Abstract
Current clinical guidelines emphasize the unmet need for technological innovations to guide physician decision-making and to transit from conventional care to personalized cardiovascular medicine. Biomarker-guided cardiovascular therapy represents an interesting approach to inform tailored treatment selection and monitor ongoing efficacy. However, results from previous publications cast some doubts about the clinical applicability of biomarkers to direct individualized treatment. In recent years, the non-coding human transcriptome has emerged as a new opportunity for the development of novel therapeutic strategies and biomarker discovery. Non-coding RNA (ncRNA) signatures may provide an accurate molecular fingerprint of patient phenotypes and capture levels of information that could complement traditional markers and established clinical variables. Importantly, ncRNAs have been identified in body fluids and their concentrations change with physiology and pathology, thus representing promising non-invasive biomarkers. Previous publications highlight the translational applicability of circulating ncRNAs for diagnosis and prognostic stratification within cardiology. Numerous independent studies have also evaluated the potential of the circulating non-coding transcriptome to predict and monitor response to cardiovascular treatment. However, this field has not been reviewed in detail. Here, we discuss the state-of-the-art research into circulating ncRNAs, specifically microRNAs and long non-coding RNAs, to support clinical decision-making in cardiovascular therapy. Furthermore, we summarize current methodological and conceptual limitations and propose future steps for their incorporation into personalized cardiology. Despite the lack of robust population-based studies and technical barriers, circulating ncRNAs emerge as a promising tool for biomarker-guided therapy.
Keywords: Biomarker , Cardiovascular disease , Long non-coding RNAs , MicroRNAs , Non-coding RNAs , Personalized medicine , Precision medicine
Biomarker-guided cardiovascular therapy
Recent decades have seen advances in the treatment of cardiovascular disease (CVD) due to the development of highly effective and well-tolerated therapeutic interventions.1,2 Despite this, CVD persists as the leading cause of death worldwide (http://www.who.int/cardiovascular_diseases). This is in part due to the interindividual variability in the response to medical treatments.3
The variability in therapeutic outcomes differs according to demographic, clinical, environmental, and genetic factors.3,4 Nevertheless, therapy is rarely tailored, as evidence-based medicine assumes a uniform patient phenotype.5 Several authors have proposed to incorporate molecular profiling into clinical decision-making to allow patients, or subgroups of patients, to receive precision medical care.6,7 Tailored therapies based on a refined understanding of patient’s molecular characteristics could be administered to those who are anticipated to benefit most, whereas simultaneously limiting ineffective or harmful interventions.5,8 This would ultimately lead to improved clinical outcomes, increased patient quality of life, and better allocation of health care resources. In this context, utilizing biomarkers to objectively assist in individualized treatment has gained attention in recent years.9 Indeed, drug-diagnostic co-development (‘companion diagnostics’) has become increasingly important in the pharmaceutical industry.10
Biomarker-guided therapy, the use of biomarkers to tailor medical treatment, could assist in: (i) selection of therapy, (ii) evaluation of efficacy, (iii) optimal dose selection, and (iv) recognition and avoidance of side-effects (Figure 1). Reflecting this, previous studies have illustrated the usefulness of circulating biomarkers to tailor cardiovascular therapy.11 Most notably, N-terminal prohormone of brain natriuretic peptide (NT-proBNP)-guided treatment has been suggested to reduce or delay cardiovascular outcomes and to improve cost-effectiveness compared to guideline-based therapy.12 Despite promising findings, other data cast doubts as to whether circulating biomarkers can support clinical decision-making.13 Genome sequence variations, including single nucleotide polymorphisms (SNPs), could act as biological markers of the response to certain drugs.14 Nevertheless, genomics alone provide an incomplete picture of the patient’s phenotype.15 Thus, there is a clinical need for innovative biomarkers to tailor cardiovascular therapy.
Figure 1.
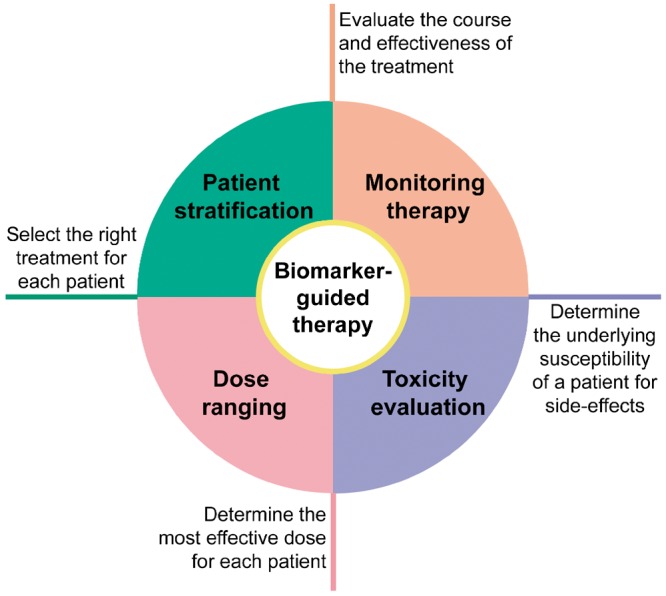
Biomarker-guided therapy.
Non-coding RNAs, an emerging concept in disease and therapy
Although 80% of the human genome is transcribed, only 1–3% of all transcripts are further translated into proteins (Figure 2).16 The vast majority of the human transcriptome represents non-coding RNAs (ncRNAs) (Figure 2),17 which play key regulatory functions.18,19 Contrary to the number of protein-coding genes, which is relatively static since the evolution of multicellularity,17 the amount of ncRNAs has continued to increase along with the complexity of the organism.20 This suggests a fundamental role of ncRNAs in the evolution of biological complexity.
Figure 2.
Coding and non-coding transcriptome. Although approximately 80% of the human genome is transcribed, only 1–3% of this corresponds to protein-coding nucleotide sequences (messenger RNAs, green circles). The remaining 97–99% corresponds to RNAs not translated into proteins: non-coding RNAs (orange), except for some specific exceptions. The non-coding RNA transcriptome includes: circular RNA (circRNAs), enhancer RNAs (eRNAs), germline small RNAs (gsRNAs), long non-coding RNAs (lncRNAs), microRNAs (miRNA), piwi-interacting RNAs (piRNAs), ribosomal RNAs (rRNAs), ribozymes, small conditional RNAs (scRNAs), small interfering RNAs (siRNAs), small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), transfer RNAs (tRNAs), and Y RNA (YRNA). The symbols used are merely indicative.
Non-coding transcripts participate in most cellular processes, and therefore, play a causative role in a number of diseases.21,22 Accordingly, the manipulation of ncRNA expression has emerged as a novel and promising therapeutic approach.23,24 Of note, clinical trials have evaluated the safety, efficacy, and tolerability of ncRNAs in the treatment of various diseases, such as chronic hepatitis C (www.clinicaltrials.gov, NCT01200420) and kidney disease (www.clinicaltrials.gov, NCT02855268). Excellent publications have recently reviewed the role of ncRNAs as potential therapeutic targets in CVD,18,19,25–27 and therefore this issue is beyond the scope of this review. Since the great majority of investigations are in pre-clinical stages, further efforts are needed to demonstrate the clinical safety and efficacy of ncRNAs in cardiovascular therapy.
