Abstract
Radiotherapy resistance is one of the major factors limiting the efficacy of radiotherapy in lung cancer patients. The extensive investigations indicate the diversity in the mechanisms underlying radioresistance. Here, we revealed that DNA damage binding protein 2 (DDB2) is a potential regulator in the radiosensitivity of non-small cell lung cancer (NSCLC) cells. DDB2, originally identified as a DNA damage recognition factor in the nucleotide excision repair, promotes the survival and inhibits the apoptosis of NSCLC cell lines upon ionizing radiation (IR). Mechanistic investigations demonstrated that DDB2 is able to facilitate IR-induced phosphorylation of Chk1, which plays a critical role in the cell cycle arrest and DNA repair in response to IR-induced DNA double-strand breaks (DSBs). Indeed, knockdown of DDB2 compromised the G2 arrest in the p53-proficient A549 cell line and reduced the efficiency of homologous recombination (HR) repair. Taken together, our data indicate that the expression of DDB2 in NSCLC could be used as a biomarker to predict radiosensitivity of the patients. Targeting Chk1 can be used to increase the efficacy of radiotherapy in patients of NSCLC possessing high levels of DDB2.
Keywords: DDB2, Chk1, Radioresistance, NSCLC, Homologous recombination
Introduction
Lung cancer is the leading cause of cancer death in the USA. It is estimated that there will be 224,390 new lung and bronchus cancer cases and 158,080 lung cancer deaths in 2016 [1]. About 85 to 90 % of lung cancers are non-small cell lung cancer (NSCLC), and radiotherapy plays a critical role in the management of inoperable NSCLC patients, particular those at advanced stages [2, 3]. However, radioresistance has been a major factor limiting the efficacy of radiotherapy. The mechanisms underlying cancer radioresistance have been extensively investigated, yet still remain unclear, probably due to multiple factors involved and tumor heterogeneity [4].
Ionizing radiation (IR) kills cancer cells mainly through the induction of DNA damage [5, 6]. Thus, enhanced DNA damage responses and DNA repair capacity in cancer cells have been considered the major contributors to chemoresistance for a long time. Upon DNA damage, the activation of a series of kinases is triggered, which include the phosphoinositide-3-kinase-related protein kinase (PIKK) family members ATM, ATR, and DNA-PKcs. While ATR activation is associated with single-stranded DNA and stalled DNA replication forks, ATM and DNA-PKcs respond mainly to DNA double-strand breaks (DSBs) [7]. ATM-Chk2 and ATR-Chk1 regulate the checkpoint pathways in the mammalian cells, resulting in cell cycle arrest, which is thought to increase the time available for DNA repair before replication or mitosis ensues. For the repair of DSBs, two principal mechanisms are used: non-homologous end-joining (NHEJ) [8] and homologous recombination (HR) [9]. NHEJ is active throughout the cell cycle. In contrast, HR-directed DSB repair is confined to the late S and G2 phases of proliferating cells when sister chromatids are available as repair templates. Thus, inhibition of DNA damage response and DNA repair capacity in cancer cells have been extensively exploited for overcoming radioresistance [10].
DNA damage binding protein 2 (DDB2) was originally identified as a damage sensor, which recognizes ultraviolet (UV) light-induced cyclobutane pyrimidine dimers (CPD) in the context of chromatin, and facilitates the nucleotide excision repair (NER) pathway [11]. Recently, new functions of DDB2 beyond its role in DNA repair have also been identified, e.g., enhancing cellular apoptosis through downregulation of Bcl-2 [12, 13], and p21 [14], suppressing colon tumor metastasis through blocking epithelial-mesenchymal transition (EMT) [15], and limiting the motility and invasiveness of invasive human breast tumor cells by regulating NF-κB activity [16], as well as mediating premature senescence [17]. DDB2 is also considered a tumor suppressor based on the finding that DDB2−/− mice were not only susceptible to UV-induced carcinogenesis but also developed spontaneous malignant tumors at a high rate [18, 19]. DDB2 is also found to be induced in the radioresistant NSCLC A549 cell line upon IR, suggesting that DDB2 might play a role in the cellular response to IR-induced DNA damage [20].
Here, we determined the role of DDB2 in the radioresistance of NSCLC cell lines A549 and H1299 and found that enhanced DDB2 expression promoted, while knockdown of DDB2 expression inhibited the radioresistance in these cells. Furthermore, we demonstrated that DDB2-facilitated, IR-induced phosphorylation of Chk1, and the subsequent cell cycle arrest and enhanced HR could be one of the contributors to the radioresistance in NSCLC cells.
Materials and methods
Cell culture and treatment
NSCLC cell lines A549 and H1299 were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). HeLa-DR13–9 cells, stably transfected with the pDR-GFP plasmid, were kindly provided by Dr. Jeff Parvin (The Ohio State University). A549 and H1299 cell lines were cultured in RPM I 1640 medium, while HeLa-DR-13–9 cells were cultured in DMEM, supplemented with 10 % of fetal bovine serum (FBS) and 50 U/ml each of penicillin and streptomycin, and maintained in a humidified atmosphere with 5 % CO2 at 37 °C. For IR treatment, X-ray was delivered to cultured cells in a RS-2000 X-ray Biological Irradiator (Rad Source Technologies, Inc., Suwanee, GA, USA).
