Abstract
Patients with deletion of chromosome 13 present with variable clinical features, and the correlation between phenotype and genomic aberration is not well established in the literature, mainly due to variable sizes of the deleted segments and inaccuracy of breakpoint mapping. In order to improve the genotype-phenotype correlation, we obtained clinical and cytogenomic data from 5 Brazilian patients with different chromosome 13 deletions characterized by G-banding and array techniques. Breakpoints were nonrecurrent, with deletion sizes ranging from 3.8 to 43.3 Mb. Our patients showed some classic features associated with 13q deletion, independent of the location and size of the deletion: hypotonia, growth delay, psychomotor developmental delay, microcephaly, central nervous system anomalies, and minor facial dysmorphism as well as urogenital and limb abnormalities. Comparisons between the literature and our patients' data allowed us to narrow the critical regions that were previously reported for microphthalmia and urogenital abnormalities, indicating that gene haploinsufficiency of ARHGEF7, PCDH9 and DIAPH3, of MIR17HG and GPC6, and of EFNB2 may contribute to microcephaly, cardiovascular disease, and urogenital abnormalities, respectively. The knowledge about genes involved in the phenotypic features found in 13q deletion patients may help us to understand how the genes interact and contribute to their clinical phenotype, improving the patient's clinical follow-up.
Keywords: Chromosome 13, Genotype-phenotype correlation, 13q deletion syndrome
Cytogenomic disorders constitute human genetic diseases caused by DNA rearrangements that result in gain, loss, or disruption of dosage-sensitive genes [Stankiewicz and Lupski, 2006]. Investigating chromosome rearrangements with similar genome imbalances is important for a better understanding of genotype-phenotype correlation, essential for diagnosis, prognosis, and genetic counseling.
Patients with chromosome 13 deletion, including ring chromosomes, constitute a rare group of patients. These patients may have overlapping clinical features making it difficult to delineate a specific clinical phenotype for each group, mainly due to variable sizes of the deleted segments and to the inaccuracy of breakpoint mapping in some cases previously described.
Individuals with 13q partial deletion present with a wide phenotypic spectrum which includes intellectual disability; growth delay; minor facial dysmorphic features; major malformations involving the brain, renal, heart, genital, and gastrointestinal systems; eye anomalies such as retinoblastoma, and distal limb abnormalities [Brown et al., 1993; Van Buggenhout et al., 1999; Alanay et al., 2005; Christofolini et al., 2006; Ballarati et al., 2007; Walczak-Sztulpa et al., 2008]. Brown et al. [1993] classified the patients with 13q deletion in 3 groups: (1) those with proximal deletions, usually not extending into 13q32, presenting with mild or moderate intellectual disability, variable minor anomalies, growth delay, and sometimes, retinoblastoma; (2) those with more distal deletions, including at least part of 13q32, with severe intellectual disability, growth delay, microcephaly, and brain, eye, gastrointestinal tract, and distal limb abnormalities, and (3) those with the most distal deletions, involving 13q33q34, presenting with severe intellectual disability but without other major malformations or growth delay. In 1995, Brown et al. defined 13q32 as the critical region for the most severe phenotypes, including brain abnormalities, severe intellectual disability, and growth delay.
Regarding ring chromosome 13, due to the different mechanisms of formation, the patients exhibit a wide spectrum of phenotype variability, in which some of the features are similar to those found in cases with del(13q), such as growth delay, intellectual disability, brain anomalies, microcephaly, facial dysmorphism, genital malformations, anal atresia, eye malformations, multiple malformations in the inner organs, and hand, foot or toe abnormalities [Schinzel, 2001]. Less frequent abnormalities associated with r(13) are hypoplasia of different organs, cardiac anomalies, pigment anomalies, and epilepsy [Brandt et al., 1992; Bedoyan et al., 2004].
Although several attempts have been made to correlate alterations in chromosome 13 with distinct phenotypes, there is still no consensus between the deletion of distinct 13q regions and specific clinical features. Here, we present 5 patients with different deletions involving chromosome 13, revealed by G-banding and array techniques.
Clinical Reports
All patients were selected and evaluated by the geneticists of the Medical Genetics Center of the Universidade Federal de São Paulo.
The clinical data from 5 patients with chromosome 13 deletions were revised by the same geneticist to ensure the correct genotype-phenotype correlation and are summarized in Table 1.
Table 1.
