Abstract
Targeting of newly synthesized membrane proteins to the endoplasmic reticulum is an essential cellular process. Most membrane proteins are recognized and targeted co-translationally by the signal recognition particle. However, nearly 5% of membrane proteins are ‘tail-anchored’ by a single carboxy-terminal transmembrane domain that cannot access the co-translational pathway. Instead, tail-anchored proteins are targeted post-translationally by a conserved ATPase termed Get3. The mechanistic basis for tail-anchored protein recognition or targeting by Get3 is not known. Here we present crystal structures of yeast Get3 in ‘open’ (nucleotide-free) and ‘closed’ (ADP·AlF 4−-bound) dimer states. In the closed state, the dimer interface of Get3 contains an enormous hydrophobic groove implicated by mutational analyses in tail-anchored protein binding. In the open state, Get3 undergoes a striking rearrangement that disrupts the groove and shields its hydrophobic surfaces. These data provide a molecular mechanism for nucleotide-regulated binding and release of tail-anchored proteins during their membrane targeting by Get3.
Eukaryotic cells have evolved sophisticated machinery to ensure high fidelity targeting and insertion of membrane proteins into intracellular organelles. For targeting to the endoplasmic reticulum (ER), the best understood mechanism is a co-translational pathway mediated by the cytosolic signal recognition particle (SRP), the ER-associated SRP receptor, and the Sec61 protein translocation channel1–3. This pathway is conserved from bacteria to humans and is essential for biosynthesis of a wide range of membrane proteins. However, many ER-targeted proteins cannot access the SRP-dependent co-translational pathway.
A particularly important example is the tail-anchored (TA) protein, defined by a single C-terminal transmembrane domain (TMD)4,5 and a cytosolic facing amino-terminal domain. TA proteins mediate numerous essential biochemical activities in nearly every cellular membrane. Well-known examples include SNARE proteins involved in vesicle trafficking, components of the mitochondrial and ER protein translocation apparatus, members of the Bcl2 family of apoptotic proteins, and several viral envelope and non-structural proteins4,6,7. The targeting information for TA proteins resides solely within the TMD. Because the TMD is still within the ribosomal tunnel when the termination codon is reached, co-translational targeting is precluded8. Thus, TA proteins are obligatorily recognized and targeted post-translationally by an SRP-independent pathway.
A central component of the TA protein pathway to the ER is a highly conserved cytosolic ATPase termed Asna1 or TRC40 (refs 9, 10). Although originally annotated Asna1 owing to ~27% sequence identity to ArsA, the catalytic subunit of the Escherichia coli arsenite resistance operon11–13, eukaryotic TRC40s have evolved different functions. Both mammalian TRC40 and its yeast homologue Get3 recognize and selectively bind the TMD of TA proteins in the cytosol. This complex then targets to the ER by membrane-bound receptors (termed Get1 and Get2 in yeast14), where the TA protein is released for insertion. This spatially restricted unidirectional targeting is regulated by ATP binding and hydrolysis, but the mechanistic basis of this process is not understood. To address this, we determined the crystal structures of yeast Get3 in the nucleotide-free and ADP·AlF 4−-bound states. Our structural and functional analysis suggests how Get3 binds selectively to TMD substrates, and how ATP hydrolysis regulates TA protein binding and release during targeting to the ER.
Overall structure of the Get3 homodimer
We determined the X-ray crystal structures of ADP·AlF 4−-bound Saccharomyces cerevisiae Get3 and nucleotide-free Schizosaccharomyces pombe Get3 (~58% identical to S. cerevisiae Get3) at 2.0 and 3.0 Å resolution, respectively (Supplementary Table 1 and Supplementary Figs 1 and 2). Both structures show a symmetric homodimer (Fig. 1a,b). Each monomer comprises a core ATPase subdomain and an α-helical subdomain (Supplementary Fig. 3a). The Get3 ATPase subdomain shows structural similarity to other members of the SIMIBI (after signal recognition particle, MinD and BioD) class of NTP-binding proteins15, including its closest sequence homologue of known structure, ArsA16, and the more distantly related nitrogenase iron protein (NifH)17, Soj18, MinD19 and the SRP GTPases20,21.
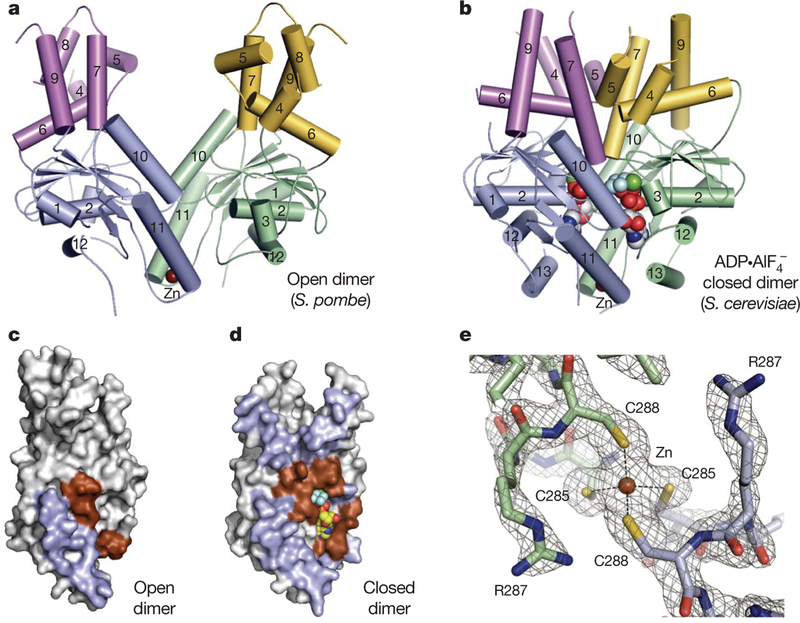
Figure 1 |. Get3 is a dynamic, metal-stabilized homodimer.
a, b, Crystal structures of Get3 in open (a) and closed (b) dimer states. Each monomer comprises a core ATPase subdomain (blue, green) and an α-helical subdomain (magenta, yellow). A tightly bound zinc atom (brown sphere) lies at the dimer interface. c, d, Residues in the homodimer interface (blue, brown) are mapped to the surface of a Get3 monomer from the open (c) and closed (d) dimer structures. Monomers are rotated ~90° about the dimer pseudo-two-fold axis, relative to a and b. Interfacial regions involving the conserved ATPase motifs are coloured brown. e, Details of the Get3 dimerization motif in the closed dimer structure, highlighting the tetrahedral coordination of zinc by the conserved CXXC sequence motif. Electron density is from a σA-weighted 2Fo − Fc map calculated at 2.0 Å resolution and contoured at 2σ.