Plasmids, siRNA, shRNA, and establishment of stable transfected cell lines
SMARTpools siRNA designed to target human DDB2 (M-011022–01), a scramble non-targeting control siRNA (5′- UUC UCC GAA CGU GUC ACG U − 3′), and TRIPZ-inducible shDDB2 plasmids (pTRIPZ-shDDB2) were purchased from GE Dharmacon (Lafayette, CO, USA). The pcDNA3.1-DDB2 vector has been described in our previous publication [21]. To construct the pTRE3G-DDB2 vector, DDB2 cDNA was amplified using pcDNA3.1-DDB2 expression construct as a template and subcloned into pTRE3G-BI-ZsGreen1 (Clontech, Mount View, CA, USA) to generate pTRE3G-DDB2. The I-SceI endonuclease expression vector pCBASce was kindly provided by Dr. Jeff Parvin.
To establish a cell line stably expressing the Tet-inducible shDDB2 plasmid, A549 cells were transfected with pTRIPZ-shDDB2 plasmids using Lipofectamine (Life Technologies, Grand Island, NY, USA), selected by Puromycin and screened for the induction of DDB2 knockdown with Doxycycline (Dox) treatment (A549-pTRIPZ-shDDB2). To establish a cell line stably expressing the Tet-inducible DDB2 overexpression plasmid, H1299 cells were first transfected with the regulator vector pCMV-Tet3G (Clontech). The stable transfected cell line H1299-Tet-On 3G was selected by G418 and screened for inducibility according to the manufacturer’s instruction. Then, H1299-Tet-On 3G cell line was further transfected with the pTRE3G-DDB2 vector and a liner Puromycin Marker (Clontech), the stably transfected cells were selected using puromycin, and validated for the Dox-induced DDB2 expression (H1299-pTRE3G-DDB2).
Clonogenic formation assay
Cells were seeded in 60-mm culture dishes, treated with IR at various doses, and further cultured for 14 days. The cells were fixed with ethanol, stained with methylene blue, and colonies of >50 cells were counted. The surviving fraction was calculated relative to the untreated cells.
Western blotting
Whole cell lysates were prepared by boiling cell pellets in the SDS lysis buffer [2 % SDS, 10 % glycerol, 62 mM Tris-HCl, pH 6.8, and a complete mini-protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN)]. After protein quantification with Bio-Rad Dc Protein Assay (Bio-Rad Laboratories, Hercules, CA), equal amount of proteins were loaded, separated on a polyacrylamide gel, and transferred to a nitrocellulose membrane. Protein bands were immunodetected with appropriate antibodies, e.g., goat anti-DDB2 (R&D Systems, Minneapolis, MN, USA. Cat. No. AF3297, 1:2000), rabbit anti-cleaved PARP (Cell Signaling Technology, Danvers, MA, USA, Cat. No. 5625, 1:1000), rabbit anti-cleaved caspase 3 (Cell Signaling Technology, Cat. No. 9603, 1:1000), mouse anti-Chk1 (Cell Signaling Technology, Cat. No. 2360, 1:1000), rabbit anti-pChk1 (Cell Signaling Technology, Cat. No. 2348, 1:1000), rabbit anti-Chk2 (Cell Signaling Technology, Cat. No. 6334, 1:1000), rabbit anti-pChk2 (Cell Signaling Technology, Cat. No. 2197, 1:1000), rabbit anti-ATM (Cell Signaling Technology, Cat. No. 2873, 1:1000), rabbit anti-pATM (Cell Signaling Technology, Cat. No. 5883, 1:1000), rabbit anti-ATR (Cell Signaling Technology, Cat. No. 13,934, 1:1000), rabbit anti-pATR (Cell Signaling Technology, Cat. No. 2853, 1:1000), mouse anti-γH2AX (Millipore, Billerica, MA, Cat. No. 05–636, 1:1000), mouse anti-actin (Santa Cruz Biotechnology, Dallas, TX, USA, Cat. No. sc-47,778, 1:1000), and goat anti-Lamin B (Santa Cruz Biotechnology, Cat. No. sc-6216, 1:1000).
Cell cycle analysis
Cells were trypsinized, washed with PBS, and resuspended in 0.5 ml of PBS to make a single-cell suspension. Cells were then fixed by drop-wise addition of 5 ml of ice-cold 70 % ethanol into the cell suspension. For propidium iodide (PI) staining, the ethanol-suspended cells were centrifuged, washed with PBS and resuspended in 0.5 ml of PI-Triton X-100 staining solution containing RNase A (0.1 % Triton X-100 in PBS, 0.2 mg/ml DNase-free RNase A, 20 μg/ml PI), and incubated at 37 °C for 15 min. DNA content analysis was carried out in a BD FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA). Cell cycle analysis was performed using the FCS Express Software (De Novo Software, Glendale, CA, USA).
