign a latent Bernoulli indicator variable zi which represents whether the SSNV is classified as a somatic mutation. The prior probability of a candidate SSNV being somatic, ϕ, is set based on the expected ratio of somatic to rare inherited germline variants, which varies by tumor type (e.g. the somatic mutation frequency in chronic lymphocytic leukemia is 1 mutation per megabase and the rate of rare germline SNPs is 10 mutations per megabase, therefore, ϕ is set to 1/11). For most sites with sufficient coverage (depth > 20) the prior has effectively no impact on the classification as somatic mutation.
To calculate the probability of each variant being somatic, we consider the probability of the observed data under 3 scenarios. (i) The variant is a somatic mutation and thus the observed allele counts are due to TiN (zv = 1, avtin = fvtCsnθhvn, rvtin = hvn − avtin); (ii) The variant is a germline polymorphism and allele fraction is determined as described above (SNP) (zv = 0, , rvhet = hvn − avhet); and (iii) The variant is an artifact and the underlying allele fractions are equal in both samples (zv = 0, avt, rvt). A priori we consider candidate variants to be equally likely to be germline variants or sequencing artifacts.
We compute the SSNV data log-likelihood for θ over all candidate variants:
Inference using SSNVs:
To estimate TiN using SSNVs, we use the EM algorithm. Briefly, θ is initialized to 0, and expectation of the variant assignments (zv) are calculated given θ. Then we find θSSNVs* which maximizes the likelihood function (over a grid [0, 0.01, 0.02, …, 1]). We repeat this procedure until the estimate on θ converges (typically in a few iterations).
Inference using the joint likelihood function:
The likelihood functions for SSNVs and aSCNAs are nearly independent since they are generated by distinct underlying processes and use different measurements. Therefore, when both data types are available, deTiN calculates the joint TiN estimate (θ*) and posterior distribution by summing and normalizing the log-likelihood functions for SSNVs and aSCNAs. Next we compare the model θ = 0 to θ = θ*:
As a final step, if the model θ = θ* is chosen we recalculate E[zv] given θ* and classify as somatic candidate variants for which E[zv] > κ (we use κ = 0.5). Finally, to remove variants that do not fit any of our models, we remove sites where the predicted normal allele fraction is unlikely given the observed normal allele counts.
Derivation of fvn as a function of Rvt and θ:
In order to estimate TiN we calculate the expected normal allele fraction of each variant given a TiN value, observed tumor allele fractions, and total copy ratio. We define the allele fractions and total copy ratios as follows, where m is the multiplicity of some variant v, α is the fraction of tumor cells in the tumor sample, β is the fraction of tumor cells in the normal sample, qv is the local total copy number in the tumor sample, τ is the ploidy of the tumor cells, and 2 is the ploidy of normal cells, the allele fractions (fvn, fvt) and copy ratios (Rvn, Rvt) of variants in each sample follow:
We then want to derive a factor Z, which allows us to translate tumor allele fractions fvt to allele fractions in the normal fvn given θ:
We can then show that Rvn = θRvt + 2(1 − θ) and thus derive Csn:
Finally we have the following expression translating a tumor allele fraction to a normal allele fraction given TiN:
Filtering of segments and SNPs:
DeTiN uses only large segments (≥ 200 capture probes) that have at least 20 balanced heterozygous SNPs (ensuring the same number of SNPs with allele fractions below and above 0.5, in the normal sample, by downsampling the more abundant allele). DeTiN ensures an equal number of SNPs above and below 0.5 in the normal sample to remove mapping artifacts. Mapping artifacts are often associated with false-positive calls at low allele fractions. Therefore, segments that cover low mappability regions accumulate reads with errors. These errors tend to be at low allele fraction and some are mis-called as germline SNPs. Accumulation of these spurious germline SNPs can cause methods that estimate allelic copy numbers to incorrectly infer allelic imbalance at these loci. It is important to account for this accumulation of low allele fraction errors since they occur equally in the tumor and normal sample and thus will negatively impact the accuracy of deTiN.
After segment and variant filtering, for each segment s in the tumor data, we calculate the average absolute shift of the allele fractions from balance, ψst, and it’s population variance, σs2;
DeTiN uses segments for which with (ψst) greater than TaSCNA (we use 0.1) and absolute allele shift variance less than 0.025 (σs2 < 0.025).