Circulating non-coding RNAs
In addition to the relevance of ncRNAs as targets or tools for therapy, a plethora of reports suggest that the transcriptomic signature, particularly from ncRNAs, could give insights into both physiological and pathological states of a patient.18 Indeed, transcriptome-originating biomarkers may provide a deep characterization of disease phenotypes and could offer information that is not captured by traditional markers or clinical characteristics.28 Despite this, only a small percentage of the non-coding transcriptome has been explored for this purpose.25 ncRNA expression exhibits high variations between cells, is highly dynamic and rapidly alters in response to stressors. Indeed, a close correlation between dysregulation of the ncRNA profile and the pathophysiology of numerous cardiovascular conditions has been demonstrated.29 Furthermore, ncRNA alterations could be indicative of complex interaction between genetic and environmental factors. Diet, exercise, biological rhythms, or other exogenous factors not routinely considered during clinical evaluation influence ncRNA expression.30 Therefore, the non-coding transcriptome provides an interesting opportunity for the identification of novel tools to assist in clinical decision-making.
Supporting their potential as easily accessible and minimally invasive biomarkers, the presence of ncRNAs has been extensively described in a large number of body fluids.31 From a clinical perspective, the non-coding transcriptome, in particular microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), have the optimal biochemical properties to be excellent biomarkers. Indeed, miRNAs and lncRNAs are stable against degradation, have a long half-life in biological samples and can be obtained by standard laboratory techniques.32,33 RNA can be quantified with high sensitivity and specificity through quantitative reverse transcription-polymerase chain reaction (RT-qPCR), a technique readily available in clinical laboratories. In comparison to peptide biomarkers, ncRNAs can be quantified even at low amounts. Furthermore, global profiles can be obtained in a single experiment using RT-qPCR panels, next-generation sequencing or microarrays.
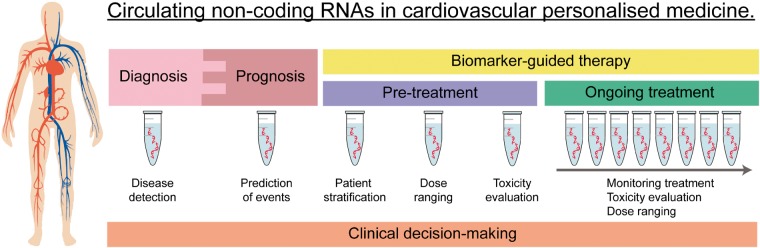
Most studies on circulating ncRNAs as cardiovascular biomarkers are focused on the potential clinical application of miRNAs and lncRNAs in the diagnosis of disease and assessment of cardiovascular risk.34–36 These approaches provide valuable information on patient selection for given therapies (Figure 3). However, the role of ncRNAs as diagnostic and prognostic biomarkers is beyond the interest of the current review and has been extensively reviewed elsewhere.37,38 Treatment selection and monitoring are key aspects of medical management (Figure 3). Based on their biological characteristics, the ncRNA profile may reflect the molecular configuration required for any intervention to exert its therapeutic effect, providing an innovative approach for treatment allocation. Additionally, the post-baseline changes in the ncRNA signature may be useful to monitor ongoing effectiveness. Despite a number of publications have addressed these issues in the cardiovascular context, this research field has not been reviewed in detail. Here, we summarize the current knowledge on ncRNAs, specifically miRNAs and lncRNAs, as biomarkers to predict and monitor the response to cardiovascular treatment.
Figure 3.
Potential clinical application of circulating non-coding RNAs in personalized cardiovascular medicine.
Circulating microRNAs in biomarker-guided cardiovascular therapy
miRNAs are a group of small ncRNAs (19–25 nucleotides in length) that regulate gene expression at the post-transcriptional level. Approximately 2500 human miRNAs have been annotated in the miRBase database (http://www.mirbase.org).39 miRNAs are predicted to target >60% of protein-coding genes,40 and play a key role in development, homeostasis, and response to stress.41,42 miRNAs are usually expressed in complex networks that enable subtile control of the cellular phenotype.43
In addition to their intracellular localization, miRNAs have also been described in the extracellular milieu. Extracellular miRNAs are protected from degradation by packaging in lipid vesicles and by binding to proteins or lipoproteins.44,45 Extracellular miRNAs participate in intercellular communication at autocrine, paracrine, and endocrine levels.46,47 They have been implicated in both physiological and adaptive responses,30 as well as in the onset and development of disease states, including in CVD.48
Results from a number of publications support the promising role of circulating miRNAs in the prediction of the therapeutic response and monitoring the effectiveness of treatment (Table 1).
Table 1.
Studies focused on circulating microRNAs in biomarker-guided therapy
| Therapy | Patients/Subjects/Model | Source | Main findings | References |
|---|---|---|---|---|
| Aspirin | Patients with T2DM and without a history of CVD | Plasma | Therapy reduces circulating miR-126 levels | 49 |
| Aspirin | Healthy subjects | Plasma | Therapy reduces the circulating levels of a panel of miRNAs, including platelet-enriched miRNAs | 50 |
| Patients with symptomatic carotid atherosclerosis | ||||
| Aspirin | Healthy volunteers | Plasma | Interindividual variability in circulating miRNA profile decreased with prolonged therapy | 51 |
| Patients with a history of ACS | Platelet-related miRNAs correlated with platelet function tests | |||
| Bruneck cohort | Platelet-related miRNAs correlated with platelet activation markers | |||
| Aspirin | Patients with intermittent claudication | Plasma | Combination of circulating miR-92a and PDW is a predictor of aspirin resistance | 52 |
| Aspirin | Healthy subjects without history of CVD | Platelets | Changes in expression levels of miR-19b-1-5p, miR-548e, miR-587, miR-1225-3p, miR-1271, and miR-1537-5p correlate with a reduction in platelet aggregation | 53 |
| Platelet miR-19b-1-5p expression is associated with aspirin insensitivity | ||||
| Aspirin | Healthy subjects | Platelets | No change in miRNA expression | 54 |
| Clopidogrel | CHD patients with NSTE-ACS | Platelets | Platelet miR-223 expression is a predictor of clopidogrel responsiveness | 55 |
| CRT | Patients with HF eligible for CRT | Serum | No differences in miRNA expression profile between responders and non-responders at baseline | 56 |
| Different changes in circulating miRNA between responders and non-responders after the therapy | ||||
| At follow-up, circulating miR-26b-5p, miR-29a-3p, miR-30e-5p, miR-92a-3p, and miR-145-5p were differentially expressed between responders and non-responders | ||||
| CRT | Patients with HF referred for CRT | Plasma | Higher levels of circulating miR-30d, miR-142-5p, and miR-766 in responders at baseline | 57 |
| Association between circulating miR-30d levels and CRT responsiveness | ||||
| Reduction in circulating miR-30d levels at 6 months in responders | ||||
| LVAD | Patients with severe advanced HF | Plasma | Circulating miR-483-3p monitors the response to LVAD therapy | 58 |
| Circulating miR-1202 predicts the response to LVAD therapy | ||||
| LVAD | Patients with end-stage HF | Plasma / Serum | Reduction in circulating levels of heart-enriched miRNAs in response to LVAD therapy | 59 |
| LVAD | Patients with end-stage HF | Plasma | Circulating miR-21 decreased at 1, 3, and 6 months after LVAD implantation | 60 |
| Antihypertensive | Hypertension-induced heart disease rat models | Plasma | Reduction of circulating levels of miR-16, miR-20b, miR-93, miR-106b, miR-223, and miR-423-5p in response to antimiR-208a and ACE inhibitor therapies | 61 |
| Circulating miR-19b could serve as a specific biomarker for antimiR-208 therapy | ||||
| CPAP | Patients with RH and OSA | Plasma | A cluster of circulating miRNAs (miR-100-5p, miR-378-3p, and miR-486-5p) predict blood pressure response | 62 |
| Different profile in miRNA changes between responders and non-responders |
ACE, angiotensin-converting enzyme; ACS, acute coronary syndrome; CPAP, continuous positive airway pressure; CHD, coronary heart disease; CRT, cardiac resynchronization therapy; CVD, cardiovascular disease; HF, heart failure; LVAD, left ventricular assist device; NSTE-ACS, non-ST elevation acute coronary syndrome; OSA, obstructive sleep apnoea; PDW, platelet distribution width; RH, resistant hypertension; T2DM, type 2 diabetes mellitus.