HR reporter assay
The effect of DDB2 on the HR activity was determined using HeLa-DR13–9 cells [22]. Briefly, HeLa-DR13–9 cells, containing the HR reporter DR-GFP, were first transfected with control or DDB2 siRNA for 2 days. Cells were then con-transfected with siCtrl or siDDB2 and I-SceI expression vector (pCBASce). Cells were harvested after 3 days, and the GFP-positive cells were analyzed using flow cytometry.
Statistical analyses
Results are presented as the mean ± SD for at least three independent experiments for each group. Statistic differences were determined by using two sample t tests for independent samples. P values less than 0.05 were defined as statistically significant.
Results
DDB2 protects NSCLC cells from IR-induced cell killing
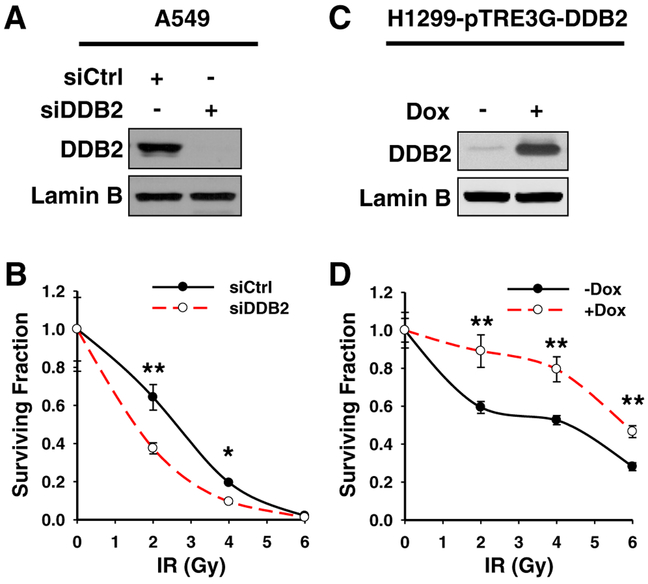
Our previous studies have revealed the role of DDB2 in mediating the sensitivity of ovarian cancer cells to cisplatin [12, 13]. As a DNA damage binding protein, DDB2 is involved in the repair of UV-induced CPD by the NER pathway, but is not required for the repair of cisplatin-induced intrastrand crosslinks. Instead, DDB2 promotes cisplatin-induced apoptosis in ovarian cancer cell lines through downregulation of anti-apoptotic protein Bcl-2 [12, 13] and cell cycle regulator p21 [14]. In order to understand whether DDB2 plays a role in regulating the sensitivity of cancer cells to radiotherapy, we knocked down DDB2 in the A549 NSCLC cell line and determined the cell survival upon IR. As shown in Fig. 1, DDB2 siRNA efficiently downregulated the expression of DDB2 in A549 cells (Fig. 1a), and downregulation of DDB2 sensitized A549 cells to IR, reflected by reduced clonogenic formation in these cells upon IR treatment at 2 and 4 Gy (Fig. 1b). To confirm this finding, we generated a Tet-inducible DDB2 overexpression cell line by using H1299 cells (H1299-pTRE3G-DDB2), in which DDB2 expression can be increased by treating cells with Dox (Fig. 1c). In contrast to DDB2 knockdown, DDB2 over-expression reduced the sensitivity of H1299 cells to IR treatment (Fig. 1d). Collectively, these data indicate that DDB2 plays a protective role in NSCLC cells in response to IR, suggesting that high expression of DDB2 in NSCLC cells might increase these cells’ resistance to radiotherapy.
Fig. 1.