The nucleotide-free structure of S. pombe Get3 shows the two subunits splayed apart (Fig. 1a) with ~900 Å2 of surface area buried in the interface (Fig. 1c). A 3.8 Å resolution structure of nucleotide-free S. cerevisiae Get3 shows the same open dimer architecture (Supplementary Table 1 and Supplementary Fig. 2), suggesting that the open form reflects a conserved conformation of nucleotide-free Get3. In contrast, the ADP·AlF 4−-bound form of S. cerevisiae Get3 (Fig. 1b) shows a large conformational change approximated by a ~37° rotation of one subunit towards the other. This movement ‘closes’ the quaternary structure into a more compact form that buries ~2,400 Å2 of surface area in an extensive dimer interface that spans both subdomains (Fig. 1d).
The open-to-closed transition in Get3 occurs about a hinge point centred on a zinc ion bound at the homodimer interface. This zinc is coordinated by the side chains of two cysteine residues from each monomer (Fig. 1e), which are conserved in eukaryotic Get3 homologues (‘CXXC motif’; Supplementary Fig. 3b). Although no exogenous zinc was added during the experiment, the tetrahedral geometry, the identity of the coordinating ligands, and the X-ray fluorescence spectrum of bacterially purified Get3 assign this ion as zinc. Variants in which these cysteines are mutated do not coordinate zinc, fail to dimerize in solution, and are unable to complement the growth defect of Δget3 yeast22. Thus, Get3 is an obligate dimer containing a structural metal ion that stabilizes functionally distinct conformations.
The composite hydrophobic groove
The α-helical subdomain of Get3 overlaps with the allosteric metalloid binding site of ArsA, but shows important differences in sequence, structure and function. Eukaryotic TRC40s lack conserved residues used by ArsA to bind As/Sb(III). Instead, this region is enriched in methionine (Supplementary Fig. 3b, red), which is observed at 2–3 times greater frequency than typically found in vertebrate proteins. Furthermore, the eukaryotic TRC40 homologues contain a unique ~20-residue insertion in the α-helical subdomain that we term the ‘TRC40-insert’ (Supplementary Fig. 3b, yellow).
In the Get3 open dimer, the α-helical subdomains are separated by more than 20 Å, creating a large cleft between the two subunits (Figs 1a and 2a). The surface of the cleft is charged rather than hydrophobic, making it unsuitable for TMD binding. In contrast, in the closed dimer state, the α-helical subdomains are in direct contact and define a continuous, solvent-exposed, hydrophobic groove that spans both monomers (Figs 1b and 2b).
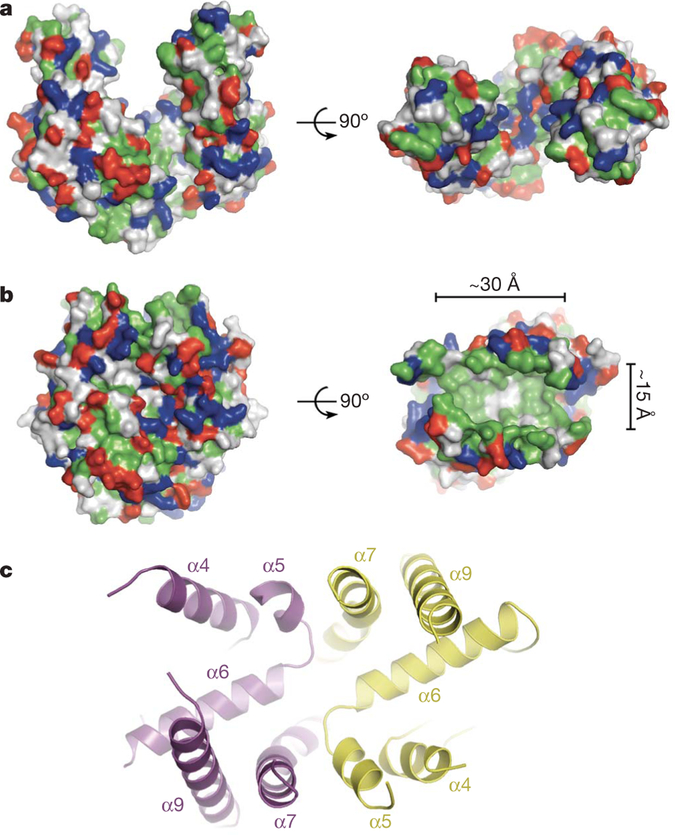
Figure 2 |. A composite hydrophobic groove at the closed dimer interface.
a, Surface representation of the Get3 open dimer with hydrophobic residues coloured green; positively and negatively charged residues are coloured blue and red, respectively. b, As in a, but for the Get3 closed dimer. The approximate dimensions of the large hydrophobic groove (right panel) are indicated. c, Architecture of the composite hydrophobic groove formed by the association of α-helical subdomains (magenta and yellow, coloured as in Fig. 1 and oriented as in b, right panel) at the homodimer interface.