Antiplatelet therapy
Antiplatelet therapy is the cornerstone treatment in patients at high risk of thrombosis.63 Since resistance to the therapy has been associated with higher incidence of cardiovascular events,63 non-responder patients may benefit from alternative antiplatelet therapies. The development of robust laboratory tests to monitor response to antiplatelet therapy is thus of clinical importance. However, the current methodology has provided varying results.64
Results from different studies suggest the potential of circulating miRNA levels as surrogate biomarkers of antithrombotic therapy efficacy. In 2013, de Boer et al.49 demonstrated that aspirin administration (300 mg/day) resulted in a significant reduction in circulating levels of miR-126 in patients with type 2 diabetes mellitus (T2DM) and without a history of CVD after 6 weeks of treatment. miR-126 levels also showed a direct correlation with platelet activation following treatment, irrespective of dose. Indeed, aspirin inhibited in vitro transfer of platelet-enriched miRNAs from the platelets to the extracellular milieu. The potential of circulating miR-126 to monitor aspirin therapy was corroborated by Willeit et al.50 who evaluated a panel of 92 miRNAs in plasma of healthy volunteers after treatment with 10 mg prasugrel, followed by combination therapy with varying aspirin doses in successive weeks. Plasma levels of 15 circulating miRNAs, including miR-126 and other platelet-enriched miRNAs (miR-191 and miR-223), decreased with further platelet inhibition. These findings were validated in patients with symptomatic carotid artery atherosclerosis under treatment with dipyridamole or clopidogrel, in addition to aspirin. The same group demonstrated that circulating miRNAs are closely correlated with platelet activation biomarkers in general population.51 The circulating signature of miRNAs was also associated with the residual platelet activity in patients with acute coronary syndrome on antiplatelet therapy.51 It is worth of noting that the authors suggested a role for the small ncRNA family, YRNAs, as novel biomarkers to monitor antiplatelet therapy.51 Interestingly, a score based on the plasma miR-92a levels and the platelet distribution width (PDW), can identify aspirin resistance post-baseline with a specificity of 97.5% and a sensitivity of 80.0% in patients with intermittent claudication on daily aspirin monotherapy (75 mg/day, 100 mg/day, or 150 mg/day).52
Results from these studies were validated in alternative sources of miRNAs. Kok et al.53 performed miRNA microarray analysis using platelet RNA obtained from a small cohort of healthy male subjects without personal history of CVD. Results showed that altered expression of miR-19b-1-5p, miR-548e, miR-587, miR-1225-3p, miR-1271, and miR-1537-5p correlated with the extent of platelet aggregation reduction after 2 weeks of aspirin therapy (100 mg/day). Lower platelet miR-19b-1-5p expression after aspirin therapy was associated with platelet aggregation in the presence of indomethacin, which mimics the effect of aspirin. As such, miR-19b-1-5p may be a suitable biomarker to identify aspirin insensitivity. It should be noted that a previous publication demonstrated that the platelet miRNA profile is remarkably stable and is not affected by a single-dose of aspirin in a small healthy cohort.54 However, single-dose treatment may not have an immediate effect on platelet miRNAs.54
Shi et al.55 explored the association between platelet miRNA expression and the responsiveness to a clopidogrel loading dose (300 mg) in a cohort of non-diabetic coronary heart disease patients with non-ST elevation acute coronary syndrome (NSTE-ACS). Logistic regression analyses demonstrated that among a number of factors potentially associated with platelet reactivity (age, obesity, smoking, genotype, and other medication), miR-223 expression was an independent predictor associated with clopidogrel responsiveness [Odds ratio (OR) 0.189, 95% Confidence Interval (CI) 0.043-0.836]. Plasma miR-223 showed a clinically acceptable discriminative value [Area Under the Curve (AUC) 0.706, 95% CI 0.521-0.891] and the association with clopidogrel responsiveness remained significant even after adjusting for CYP2C19*2 (OR 0.193, 95% CI 0.044–0.855), a genetic polymorphism that influences the response to clopidogrel.
These observations suggest that miRNAs are able to reveal measurable changes in platelet activation and aggregation after antiplatelet therapies and may serve as indicators of compliance in both general and specific patient populations. However, due to the high incidence of suboptimal responses63 and the number of alternatives in antiplatelet therapy, the further development of novel biomarkers to predict the response to the treatment seems fundamental.
Cardiac resynchronization therapy
Cardiac resynchronization therapy (CRT) has emerged as a well-established therapy for advanced heart failure (HF).65 However, a significant number of patients do not show objective evidence of a clinical benefit.66 Implantation of a CRT device is invasive and has high costs. Therefore, there is great interest in identifying patients who are most likely to benefit. Melman et al.57 evaluated the role of circulating miRNAs as biomarkers to predict the response to CRT in a cohort of patients referred for CRT pacemaker implantation with and without subsequent echocardiographic improvement after 6 months. Responders were defined as individuals with an increase in left ventricular ejection fraction (LVEF) ≥ 10% between baseline and follow-up transthoracic echocardiography. After an initial discovery phase using a 766 miRNA array and a subsequent validation phase, plasma miR-30d was identified as a predictor of therapeutic response in multivariate models, even after adjusting for serum creatinine, previous revascularization, preimplantation QRS duration, and LVEF (OR 2.52, 95% CI 1.07–5.94). The discriminative ability was superior to clinical variables such as QRS duration in univariate logistic regression analyses (OR 2.32, 95% CI 1.16–4.67 vs. OR 1.00, 95% CI 0.82–1.34). Thus, preimplantation circulating miR-30d could be a biomarker for the selection of CRT or alternative medical interventions. Interestingly, using in vitro and in vivo models, the authors demonstrated that miR-30d mediates cardiac hypertrophy and protection against inflammation-induced cardiomyocyte apoptosis.
Marfella et al.56 evaluated 84 miRNAs previously associated with the pathogenesis of the failing heart in 81 patients with HF eligible for CRT. At 12 months, patients were categorized as responders if they exhibited LV reverse remodelling [reduction >10% in LV end-systolic volume (LVESV) index and an increment of >10% in LVEF] or non-responders (reduction <10% in LVESV index and an increment of <10% in LVEF). In contrast to previous findings, similar levels of circulating miRNAs were observed between responders and non-responders at baseline, which makes the potential for patient stratification by preimplantation miRNA levels unclear. However, the authors reported higher expression levels of circulating miR-26b-5p, miR-29a-3p, miR-30e-5p, miR-92a-3p, and miR-145-5p after 12 months of CRT in responders (fold-changes from 15.2 to 6.2) compared to non-responders (fold-changes from 5.6 to 2.1). These miRNAs were directly associated with changes in LVEF and NT-proBNP, even after adjusting for age, gender, functional class, and treatments. Thus, post-baseline circulating miRNA levels may be useful to assist in monitoring of efficacy of CRT.