DDB2 reduces IR sensitivity in lung cancer cells. a, b A549 cells were transfected with either control or DDB2 siRNA. Immunoblotting was conducted to show the knockdown efficiency of DDB2 by siRNA. Lamin B was detected as a loading control (a); the clonogenic formation assay was conducted to show the sensitivity of these cells to IR (b). c, d H1299 cells containing Tet-inducible DDB2 were cultured in the presence of Dox to induce DDB2 overexpression. Immunoblotting was conducted to show the induction of DDB2 by Dox; Lamin B was detected as a loading control (c). The clonogenic formation assay was conducted to show the sensitivity of these cells to IR (d). N = 4, bar: SD, *p < 0.05, **p < 0.01
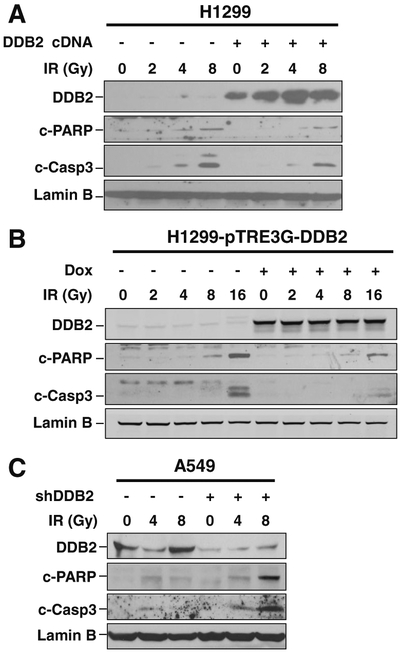
We have shown in our previous study that DDB2 facilitates cisplatin-induced apoptosis [12, 13]. Given our finding that DDB2 inhibits IR-induced cell killing, we wanted to know whether DDB2 is involved in IR-induced apoptosis in NSCLC cells. We first transiently transfected DDB2-expressing constructs into H1299 cell and analyzed cellular apoptosis upon IR. As shown in Fig. 2a, IR treatment could induce apoptosis in H1299 cells at a dose as low as 2 Gy, reflected by the detection of cleaved PARP and cleaved caspase 3. However, when DDB2 was overexpressed in these cells, the lowest dose of IR to induce apoptosis was 4 Gy, indicating that DDB2 is able to protect H1299 cells from IR-induced apoptosis. In addition, we confirmed this finding in H1299 cells containing Tet-inducible DDB2 (Fig. 2b). It is very clear that Dox-induced DDB2 expression dramatically reduced the amount of cleaved-PARP and cleaved-caspase 3 in cells treated with IR at 8 and 16 Gy. In addition, we knocked down DDB2 expression in A549 cells and found that downregulation of DDB2 promoted IR-induced cellular apoptosis, as reflected by increased cleaved PARP and cleaved caspase 3 (Fig. 2c). Taken together, our data indicate that DDB2 is able to protect NSCLC cells from IR-induced apoptosis, resulting in an inefficient killing of cancer cells and subsequent radiotherapy.
Fig. 2.
DDB2 inhibits IR-induced apoptosis in lung cancer cells. a, b DDB2 was overexpressed in H1299 cells by transfecting with DDB2-expressing plasmids (a) or in H1299-pTRE3G-DDB2 cells by treating with Dox (b). Cells were irradiated with X-ray at various doses and further cultured for 48 h. Whole cell lysates were prepared and subjected to immunoblotting to detect cleaved PARP (c-PARP) and cleaved caspase-3 (c-Casp3) to reflect cellular apoptosis. Lamin B was also detected to serve as a loading control. c DDB2 was downregulated in A549 cells by transfecting with DDB2 siRNA. Cells were treated with X-ray, and cellular apoptosis was detected as in a and b
DDB2 promotes DNA damage responses upon IR
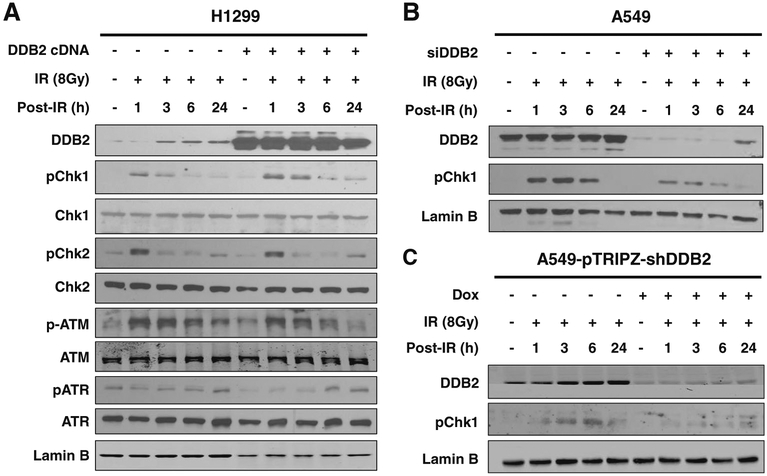
Prompt activation of DNA damage response and enhanced DNA repair capacity can promote cells to survive DNA-damaging agents. DDB2 has been suggested to be involved in a general cellular response to DNA damage [23]. Therefore, to understand the mechanism by which DDB2 facilitates cell survival upon IR, we determined the effect of DDB2 expression level on the activation of various DNA damage response proteins. As shown in Fig. 3a, transient transfection of DDB2 into H1299 cells enhanced the IR-induced phosphorylation of Chk1, but not ATM, ATR, and Chk2. We then downregulated the expression of DDB2 in A549 cells through transient transfection of DDB2 siRNA and found a reduced phosphorylation of Chk1 after IR treatment (Fig. 3b). This result was further confirmed in an A549 cell line containing a Tet-inducible DDB2 shRNA (Fig. 3c). Taken together, these data indicate that DDB2 is able to promote DNA damage checkpoint signaling by facilitating phosphorylation of Chk1 upon IR.
Fig. 3.