Filtering of SSNVs:
DeTiN uses candidate SSNVs which are labeled somatic or rejected solely due to observing evidence in the normal. When using MuTect, SSNVs are considered candidates if and only if the judgement column is “KEEP” or the failure reasons column contains only “normal_lod” or “alt_allele_in_normal” or both. Next, we annotate each variant as representing a likely germline SNP or a potential SSNV based on its allele frequency in the ExAC database22. Variants with an ExAC population frequency ≥ 0.01 are considered germline SNPs and variants with < 0.01 allele frequency are considered candidate SSNVs. Variants with less then 15 reads in either sample or below 15% allele fraction in the tumor are not used for TiN estimation but are considered for SSNV recovery.
Clustering of aSCNA data:
In order to identify multiple modes of TiN contamination, deTiN perform’s K means clustering on the posterior TiN distributions of the aSCNAs. DeTiN considers K ∈ {1, 2, 3} clusters and then performs model selection using the bayesian information criterion (BIC). When N is the total number of segments, Nk is the number of segments assigned to cluster k, ns is the number of variants (v) in segment (s), θv refers to the MAP TiN estimate for a SNP, μk is the cluster center, and RSSk is the residual sum of squares for k number of clusters. We determine the BIC score for each number of clusters:
We disregard values of k for which the minimum distance between clusters is less then 2σk, where σk represents the within cluster standard deviation for solution k. We then select the number of clusters (K*) with the minimal BIC, and ensure that BICk * − 1 − BICk* > 10.
Role of tumor derived phasing in deTiN:
Phasing information derived from the tumor sample is important because it reduces the uncertainty on the estimate of allele shift. Given a segment which has an allele shift in the tumor data, one would require two steps in order to estimate the allele imbalance in the normal: (i) comparing the evidence for allele shift with the evidence for balance (the null hypothesis); and (ii) estimating allele shift using the count data. Using the phasing data we can directly compute the best estimate of the allele shift. Without the phasing data, there is an additional step of accounting for the uncertainty of the phase of each SNP. In this scenario, each SNP has a probability, which depends on its allele counts, of representing the higher (allele fraction > 50%) or lower allele (allele fraction < 50%). For example, a SNP with 20 alternate reads and 20 reference reads has equal probability of belonging to each allele, but a SNP with 30 alternate reads and 10 reference reads is more likely to represent the higher allele. In the case of a small allele shift in the normal (ie. most SNPs are close to balance) or in cases of low coverage there is more uncertainty in the phase of the SNP. The uncertainty in the phasing yields greater uncertainty in the estimate of the allele shift in the normal because for each SNP we need to account for the probability of it being generated by each allele. Ignoring the phase information coming from the tumor sample produces less accurate results.
Data Generation:
In-silico simulations:
We selected tumor-normal pairs, for in-silico simulations, from TCGA. We applied the following criteria to select samples: high coverage (200x in the tumor and 80x in the normal), high purity (ABSOLUTE21 purity estimate > 95%), somatic mutation frequency > 1 mutation / Mb, and at least one arm-level aSCNA. Applying this criteria resulted in 5 tumor - normal peripheral blood sample pairs from three tumor types (bladder cancer, glioblastoma multiforme (x3), and a malignant melanoma; Supplementary Table 1).
To create the simulations, we first down-sampled each bam file using SAMtools23 to establish uniform coverage (120x in tumors and 60x in the normals). Then, we down-sampled the normals and tumors in ratios corresponding to the TiN mixtures and mixed each of the resulting bam files and fixed read groups using picard tools. For example, to generate a 0.5 TiN simulation, we down-sampled a normal to 0.5 (30x) and down-sampled the matched tumor to 0.25 (30x), and then mixed them together to generate a 50% TiN mixture (at 60x).
In-vitro simulations:
To evaluate the performance of deTiN on experimentally derived sequencing data, we mixed tumor and normal cell lines in various ratios. For the tumor sample we selected the cell line CRL-2321D and for the normal CRL-2362D. DNA from these samples was mixed in equal amounts to generate a 0.5 TiN pool with total mass of 500ng. We then mixed pure tumor and pure normal with this pool to generate the other mixtures. Samples were volume checked using nanodrop to ensure we achieved the desired mixtures.