Left ventricular assist devices
The use of LV assist devices (LVAD) in patients with advanced HF is limited by high rates of adverse events and cost.67,68 Various clinical variables have been studied to select patients based on anticipated therapeutic response.68 However, appropriate patient stratification remains challenging.69
The potential of circulating miRNAs to predict the response to LVAD has been previously evaluated in patients with severe advanced HF. Morley-Smith et al.58 classified the clinical response to LVAD therapy based on NT-proBNP concentration at 3 months. Selected patients whose change in NT-proBNP was above the 50th percentile were identified as ‘poor responders’ and patients below the 50th percentile as ’good responders’. Using a screening approach including 1113 miRNAs, the authors demonstrated that miR-1202 was a useful biomarker to predict the response to LVAD therapy. Baseline circulating miR-1202 levels stratified the cohort into poor and good responders with greater accuracy than baseline NT-proBNP (AUC 0.976, 95% CI 0.904–1.000 vs. AUC 0.071, 95% CI 0.000–0.210). Thus, miR-1202 emerges as an interesting biomarker to select LVAD therapy or alternative interventions, such as urgent cardiac transplantation.
Within the same study, circulating miR-483-3p levels were upregulated following intervention, with >2-fold-change from baseline even after 3 months. This up-regulation mirrored the improvement in LV function. Other studies similarly support the alteration in the circulating miRNA signature after LVAD implantation.59,60 Since the evaluation of the changes in ventricular function after LVAD implantation has limitations,58 serial measurements of the circulating miRNA signature could be an interesting and easily accessible tool to complement the current methodology for monitoring the therapeutic response to LVAD.
Antihypertensive therapy
Efforts have been made to predict and monitor antihypertensive therapy using circulating miRNAs. Sánchez-de-la-Torre et al.62 analysed a panel of 84 miRNAs functionally associated with the cardiovascular system in plasma samples from 38 patients with resistant hypertension (RH) and obstructive sleep apnoea (OSA) at baseline and after 3 months of adherent continuous positive airway pressure (CPAP) use. Pre-treatment levels of a cluster of miRNAs, miR-100-5p, miR-378-3p, and miR-486-5p, discriminated between non-responders and patients with a favourable decrease in blood pressure (AUC 0.92, 95% CI 0.79–0.99). Supporting previous results from different medical treatments, the changes in plasma miRNA profile after the treatment differed between responders and non-responders. Notably, the authors developed a predictive screening algorithm (HIPARCOScore) based on the specific cluster of miRNAs to identify those responders.
Dickinson et al.61 demonstrated that circulating miRNAs are biomarkers of therapeutic efficacy in different hypertension-induced heart disease rat models. Although the clinical application of the results is limited due to the lack of patient-based data, this study provides interesting conceptual implications. Different miRNA profiles were observed in response to established and experimental therapies; angiotensin-converting enzyme (ACE) inhibitor (captopril) and antimiR-208a treatment. The results support the potential of circulating miRNAs as specific treatment biomarkers. Indeed, circulating levels of miR-19b only changed in response to antimiR-208a therapy. Furthermore, levels of circulating miRNAs correlated with the expression of the myosin isoform Myh7, suggesting that miRNAs could be an interesting tool to monitor the improvement in cardiac function.
Circulating long non-coding RNAs in biomarker-guided cardiovascular therapy
lncRNAs comprise a vast family of ncRNAs longer than 200 nucleotides that participate in multiple biological processes, including imprinting, scaffold/guide for epigenetic and transcription factors, transcription activation, RNA splicing, nuclear trafficking, and regulation of the miRNA function.29,70 Several lines of evidence indicate that lncRNAs play an essential role in development, homeostasis, and disease.18 lncRNAs exhibit cell type-specific expression, precise subcellular localization, and dysregulated expression in pathological conditions.71 Around 28 000 human lncRNAs have been annotated according to the GENCODE project.72
Compared to miRNAs, the characterization of extracellular lncRNAs is poor. The precise mechanism of lncRNA release into the extracellular space and their role in intercellular communication is not yet understood. Despite this, lncRNAs have also been detected in stable form in extracellular fluids.32
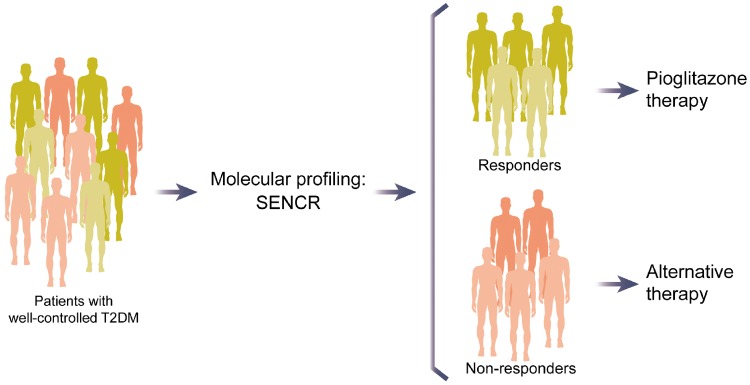
Circulating lncRNAs, such as the long intergenic non-coding RNA predicting cardiac remodelling (LIPCAR), are robust biomarkers of cardiovascular conditions.32,73 However, the literature pertaining to the role of circulating lncRNAs as biomarkers to tailor cardiovascular therapy is scarce. In contrast, oncological research supports the use of circulating lncRNAs in biomarker-guided therapy.74 Our group has recently proposed the potential of circulating lncRNA as blood-based biomarkers to guide pioglitazone therapy.75 After analysing a panel of lncRNAs related to cardiovascular pathology in a cohort of patients with well-controlled T2DM, we reported that pre-treatment levels of the circulating lncRNA smooth muscle and endothelial cell-enriched migration/differentiation-associated long non-coding RNA (SENCR) are positively associated with the rate of change in diastolic function after 24 weeks of pioglitazone therapy. Of note, levels of circulating SENCR before pioglitazone therapy allowed the discrimination of responders from non-responders (AUC 0.763, 95% CI 0.558–0.967). Thus, pre-treatment levels of circulating SENCR may constitute a clinically useful biomarker for the identification of patients that will benefit from pioglitazone administration (Figure 4).
Figure 4.
Potential clinical application of the long non-coding RNA smooth muscle and endothelial cell-enriched migration/differentiation-associated long non-coding RNA (SENCR) in patient stratification to pioglitazone therapy.