Summary of clinical findings in the patients with 13q deletion
| P1 | P2 | P3 | P4 | P5 | |
|---|---|---|---|---|---|
| Genomic imbalance | del(13)(q34) | del(13)(q33.1q34) | del(13)(q33.1q34) | del(13)(q21.33q31.3) | del(13)(q21.1q32.2) |
| Sex | M | M | M | F | F |
| Age at evaluation | 3 yrs 5 mo | 9 mo | 2 yrs 9 mo | 17 yrs 4 mo | 1 y 3 mo |
| Birth age | Term | Preterm (32 w) | Preterm (37 w) | Term | Term |
| Birth weight, g | 2,880 (−1.54 SD) | 1,730 (−0.17 SD) | 1,520 (−3.46 SD) | 2,850 (−1.22 SD) | 2,590 (−1.84 SD) |
| Birth length, cm | 45 (−2.75 SD) | 42 (−0 SD) | 41 (−3.03 SD) | 45 (−2.39 SD) | 44 (−2.83 SD) |
| Birth head circumference, cm | 29 (−4.25 SD) | 29 (−0.26 SD) | 28 (−3.61 SD) | NE | 32 (−1.96 SD) |
| Hypotonia | + | + | + | + | + |
| Growth delay | + | + | + | + | + |
| Psychomotor developmental delay | + | + | + | + | + |
| Microcephaly | + | + | + | + | + |
| Central nervous system anomalies | + | NE | + | NE | NE |
| Hearing loss | − | NE | + | + | − |
| Microphthalmia | − | − | − | − | − |
| Ocular hypertelorism | + | + | + | + | + |
| Upslanting palpebral fissures | + | + | − | + | + |
| Strabismus | + | − | + | + | + |
| Bilateral epicanthus | + | + | + | + | + |
| Large ears | − | + | + | − | − |
| Retrognathia | + | − | − | + | + |
| Short neck | + | − | + | + | + |
| Congenital heart defect | − | NE | NE | − | ASD |
| Urogenital abnormalities | − | + | + | + | − |
| Limb abnormalities | − | + | + | + | + |
ASD, atrial septal defect; del, deletion; dup, duplication; mo, months; NE, not evaluated; w, weeks; yrs, years; +, present; –, absent.
Patient 1
Patient 1 is the first child of healthy nonconsanguineous parents. At 3 years and 9 months, the boy's measurements were: height 97.5 cm (−0.73 SD), weight 17.2 kg (0.74 SD), and OFC 46.5 cm (< −3 SD). He presented with hypotonia, mild psychomotor developmental delay, speech delay, microcephaly, a narrow and sloping forehead, ocular hypertelorism, mild upslanting palpebral fissures, bilateral epicanthus, strabismus, a prominent and wide nasal bridge, retrognathia, thin upper lip, prominent upper incisors, a short neck, and wide-spaced inverted nipples. The brain MRI revealed cerebellar hypoplasia and mastoiditis.
Patient 2
The boy is the second child of a healthy nonconsanguineous couple. At 3 years and 4 months, he presented with hypotonia, growth and speech delay, severe psychomotor developmental delay, microcephaly, an elongated face, narrow and sloping forehead, ocular hypertelorism, upslanting palpebral fissures, bilateral epicanthus, prominent nasal bridge with hypoplastic nares, large ears, prominent upper incisors, a high palate, renal ectopia, penoscrotal transposition, scrotal hypoplasia, and bilateral large halluces.
Patient 3
This patient is the first child of a healthy nonconsanguineous couple. At birth, the boy showed atypical genitalia with hypospadias, penoscrotal transposition and scrotal hypoplasia (surgically corrected at age 2 years), hypotonia, moderate psychomotor developmental delay, speech delay, microcephaly, brachycephaly, narrow and sloping forehead, ocular hypertelorism, strabismus, bilateral epicanthus, large ears, flat nasal root, anteverted nares, short columella, a triangular mouth with downturned corners, a short neck, left testicle in inguinal canal, right transverse palmar crease, and bilateral proximally placed second toes. The brain MRI showed right cerebellar hypoplasia (Dandy-Walker variant) and slight enlargement of the 4th and lateral ventricles. At 2 years and 7 months, his measurements were: height 91.5 cm (−0.05 SD), weight 13 kg (−0.44 SD), and OFC 46 cm (< −3 SD).
Patient 4
Patient 4 is the third child of healthy nonconsanguineous parents. At 5 years and 7 months, the girl's measurements were: height 90 cm (−4.94 SD), weight 10.7 kg (−6.10 SD), and OFC 47 cm (< −3 SD). She presented with the following main dysmorphic features: short stature, microcephaly, a triangular face, strabismus, ocular hypertelorism, upslanting palpebral fissures, bilateral epicanthus, bilateral low-set ears, retrognathia, high palate, a short neck, genital hypoplasia, bilateral clinodactyly of the 5th fingers, and a short foot as well as axial hypotonia and moderate psychomotor developmental delay. At age 17 years and 4 months, her height was 143.5 cm (−3.01 SD), weight 36 kg (−4.13 SD), and her OFC was 53 cm (2 SD). In addition to the dysmorphic features described above, she had a retinal detachment treated surgically by laser at 15 years of age.