We then performed library preparation. Briefly, dsDNA was quantified by Picogreen fluorescence assay using provided DNA standards, 100ng of DNA were fragmented to obtain 150bp pieces by sonication using a Covaris E210 instrument. Solid phase reversible immobilization purification and library construction were performed using AMPure XP Beads, KAPA Library Preparation and KAPA Library Amplification Kits. Library preparation was performed in 96-well plates on an Agilent Bravo Liquid Handler.
Finally we performed hybrid selection, capture and sequencing. DNA was processed through two hybridization events using the Illumina Content Exome Rapid Capture Kit. Samples were normalized to 2ng/uL and pooled. Quantitative PCR (qPCR) was then performed on the pool in order to normalize it to 2nM, before using 0.1M NaOH to denature. Samples were sequenced on Illumina HiSeq2500 machines in Rapid Run mode using 76 base-pair, paired-end reads. The bam files generated by these experiments are publicly available on google cloud, bucket id: fc-070aec01-a599-4fe3-9ed0-2f39288f912e, firecloud: https://portal.firecloud.org/#workspaces/broad-firecloud-testing/deTiN_release_data and the Sequencing Read Archive (PRJNA422575).
Alignment/assembly and Quality control:
Exome sequence processing was performed using established analytical pipelines at the Broad Institute. A BAM file was produced with the Picard pipeline (http://picard.sourceforge.net/), which aligns the tumor and normal sequences to the hg19 human genome build using Illumina sequencing reads. The BAM was uploaded into the Firehose pipeline (http://www.broadinstitute.org/cancer/cga/Firehose), which manages input and output files to be executed.
Quality control modules for assessment of genotype concordance and cross contamination using ContEst24 were applied within Firehose.
Mutation calling and copy number analysis:
MuTect1, Strelka2, and Varscan23 were applied to identify somatic single-nucleotide variants. Strelka2 was applied to identify small insertions or deletions. Variants were filtered by a panel of normal samples to remove sequencing variants as previously described9. Annotation of identified variants was done using Oncotator25.
Copy-ratios and germline SNPs were inferred using GATK’s CNV analysis suite (https://github.com/broad institute/gatk). Briefly, read depth at capture probes in tumor samples was normalized using tangent normalization against a panel of normal samples. The resulting normalized coverage ratios are then segmented using the circular binary segmentation (CBS) algorithm. This data was then transformed into allelic copy number data via integration of data from informative inherited SNPs. MuTect’s “call-stats” raw variant file, allelic copy number data, and inherited SNPs are the required inputs to deTiN. See below.
Statistics and data analysis:
For in-silico simulation data points in Figure 1a, Figure 1c, and Supplementary Figure 3a show the weighted mean TiN estimate from 5 independent experiments (n=5 for each TiN level). Error bars in these figures show the standard error on the weighted mean. For in-vitro simulation data points in Figure 1b, Figure 1d, Supplementary Figure 2 a–c, and Supplementary Figure 3b, panels show results from a single experiment (n=1 for each TiN level). Error bars show the 95% confidence interval on the TiN estimate in Figure 1b and show the 95% confidence interval on the sensitivity calculated using the beta distribution (MATLAB function “betapdf”) in Figure 1d,Supplementary Figure 2 a–c, and Supplementary Figure 3b. TiN estimates and sensitivities are reported in Supplementary Tables 1 and 2. ROC curves and AUCs in Supplementary Figure 3e–f were calculated using the in-vitro sequencing experiment and the python package scikit-learn function “roc_auc_score”. Error bars in Supplementary Figure 3f show the 95% confidence interval generated via bootstrapping (n=100 iterations). Error bars shown in Supplementary Figure 4a–b,d are based on 100 iterations of downsampling. Error bars shown in Supplementary Figure 4c and e indicate 95% confidence interval on TiN estimate calculated using the in-vitro sequencing mixture.
Comparisons of TiN estimates and mutation rates shown in Figure 2a and Figure 2b were performed using a two-tailed Mann-Whitney Test (MATLAB function “ranksum”). For each panel n=257. Error bars shown in Supplementary Figure 6b (red) and Figure 3c show one standard deviation on the allele fraction calculated using the beta distribution. Estimates and mutations are reported in Supplementary Table 3. Error bars in Figure 3b show standard error on mean sensitivities (for TiN = 0:n=230; TiN=0.01:n=9; TiN = 0.03:n=9; TiN=0.07:n=4; otherwise no error bar is shown). Normal blood samples were used to generate “truth set” variants. Calls with lower than 10x coverage in tumor or normal samples and lower than 10% allele fraction in the tumor were excluded from this analysis.