Each α-helical subdomain contributes six amphipathic helices to the composite groove. The bottom of the groove is formed by two ‘crossing’ helices (α6), whereas eight further helices (α4, α5, α7 and α9), that are roughly orthogonal to the crossing helices, form the sides of an extended groove (Fig. 2c). Electron density in this region is generally weaker than in the rest of the protein, and this is reflected in higher B-factors; indeed, the regions connecting helices α4–α5 and α7–α9 (including helix α8 of the TRC40-insert) are disordered in electron density maps of the closed dimer (Supplementary Fig. 1b).
The Get3 composite groove exposes more than 3,000 Å2 of hydrophobic surface area to solvent. In contrast, the signal sequence binding groove in the M-domain of Thermus aquaticus Ffh (the homolog of eukaryotic SRP54) exposes only ~1,500 Å2 of hydrophobic surface area23. The non-polar character of the Get3 groove is conserved across eukaryotic TRC40s, but the sequence identity of residues lining the groove is not (Supplementary Fig. 3b). Polar and charged residues are almost completely excluded from the groove, whereas the ends, which are open to solvent, are delineated by two conserved, positively charged surface patches (Lys 147, Lys 150 and Lys 215). The hydrophobic region of the groove is approximately 30 Å long, 15 Å wide, and 15 Å deep (Fig. 2b), which is sufficient to accommodate an α-helical TMD of ~20 residues, and could therefore form the TA substrate-binding site.
To test this, 24 Get3 variants with single aspartate substitutions for hydrophobic residues on the surface of the groove were analysed for substrate interaction in vitro and functional complementation of a Δget3 yeast strain in vivo. All but one of these expressed at wild-type levels in E. coli, were soluble after purification, and possessed detectable levels of ATPase activity (Supplementary Table 2). A native pull-down assay showed that wild-type Get3 bound to in-vitro-synthesized SEC61β (a model TA protein) in a TMD-dependent manner (Supplementary Fig. 4). Among the variant Get3 proteins, reduced TA substrate binding was seen for mutations clustered along helices α7 and α8 (Fig. 3a, c, green and magenta). Notably, these regions include the C-terminal end of Switch II (helix α7), and the N-terminal end of the TRC40-insert (helix α8). In qualitative agreement with the substrate pull-down assay, the strongest in vivo defects clustered along helices α7 and α8 (Supplementary Fig. 5).
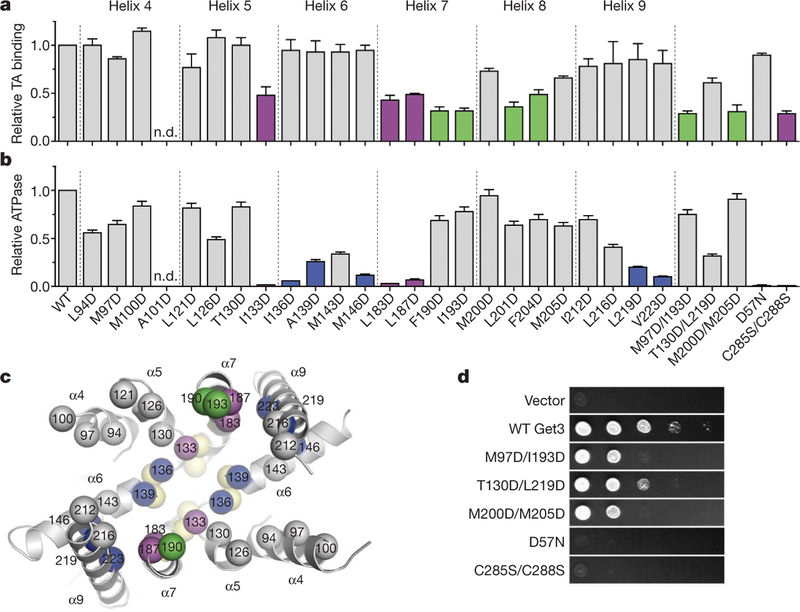
Figure 3 |. Functional analysis of the hydrophobic groove.
a, The effect of site-specific Get3 mutations on binding to full-length human SEC61β was measured by native immunoprecipitation. Each value is the average of between three and six independent measurements, performed on different days, relative to wild type. Error bars denote s.e.m. Variants showing less than ~50% of wild-type binding are highlighted (green, magenta). b, ATPase activity was determined in triplicate, and values are Vmax relative to wild type. Error bars denote s.e.m. Variants showing less than ~25% of wild-type ATPase activity are highlighted (blue, magenta). c, Mutations showing the strongest defects in TA substrate binding (green), ATP hydrolysis (blue), or both (magenta) are mapped onto the composite hydrophobic groove (oriented as in Fig. 2c). ATPase mutants localize to the base of the groove, adjacent to the Get3 nucleotide sensor (yellow; see Fig. 4b). Helix α8 is disordered in the Get3 closed dimer structure and is therefore not visible here. d, In vivo analysis of Get3 mutants. WT, wild type.
As expected, a set of double aspartate mutants, as well as the dimerization-deficient Cys285Ser/Cys288Ser double mutant, showed stronger growth defects that correlated well with in vitro TA substrate binding activity (Fig. 3a, d). Notably, two single mutations (Met200Asp and Met205Asp) that showed relatively modest in vitro and in vivo defects had a much stronger effect when combined. The double mutants expressed to high levels in E. coli, were soluble, and showed ATPase activities that are within ~2–3-fold of wild-type Get3 (Fig. 3b and Supplementary Table 2), suggesting that the observed defects were not due to improper folding or impaired catalysis. Considered together with the crystallographic analysis, these functional data suggest that newly synthesized TA substrates bind to the composite hydrophobic groove of Get3 during targeting to the ER.
The nucleotide sensor
In the ADP·AlF 4−-bound Get3 crystal structure, two nucleotides are buried in a head-to-head conformation at the homodimer interface (Fig. 1b). Nearly 30% of the surface area buried in this interface involves the conserved ATPase sequence motifs (Fig. 1d). Although each nucleotide is principally associated with one monomer, key contacts are also made to the second monomer, explaining why dimerization-deficient mutants of Get3, including Cys285Ser/Cys288Ser (Fig. 3a, b, d), are functionally inactive22.