Patient 5
This patient is the second child of a healthy, young nonconsanguineous couple. At 15 months, the girl's height was 66.5 cm (−3.39 SD), weight 6.3 kg (−5.07 SD), and her OFC was 40 cm (−4.65 SD). She showed signs of Feingold syndrome type 2 with the following main dysmorphic features: proportionate short stature, microcephaly, ocular hypertelorism, upslanting palpebral fissures, bilateral epicanthus, long and thick eyelashes, bilateral nystagmus, strabismus, both ears with prominent antihelix, downturned corners of the mouth, retrognathia, high palate, a short neck, wide-spaced nipples, genital hypoplasia, brachydactyly of fingers and toes, bilateral clinodactyly, camptodactyly of the 2nd right digit, a short foot, anteriorly placed anus, axial hypotonia, and moderate psychomotor developmental delay. An echocardiogram revealed an atrial septal defect.
Methods
Cytogenetic Analysis
G-banding [Moorhead et al., 1960; Sanchez et al., 1973] was performed on peripheral blood lymphocyte cultures according to standard procedures. At least 20 G-banded metaphases were analyzed for each patient. The metaphase chromosomes, with a 550-band resolution, were classified according to the International System for Human Cytogenomic Nomenclature [ISCN, 2016].
Array Analysis
DNA was isolated from peripheral blood using the Gentra Puregene Kit (Qiagen Sciences Inc., Germantown, MD, USA). Array was performed using Human610-Quad and BeadStation (Illumina, San Diego, CA, USA), Genome-Wide Human SNP Array 6.0 or CytoScan HD Array (Affymetrix, Santa Clara, CA, USA), according to the manufacturer's instructions. All cases were referred using annotation GRCh37/hg19.
Results
G-banding analysis revealed a ring chromosome 13 in 2 patients (P1 and P2), 1 patient with terminal 13q deletion (P3), and 2 patients with interstitial 13q deletions (P4 and P5).
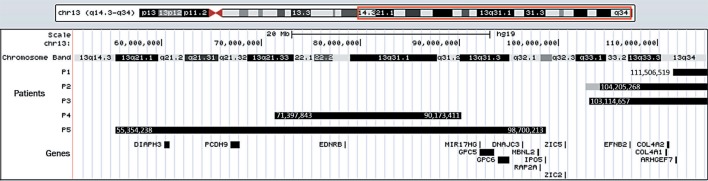
The array analysis determined the breakpoints and the genomic coordinates in all cases and revealed a concomitant 1.5-Mb duplication in r(13) for patient 2 (Fig. 1). Table 2 shows the karyotype and chromosomal microarray results for all patients.
Fig. 1.
Genome map of the distal part of the long arm of chromosome 13, showing the chromosome bands, the deleted (black bars) and the duplicated (gray bar) regions in the patients (P1-P5) as well as the genes of interest discussed in the text. The genomic coordinates of the deleted regions are indicated.
Table 2.
Cytogenomic results of the patients
| Patient | Cytogenomic data | Genomic imbalance |
|---|---|---|
| P1 | 46,XY,r(13)(p13q34).arr 13q34(111506519_115105020)×1 | del 13q (3.8 Mb) |
| P2 | 46,XY,r(13)(p13q33.1).arr 13q33.1(102745509_104203461)×3,13q33.1q34(104205268_115109878)×1 | dup 13q (1.5 Mb) |
| del 13q (11.1 Mb) | ||
| P3 | 46, XY, del(13)(q33.1).arr 13q33.1q34(103114657_115108385)×1 | del 13q (l1.9 Mb |
| P4 | 46, XX, del(l3)(q21.33q31.3).arr 13q21.33q31.3(71397843_90173411)×1 | del 13q (l8.8 Mb) |
| P5 | 46, XX, del(l3)(q21.1q32.2).arr 13q21.1q32.2(55354238_98700213)×1 | del 13q (43.3 Mb) |
del, deletion; dup, duplication.
Discussion
After the precise determination of the breakpoints, we attempted to correlate the clinical findings with the location of the 13q deletion and with the genes deleted in these patients.
Brown et al. [1993] classified the patients in 3 groups based on the correlations of 13q deletion intervals with specific phenotypic signs. According to this classification, our patient 4 belongs to group 1, patient 5 belongs to group 2, and patients 1, 2, and 3, to group 3.