DDB2 promotes IR-induced phosphorylation of ChK1 in lung cancer cells. a H1299 cells were transfected with DDB2-expressing vectors, treated with IR and further cultured for various time periods. Whole cell lysates were prepared and subjected to immunoblotting to detect various checkpoint proteins. b, c DDB2 was downregulated in A549 cells by transfecting with DDB2-specific siRNA (b) or in A549-pTRIPZ-shDDB2 cells by treating with Dox (c). Cells were treated with IR and further cultured for various time periods. Whole cell lysates were prepared and subjected to immunoblotting to detect phosphorylated Chk1. Lamin B was also detected as a loading control
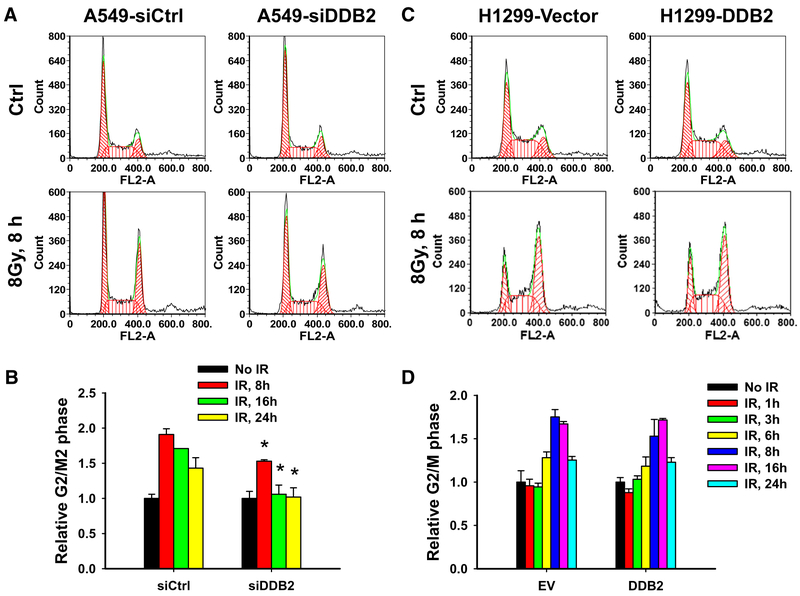
Given that Chk1 phosphorylation plays an important role in the regulation of G2 arrest in response to DNA damage, we sought to understand whether the DDB2 expression level has influence on G2 arrest upon IR. A549 cells were transfected with the DDB2 siRNA to downregulate DDB2 expression or transfected with a control siRNA. Cells were IR treated and further cultured for up to 24 h, and the cell cycle phase distribution was detected at different time points. As shown in Fig. 4a, b, IR treatment induced G2 arrest in siCtrl-transfected A549 cells, and the G2 arrest was kept for up to 24 h. However, IR-induced G2 arrest was significantly compromised in siDDB2-transfected A549 cells, particularly at 16 and 24 h post-IR. Given that ATM-Chk2 signaling initiates cellular G2 arrest, and the maintenance of this arrest is performed by ATR-Chk1 signaling, in response to IR-induced DSBs [24, 25], the reduced G2 arrest in DDB2 knockdown cells upon IR can be attributed to the decrease in Chk1 activation.
Fig. 4.
DDB2 affects IR-induced G2 arrest differentially in p53-proficient and p53-deficient cells. DDB2 was downregulated in A549 cells by transfecting with DDB2 siRNA (a, b) or overexpressed in H1299 cells by transfecting with DDB2-expressing plasmids (c, d). Cells were IR treated and further cultured for different time periods. Cells were harvested and stained with PI, cell cycle distribution was analyzed by flow cytometry. N = 3, bar: SD, *p < 0.05 compared with control siRNA (siCtrl) or empty vector transfected cells, respectively
We then overexpressed DDB2 in H1299 cells, and analyzed the cell cycle phase distribution in these cells upon IR. Similar to A549 cells, we also found IR-induced G2 arrest in both empty vector and DDB2 cDNA transfected H1299 cells after 6 h post-IR. However, we failed to see the effect of DDB2 overexpression on this IR-induced G2 arrest (Fig. 4c,d). Given that H1299 cells do not express p53 protein due to homozygous partial deletion of the TP53 gene, and p53 is not required for the initial arrest of cells in G2 but is essential for the long-term maintenance of the arrest [26], it is possible that DDB2 knockdown-induced inhibition of G2 arrest in response to IR requires functional p53.