The active sites of both Get3 monomers are locked in virtually identical conformations mimicking the transition state (Fig. 4a). This contrasts with ArsA, in which the nucleotide-binding sites seem to be structurally and functionally non-equivalent24. In Get3, the square-planar AlF 4− and α- and β-phosphates of ADP interact with the P-loop through backbone amide nitrogens of residues 28–33 and the side chains of Thr 32 and Thr 33. Mg2+ is octahedrally coordinated by three water molecules, a β-phosphate oxygen, one of the fluoride ligands of AlF 4−, and by the side chain of Thr 32. Furthermore, the Switch II residues Asp 166 and Thr 167 each make second shell interactions with Mg2+. A well-ordered water molecule, coordinated by the side chain of Asp 57 (in Switch I) and only 3.6 Å from Pro 169 (in Switch II), is positioned for in-line nucleophilic attack on the γ-phosphate of ATP. This interaction is essential because an Asp57Asn mutant is defective for ATP hydrolysis and unable to rescue the hygromycin B-sensitive phenotype of Δget3 yeast (Fig. 3b, d). In addition, the conserved P-loop residue Lys 26 reaches across the dimer interface to contact the α- and β-phosphates and AlF 4− in the opposing active site. This intersubunit interaction is essential for catalysis in related systems, including NifH25 and Soj18. In Get3, Lys 26 probably functions analogously to stabilize the transition state by neutralization of the negative charge at the γ-phosphate.
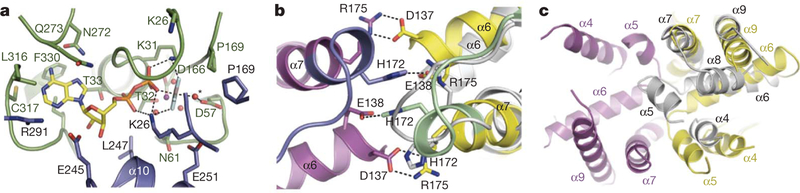
Figure 4 |. The Get3 nucleotide sensor.
a, Key interactions within the composite ATP-binding site of one subunit (green) of the closed dimer. The essential catalytic residue Asp 57 coordinates the putative nucleophilic water molecule (red sphere, asterisk), adjacent to AlF4−. The nucleotide makes further interactions with residues in the second subunit (blue), including the P-loop residue Lys 26. b, A coil-to-helix transition in the Switch II region (green and blue) is observed in the presence of ADP·AlF 4− relative to the nucleotide free state (grey subunit). Viewed ~180° from the orientation in c, looking along the dimer pseudo-two-fold axis, and coloured as in Fig. 1. Conserved, cross-monomer interactions between Switch II/α7 and the α-helical subdomain are disrupted in the open dimer (the second subunit of the nucleotide-free dimer is not visible here). c, The α-helical subdomains move apart in the open dimer (grey), and helices within each of the resulting ‘half-sites’ rearrange; helix α8, disordered in the closed dimer, inserts into the hydrophobic half-site to shield it from solvent.
The observation that ATP hydrolysis is required for efficient release of the TA substrate at the ER membrane10 suggests that the ATPase and α-helical subdomains of Get3 are functionally linked. Consistent with this, we found that a subset of aspartate point mutations within the α-helical subdomain reduced the rate of ATP hydrolysis in vitro (Fig. 3b, c, blue and magenta). These mutations are adjacent to the conserved Switch II region (Fig. 3c, yellow), which undergoes nucleotide-mediated conformational changes in related systems, including NifH25,26. In the Get3 crystal, the transition from an open to a closed state is accompanied by a coil-to-helix transition in the N-terminal end of helix α7, part of Switch II (Fig. 4b). In the helical conformation, the side chain of the Switch II residue His 172 (which binds to metalloid substrates in ArsA) stacks against the His 172 side chain from the opposing subunit. Each His 172 makes an additional cross-monomer interaction with Glu 138 (part of helix α6), and Arg 175 (located in helix α7, immediately C-terminal to Switch II) forms an inter-monomer salt bridge to Asp 137 (part of helix α6). Remarkably, these residues are conserved from yeast to human
The network of Switch-II-mediated cross-monomer interactions between the ATPase and α-helical subdomains in the closed dimer provides a direct link between nucleotide switching and formation of the composite TA binding site. Consistent with this, the nucleotide-free crystal structure shows Switch II/α7 in a coiled conformation, and the network of inter-subunit interactions observed in the closed dimer is completely disrupted (Fig. 4b, grey). In this open state, the α-helical subdomains are separated by more than 20Å (Fig. 1a), and the composite hydrophobic groove is disrupted. Helices within the isolated α-helical subdomains are rearranged (Fig. 4c, grey); most notably, helix α8, which is disordered in the closed dimer structure, packs against the α-helical subdomain, where it shields the hydrophobic surface from solvent. Taken together, and by analogy to other SIMIBI proteins25,27,28, the Get3 crystal structures suggest that Switch II functions to relay structural changes induced by ATP binding and hydrolysis to the homodimer interface and the TA substrate binding site.
Implications for TA protein binding
We propose that the composite hydrophobic groove observed in the closed configuration of the Get3 homodimer forms the TMD binding site. This assignment is consistent with our biochemical, genetic and structural analysis. The dimensions of the groove are well suited for binding to an α-helical TMD of ~20 residues (Fig. 5a). The conserved TRC40-insert, which is disordered in the closed dimer structure (presumably reflecting the absence of a bound TA substrate), is a dynamic feature of the TA binding site. It may serve as a ‘lid’ to shield the exposed face of a bound TMD from solvent during targeting, and to protect the hydrophobic surface of the isolated α-helical subdomain in the absence of substrate.