Concerning group 1, our patient meets the criteria established by Brown et al. [1993] since she presents with moderate psychomotor developmental delay, minor anomalies, and growth delay. However, the patient also has genital hypoplasia, bilateral clinodactyly, and retinal detachment. Patient 5 has the largest deletion (43.3 Mb) including part of 13q32 with moderate intellectual disability, not meeting Brown's criteria that describe these patients with severe intellectual disability. She also presents with proportionate short stature, microcephaly, atrial septal defect, anteriorly placed anus, genital hypoplasia, and brachydactyly of fingers and toes, corroborating the fact that patients with deletions including the 13q32 region show the most severe phenotypes [Brown et al., 1995]. According to the classification of Brown et al. [1993], patients in group 3 present with severe intellectual disability but without other major malformations or growth delay. Our 3 patients with distal deletions present with microcephaly, hypotonia, and growth/psychomotor developmental delay. Two of them also show central nervous system anomalies (P1 and P3) and urogenital/distal limb anomalies (P2 and P3) - major malformations that do not fit to Brown's criteria.
As shown in Table 1, our 5 patients demonstrate some classic features associated with 13q deletion independent of the location and size of the deletion: hypotonia (5/5), growth delay (5/5), psychomotor developmental delay (5/5), microcephaly (5/5), hearing loss (2/4), facial minor dysmorphic features (including ocular hypertelorism (5/5), upslanting palpebral fissures (4/5), strabismus (4/5), bilateral epicanthus (5/5), retrognathia (3/5), short neck (4/5), urogenital anomalies (3/5), limb abnormalities (4/5), whereas central nervous system anomalies were noted in 2 patients with terminal deletion, and congenital heart defect was found only in the patient with interstitial deletion that includes the 13q32 region.
Despite some common phenotypes, it is difficult to group patients and compare them without considering the genes included in the deleted region in each case. So, we attempted to correlate the deleted candidate genes in our patients with the main features described in 13q deletions. However, no specific phenotypic effect due to the presence of the ring chromosome or the small region duplicated in patient 2 could be observed.
Microcephaly
A common feature in individuals with 13q deletion is microcephaly which was found in all of our patients. Kirchhoff et al. [2009] correlated the microcephaly phenotype to the ∼6-Mb terminal 13q region, between 13q33.3 and 13q34. In most patients reported by Ballarati et al. [2007] and Quelin et al. [2009] who had terminal deletions, microcephaly was observed. Mapped in 13q34, the ARHGEF7 gene, known to play a role in the development of human cerebral cortex [Sheen et al., 2004], was suggested as a candidate gene for psychomotor developmental delay and microcephaly [Walczak-Sztulpa et al., 2008]. In 3 out of 5 patients (P1, P2, and P3) with microcephaly, the terminal deletions include the ARHGEF7 gene, reinforcing this region as a good candidate for this feature.
Mitter et al. [2011] suggested another region (13q21.32q21.33) as critical for microcephaly and the PCDH9 gene as a good candidate. The PCDH9 gene has a putative role in specific neuronal connections and signal transduction [Strehl et al., 1998]. Our patient 5 with microcephaly has a 43.3-Mb interstitial deletion that includes the candidate PCDH9 gene, suggesting that this gene may also be involved in microcephaly. Another gene, DIAPH3, located in 13q21.2, was recently associated with microcephaly. Inactivation of the Diaph3 gene causes a loss of cortical progenitor cells in mice, resulting in microcephaly [Damiani et al., 2016]. Additionally, a mutation in another gene of the same family, DIAPH1, located in 5q31.3, has already been described to play a role in human microcephaly [Ercan-Sencicek et al., 2015]. The interstitial deleted region of our patient 5 also includes the DIAPH3 gene, so the involvement of a second gene in this case cannot be ruled out. Interestingly, patient 4, who has a deletion not involving any candidate gene, showed microcephaly during infancy, and at age 17 years, the OFC was normal.
Central Nervous System Abnormalities
Central nervous system abnormalities are often described in 13q deletion patients including holoprosencephaly, neural tube defects, agenesis/dysgenesis of the corpus callosum, cortical dysplasia, and posterior fossa anomaly [McCormack et al., 2002; Ballarati et al., 2007]. Ballarati et al. [2007] and Kirchhoff et al. [2009] reported the association between the 13q32.2q33.2 region and Dandy-Walker malformation or cerebellar hypoplasia and proposed the ZIC2 and ZIC5 genes as the cause of posterior fossa anomaly. ZIC2 is important for normal development of the forebrain [McCormack et al., 2002] and its mutations cause holoprosencephaly [Brown et al., 1998]. All the patients described in this study show psychomotor developmental delay, and in 2 patients (P1 and P3) posterior fossa anomaly was present, but the abovementioned genes were not deleted. Patient 3, with right cerebellar hypoplasia (Dandy-Walker malformation variant) and slight enlargement of the 4th and lateral ventricles, carries an 11.9-Mb deletion in the 13q33.1q34 region, and patient 1 with cerebellar hypoplasia, carries the smallest 13q deletion (3.8 Mb) in 13q34. In both cases, the affected region does not correspond with the previously defined critical region. Myers et al. [2017] suggested that posterior fossa anomaly in distal 13q deletion syndrome can occur due to disruption of multiple genes, other than haploinsufficiency of ZIC2 and ZIC5, and our patients 1 and 3 support this hypothesis.
