Abstract
The level of microRNA-93 (miR-93) in tumors has been recently reported to be negatively correlated with survival of lung cancer patients. Considering that the most devastating aspect of lung cancer is metastasis, which can be promoted by transforming growth factor-β (TGF-β)-induced epithelial- to-mesenchymal transition (EMT), we sought to determine whether miR-93 is involved in this process. Here, we report that a previously unidentified target of miR-93, neural precursor cell expressed developmentally downregulated gene 4-like (NEDD4L), is able to mediate TGF-β-mediated EMT in lung cancer cells. miR-93 binds directly to the 3′-UTR of the NEDD4L messenger RNA (mRNA), leading to a downregulation of NEDD4L expression at the protein level. We next demonstrated that the downregulation of NEDD4L enhanced, while overexpression of NEDD4L reduced TGF-β signaling, reflected by increased phosphorylation of SMAD2 in the lung cancer cell line after TGF-β treatment. Furthermore, overexpression of miR-93 in lung cancer cells promoted TGF-β- induced EMT through downregulation of NEDD4L. The analysis of publicly available gene expression array datasets indicates that low NEDD4L expression correlates with poor outcomes among patients with lung cancer, further supporting the oncogenic role of miR-93 in lung tumorigenesis and metastasis.
Keywords: Lung cancer, NEDD4L, miR-93, TGF-β signaling, EMT
Introduction
Lung cancer is the leading cause of cancer death worldwide. In 2015, an estimated 221,210 new cases of lung and bronchial cancer will be diagnosed, and 158,040 deaths are estimated to occur. Despite intensive research over the past decade, the 5-year survival of patients with lung cancer is only 17 % [1]. Histologically, the majority of lung cancers (75–85 %) are classified as non-small cell lung cancer (NSCLC), in which adenocarcinoma (40 %) and squamous cell carcinoma (30–35 %) are the two most common subtypes of NSCLC [2]. The most devastating aspect of lung cancer is metastasis, which is a complicated and continuous process involving multiple steps [3]. It has been reported that more than 90 % of lung cancer patients had metastasis by the time of their death [4]. Transforming growth factor-β (TGF-β)-induced epithelial-to-mesenchymal transition (EMT) is a major contributor to lung cancer metastasis [2]. Therefore, understanding the molecular mechanisms of TGF-β signaling transduction might facilitate the identification of new diagnostic markers and the development of novel therapeutic strategies in the treatment of lung cancer.
miR-93 belongs to the miR106b-25 cluster that has been reported to be overexpressed in different types of cancer, such as gastric cancer, prostate cancer, pancreatic neural endocrine tumor, neuroblastoma, and multiple myeloma [5]. It has recently been reported that miR-93 expression was upregulated in NSCLC cells both in vitro and in vivo studies [6]. miR-93 overexpression has an important role in promoting lung cancer cell growth, and high levels of miR-93 are correlated with poor survival of lung cancer patients, suggesting that miR-93 may play an important role in the tumor progression of lung cancer [7–9].
Neural precursor cell expressed developmentally downregulated gene 4-like (NEDD4L) is one of the nine NEDD4-like E3 ubiquitin ligases [10]. The best known and most studied role of NEDD4L is its function as a regulator of ion channel receptors and transporters [11, 12]. Recent studies have shown that NEDD4L also has potential functions in regulating key signaling pathways, such as EGFR, TGF-β, and Wnt pathways [13–15]. NEDD4L is able to limit the half-life of TGF-β-activated SMADs by regulating the turnover rate of two key mediators, SMAD 2 and SMAD 3 [16]. However, a reduced level of NEDD4L messenger RNA (mRNA) has been reported in lung cancer tissues compared with normal lung tissues [13], and this low NEDD4L expression is associated with clinical-pathological factors, such as excessive smoking history, histological grade (moderately and poorly), T stage (T2–4), lymph node metastasis, and pathological stage (stages II–IV) [13], suggesting that NEDD4L may play a role in preventing tumor progression.
In this study, we identified NEDD4L as a direct target of miR-93. Overexpression of miR-93 promoted TGF-β- induced EMT through downregulation of NEDD4L in lung cancer cells, unveiling a novel mechanism for miR-93- promoted lung cancer progression.