Figure 5 |. Model for TA protein targeting.
a, The 20-residue α-helical TMD of the Methanococcus jannaschii TA protein Sec61β (PDB accession 1RHZ)38 can be modelled into the Get3 hydrophobic groove with good physiochemical complementarity. b, The Get3 open dimer binds ATP and newly synthesized TA proteins destined for the ER (step 1). The Get3-substrate complex is targeted to the ER by an interaction with the membrane bound receptor, Get1/2 (step 2). After ATP hydrolysis, conformational changes in the nucleotide sensor destabilize the composite groove, driving TA substrate release (step 3). Membrane insertion may be spontaneous, or facilitated by a dedicated integrase (not shown). Disruption of the closed dimer following ATP hydrolysis and TA substrate insertion drives release from the membrane and restores Get3 to its open dimer configuration (step 4).
The extensive hydrophobic surface area (>3,000 Å2) within the groove suggests that Get3 binds to a wide range of ER-directed targeting signals with high affinity. Consistent with this, many of the single aspartate Get3 mutants remain capable of interacting with TA substrates despite the presence of charge within the hydrophobic groove (Fig. 3a and Supplementary Fig. 5). Phylogenetic analysis of eukaryotic TRC40 sequences indicates that methionine is unusually abundant within the groove. Similar to previous proposals for signal sequence binding by the methionine-rich M-domain of SRP54 and Ffh23,29, the intrinsic side- and main-chain dynamics of the groove probably contribute to the ability of Get3 to accommodate diverse TA protein targeting signals.
The structures also suggest how Get3 distinguishes between closely related ER and mitochondrial outer membrane (MOM)-directed targeting signals. ER-targeted TA proteins typically possess longer and more hydrophobic TMDs than TA proteins destined for the MOM30. Thus, the composite groove in Get3 may function as a molecular ruler, preferentially binding to ER-directed TA proteins of sufficient length (~30 Å) and hydrophobicity. Similarly, MOM-directed targeting signals generally contain positive charge immediately C-terminal to the TMD31,32. Thus, further discrimination may occur at the positively charged ends of the Get3 hydrophobic groove (Fig. 2b), where charge repulsion biases Get3 against binding to MOM targeting signals. Together, these structural features may allow selective binding to ER-directed targeting signals.
A model for TA protein targeting
Our structural and functional data allow us to propose a physically plausible model by which ATP binding and hydrolysis regulate TA substrate binding and release during targeting to the ER (Fig. 5b). Targeting is initiated in the cytosol when ATP binding drives Get3 towards the closed dimer state, facilitating recognition of newly synthesized TA membrane proteins in a TMD-dependent manner9,10,14. To minimize unproductive ATP hydrolysis, active site residues in the closed dimer interface (for example, Lys 26) are held in non-catalytic conformations until the pre-targeting complex is delivered to the Get1/2 receptor at the ER membrane14. Because ATP hydrolysis is required for insertion10, association with the receptor, the lipid bilayer and/or a putative integrase drives the ‘loose’ pretargeting complex into a catalytically competent conformation. After ATP hydrolysis and dissociation of ADP and/or inorganic phosphate, the Switch II region collapses, thereby disrupting the network of conserved cross-monomer interactions that stabilize the composite groove (Fig. 4b). Initially, these conformational changes may be propagated through helix a7 to ‘pop’ open the TRC40-insert lid, as observed in the closed dimer, facilitating TA substrate release. An intriguing possibility is that the conserved TRC40-insert transiently partitions into the outer leaflet of the ER lipid bilayer to drive TA substrate insertion into the membrane. Subsequently, Get3 reverts towards the open dimer state, lowering its affinity for the Get1/2 receptor, and returning it to the cytosol to initiate a new round of targeting.
METHODS SUMMARY
Wild-type and selenomethionyl proteins were expressed in E. coli and purified using Ni-NTA and size-exclusion chromatography. Crystals were grown in hanging drop, vapour diffusion format. Diffraction data were collected from cryo-protected crystals at beamline 21-IDG of the Advanced Photon Source (Argonne National Laboratories). The structure of S. cerevisiae Get3 complexed with ADP·AlF 4− was determined by single-wavelength anomalous dispersion (SAD) from selenomethionine-containing protein using PHENIX33. The structures of nucleotide-free S. cerevisiae and S. pombe Get3 were solved by molecular replacement in PHASER34. A monomer of the S. cerevisiae Get3–ADP·AlF 4− complex (with nucleotide and the α-helical subdomain removed) was used as the search model. Refinement and model building were carried out with PHENIX33 and COOT35.
A series of GET3 genes containing site-specific mutations were generated by Quik-Change mutagenesis. The identity of each mutant was confirmed by DNA sequencing. Proteins were expressed in E. coli and purified by Ni-NTA chromatography. TA substrate binding was monitored using a native pull-down assay in which full-length 35S-labelled human SEC61β was translated in a TRC40-depleted reticulocyte lysate translation extract with or without recombinant wild-type or mutant Get3 protein. After translation, Get3 was immunoprecipitated under native conditions, analysed by SDS–PAGE, and quantified by phosphorimaging. The ATPase activity of Ni-NTA-purified protein was determined using an NADH-coupled microplate photometric assay36,37
Wild-type and mutant GET3 genes were subcloned into a low copy number URA plasmid under the control of a medium-strength, constitutive ACT1 promoter, and transformed into a Δget3 strain (Open Biosystems); serial dilutions of each transformant were spotted (along with wild type and vector only controls) onto synthetic defined medium (−uracil) supplemented with 100 μg ml−1 hygromycin B. Plates were photographed after 2 days at 37 °C.
METHODS
Protein cloning, expression and purification.