Life Sciences Reporting Summary:
Further information on experimental design is available in the Life Sciences Reporting Summary.
Code availability:
DeTiN is available for use https://www.broadinstitute.org/cancer/cga/deTiN and source code is available at https://github.com/broadinstitute/deTiN. Furthermore deTiN is accessible using the Broad Institute’s genomics analysis platform firecloud. Module: broadinstitute_cga/detin_v1.0. Data in this paper was generated using a MATLAB implementation of deTiN (https://hub.docker.com/r/broadinstitute/detin_matlab) which is available upon request but no longer being supported.
Data availability:
Additionally, the in-vitro validation sequencing data is available on the Sequencing Read Archive (PRJNA422575)
Supplementary Material
Acknowledgements:
G.G. was partially funded by the NIH TCGA Genome Data Analysis Center (U24CA143845) and the Paul C. Zamecnick, MD, Chair in Oncology at MGH Cancer Center. A.T.W was funded in part by T32 HG002295 from the National Human Genome Research Institute, NIH. A.T.W, C.S and C.J.W. were partially funded by grants from the National Institutes of Health (NCI P01CA206978–01, R01CA182461–01, U10CA180861–01, R01CA184922–02). C.J.W. is a Scholar of the Leukemia and Lymphoma Society.
Footnotes
Competing Financial Interests Statement:
C.J.W. is a co-founder of Neon Therapeutics and a member of its scientific advisory board.
References for main text
- 1.Cibulskis K et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples 31, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saunders CT et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 28, 1811–7 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Koboldt DC et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22, 568–76 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stieglitz E et al. The genomic landscape of juvenile myelomonocytic leukemia. Nat. Genet 47, 1326–1333 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei L et al. Pitfalls of improperly procured adjacent non-neoplastic tissue for somatic mutation analysis using next-generation sequencing. BMC Med. Genomics 9, 64 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genomic and Epigenomic Landscapes of Adult De Novo Acute Myeloid Leukemia. N. Engl. J. Med 368, 2059–2074 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor-Weiner A et al. Genomic evolution and chemoresistance in germ-cell tumours. Nature 540, 114–118 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welch JS et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 150, 264–78 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landau DA et al. Mutations driving CLL and their evolution in progression and relapse. Nature 526, 525–530 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng G, Lu Y, Zlotnikov G, Thor AD & Smith HS Loss of Heterozygosity in Normal Tissue Adjacent to Breast Carcinomas doi: 10.1126/science.274.5295.2057 [DOI] [PubMed] [Google Scholar]
- 11.Försti A et al. Loss of heterozygosity in tumour-adjacent normal tissue of breast and bladder cancer. Eur. J. Cancer 37, 1372–1380 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Leung WK et al. Concurrent hypermethylation of multiple tumor-related genes in gastric carcinoma and adjacent normal tissues. Cancer 91, 2294–2301 (2001). [PubMed] [Google Scholar]
- 13.Braakhuis BJM, Tabor MP, Kummer JA, Leemans CR & Brakenhoff RH A Genetic Explanation of Slaughter’s Concept of Field Cancerization: Evidence and Clinical Implications. CANCER Res 63, 1727–1730 (2003). [PubMed] [Google Scholar]
- 14.Forbes SA et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res 43, D805–11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rheinbay E et al. Recurrent and functional regulatory mutations in breast cancer. Nat. Publ. Gr 547, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Allen EM et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat. Med 20, 682–688 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giannakis M et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep (2016). doi: 10.1016/j.celrep.2016.03.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanda M et al. Presence of Somatic Mutations in Most Early-Stage Pancreatic Intraepithelial Neoplasia doi: 10.1053/j.gastro.2011.12.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bettegowda C et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med 6, 224ra24 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwarzenbach H, Hoon DSB & Pantel K Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 11, 426–437 (2011). [DOI] [PubMed] [Google Scholar]
Supplementary References:
- 21.Carter SL et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol 30, 413–421 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lek M et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cibulskis K et al. ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics 27, 2601–2602 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramos AH et al. Oncotator Cancer Variant Annotation Tool 36, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additionally, the in-vitro validation sequencing data is available on the Sequencing Read Archive (PRJNA422575)