DDB2 enhances homologous recombination repair upon IR treatment
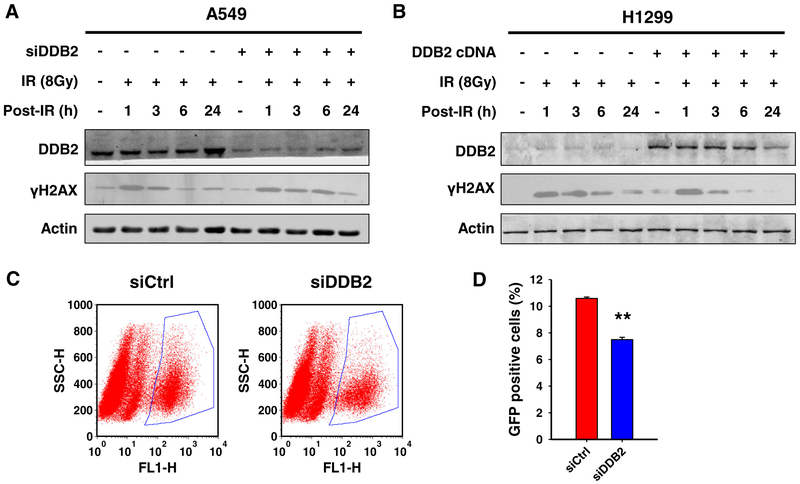
Although DDB2 does not affect IR-induced G2 arrest in H1299 cells, it does increase the resistance of H1299 cells to IR-induced cell killing and apoptosis (Figs. 1 and 2). Thus, additional mechanisms underlying DDB2-mediated radioresistance in NSCLC cells might exist. To investigate whether DDB2 is involved in the repair of IR-induced DNA DSBs, we either knocked down or overexpressed DDB2 in A549 or H1299 cells, respectively, and detected the kinetics of γH2AX amounts, a marker for DSBs, with the time of post-IR. As shown in Fig. 5a, the amount of γH2AX reduced dramatically at 6 h post-IR in A549 cells transfected with control siRNA, while still remained at a high level at the same time point post-IR in A549 cells transfected with DDB2 siRNA, indicating that downregulation of DDB2 halted the repair of IR-induced DSBs. Conversely, a considerable amount of γH2AX can be detected in H1299 cells, which exhibit low levels of DDB2, at both 6 and 24 h post-IR. Overexpression of DDB2 in these cells promoted the decrease in the amount of γH2AX (Fig. 5b), further confirming that DDB2 facilitates the repair of DSBs following IR.
Fig. 5.
DDB2 facilitates homologous recombination to repair IR-induced DSBs. a, b A549 and H1299 cells were transfected with DDB2 siRNA (a) and DDB2-expressing vectors (b), respectively. Cell were treated with IR and further cultured for the indicated time periods. Whole cell lysates were prepared and subjected to immunoblotting to detect DDB2 and phosphorylated H2AX (γH2AX). Actin was also detected as a loading control. c, d The HeLa cells stably transfected with pDR-GFP (HeLa-DR13–9 cells) were transfected with either control siRNA (siCtrl) or DDB2 siRNA (siDDB2) along with pCBASce plasmids for 3 days. GFP-positive cells were detected by flow cytometry. N = 3, bar: SD, **p < 0.01
HR is one of the major DSB repair pathways. We thus determined whether DDB2 affects the efficiency of HR by using a HR reporter assay. HeLa cells containing integrated DR-GFP reporter constructs were transfected with either control or DDB2 siRNA, along with I-SceI to induce DSBs. The GFP+ cells, representing HR-repaired cells, were analyzed using FACS. As shown in Fig. 5c, d, knockdown of DDB2 in these cells significantly reduced the HR efficiency, reflected by about 30 % decrease in GFP+ cells in cells with DDB2 downregulation. These data indicate that DDB2 is able to promote the repair of DSBs through the HR pathway.
Discussion
In this study, we reported for the first time that DDB2 plays a role in the activation of Chk1 upon IR and facilitates HR repair, thus, increases radioresistance of NSCLC cells.
Upon DNA damage, ATM and ATR, two of DNA damage sensors, can be activated by phosphorylation, and further phosphorylate Chk2 and Chk1, respectively, to transduce the signal for cell cycle checkpoint and DNA repair [23]. The ATM-Chk2 pathway is primarily activated in response to DSBs, while the ATR-Chk1 pathway is mainly activated by UV light and DNA replication stress. In addition, the ATR-Chk1 pathway can also be activated upon IR-induced DSBs [27, 28], through the generation of structures containing single-stranded DNA (ssDNA) adjacent to dsDNA, and the coating of replication protein A (RPA) [29]. In our studies, we have demonstrated that both ATM-Chk2 and ATR-Chk1 pathways are activated in response to IR in NSCLC cell lines. Interestingly, among these four proteins, only IR-induced phosphorylation of Chk1 can be regulated by the DDB2 expression level, indicating that DDB2 might affect the formation of ssDNA-RPA platform for ATR recruitment. It has been reported that many proteins are involved in this process, including Rad17, TopBP1, the 9-1-1 complex, BRCA1, Claspin, and CBP/p300 [29, 30]. Interestingly, DDB2 has been shown to be associated with Claspin [31] and CBP/p300 [32]. Thus, it is very likely that DDB2 promotes IR-induced Chk1 phosphorylation by bringing Claspin and p300 to the damage sites for facilitating platform formation and ATR activity stimulation. However, we could not exclude the possibility that DDB2 may enhance IR-induced Chk1 phosphorylation by inhibiting Chk1 dephosphorylation.
Another novel finding in this study is that DDB2 promotes HR in response of IR-induced DSBs. Although DDB2 has been shown to bind to a broad spectrum of different lesions, such as apurinic/apyrimidinic sites and interstrand crosslinks [33–35], and play an important role in NER [36], its role in other DNA repair pathways has not yet been investigated. DDB2 expression can be induced by IR in a p53-dependent manner [37]. Given that DDB2 is able to bind to abasic sites [35], induction of DDB2 by IR indicates that DDB2 might be involved in the repair of IR-induced oxidative DNA damage [38]. In addition, given that Chk1 positively regulates mammalian HR repair through interaction with and phosphorylation of RAD51 [39], DDB2 could enhance HR through pro-motion of Chk1 activity, as we revealed in this study.