Microphthalmia
Ballarati et al. [2007] suggested a relationship between microphthalmia, blindness, and the 13q33.2q33.3 region, which includes the EFNB2 gene, whose haploinsufficiency was postulated to be responsible for micro/anophthalmia. Another critical region for microphthalmia was defined by Kirchhoff et al. [2009] between 13q31.3 and 13qter and narrowed to a smaller region of 18 Mb between 13q32.1 (∼97.3 Mb) and 13qter by Quelin et al. [2014].
Three of our patients have distal deletions (P1, P2, and P3) and 2 of them (P2 and P3) have deletions comprising the EFNB2 gene, but none with microphthalmia. In 2 of our patients with interstitial deletions (P4 and P5), eye abnormalities do not occur.
Therefore, our data suggest that deletions of the 13q33.1qter region and the EFNB2 gene haploinsufficiency are probably not causative of micro/anophthalmia.
Moreover, the breakpoint of patient 5 allowed us to narrow the critical region for microphthalmia to 13q32.2q33.1 (between 98.7 and 103.1 Mb). Interestingly, the 13q32.2q33.1 region partially overlaps the keratoconus (KTCN) type 7 locus. KTCN is a noninflammatory thinning of the cornea that results in distortion of the corneal surface, altered refractive powers of the eye, and reduced visual acuity. Gajecka et al. [2009] reported a KTCN locus in 13q32 and recently, a patient carrying a 17.2-Mb deletion in 13q31.1q32.2 with Feingold syndrome type 2 and KTCN was described by Sirchia et al. [2017]. The authors propose IPO5, DNAJC3, MBNL2, and RAP2A as best candidate genes. Our patient 5 has a deletion that encompasses these genes, however, without KTCN. Since KTCN generally occurs during puberty, we cannot exclude the involvement of deleted genes, but IPO5 seems to be a good candidate.
Heart Defects
Concerning congenital heart defects, Huang et al. [2012] proposed that 13q33.1q34 may be one of the responsible regions. Wang et al. [2017] reported that the MIR17HG cluster host gene and the GPC6 gene, located in the 13q31.3 region, as well as EFNB2, COL4A1 and COL4A2 in the 13q33.1q34 region, may contribute to cardiovascular disease development. Since cardiac defects could not be evaluated in our 2 patients (P2 and P3), whose deletions involve these last 3 genes, we cannot confirm them as causative of congenital heart defects.
Mutations in GPC6 are associated with Omodysplasia 1 [Campos-Xavier et al., 2009], a rare autosomal recessive skeletal dysplasia. Patients with GPC6 mutations may additionally present with congenital heart defects, but it is not clear if haploinsufficiency has an impact on heart development. Deletions in MIR17HG, a miRNA encoding gene, are associated with Feingold syndrome type 2, an autosomal dominant disorder associated with microcephaly, short stature, digital anomalies, learning disabilities, hearing loss, and occasionally congenital heart defect [de Pontual et al., 2011; Grote et al., 2015]. Since our patient 5 has an atrial septal defect and a 13q21.1q32.2 deletion that includes both MIR17HG and GPC6, we could confirm the possible involvement of these genes in cardiovascular disease.
Urogenital Abnormalities
Urogenital abnormalities in male patients, including penoscrotal transposition, have a well-established association with 13q deletion. Deletions in the 13q32.2q34 region were previously described as critical for anal atresia, penoscrotal transposition, hypospadias, anteriorly placed anus, imperforate anus, and atypical genitalia [Bartsch et al., 1996; Garcia et al., 2006; Kirchhoff et al., 2009]. In females, the 13q distal deletion is associated with an absent or bicornuate uterus, imperforate anus with vaginal fistula, or cloacal abnormality [Garcia et al., 2006]. Our male patients 2 and 3 have urogenital abnormalities, while the male patient 1, with a deletion that involves only part of the 13q34 band, has no urogenital abnormality, suggesting that the region responsible for the abnormality is within a 7.3-Mb region between positions 104.2 and 111.5 Mb in 13q33.1q34.