Limitations and perspectives
The reproduction of current results in independent and multicentre studies with large population sizes will be a key step to obtain convincing evidence for clinical applicability. Only few overlapping ncRNAs on similar therapies have been described, possibly due to the great heterogeneity in the selection of patients and the evaluation of numerous therapeutic responses. Another aspect to be considered is the inadequate control for confounders. Circulating levels of ncRNAs are affected by factors such as age, sex, cardiovascular risk factors, and pharmacological treatments. These should be considered in the interpretation of results. The effects of disease stage and subtype on ncRNA expression are additional sources of variation that should be similarly considered. The use of ncRNA clustering may provide higher accuracy than single ncRNAs. The inclusion of novel members of the non-coding transcriptome that exhibit a great potential to be used as biomarkers, such as circular RNAs (circRNAs),76 is also required. Unfortunately, a remarkable proportion of published studies have not compared patterns of circulating ncRNAs with established indicators (e.g. peptide biomarkers). Furthermore, most studies have evaluated the univariate association between ncRNAs and treatment response. It is fundamental to explore whether the addition of specific ncRNAs, or a signature of diverse ncRNAs, to composite scores based on multiple parameters from different methodologies provide greater characterization and prediction of patient responses. Advances in ‘Big Data’ should be utilized to combine the complexity of clinical, biochemical, genomic, transcriptomic variables, in addition to novel pieces of information such as the environment and the microbiome, to maximize prediction.26 Ongoing European consortia such as HOMAGE (http://www.homage-hf.eu/) will together identify the role of circulating ncRNAs in thousands of CVD patients. Ischaemic heart disease deserves particular attention. Despite myocardial ischaemia/reperfusion injury and ischaemic HF being the main causes of morbidity and mortality in industrialized societies,77 the number of publications that have addressed the potential clinical application of ncRNAs in biomarker-guided therapy is scarce, and merely focused on platelet activation and aggregation.51,55 Further studies, should evaluate whether ncRNAs are useful tools to inform tailored treatment selection and monitor ongoing efficacy in alternative treatments. This is particularly important in the context of the new expensive monoclonal antibodies targeting PCSK9 (evolocumab and alirocumab) and interleukin-1β (canakinumab).78–80
Despite having promising biochemical properties for use as biomarkers, implementation of circulating ncRNAs is currently not feasible in all clinical laboratories. ncRNA quantification is laborious with current technology and is highly dependent on specimen collection, storage, and handling. As such, ncRNAs quantification and data processing is highly variable between studies. There is a crucial need to develop guidelines of best practice and standard operating procedures in order to increase consistency among results and to make studies comparable and reproducible. Furthermore, the addition of ncRNA testing to clinical practice would currently be time-consuming and may delay commencement of therapy. The development of automated and standardized assays for clinical application should thus be intensified. Finally, evaluation of the cost-effectiveness of circulating ncRNAs in comparison to current methods must be undertaken to assess future viability.
Concluding remarks
A blood test based on circulating ncRNAs would constitute a promising tool to assist physician decision-making in cardiovascular therapy (Figure 3). Nevertheless, since most studies are still in pre-clinical phases, additional research is needed to clinically adopt the non-coding transcriptome for personalized medicine.
Acknowledgements
DdG-C and VLl-C are members of the CardiolincTM network.
Funding
Sara Borrell grant from the Instituto de Salud Carlos III Grant (CD14/00109) and Juan de la Cierva-Incorporación grant from the Ministerio de Economía y Competitividad (IJCI-2016-29393) to D.d.G.-C.; CIBER Cardiovascular (CB16/11/00403 to D.d.G.-C. and V.L.-C.) is a project of the Instituto de Salud Carlos III; EU, project HOMAGE, the ERANET-CVD LIPCAR-HF, and the DFG (TH903/18-1) to T.T.
Conflict of interest: T.T. is founder of Cardior Pharmaceuticals GmbH and holds and licenced patents about the therapeutic and diagnostic use of ncRNAs. There is no conflict of interest for other authors.
References
- 1. Catapano AL, Graham I, De Backer G, Wiklund O, Chapman MJ, Drexel H, Hoes AW, Jennings CS, Landmesser U, Pedersen TR, Reiner Z, Riccardi G, Taskinen MR, Tokgozoglu L, Verschuren WM, Vlachopoulos C, Wood DA, Zamorano JL.. 2016 ESC/EAS guidelines for the management of dyslipidaemias. Eur Heart J 2016;37:2999–3058. [DOI] [PubMed] [Google Scholar]
- 2. Piepoli MF, Hoes AW, Agewall S, Albus C, Brotons C, Catapano AL, Cooney MT, Corra U, Cosyns B, Deaton C, Graham I, Hall MS, Hobbs FDR, Lochen ML, Lollgen H, Marques-Vidal P, Perk J, Prescott E, Redon J, Richter DJ, Sattar N, Smulders Y, Tiberi M, van der Worp HB, van Dis I, Verschuren WMM, Binno S; ESC Scientific Document Group. 2016 European Guidelines on cardiovascular disease prevention in clinical practice: The Sixth Joint Task Force of the European Society of Cardiology and Other Societies on Cardiovascular Disease Prevention in Clinical Practice (constituted by representatives of 10 societies and by invited experts) Developed with the special contribution of the European Association for Cardiovascular Prevention & Rehabilitation (EACPR). Eur Heart J 2016;37:2315–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weeke P, Roden DM.. Applied pharmacogenomics in cardiovascular medicine. Annu Rev Med 2014;65:81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Simon JA, Lin F, Hulley SB, Blanche PJ, Waters D, Shiboski S, Rotter JI, Nickerson DA, Yang H, Saad M, Krauss RM.. Phenotypic predictors of response to simvastatin therapy among African-Americans and Caucasians: the cholesterol and pharmacogenetics (cap) study. Am J Cardiol 2006;97:843–850. [DOI] [PubMed] [Google Scholar]
- 5. van der Leeuw J, Ridker PM, van der Graaf Y, Visseren FL.. Personalized cardiovascular disease prevention by applying individualized prediction of treatment effects. Eur Heart J 2014;35:837–843. [DOI] [PubMed] [Google Scholar]
- 6. Antman EM, Loscalzo J.. Precision medicine in cardiology. Nat Rev Cardiol 2016;13:591–602. [DOI] [PubMed] [Google Scholar]
- 7. Hall MA, Moore JH, Ritchie MD.. Embracing complex associations in common traits: critical considerations for precision medicine. Trends Genet 2016;32:470–484. [DOI] [PubMed] [Google Scholar]