DDB2 has been shown to facilitate apoptosis in response to UV and cisplatin. [12–14]. However, we found that DDB2 inhibits IR-induced apoptosis in NSCLC cells in this study. It is well known that upon DNA damage, the DNA repair machinery starts to remove DNA lesions. The unrepaired DNA lesions trigger cellular apoptosis to eliminate heavily damaged or seriously deregulated cells [23]. Given that DDB2 plays a role in both DNA repair and apoptosis, if the contribution of DDB2 to DNA repair is less than that to apoptosis, cells will exhibit increased apoptosis in the presence of DDB2. In contrast, if the contribution of DDB2 to DNA repair is more than that to apoptosis, cells will exhibit decreased apoptosis in the presence of DDB2 because DNA damage is efficiently repaired. We believe that DDB2 facilitates the HR pathway to repair IR-induced DSBs in NSCLC cells, thus reduces the amount of triggers of apoptosis, and results in a decreased apoptosis in these cells when DDB2 expression is enhanced.
Chk1 has been shown to contribute to therapy resistance in many cancers. Thus, targeting Chk1 has been exploited for cancer therapy, particularly for improving the efficacy of radiotherapy and chemotherapies that induce DNA damage [29]. Given our finding that DDB2 facilitate the activation of Chk1 upon IR, targeting Chk1 could be an efficient strategy to overcome radioresistance in DDB2 highly expressing NSCLC.
Acknowledgment
We thank Dr. Jeff Parvin (The Ohio State University) for providing HeLa-DR cells and pCBASce plasmids. This work was supported by National Institute of Health (CA151248 to Q.E.W.).
Footnotes
Conflicts of interest None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. [DOI] [PubMed] [Google Scholar]
- 2.Sause WT. The role of radiotherapy in non-small cell lung cancer. Chest. 1999;116:504S–8S. [DOI] [PubMed] [Google Scholar]
- 3.Koh PK, Faivre-Finn C, Blackhall FH, De RD. Targeted agents in non-small cell lung cancer (NSCLC): clinical developments and rationale for the combination with thoracic radiotherapy. Cancer Treat Rev. 2012;38:626–40. [DOI] [PubMed] [Google Scholar]
- 4.Chang L, Graham P, Hao J, Ni J, Deng J, Bucci J, Malouf D, Gillatt D, Li Y: Cancer stem cells and signaling pathways in radioresistance. Oncotarget 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bedford JS. Sublethal damage, potentially lethal damage, and chromosomal aberrations in mammalian cells exposed to ionizing radiations. Int J Radiat Oncol Biol Phys. 1991;21:1457–69. [DOI] [PubMed] [Google Scholar]
- 6.Frankenberg-Schwager M, Frankenberg D, Blocher D, Adamczyk C. Effect of dose rate on the induction of DNA double-strand breaks in eucaryotic cells. Radiat Res. 1981;87:710–7. [PubMed] [Google Scholar]
- 7.Durocher D, Jackson SP. DNA-PK, ATM and ATR as sensors of DNA damage: variations on a theme? Curr Opin Cell Biol. 2001;13:225–31. [DOI] [PubMed] [Google Scholar]
- 8.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–57. [DOI] [PubMed] [Google Scholar]
- 10.O’Connor MJ. Targeting the DNA damage response in cancer. Mol Cell. 2015;60:547–60. [DOI] [PubMed] [Google Scholar]
- 11.Dualan R, Brody T, Keeney S, Nichols AF, Admon A, Linn S. Chromosomal localization and cDNA cloning of the genes (DDB1 and DDB2) for the p127 and p48 subunits of a human damage-specific DNA binding protein. Genomics. 1995;29:62–9. [DOI] [PubMed] [Google Scholar]
- 12.Barakat BM, Wang QE, Han C, Milum K, Yin DT, Zhao Q, Wani G, Arafa ES, El-Mahdy MA, Wani AA. Overexpression of DDB2 enhances the sensitivity of human ovarian cancer cells to cisplatin by augmenting cellular apoptosis. Int J Cancer. 2009;127:977–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao R, Han C, Eisenhauer E, Kroger J, Zhao W, Yu J, Selvendiran K, Liu X, Wani AA, Wang QE. DNA damage-binding complex recruits HDAC1 to repress Bcl-2 transcription in human ovarian cancer cells. Mol Cancer Res. 2014;12:370–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoyanova T, Roy N, Kopanja D, Bagchi S, Raychaudhuri P. DDB2 decides cell fate following DNA damage. Proc Natl Acad Sci U S A. 2009;106:10690–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roy N, Bommi PV, Bhat UG, Bhattacharjee S, Elangovan I, Li J, Patra KC, Kopanja D, Blunier A, Benya R, Bagchi S, Raychaudhuri P. DDB2 suppresses epithelial-to-mesenchymal transition in colon cancer. Cancer Res. 2013;73:3771–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ennen M, Klotz R, Touche N, Pinel S, Barbieux C, Besancenot V, Brunner E, Thiebaut D, Jung AC, Ledrappier S, Domenjoud L, Abecassis J, Plenat F, Grandemange S, Becuwe P. DDB2: a novel regulator of NF-kappaB and breast tumor invasion. Cancer Res. 2013;73:5040–52. [DOI] [PubMed] [Google Scholar]