Located in 13q33.3, the EFNB2 gene has been suggested as a candidate to play a role in male urogenital abnormalities in several studies [Dravis et al., 2004; Garcia et al., 2006; Walczak-Sztulpa et al., 2008; Kirchhoff et al., 2009; Andresen et al., 2010; Wang et al., 2017]. In fact, EFNB2 haploinsufficiency may provide a causative explanation for the observed urogenital abnormalities in our 13q deletion male patients 2 and 3.
Wang et al. [2017] suggested the EDNRB gene, located in the 13q22.3 region, as another candidate for urogenital abnormalities. Although both our female patients carry deletions involving EDNRB, only patient 5 presents external genital hypoplasia, suggesting that this correlation is not so definitive.
Limb Abnormalities
Concerning limb abnormalities, absent or small thumbs were associated with 2 regions: 13q31.3q33.1 and 13q33.3q34 [Kirchhoff et al., 2009]. Quelin et al. [2009] suggested the GPC5 gene, which maps in 13q31.3, as a candidate gene for finger abnormalities. De Pontual et al. [2011] also proposed a possible contribution of GPC5 haploinsufficiency in limb abnormalities, supported by studies in mice [Saunders et al., 1997] that demonstrated the involvement of this gene in growth and morphogenesis within the limbs, central nervous system, and kidneys. However, Quelin et al. [2014] narrowed the proximal critical region to 13q32.2q33.1, excluding this gene.
Our patients 2 and 3, who present bilateral large halluces and bilateral proximally placed second toes, respectively, had deletions including only a small part of the 13q33.1 region. Patient 5, who had bilateral clinodactyly, camptodactyly of the 2nd right digit and brachydactyly of fingers and toes, carries a deletion that includes the GPC5 gene, proposed as a candidate gene by Quelin et al. [2009] and de Pontual et al. [2011]. However, our patient 4 has bilateral clinodactyly of the 5th fingers and a deletion not including the previously proposed critical region, and patient 1, who has a 13q34 deletion, shows no limb abnormalities. Thus, according to our data, limb abnormalities could not be associated with a specific genomic region in chromosome 13.
Conclusion
The data from the literature combined with our data allow an update of the genotype-phenotype correlation for partial 13q deletions. The literature shows a wide variability in the phenotypic features of patients even with deletions in the same chromosomal regions. We pointed out that defining the precise breakpoints narrowing the critical regions is essential for a better phenotype-genotype correlation. The knowledge about genes involved in the phenotypic features found in 13q deletion patients may help us to understand how the genes interact and contribute to the clinical phenotype.
Statement of Ethics
This study was approved by the Ethics Commission of the Universidade Federal de São Paulo and informed written consent was obtained.
Disclosure Statement
All authors declare no conflict of interests.
Funding Sources
This work was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Brazil (grant to M.I.M. 2014/11572-8).
References
- 1.Alanay Y, Aktas D, Utine E, Talim B, Onderoğlu L, et al. Is Dandy-Walker malformation associated with “distal 13q deletion syndrome”? Findings in a fetus supporting previous observations. Am J Med Genet A. 2005;136:265–268. doi: 10.1002/ajmg.a.30808. [DOI] [PubMed] [Google Scholar]
- 2.Andresen JH, Aftimos S, Doherty E, Love DR, Battin M. 13q33.2 deletion: a rare cause of ambiguous genitalia in a male newborn with growth restriction. Acta Paediatr. 2010;99:784–786. doi: 10.1111/j.1651-2227.2010.01683.x. [DOI] [PubMed] [Google Scholar]
- 3.Ballarati L, Rossi E, Bonati MT, Gimelli S, Maraschio P, et al. 13q Deletion and central nervous system anomalies: further insights from karyotype-phenotype analyses of 14 patients. J Med Genet. 2007;44:e60. doi: 10.1136/jmg.2006.043059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartsch O, Kuhnle U, Wu LL, Schwinger E, Hinkel GK. Evidence for a critical region for penoscrotal inversion, hypospadias, and imperforate anus within chromosomal region 13q32.2q34. Am J Med Genet. 1996;65:218–221. doi: 10.1002/(SICI)1096-8628(19961028)65:3<218::AID-AJMG9>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 5.Bedoyan JK, Flore LA, Alkatib A, Ebrahim SA, Bawle EV. Transmission of ring chromosome 13 from a mother to daughter with both having a 46,XX,r(13)(p13q34) karyotype. Am J Med Genet A. 2004;129A:316–320. doi: 10.1002/ajmg.a.30242. [DOI] [PubMed] [Google Scholar]