- 8. Jameson JL, Longo DL.. Precision medicine—personalized, problematic, and promising. N Engl J Med 2015;372:2229–2234. [DOI] [PubMed] [Google Scholar]
- 9. Lyman GH, Moses HL.. Biomarker tests for molecularly targeted therapies—the key to unlocking precision medicine. N Engl J Med 2016;375:4–6. [DOI] [PubMed] [Google Scholar]
- 10. Drucker E, Krapfenbauer K.. Pitfalls and limitations in translation from biomarker discovery to clinical utility in predictive and personalised medicine. EPMA J 2013;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ridker PM, Danielson E, Fonseca FA, Genest J, Gotto AM Jr, Kastelein JJ, Koenig W, Libby P, Lorenzatti AJ, MacFadyen JG, Nordestgaard BG, Shepherd J, Willerson JT, Glynn RJ; JUPITER Study Group. Rosuvastatin to prevent vascular events in men and women with elevated c-reactive protein. N Engl J Med 2008;359:2195–2207. [DOI] [PubMed] [Google Scholar]
- 12. Chow SL, Maisel AS, Anand I, Bozkurt B, de Boer RA, Felker GM, Fonarow GC, Greenberg B, Januzzi JL Jr, Kiernan MS, Liu PP, Wang TJ, Yancy CW, Zile MR.. Role of biomarkers for the prevention, assessment, and management of heart failure: a scientific statement from the American Heart Association. Circulation 2017;135:e1054–e1091. [DOI] [PubMed] [Google Scholar]
- 13. Felker GM, Anstrom KJ, Adams KF, Ezekowitz JA, Fiuzat M, Houston-Miller N, Januzzi JL, Mark DB, Piña IL, Passmore G, Whellan DJ, Yang H, Cooper LS, Leifer ES, Desvigne-Nickens P, O’Connor CM.. Effect of natriuretic peptide-guided therapy on hospitalization or cardiovascular mortality in high-risk patients with heart failure and reduced ejection fraction. A randomized clinical trial. JAMA 2017;318:713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Epstein RS, Moyer TP, Aubert RE, O’Kane DJ, Xia F, Verbrugge RR, Gage BF, Teagarden JR.. Warfarin genotyping reduces hospitalization rates results from the MM-WES (Medco-Mayo Warfarin Effectiveness Study). J Am Coll Cardiol 2010;55:2804–2812. [DOI] [PubMed] [Google Scholar]
- 15. MacRae CA, Vasan RS.. The future of genetics and genomics: closing the phenotype gap in precision medicine. Circulation 2016;133:2634–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pennisi E. Genomics. Encode project writes eulogy for junk DNA. Science 2012;337:1159–1161.. [DOI] [PubMed] [Google Scholar]
- 17. Liu G, Mattick JS, Taft RJ.. A meta-analysis of the genomic and transcriptomic composition of complex life. Cell Cycle 2013;12:2061–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beermann J, Piccoli MT, Viereck J, Thum T.. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev 2016;96:1297–1325. [DOI] [PubMed] [Google Scholar]
- 19. Boon RA, Jae N, Holdt L, Dimmeler S.. Long noncoding RNAs: from clinical genetics to therapeutic targets? J Am Coll Cardiol 2016;67:1214–1226. [DOI] [PubMed] [Google Scholar]
- 20. Shabalina SA, Spiridonov NA.. The mammalian transcriptome and the function of non-coding DNA sequences. Genome Biol 2004;5:105.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bar C, Chatterjee S, Thum T.. Long noncoding RNAs in cardiovascular pathology, diagnosis, and therapy. Circulation 2016;134:1484–1499. [DOI] [PubMed] [Google Scholar]
- 22. Xu Z, Yan Y, Zeng S, Dai S, Chen X, Wei J, Gong Z.. Circular RNAs: clinical relevance in cancer. Oncotarget 2018;9:1444–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JT, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J, Engelhardt S.. MicroRNA-21 contributes to myocardial disease by stimulating map kinase signalling in fibroblasts. Nature 2008;456:980–984. [DOI] [PubMed] [Google Scholar]
- 24. Ucar A, Gupta SK, Fiedler J, Erikci E, Kardasinski M, Batkai S, Dangwal S, Kumarswamy R, Bang C, Holzmann A, Remke J, Caprio M, Jentzsch C, Engelhardt S, Geisendorf S, Glas C, Hofmann TG, Nessling M, Richter K, Schiffer M, Carrier L, Napp LC, Bauersachs J, Chowdhury K, Thum T.. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat Commun 2012;3:1078.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Poller W, Dimmeler S, Heymans S, Zeller T, Haas J, Karakas M, Leistner DM, Jakob P, Nakagawa S, Blankenberg S, Engelhardt S, Thum T, Weber C, Meder B, Hajjar R, Landmesser U.. Non-coding RNAs in cardiovascular diseases: diagnostic and therapeutic perspectives. Eur Heart J 2017;doi:10.1093/eurheartj/ehx165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Perrino C, Barabasi AL, Condorelli G, Davidson SM, De Windt L, Dimmeler S, Engel FB, Hausenloy DJ, Hill JA, Van Laake LW, Lecour S, Leor J, Madonna R, Mayr M, Prunier F, Sluijter JP, Schulz R, Thum T, Ytrehus K, Ferdinandy P.. Epigenomic and transcriptomic approaches in the post-genomic era: path to novel targets for diagnosis and therapy of the ischemic heart? Cardiovasc Res 2017;113:725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vegter EL, van der Meer P, de Windt LJ, Pinto YM, Voors AA.. MicroRNAs in heart failure: from biomarker to target for therapy. Eur J Heart Fail 2016;18:457–468. [DOI] [PubMed] [Google Scholar]
- 28. Mayr M, Zampetaki A, Willeit P, Willeit J, Kiechl S.. MicroRNAs within the continuum of postgenomics biomarker discovery. Arterioscler Thromb Vasc Biol 2013;33:206–214. [DOI] [PubMed] [Google Scholar]
- 29. Thum T, Condorelli G.. Long noncoding RNAs and microRNAs in cardiovascular pathophysiology. Circ Res 2015;116:751–762. [DOI] [PubMed] [Google Scholar]
- 30. de Gonzalo-Calvo D, Dávalos A, Montero A, García-González Á, Tyshkovska I, González-Medina A, Soares SMA, Martínez-Camblor P, Casas-Agustench P, Rabadán M, Díaz-Martínez ÁE, Úbeda N, Iglesias-Gutiérrez E.. Circulating inflammatory miRNA signature in response to different doses of aerobic exercise. J Appl Physiol (1985) 2015;119:124–134. [DOI] [PubMed] [Google Scholar]
- 31. Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K.. The microRNA spectrum in 12 body fluids. Clin Chem 2010;56:1733–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kumarswamy R, Bauters C, Volkmann I, Maury F, Fetisch J, Holzmann A, Lemesle G, de Groote P, Pinet F, Thum T.. Circulating long noncoding RNA, lipcar, predicts survival in patients with heart failure. Circ Res 2014;114:1569–1575. [DOI] [PubMed] [Google Scholar]
- 33. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M.. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 2008;105:10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. de Gonzalo-Calvo D, van der Meer RW, Rijzewijk LJ, Smit JW, Revuelta-Lopez E, Nasarre L, Escola-Gil JC, Lamb HJ, Llorente-Cortes V.. Serum microRNA-1 and microRNA-133a levels reflect myocardial steatosis in uncomplicated type 2 diabetes. Sci Rep 2017;7:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Masson S, Batkai S, Beermann J, Bar C, Pfanne A, Thum S, Magnoli M, Balconi G, Nicolosi GL, Tavazzi L, Latini R, Thum T.. Circulating microRNA-132 levels improve risk prediction for heart failure hospitalization in patients with chronic heart failure. Eur J Heart Fail 2018;20:78–85. [DOI] [PubMed] [Google Scholar]