Materials and methods
Cell culture and TGF-β treatment
Human lung adenocarcinoma epithelial cell lines A549 and H1650 were purchased from ATCC (Manassas, VA). Cells were grown in RPMI 1640 medium supplemented with 50 U/ml each of penicillin and streptomycin and 10 % (v/v) fetal bovine serum (FBS) and maintained in a humidified atmosphere with 5 % CO2 at 37 °C.
Plasmids, shRNA, and miRNA
To construct wild-type psiCHECK-2-NEDD4L-3′-UTR, a 460-base pair fragment of the 3′-UTR of the NEDD4L gene (positions 1045–1051) that contains the putative miR-93 binding site was cloned into the psiCHECK-2 reporter vector (Promega, Madison, WI), downstream of the Renilla luciferase gene (WT-NEDD4L). To construct mutant plasmids, the putative miR-93 binding site in NEDD4L 3′-UTR was mutated (Mut-NEDD4L) using QuikChange Site-Directed Mutagenesis Kit (Stratagene, Santa Clara, CA). The insert was sequenced to verify the mutation. NEDD4L short hairpin RNA (shRNA) was purchased from Sigma (St. Louis, MO, SHCLNG-015277). miR-93 mirVana mimics (miR-93-M) (Cat No. 4464066) and miR-93 inhibitors (miR-93-I) (Cat No. 4464084) were purchased from Life Technologies (Carlsbad, CA). The pCMV-NEDD4L plasmid was purchased from transOMIC technologies (Huntsville, AL). The plasmids shRNA and microRNA (miRNA) were transfected into cells by using Lipofectamine 2000 reagents (Life Technologies) according to the manufacturer’s instructions.
Luciferase reporter assay
Of A549 cells, 2×105 were seeded in 12-well plates. After 24 h, the cells were co-transfected with 200 ng of psiCHECK-2-NEDD4L-3′-UTR vectors and 20 μM of miR-93 mimics. The cells were harvested 48 h after transfection. Firefly and Renilla luciferase activities were measured by the Dual-Luciferase Reporter Assay Kit (Promega) on a Veritas Microplate Luminometer (Turner Biosystems, Sunnyvale, CA, USA). Renilla luciferase activity was normalized to Firefly luciferase activity to evaluate the effect of miR-93 on the expression of Renilla luciferase.
Western blot analysis
Cells were lysed and subjected to western blot analysis as previously described [17]. Briefly, cells were lysed in sodium dodecyl sulfate (SDS) lysis buffer (2 % SDS, 10 % glycerol, 62 mM Tris-HCl, pH 6.8, and a complete mini-protease inhibitor cocktail [Roche Life Science]) and boiled for 10 min. After protein quantification, equal amount of protein was loaded, separated on a polyacrylamide gel, and transfering to a nitrocellulose membrane. Protein bands were immunodetected with appropriate antibodies, e.g., rabbit-anti- NEDD4L (Cell Signaling, Danvers, MA, Cat. No. 4013, 1:1000), mouse anti-E-cadherin (BD Transduction Laboratories, San Jose, CA, Cat. No. 610181, 1:1000), mouse antivimentin (Cell Signaling, Cat. No. 3390, 1:1000), rabbitanti-pSMAD2 (Cell Signaling, Cat. No. 9101, 1:1000), mouse anti-GAPDH (Santa Cruz Biotechnology, Dallas, TX, Cat. No. sc-47724, 1:4000), goat anti-actin (Santa Cruz Biotechnology, Cat. No. sc-1616, 1:1000), and their corresponding secondary antibodies (Santa Cruz Biotechnology).