- 17.Roy N, Stoyanova T, Dominguez-Brauer C, Park HJ, Bagchi S, Raychaudhuri P. DDB2, an essential mediator of premature senescence. Mol Cell Biol. 2010;30:2681–92. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Yoon T, Chakrabortty A, Franks R, Valli T, Kiyokawa H, Raychaudhuri P. Tumor-prone phenotype of the DDB2-deficient mice. Oncogene. 2005;24:469–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itoh T, Iwashita S, Cohen MB, Meyerholz DK, Linn S. Ddb2 is a haploinsufficient tumor suppressor and controls spontaneous germ cell apoptosis. Hum Mol Genet. 2007;16:1578–86. [DOI] [PubMed] [Google Scholar]
- 20.Yang HJ, Kim N, Seong KM, Youn H, Youn B. Investigation of radiation-induced transcriptome profile of radioresistant non-small cell lung cancer A549 cells using RNA-seq. PLoS One. 2013;8: e59319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Wang QE, Zhu Q, El-Mahdy MA, Wani G, Praetorius-Ibba M, Wani AA. DNA damage binding protein component DDB1 participates in nucleotide excision repair through DDB2 DNA-binding and cullin 4A ubiquitin ligase activity. Cancer Res. 2006;66:8590–7. [DOI] [PubMed] [Google Scholar]
- 22.Parvin J, Chiba N, Ransburgh D: Identifying the effects of BRCA1 mutations on homologous recombination using cells that express endogenous wild-type BRCA1. J Vis Exp 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. [DOI] [PubMed] [Google Scholar]
- 24.Xu B, Kim ST, Lim DS, Kastan MB. Two molecularly distinct G(2)/M checkpoints are induced by ionizing irradiation. Mol Cell Biol. 2002;22:1049–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown EJ, Baltimore D. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 2003;17: 615–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene. 2001;20:1803–15. [DOI] [PubMed] [Google Scholar]
- 27.Myers JS, Cortez D. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J Biol Chem. 2006;281:9346–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Hunter T. Roles of Chk1 in cell biology and cancer therapy. Int J Cancer. 2014;134:1013–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stauffer D, Chang B, Huang J, Dunn A, Thayer M. p300/CREB-binding protein interacts with ATR and is required for the DNA replication checkpoint. J Biol Chem. 2007;282:9678–87. [DOI] [PubMed] [Google Scholar]
- 31.Praetorius-Ibba M, Wang QE, Wani G, El-Mahdy MA, Zhu Q, Qin S, Wani AA. Role of Claspin in regulation of nucleotide excision repair factor DDB2. DNA Repair (Amst). 2007;6:578–87. [DOI] [PubMed] [Google Scholar]
- 32.Datta A, Bagchi S, Nag A, Shiyanov P, Adami GR, Yoon T, Raychaudhuri P. The p48 subunit of the damaged-DNA binding protein DDB associates with the CBP/p300 family of histone ace-tyltransferase. Mutat Res. 2001;486:89–97. [DOI] [PubMed] [Google Scholar]
- 33.Payne A, Chu G. Xeroderma pigmentosum group E binding factor recognizes a broad spectrum of DNA damage. Mutat Res Fundam Mol Mech Mutagen. 1994;310:89–102. [DOI] [PubMed] [Google Scholar]
- 34.Fujiwara Y, Masutani C, Mizukoshi T, Kondo J, Hanaoka F, Iwai S. Characterization of DNA recognition by the human UV-damaged DNA-binding protein. J Biol Chem. 1999;274:20027–33. [DOI] [PubMed] [Google Scholar]
- 35.Wittschieben BO, Iwai S, Wood RD. DDB1-DDB2 (xeroderma pigmentosum group E) protein complex recognizes a cyclobutane pyrimidine dimer, mismatches, apurinic/apyrimidinic sites, and compound lesions in DNA. J Biol Chem. 2005;280:39982–9. [DOI] [PubMed] [Google Scholar]
- 36.Wittschieben BO, Wood RD. DDB complexities. DNA Repair (Amst). 2003;2:1065–9. [DOI] [PubMed] [Google Scholar]
- 37.Hwang BJ, Toering S, Francke U, Chu G. p48 activates a UV-damaged-DNA binding factor and is defective in xeroderma pigmentosum group E cells that lack binding activity. Mol Cell Biol. 1998;18:4391–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cramers P, Filon AR, Pines A, Kleinjans JC, Mullenders LH, Van Zeeland AA. Enhanced nucleotide excision repair in human fibroblasts pre-exposed to ionizing radiation. Photochem Photobiol. 2012;88:147–53. [DOI] [PubMed] [Google Scholar]
- 39.Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, Helleday T. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol. 2005;7:195–201. [DOI] [PubMed] [Google Scholar]