- 36. Vausort M, Wagner DR, Devaux Y.. Long noncoding RNAs in patients with acute myocardial infarction. Circ Res 2014;115:668–677. [DOI] [PubMed] [Google Scholar]
- 37. Romaine SP, Tomaszewski M, Condorelli G, Samani NJ.. MicroRNAs in cardiovascular disease: an introduction for clinicians. Heart 2015;101:921–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Viereck J, Thum T.. Circulating noncoding RNAs as biomarkers of cardiovascular disease and injury. Circ Res 2017;120:381–399. [DOI] [PubMed] [Google Scholar]
- 39. Kozomara A, Griffiths-Jones S.. Mirbase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 2014;42:D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Friedman RC, Farh KK, Burge CB, Bartel DP.. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2008;19:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mendell JT, Olson EN.. MicroRNAs in stress signaling and human disease. Cell 2012;148:1172–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Small EM, Olson EN.. Pervasive roles of microRNAs in cardiovascular biology. Nature 2011;469:336–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Matkovich SJ, Hu Y, Dorn GW 2nd. Regulation of cardiac microRNAs by cardiac microRNAs. Circ Res 2013;113:62–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M.. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA 2011;108:5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. de Gonzalo-Calvo D, Cenarro A, Garlaschelli K, Pellegatta F, Vilades D, Nasarre L, Camino-Lopez S, Crespo J, Carreras F, Leta R, Catapano AL, Norata GD, Civeira F, Llorente-Cortes V.. Translating the microRNA signature of microvesicles derived from human coronary artery smooth muscle cells in patients with familial hypercholesterolemia and coronary artery disease. J Mol Cell Cardiol 2017;106:55–67. [DOI] [PubMed] [Google Scholar]
- 46. Hergenreider E, Heydt S, Treguer K, Boettger T, Horrevoets AJ, Zeiher AM, Scheffer MP, Frangakis AS, Yin X, Mayr M, Braun T, Urbich C, Boon RA, Dimmeler S.. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat Cell Biol 2012;14:249–256. [DOI] [PubMed] [Google Scholar]
- 47. Shan Z, Qin S, Li W, Wu W, Yang J, Chu M, Li X, Huo Y, Schaer GL, Wang S, Zhang C.. An endocrine genetic signal between blood cells and vascular smooth muscle cells: role of microRNA-223 in smooth muscle function and atherogenesis. J Am Coll Cardiol 2015;65:2526–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bang C, Batkai S, Dangwal S, Gupta SK, Foinquinos A, Holzmann A, Just A, Remke J, Zimmer K, Zeug A, Ponimaskin E, Schmiedl A, Yin X, Mayr M, Halder R, Fischer A, Engelhardt S, Wei Y, Schober A, Fiedler J, Thum T.. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J Clin Invest 2014;124:2136–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. de Boer HC, van Solingen C, Prins J, Duijs JM, Huisman MV, Rabelink TJ, van Zonneveld AJ.. Aspirin treatment hampers the use of plasma microRNA-126 as a biomarker for the progression of vascular disease. Eur Heart J 2013;34:3451–3457. [DOI] [PubMed] [Google Scholar]
- 50. Willeit P, Zampetaki A, Dudek K, Kaudewitz D, King A, Kirkby NS, Crosby-Nwaobi R, Prokopi M, Drozdov I, Langley SR, Sivaprasad S, Markus HS, Mitchell JA, Warner TD, Kiechl S, Mayr M.. Circulating microRNAs as novel biomarkers for platelet activation. Circ Res 2013;112:595–600. [DOI] [PubMed] [Google Scholar]
- 51. Kaudewitz D, Skroblin P, Bender LH, Barwari T, Willeit P, Pechlaner R, Sunderland NP, Willeit K, Morton AC, Armstrong PC, Chan MV, Lu R, Yin X, Gracio F, Dudek K, Langley SR, Zampetaki A, de Rinaldis E, Ye S, Warner TD, Saxena A, Kiechl S, Storey RF, Mayr M.. Association of microRNAs and YRNAs with platelet function. Circ Res 2016;118:420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Binderup HG, Houlind K, Madsen JS, Brasen CL.. Aspirin resistance may be identified by miR-92a in plasma combined with platelet distribution width. Clin Biochem 2016;49:1167–1172. [DOI] [PubMed] [Google Scholar]
- 53. Kok MG, Mandolini C, Moerland PD, de Ronde MW, Sondermeijer BM, Halliani A, Nieuwland R, Cipollone F, Creemers EE, Meijers JC, Pinto-Sietsma SJ.. Low miR-19b-1-5p expression in isolated platelets after aspirin use is related to aspirin insensitivity. Int J Cardiol 2016;203:262–263. [DOI] [PubMed] [Google Scholar]
- 54. Stratz C, Nuhrenberg TG, Binder H, Valina CM, Trenk D, Hochholzer W, Neumann FJ, Fiebich BL.. Micro-array profiling exhibits remarkable intra-individual stability of human platelet micro-RNA. Thromb Haemost 2012;107:634–641. [DOI] [PubMed] [Google Scholar]
- 55. Shi R, Ge L, Zhou X, Ji WJ, Lu RY, Zhang YY, Zeng S, Liu X, Zhao JH, Zhang WC, Jiang TM, Li YM.. Decreased platelet miR-223 expression is associated with high on-clopidogrel platelet reactivity. Thromb Res 2013;131:508–513. [DOI] [PubMed] [Google Scholar]
- 56. Marfella R, Di Filippo C, Potenza N, Sardu C, Rizzo MR, Siniscalchi M, Musacchio E, Barbieri M, Mauro C, Mosca N, Solimene F, Mottola MT, Russo A, Rossi F, Paolisso G, D’Amico M.. Circulating microRNA changes in heart failure patients treated with cardiac resynchronization therapy: responders vs. non-responders. Eur J Heart Fail 2013;15:1277–1288. [DOI] [PubMed] [Google Scholar]
- 57. Melman YF, Shah R, Danielson K, Xiao J, Simonson B, Barth A, Chakir K, Lewis GD, Lavender Z, Truong QA, Kleber A, Das R, Rosenzweig A, Wang Y, Kass DA, Singh JP, Das S.. Circulating microRNA-30d is associated with response to cardiac resynchronization therapy in heart failure and regulates cardiomyocyte apoptosis: a translational pilot study. Circulation 2015;131:2202–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Morley-Smith AC, Mills A, Jacobs S, Meyns B, Rega F, Simon AR, Pepper JR, Lyon AR, Thum T.. Circulating microRNAs for predicting and monitoring response to mechanical circulatory support from a left ventricular assist device. Eur J Heart Fail 2014;16:871–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Akat KM, Moore-McGriff D, Morozov P, Brown M, Gogakos T, Correa Da Rosa J, Mihailovic A, Sauer M, Ji R, Ramarathnam A, Totary-Jain H, Williams Z, Tuschl T, Schulze PC.. Comparative RNA-sequencing analysis of myocardial and circulating small RNAs in human heart failure and their utility as biomarkers. Proc Natl Acad Sci USA 2014;111:11151–11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lok SI, de Jonge N, van Kuik J, van Geffen AJ, Huibers MM, van der Weide P, Siera E, Winkens B, Doevendans PA, de Weger RA, da Costa Martins PA.. MicroRNA expression in myocardial tissue and plasma of patients with end-stage heart failure during lvad support: comparison of continuous and pulsatile devices. PLoS One 2015;10:e0136404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dickinson BA, Semus HM, Montgomery RL, Stack C, Latimer PA, Lewton SM, Lynch JM, Hullinger TG, Seto AG, van Rooij E.. Plasma microRNAs serve as biomarkers of therapeutic efficacy and disease progression in hypertension-induced heart failure. Eur J Heart Fail 2013;15:650–659. [DOI] [PubMed] [Google Scholar]