- 6.Brandt CA, Hertz JM, Petersen MB, Vogel F, Noer H, Mikkelsen M. Ring chromosome 13: lack of distinct syndromes based on different breakpoints on 13q. J Med Genet. 1992;29:704–708. doi: 10.1136/jmg.29.10.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown S, Gersen S, Anyane-Yeboa K, Warburton D. Preliminary definition of a “critical region” of chromosome 13 in q32: report of 14 cases with 13q deletions and review of the literature. Am J Med Genet. 1993;45:52–59. doi: 10.1002/ajmg.1320450115. [DOI] [PubMed] [Google Scholar]
- 8.Brown S, Russo J, Chitayat D, Warburton D. The 13q-syndrome: the molecular definition of a critical deletion region in band 13q32. Am J Hum Genet. 1995;57:859–866. [PMC free article] [PubMed] [Google Scholar]
- 9.Brown SA, Warburton D, Brown LY, Yu CY, Roeder ER, et al. Holoprosencephaly due to mutations in ZIC2, a homologue of Drosophila odd-paired. Nat Genet. 1998;20:180–183. doi: 10.1038/2484. [DOI] [PubMed] [Google Scholar]
- 10.Campos-Xavier AB, Martinet D, Bateman J, Belluoccio D, Rowley L, et al. Mutations in the heparan-sulfate proteoglycan glypican 6 (GPC6) impair endochondral ossification and cause recessive omodysplasia. Am J Hum Genet. 2009;84:760–770. doi: 10.1016/j.ajhg.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christofolini DM, Yoshimoto M, Squire JA, Brunoni D, Melaragno MI, Carvalheira G. Hydrocephaly, penoscrotal transposition, and digital anomalies associated with de novo pseudodicentric rearranged chromosome 13 characterized by classical cytogenetic methods and mBAND analysis. Am J Med Genet A. 2006;140:1321–1325. doi: 10.1002/ajmg.a.31269. [DOI] [PubMed] [Google Scholar]
- 12.Damiani D, Goffinet AM, Alberts A, Tissir F. Lack of Diaph3 relaxes the spindle checkpoint causing the loss of neural progenitors. Nat Commun. 2016;13509:2016. doi: 10.1038/ncomms13509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Pontual L, Yao E, Callier P, Faivre L, Drouin V, et al. Germline deletion of the miR-17 ∼92 cluster causes skeletal and growth defects in humans. Nat Genet. 2011;43:1026–1030. doi: 10.1038/ng.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dravis C, Yokoyama N, Chumley MJ, Cowan CA, Silvany RE, et al. Bidirectional signaling mediated by ephrin-B2 and EphB2 controls urorectal development. Dev Biol. 2004;271:272–290. doi: 10.1016/j.ydbio.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 15.Ercan-Sencicek AG, Jambi S, Franjic D, Nishimura S, Li M, et al. Homozygous loss of DIAPH1 is a novel cause of microcephaly in humans. Eur J Hum Genet. 2015;23:165–172. doi: 10.1038/ejhg.2014.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gajecka M, Radhakrishna U, Winters D, Nath SK, Rydzanicz M, et al. Localization of a gene for keratoconus to a 5.6-Mb interval on 13q32. Invest Ophthalmol Vis Sci. 2009;50:1531–1539. doi: 10.1167/iovs.08-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia NM, Allgood J, Santos LJ, Lonergan D, Batanian JR, et al. Deletion mapping of critical region for hypospadias, penoscrotal transposition and imperforate anus on human chromosome 13. J Pediatr Urol. 2006;2:233–242. doi: 10.1016/j.jpurol.2006.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grote LE, Repnikova EA, Amudhavalli SM. Expanding the phenotype of Feingold syndrome-2. Am J Med Genet A. 2015;167A:3219–3225. doi: 10.1002/ajmg.a.37368. [DOI] [PubMed] [Google Scholar]
- 19.Huang C, Yang YF, Yin N, Chen JL, Wang J, et al. Congenital heart defect and mental retardation in a patient with a 13q33.1–34 deletion. Gene. 2012;498:308–310. doi: 10.1016/j.gene.2012.01.083. [DOI] [PubMed] [Google Scholar]
- 20.ISCN 2016. An International System for Human Cytogenomic Nomenclature; McGowan-Jordan J, Simons A, Schmid M (eds) Cytogenet Genome Res. 2016;149:1–140. [Google Scholar]