The genes encoding full-length S. cerevisiae and S. pombe Get3 were amplified by PCR using genomic DNA, and subcloned into a pET28 derivative (Novagen) modified to incorporate a tobacco etch virus (TEV) protease cleavage site between an N-terminal 6×His tag and the polylinker. Proteins were expressed in E. coli Rosetta2(DE3)/pLysS (Novagen) at 37 °C for 3 h by induction with 0.1 mM IPTG after the cells reached an A600 of ~0.6. Cells were disrupted in lysis buffer (50 mM Tris, pH 7.5, 500 mM NaCl, 5 mM β-mercaptoethanol, 10 mM imidazole, 10% glycerol and 1 mM PMSF) using a high-pressure microfluidizer (Avestin). After clearing by centrifugation, the supernatant was batch-purified by nickel-affinity chromatography (Ni-NTA His Bind Resin, Novagen) and dialysed into 10 mM Tris, pH 7.5, 100 mM NaCl, 2 mM dithiothreitol (DTT), 40% glycerol. This was followed optionally by cleavage with 6×His-tagged TEV protease and removal of residual uncleaved Get3 and the 6×His-tagged TEV protease by subtractive Ni-NTA purification. For crystallization, protein was further purified by gel filtration (Superdex 200 10/300 GL, GE Healthcare). Fractions were pooled, concentrated to ~10 mg ml−1 in10 mM Tris, pH 7.5, 100 mM NaCl and 2 mM DTT, and stored at −80 °C. Selenomethionyl S. cerevisiae Get3 protein was prepared by feedback inhibition39 in BL21(DE3) cells (Novagen) using the Overnight Express Autoinduction System 2 (Novagen). Incorporation of selenomethionine was confirmed by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. Selenomethionyl Get3 was purified in the same way as the native protein.
Crystallization.
Crystals of 6×His-tagged S. cerevisiae Get3 complexed with ADP·AlF4− were grown at room temperature using hanging drop vapour diffusion by mixing equal volumes of a protein solution containing 2 mM ADP, 2 mM MgCl2, 2 mM AlCl3 and 8 mM NaF with a reservoir solution containing 33% PEG 3350, 0.1 M Tris, pH 8.4, 0.2 M ammonium acetate and 75 mM Na/K tartrate. Selenomethionine-containing crystals were grown under similar conditions by mixing equal volumes of protein solution with 31% PEG 3350, 0.1 M Tris, pH 8.4, 0.2 M ammonium acetate and 50 mM glycine. Crystals were cryoprotected in 30% PEG3350, 0.1 M Tris, pH 8.4, 0.2 M ammonium acetate, 75 mM Na/K tartrate, 2 mM ADP, 2 mM MgCl2, 2 mM AlCl3, 8 mM NaF and 20% ethylene glycol (native) or 31% PEG3350, 0.1 M Tris, pH 8.4, 0.2 M ammonium acetate, 50 mM glycine, 2 mM ADP, 2 mM MgCl2, 2 mM AlCl3, 8 mM NaF and 20% glycerol (selenomet) and flash frozen in liquid nitrogen.
Crystals of S. pombe Get3 (cleaved) were grown at room temperature using hanging drop vapour diffusion by mixing equal volumes of protein solution with a reservoir solution containing 25% PEG 3350, 0.1 M MES, pH 6.5 and 0.4 M MgCl2. Crystals were collected directly and flash-frozen in liquid nitrogen
Crystals of 6×His-tagged S. cerevisiae Get3 (apo) were grown at room temperature using hanging drop vapour diffusion by mixing equal volumes of protein solution with a reservoir solution containing 1.5 M ammonium sulphate, 0.1 M MES, pH 6.5, 0.1 M NaCl, 5 mM proline and 0.2 mM C12E8. Crystals were cryoprotected in 2 M ammonium sulphate, 0.1 M MES, pH 6.5,0.1 M NaCl, 5 mM proline and 25% glycerol, and flash-frozen in liquid nitrogen.
Structure determination and refinement.
Native and selenium SAD data were collected at 100K at APS beamline 21-IDG (λ=0.97856), and processed using HKL2000 (HKL Research). Data collection statistics are listed in Supplementary Table 1.
The structure of S. cerevisiae Get3 in complex with ADP·AlF4− was determined by SAD. The positions of 38 out of 52 selenium sites in the asymmetric unit were located using PHENIX33. After phasing and density modification the resulting electron density maps were of high quality, allowing us to manually place four copies of Get3 (using the core ATPase region of ArsA as a starting model) and revealing clear density for nucleotide in the active site. A native data set to 2.0 Å was used for model rebuilding and refinement with COOT35 and PHENIX. The final model contains two Get3 homodimers, four Mg2+ ADP·AlF4− complexes, two zinc atoms and 507 water molecules, and was refinedto an R-factor (Rfree) of 17.6% (21.3%). No electron density was observed for residues 1–4, 101–120, 191–211, 279–284, 353 and 354 in chain A; 1–3, 106–125, 154–158, 194–209, 280–284 and 351–354 in chain B; 1–4, 100–125, 153–158, 189–212, 278–284 and 352–354 in chain C; and 1–4, 105–117, 152–159, 192–211, 282–284, 353 and 354 in chain D.
The structure of nucleotide-free S. pombe Get3 was determined to 3.0 Å by molecular replacement with PHASER34 using a monomer of S. cerevisiae Get3 (with the α-helical subdomain removed) as the search model (58% sequence identity to S. pombe). Although these crystals diffract relatively poorly, sixfold NCS averaging yielded interpretable electron density maps. Model building and refinement (with NCS restraints) were carried out with COOT and PHENIX. The final model contains three Get3 homodimers and three zinc atoms and was refined with tight NCS restraints to an Rfree of 23.7% (28.8%). Side-chain density is generally weakest in the α-helical subdomains, and no interpretable electron density was observed for residues 1–6, 101–108, 191–195 and 323–329 in each chain.