- 62. Sánchez-de-la-Torre M, Khalyfa A, Sánchez-de-la-Torre A, Martinez-Alonso M, Martinez-García MÁ, Barceló A, Lloberes P, Campos-Rodriguez F, Capote F, Diaz-de-Atauri MJ, Somoza M, González M, Masa J-F, Gozal D, Barbé F; Spanish Sleep Network . Precision medicine in patients with resistant hypertension and obstructive sleep apnea: blood pressure response to continuous positive airway pressure treatment. J Am Coll Cardiol 2015;66:1023–1032. [DOI] [PubMed] [Google Scholar]
- 63. Kuliczkowski W, Witkowski A, Polonski L, Watala C, Filipiak K, Budaj A, Golanski J, Sitkiewicz D, Pregowski J, Gorski J, Zembala M, Opolski G, Huber K, Arnesen H, Kristensen SD, De Caterina R.. Interindividual variability in the response to oral antiplatelet drugs: a position paper of the Working Group on antiplatelet drugs resistance appointed by the Section of Cardiovascular Interventions of the Polish Cardiac Society, endorsed by the Working Group on Thrombosis of the European Society of Cardiology. Eur Heart J 2008;30:426–435. [DOI] [PubMed] [Google Scholar]
- 64. Gremmel T, Koppensteiner R, Panzer S.. Comparison of aggregometry with flow cytometry for the assessment of agonists-induced platelet reactivity in patients on dual antiplatelet therapy. PLoS One 2015;10:e0129666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Brignole M, Auricchio A, Baron-Esquivias G, Bordachar P, Boriani G, Breithardt OA, Cleland J, Deharo JC, Delgado V, Elliott PM, Gorenek B, Israel CW, Leclercq C, Linde C, Mont L, Padeletti L, Sutton R, Vardas PE, Zamorano JL, Achenbach S, Baumgartner H, Bax JJ, Bueno H, Dean V, Deaton C, Erol C, Fagard R, Ferrari R, Hasdai D, Hoes AW, Kirchhof P, Knuuti J, Kolh P, Lancellotti P, Linhart A, Nihoyannopoulos P, Piepoli MF, Ponikowski P, Sirnes PA, Tamargo JL, Tendera M, Torbicki A, Wijns W, Windecker S, Document R, Kirchhof P, Blomstrom-Lundqvist C, Badano LP, Aliyev F, Bansch D, Baumgartner H, Bsata W, Buser P, Charron P, Daubert JC, Dobreanu D, Faerestrand S, Hasdai D, Hoes AW, Le Heuzey JY, Mavrakis H, McDonagh T, Merino JL, Nawar MM, Nielsen JC, Pieske B, Poposka L, Ruschitzka F, Tendera M, Van Gelder IC, Wilson CM.. 2013 ESC Guidelines on cardiac pacing and cardiac resynchronization therapy: the Task Force on cardiac pacing and resynchronization therapy of the European Society of Cardiology (ESC). Developed in collaboration with the European Heart Rhythm Association (EHRA). Eur Heart J 2013;34:2281–2329. [DOI] [PubMed] [Google Scholar]
- 66. Daubert C, Behar N, Martins RP, Mabo P, Leclercq C.. Avoiding non-responders to cardiac resynchronization therapy: a practical guide. Eur Heart J 2017;38:1463–1472. [DOI] [PubMed] [Google Scholar]
- 67. Baras Shreibati J, Goldhaber-Fiebert JD, Banerjee D, Owens DK, Hlatky MA.. Cost-effectiveness of left ventricular assist devices in ambulatory patients with advanced heart failure. JACC Heart Fail 2017;5:110–119. [DOI] [PubMed] [Google Scholar]
- 68. Gustafsson F, Rogers JG.. Left ventricular assist device therapy in advanced heart failure: patient selection and outcomes. Eur J Heart Fail 2017;19:595–602. [DOI] [PubMed] [Google Scholar]
- 69. Feldman D, Pamboukian SV, Teuteberg JJ, Birks E, Lietz K, Moore SA, Morgan JA, Arabia F, Bauman ME, Buchholz HW, Deng M, Dickstein ML, El-Banayosy A, Elliot T, Goldstein DJ, Grady KL, Jones K, Hryniewicz K, John R, Kaan A, Kusne S, Loebe M, Massicotte MP, Moazami N, Mohacsi P, Mooney M, Nelson T, Pagani F, Perry W, Potapov EV, Eduardo Rame J, Russell SD, Sorensen EN, Sun B, Strueber M, Mangi AA, Petty MG, Rogers J.. The 2013 International Society for Heart and Lung Transplantation guidelines for mechanical circulatory support: executive summary. J Heart Lung Transplant 2013;32:157–187. [DOI] [PubMed] [Google Scholar]
- 70. Uchida S, Dimmeler S.. Long noncoding RNAs in cardiovascular diseases. Circ Res 2015;116:737–750. [DOI] [PubMed] [Google Scholar]
- 71. Ounzain S, Micheletti R, Beckmann T, Schroen B, Alexanian M, Pezzuto I, Crippa S, Nemir M, Sarre A, Johnson R, Dauvillier J, Burdet F, Ibberson M, Guigo R, Xenarios I, Heymans S, Pedrazzini T.. Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs. Eur Heart J 2015;36:353–368a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mudge JM, Harrow J.. Creating reference gene annotation for the mouse c57bl6/j genome assembly. Mamm Genome 2015;26:366–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. de Gonzalo-Calvo D, Kenneweg F, Bang C, Toro R, van der Meer RW, Rijzewijk LJ, Smit JW, Lamb HJ, Llorente-Cortes V, Thum T.. Circulating long-non coding RNAs as biomarkers of left ventricular diastolic function and remodelling in patients with well-controlled type 2 diabetes. Sci Rep 2016;6:37354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y, Chen W, Liu F, Sun W, Li XF, Wang X, Wang Y, Xu ZY, Gao L, Yang Q, Xu B, Li YM, Fang ZY, Xu ZP, Bao Y, Wu DS, Miao X, Sun HY, Sun YH, Wang HY, Wang LH.. Exosome-transmitted lncarsr promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell 2016;29:653–668. [DOI] [PubMed] [Google Scholar]
- 75. de Gonzalo-Calvo D, Kenneweg F, Bang C, Toro R, van der Meer RW, Rijzewijk LJ, Smit JW, Lamb HJ, Llorente-Cortes V, Thum T.. Circulating long noncoding RNAs in personalized medicine: response to pioglitazone therapy in type 2 diabetes. J Am Coll Cardiol 2016;68:2914–2916. [DOI] [PubMed] [Google Scholar]
- 76. Devaux Y, Creemers EE, Boon RA, Werfel S, Thum T, Engelhardt S, Dimmeler S, Squire I.. Circular RNAs in heart failure. Eur J Heart Fail 2017;19:701–709. [DOI] [PubMed] [Google Scholar]
- 77. Wong ND. Epidemiological studies of CHD and the evolution of preventive cardiology. Nat Rev Cardiol 2014;11:276–289 [DOI] [PubMed] [Google Scholar]
- 78. Sabatine MS, Giugliano RP, Keech AC, Honarpour N, Wiviott SD, Murphy SA, Kuder JF, Wang H, Liu T, Wasserman SM, Sever PS, Pedersen TR; FOURIER Steering Committee and Investigators. Evolocumab and clinical outcomes in patients with cardiovascular disease. N Engl J Med 2017;376:1713–1722. [DOI] [PubMed] [Google Scholar]
- 79. Ridker PM, Everett BM, Thuren T, MacFadyen JG, Chang WH, Ballantyne C, Fonseca F, Nicolau J, Koenig W, Anker SD, Kastelein JJP, Cornel JH, Pais P, Pella D, Genest J, Cifkova R, Lorenzatti A, Forster T, Kobalava Z, Vida-Simiti L, Flather M, Shimokawa H, Ogawa H, Dellborg M, Rossi PRF, Troquay RPT, Libby P, Glynn RJ, Group CT.. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med 2017;377:1119–1131. [DOI] [PubMed] [Google Scholar]
- 80. Hlatky MA, Kazi DS.. Pcsk9 inhibitors: economics and policy. J Am Coll Cardiol 2017;70:2677–2687. [DOI] [PubMed] [Google Scholar]