RNA extraction and qRT-PCR
Total RNA was extracted using TRIzol reagent (Life Technologies), and the first-strand complementary DNA (cDNA) was generated by the Reverse Transcription System (Promega) in a 20-μl reaction containing 1 μg of total RNA. A 0.5-μl aliquot of cDNA was amplified by Fast SYBR Green PCR Master Mix (Life Technologies) in each 20-μl reaction. PCR reactions were run on the ABI 7900 Fast Real-Time PCR system in the OSUCCC Nucleic Acid Core Facility with the following primers: NEDD4L, forward, 5′-AGA AAC TGC CCA GAG CTC AC-3′, reverse, 5′-TCG CCT CTG CAA AAG TCT GT-3′; GAPDH, forward, 5′-GAAGGTGAAGGTCGGAGT-3′, reverse, 5′-GAAGATGGTGATGGGATTTC-3′. For miRNA detection, the TaqMan MicroRNA Assay Kit (Life Technologies) and miR-93 detection kit (Assay ID: 001090) were used. All quantitative real-time PCR analyses were carried out according to the manufacturer’s instructions. The relative expression values of NEDD4L and miR-93 were calculated and normalized to GAPDH and RNU6B (Assay ID: 001093), respectively, in each sample and compared. The experiments were performed in triplicates.
EMT induction
Of A549 or H1650 cells, 1×105 were seeded in 60-mm dishes. After 24 h, the culture medium was replaced with RPMI1640 medium supplemented with 1 % FBS and then treated with 0.5 or 5.0 ng/ml TGF-β (R&D Systems, Minneapolis, MN), respectively, for 3 days. Cells were photographed under microscope and then trypsinized for cell lysate preparation for immunoblotting analysis.
Statistical analysis
Results are presented as the mean±SD for at least three independent experiments for each group. Statistic differences were determined by using ANOVA or two sample t tests for independent samples. P values less than 0.05 were defined as statistically significant.
Results
miR-93 targets NEDD4L by directly binding to the 3′-UTR of the NEDD4L mRNA
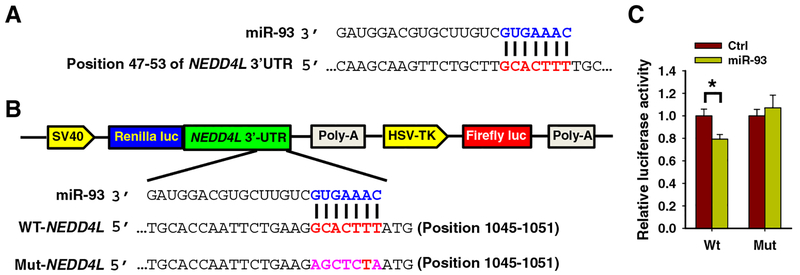
Since the miR-93 expression level is upregulated in NSCLC tissues compared to the corresponding noncancerous lung tissues [6, 18] and higher miR-93 expression correlates with poor outcome of lung cancer patients, we sought to investigate the molecular mechanisms through which miR-93 exerts its oncogenic effects. We screened its potential targets using several bioinformatics tools, including miRDB, miRNA, and TargetScan. Based on the sequence complementarity to miR-93 seed sequence, we chose NEDD4L as our interest of research (Fig. 1a), because NEDD4L has been indicated to regulate TGF-β signal transduction [16, 19], which plays a critical role in promoting lung cancer metastasis. We then determined whether miR-93 is the direct upstream regulator for NEDD4L by using the Dual-Luciferase reporter assay. NEDD4L 3′-UTR with predicted miR-93 binding site or with the mutant binding site was constructed into the psiCHECK2 vector (Fig. 1b). Co-transfection of A549 cells with miR-93 mimics and psiCHECK-2 containing WT-NEDD4L significantly repressed the expression of Renilla luciferase. When cells were transfected with miR-93 mimics and psiCHECK-2 containing Mut-NEDD4L, the expression of Renilla luciferase was not influenced (Fig. 1c). These results demonstrated that miR-93 is able to bind to the 3′-UTR of the NEDD4L mRNA for downregulation of gene expression.
Fig. 1.