The structure of nucleotide-free S. cerevisiae Get3 was determined by molecular replacement using PHASER. Identical solutions were found using either a monomer of S. cerevisiae Get3 or the S. pombe nucleotide-free open dimer as the search model (with the α-helical subdomain removed). Initial electron density maps calculated after fourfold NCS averaging clearly defined the orientation of the core ATPase domains of each monomer, and difference maps were used to confirm the presence of zinc at the dimer interface (Supplementary Fig. 2b). Helical features, including portions of α6, α7 and α9, are also observed in the α-helical subdomains. Although the S. cerevisiae apo structure shows the same open dimer architecture observed in the S. pombe apo structure (Supplementary Fig. 2c), because these crystals diffract weakly (to 3.8 Å) and anisotropically, no attempt was made to refine the model.
Refinement statistics are listed in Supplementary Table 1. All structure figures in the manuscript were generated using Pymol40 and COOT.
Preparation of site-directed mutants.
Site-directed S. cerevisiae Get3 mutants were generated using the QuikChange mutagenesis kit (Stratagene), and verified by DNA sequencing. Protein expression was carried out in E. coli as described earlier, except cells were grown at 25 °C and induced overnight. All mutants expressed to high levels. With the exception of Ala101Asp (which was insoluble), wild-type and mutant proteins were purified under non-denaturing conditions (as above) in a single step by nickel-affinity chromatography and dialyzed into 10 mM Tris, pH 7.5, 100 mM NaCl, 2 mM DTT and 40% glycerol. Protein concentrations were determined using the Bradford protein assay (Bio-Rad) standardized by calculated A280 extinction coefficients.
Get3 pull-down assay for TA substrate interaction.
Full-length 35S-labelled human SEC61β was synthesized in phenyl-depleted rabbit reticulocyte lysate translation extract (see later) with or without Get3 (or mutant Get3) protein at 50 μg ml−1. After translation for 15 min at 32 °C, the reactions were placed immediately on ice. To each reaction, 2.5 μl of anti-Get3 serum was added, incubated for 30 min on ice, and diluted to 1 ml with ice-cold pull-down buffer (50 mM HEPES, pH 7.4, 150 mM potassium acetate, and 2 mM magnesium acetate). Ten microlitres of Protein-A agarose (BioRad) was added, incubated with end-over-end mixing for 90 min at 4 °C, and washed 3 × 1 ml with pull-down buffer. Immunoprecipitated products were analysed by SDS–PAGE and quantified by phosphorimaging.
Each experiment included a reaction without Get3 to assess the extent of background pull-down. This value was subtracted from all samples. Wild-type Get3 was arbitrarily set at 100% pull-down. Each value is the average of between three and six independent measurements performed on different days. The error bars denote s.e.m. Coomassie staining verified that the polyclonal anti-Get3 antibody pulled down equal amounts of all Get3 mutants, indicating that the site-specific mutations did not affect antibody binding.
Preparation of phenyl-depleted translation extract.
Rabbit reticulocyte lysate supplemented with all components needed for translation was passed by gravity over a column of highly substituted phenyl-sepharose (GE/Amersham). The column volume was one-third of the lysate volume (typically 200 and 600 ml, respectively). The leading and trailing edges of the flow-through were not collected. Only the peak flow-through fractions (,~400 μl) containing undiluted lysate were collected and frozen in aliquots in liquid nitrogen and stored at −80 °C. Prepared in this manner, the lysate was more than 98% depleted of TRC40, but retained full translation capacity. By Coomassie staining, 95%or more of the proteins were not affected by the depletion.
ATPase assay.
ATPase activity was determined at 30 °C using a microplate photometricassay in which ATP hydrolysis is coupled to NADH oxidation36,37. The assay buffer contained 50 mM Tris, pH 7.5, 20 mM NaCl, 5 mM MgCl2, 1 mM DTT, 5% glycerol, 0.02% n-dodecyl-β-D-maltopyranoside, 4.5 mM phosphoenolpyruvate, 8.0 U lactate dehydrogenase (Sigma), 6.3 U pyruvate kinase (Sigma), 0.3 mM NADH and 2 μM Get3 (wild-type or mutant), and reactions were carried out in a final reaction volume of 200 ml. The reactions were initiated by adding ATP and the decrease in NADH concentration was followed spectrophotometrically at 340 nm. Linear steady state rates between 100 and 400 s were used to calculate Vmax and apparent Km,ATP values using a pathlength-corrected molar extinction coefficient for NADH. Kinetic parameters were determined by fitting the data to the Michaelis–Menten equation by nonlinear regression. All measurements were carried out in triplicate.
Yeast growth assay.
Wild-type and mutant GET3 genes were subcloned into a low copy number URA plasmid under the control of a medium-strength, constitutive ACT1 promoter, and transformed into a Δget3 strain (Open Biosystems). Serial dilutions of each transformant were spotted (along with wild type and vector only controls) onto synthetic defined medium (−uracil) supplemented with 100 μg ml −1 hygromycin B. Plates were photographed after 2 days at 37 °C.
Supplementary Material
Acknowledgements
Data were collected at beamlines 21-IDG and 23-IDD at the Advanced Photon Source (APS), Argonne National Laboratory, and we thank the beamline staff for support. Use of the APS was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. DE-AC02–06CH11357. We thank B. Glick and R. Strack for reagents and advice, X. Li for assay characterization, and A. Shiau and T. Steck for comments on the manuscript. This work was supported by a grant from the Edward Mallinckrodt, Jr. Foundation (to R.J.K.) and by the Intramural Research Program of the National Institutes of Health (to R.S.H.).