- 21.Kirchhoff M, Bisgaard AM, Stoeva R, Dimitrov B, Gillessen-Kaesbach G, et al. Phenotype and 244k array-CGH characterization of chromosome 13q deletions: an update of the phenotypic map of 13q21.1-qter. Am J Med Genet A. 2009;149A:894–905. doi: 10.1002/ajmg.a.32814. [DOI] [PubMed] [Google Scholar]
- 22.McCormack WM, Jr, Shen JJ, Curry SM, Berend SA, Kashork C, et al. Partial deletions of the long arm of chromosome 13 associated with holoprosencephaly and the Dandy-Walker malformation. Am J Med Genet. 2002;112:384–389. doi: 10.1002/ajmg.10659. [DOI] [PubMed] [Google Scholar]
- 23.Mitter D, Ullmann R, Muradyan A, Klein-Hitpass L, Kanber D, et al. Genotype-phenotype correlations in patients with retinoblastoma and interstitial 13q deletions. Eur J Hum Genet. 2011;19:947–958. doi: 10.1038/ejhg.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moorhead PS, Nowell PC, Mellman WJ, Battips DM, Hungerford DA. Chromosome preparations of leukocytes cultured from human peripheral blood. Exp Cell Res. 1960;20:613–616. doi: 10.1016/0014-4827(60)90138-5. [DOI] [PubMed] [Google Scholar]
- 25.Myers KA, Wallis MJ, Fitt GJ, Sarnat HB, Newton MR. Blake's pouch cyst in 13q deletion syndrome: posterior fossa malformations may occur due to disruption of multiple genes. Am J Med Genet A. 2017;173:2442–2445. doi: 10.1002/ajmg.a.38346. [DOI] [PubMed] [Google Scholar]
- 26.Quelin C, Bendavid C, Dubourg C, de la Rochebrochard C, Lucas J, et al. Twelve new patients with 13q deletion syndrome: genotype-phenotype analyses in progress. Eur J Med Genet. 2009;52:41–46. doi: 10.1016/j.ejmg.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Quelin C, Spaggiari E, Khung-Savatovsky S, Dupont C, Pasquier L, et al. Inversion duplication deletions involving the long arm of chromosome 13: phenotypic description of additional three fetuses and genotype-phenotype correlation. Am J Med Genet A. 2014;164A:2504–2509. doi: 10.1002/ajmg.a.36658. [DOI] [PubMed] [Google Scholar]
- 28.Sanchez O, Escobar JI, Yunis JJ. A simple G-banding technique. Lancet. 1973;269:1973. doi: 10.1016/s0140-6736(73)93180-2. [DOI] [PubMed] [Google Scholar]
- 29.Saunders S, Saunders S, Lander AD. Expression of the cell surface proteoglycan glypican-5 is developmentally regulated in kidney, limb, and brain. Dev Biol. 1997;190:78–93. doi: 10.1006/dbio.1997.8690. [DOI] [PubMed] [Google Scholar]
- 30.Schinzel A. ed 2. Berlin: Walter de Gruyter; 2001. Catalogue of Unbalanced Chromosome Aberrations in Man. [Google Scholar]
- 31.Sheen VL, Ganesh VS, Topcu M, Sebire G, Bodell A, et al. Mutations in ARFGEF2 implicate vesicle trafficking in neural progenitor proliferation and migration in the human cerebral cortex. Nat Genet. 2004;36:69–76. doi: 10.1038/ng1276. [DOI] [PubMed] [Google Scholar]
- 32.Sirchia F, Di Gregorio E, Restagno G, Grosso E, Pappi P, et al. A case of Feingold type 2 syndrome associated with keratoconus refines keratoconus type 7 locus on chromosome 13q. Eur J Med Genet. 2017;60:224–227. doi: 10.1016/j.ejmg.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 33.Stankiewicz P, Lupski JR. The genomic basis of disease, mechanisms and assays for genomic disorders. Genome Dyn. 2006;1:1–16. doi: 10.1159/000092496. [DOI] [PubMed] [Google Scholar]
- 34.Strehl S, Glatt K, Liu QM, Glatt H, Lalande M. Characterization of two novel protocadherins (PCDH8 and PCDH9) localized on human chromosome 13 and mouse chromosome 14. Genomics. 1998;53:81–89. doi: 10.1006/geno.1998.5467. [DOI] [PubMed] [Google Scholar]
- 35.Van Buggenhout G, Trommelen J, Hamel B, Fryns JP. 13q deletion syndrome in an adult mentally retarded patient. Genet Couns. 1999;10:177–181. [PubMed] [Google Scholar]
- 36.Walczak-Sztulpa J, Wisniewska M, Latos-Bielenska A, Linné M, Kelbova C, et al. Chromosome deletions in 13q33-34: report of four patients and review of the literature. Am J Med Genet A. 2008;146A:337–342. doi: 10.1002/ajmg.a.32127. [DOI] [PubMed] [Google Scholar]
- 37.Wang YP, Wang DJ, Niu ZB, Cui WT. Chromosome 13q deletion syndrome involving 13q31qter: a case report. Mol Med Rep. 2017;15:3658–3664. doi: 10.3892/mmr.2017.6425. [DOI] [PMC free article] [PubMed] [Google Scholar]