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Egea PF, Stroud RM & Walter P Targeting proteins to membranes: structure of the signal recognition particle. Curr. Opin. Struct. Biol 15, 213–220 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Keenan RJ, Freymann DM, Stroud RM & Walter P The signal recognition particle. Annu. Rev. Biochem 70, 755–775 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Rapoport TA Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450, 663–669 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Beilharz T, Egan B, Silver PA, Hofmann K & Lithgow T Bipartite signals mediate subcellular targeting of tail-anchored membrane proteins in Saccharomyces cerevisiae. J. Biol. Chem 278, 8219–8223 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Kalbfleisch T, Cambon A & Wattenberg BW A bioinformatics approach to identifying tail-anchored proteins in the human genome. Traffic 8, 1687–1694 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Borgese N, Brambillasca S & Colombo S How tails guide tail-anchored proteins to their destinations. Curr. Opin. Cell Biol 19, 368–375 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Wattenberg B & Lithgow T Targeting of C-terminal (tail)-anchored proteins: understanding how cytoplasmic activities are anchored to intracellular membranes. Traffic 2, 66–71 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Kutay U, Ahnert-Hilger G, Hartmann E, Wiedenmann B & Rapoport TA Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J 14, 217–223 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Favaloro V, Spasic M, Schwappach B & Dobberstein B Distinct targeting pathways for the membrane insertion of tail-anchored (TA) proteins. J. Cell Sci 121, 1832–1840 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stefanovic S & Hegde RS Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell 128, 1147–1159 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Kurdi-Haidar B et al. Isolation of the ATP-binding human homolog of the arsA component of the bacterial arsenite transporter. Genomics 36, 486–491 (1996). [DOI] [PubMed] [Google Scholar]
- 12.Kurdi-Haidar B, Heath D, Aebi S & Howell SB Biochemical characterization of the human arsenite-stimulated ATPase (hASNA-I). J. Biol. Chem 273, 22173–22176 (1998). [DOI] [PubMed] [Google Scholar]
- 13.Rosen BP, Bhattacharjee H, Zhou T & Walmsley AR Mechanism of the ArsA ATPase. Biochim. Biophys. Acta 1461, 207–215 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Schuldiner M et al. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell 134, 634–645 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leipe DD, Wolf YI, Koonin EV & Aravind L Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol 317, 41–72 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Zhou T, Radaev S, Rosen BP & Gatti DL Structure of the ArsA ATPase: the catalytic subunit of a heavy metal resistance pump. EMBO J 19, 4838–4845 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Georgiadis MM et al. Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii. Science 257, 1653–1659 (1992). [DOI] [PubMed] [Google Scholar]
- 18.Leonard TA, Butler PJ & Lowe J Bacterial chromosome segregation: structure and DNA binding of the Soj dimer—a conserved biological switch. EMBO J 24, 270–282 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayashi I, Oyama T & Morikawa K Structural and functional studies of MinD ATPase: implications for the molecular recognition of the bacterial cell division apparatus. EMBO J 20, 1819–1828 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freymann DM, Keenan RJ, Stroud RM & Walter P Structure of the conserved GTPase domain of the signal recognition particle. Nature 385, 361–364 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Montoya G,Svensson C, Luirink J & Sinning I Crystal structure of the NG domain from the signal-recognition particle receptor FtsY. Nature 385, 365–368 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Metz J, Wachter A, Schmidt B, Bujnicki JM & Schwappach B The yeast Arr4p ATPase binds the chloride transporter Gef1p when copper is available in the cytosol. J. Biol. Chem 281, 410–417 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Keenan RJ, Freymann DM, Walter P & Stroud RM Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell 94, 181–191 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Jiang Y et al. Nonequivalence of the nucleotide binding domains of the ArsA ATPase. J. Biol. Chem 280, 9921–9926 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Schindelin H, Kisker C, Schlessman JL, Howard JB & Rees DC Structure of ADP·AIF 4−-stabilized nitrogenase complex and its implications for signal transduction. Nature 387, 370–376 (1997). [DOI] [PubMed] [Google Scholar]
- 26.Sprang SR G protein mechanisms: insights from structural analysis. Annu. Rev. Biochem 66, 639–678 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Gasper R, Meyer S, Gotthardt K, Sirajuddin M & Wittinghofer A It takes two to tango: regulation of G proteins by dimerization. Nature Rev. Mol. Cell Biol 10, 423–429 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Zhou T, Radaev S, Rosen BP & Gatti DL Conformational changes in four regions of the Escherichia coli ArsA ATPase link ATP hydrolysis to ion translocation. J. Biol. Chem 276, 30414–30422 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Bernstein HD et al. Model for signal sequence recognition from amino-acid sequence of54Ksubunitofsignal recognition particle. Nature 340,482–486(1989). [DOI] [PubMed] [Google Scholar]
- 30.Horie C, Suzuki H, Sakaguchi M & Mihara K Characterization of signal that directs C-tail-anchored proteins to mammalian mitochondrial outer membrane. Mol. Biol. Cell 13, 1615–1625 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaufmann T et al. Characterization of the signal that directs Bcl-xL, but not Bcl-2, to the mitochondrial outer membrane. J. Cell Biol 160, 53–64 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuroda R et al. Charged amino acids at the carboxyl-terminal portions determine the intracellular locations of two isoforms of cytochrome b5. J. Biol. Chem 273, 31097–31102 (1998). [DOI] [PubMed] [Google Scholar]
- 33.Adams PD et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002). [DOI] [PubMed] [Google Scholar]
- 34.McCoy AJ, Grosse-Kunstleve RW, Adams PD & Winn MD Phaser crystallographic software. J. Appl. Crystallogr 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Kiianitsa K, Solinger JA & Heyer WD NADH-coupled microplate photometric assay for kinetic studies of ATP-hydrolyzing enzymes with low and high specific activities. Anal. Biochem 321, 266–271 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Vogel G & Steinhart R ATPase of Escherichia coli: purification, dissociation, and reconstitution of the active complex from the isolated subunits. Biochemistry 15, 208–216 (1976). [DOI] [PubMed] [Google Scholar]
References
- 38.Van den Berg B et al. X-ray structure of a protein-conducting channel. Nature 427, 36–44 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL & Clardy J Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol 229, 105–124 (1993). [DOI] [PubMed] [Google Scholar]
- 40.Delano WL The PyMOL Molecular Graphics System 〈http://www.pymol.org〉 (2002).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.