miR-93 targets the NEDD4L gene directly by binding to NEDD4L 3′-UTR. a Predicted binding of miR-93 to the 3′-UTR of NEDD4L. b The schematic diagram of the psiCHECK-2-NEDD4L-3′-UTR construct. Sequences were compared between mature miR-93 and the wild-type (WT) or mutant (Mut) putative target sties in the 3′-UTR of NEDD4L. c Dual-luciferase assays in A549 cells. Cells were transfected with psiCHECK-2 containing the WT or Mut target site of the NEDD4L3′- UTR plus miR-93 or non-targeting control (NC) mimic for 48 h. The luciferase activity was normalized to the Firefly activity and presented as relative activity to the corresponding NC. N=3; Bar: SD; *P<0.05
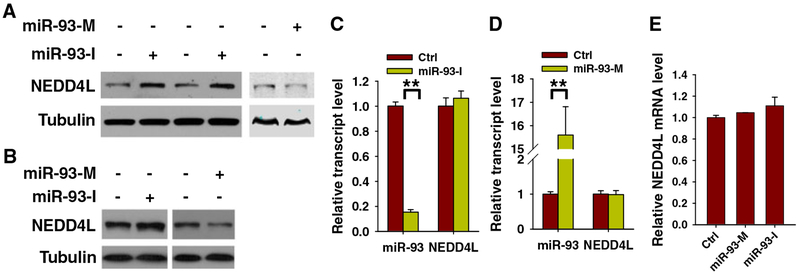
We further determined whether miR-93 downregulates the expression of NEDD4L at both the protein level and mRNA level. To this end, we transfected A549 and H1650 cells with either miR-93 inhibitors (miR-93-I) or miR-93 mimics (miR-93-M). Immunoblotting analyses demonstrated an increase of NEDD4L protein level after inhibition of miR-93, while overexpression of miR-93 decreased NEDD4L protein level in both lung cancer cell lines (Fig. 2a, b). However, although miR-93-M and miR-93-I significantly changed the expression level of miR-93 in lung cancer cells, no change was found in NEDD4L at the mRNA level in both cell lines (Fig. 2c–e). These data indicate that miR-93 may regulate the expression of NEDD4L by interfering with its mRNA translation, but not mRNA stability [20].
Fig. 2.
miR-93 downregulates NEDD4L expression through translational repression. a, bmiR-93 downregulates the NEDD4L protein level. A549 (a) and H1650 (b) cells were transfected with either miR-93 inhibitor (miR-93-I) or miR-93 mimic (miR-93-M) for 48 h, and western blot analyses were carried out to detect the protein level of NEDD4L in these cells. c–e miR-93 does not affect the NEDD4L mRNA level. A549 (c, d) and H1650 (e) cells were transfected with either miR-93 inhibitor or miR-93 mimic for 48 h, and quantitative RT-PCR was conducted to detect the mRNA level of NEDD4L in these cells. N=3; Bar: SD; **P<0.01
Downregulating NEDD4L expression enhanced TGF-β signaling in the lung cancer cell line
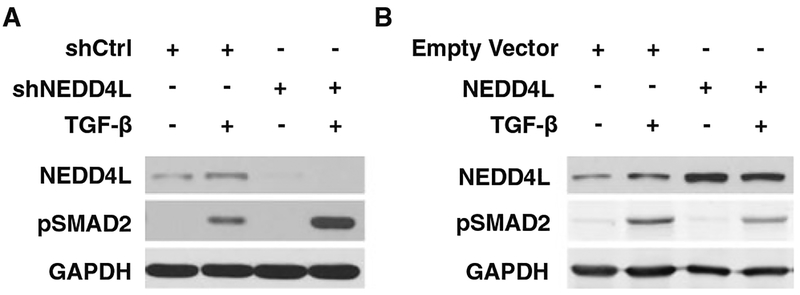
It has been reported that downregulation of NEDD4L promoted TGF-β-induced phosphorylation of SMAD2 in human HaCaT keratinocytes. To determine whether NEDD4L also inhibits the TGF-β signaling in human lung cancer cells, we knocked down or overexpressed NEDD4L in A549 cell, treated with TGF-β, and detected the phosphorylation of SMAD2, a downstream signaling in the TGF-β pathway. As shown in Fig. 3, TGF-β treatment significantly increased the phosphorylation of SMAD2. Downregulation of NEDD4L expression further magnified this TGF-β-induced SMAD2 phosphorylation (Fig. 3a), while overexpression of NEDD4L compromised such TGF-β-induced SMAD2 phosphorylation (Fig. 3b), indicating that NEDD4L plays an inhibitory role in the TGF-β signaling in lung cancer cells.
Fig. 3.
NEDD4L inhibits the TGF-β signaling pathway in lung cancer cells. A549 cells were transfected with either shNEDD4L (a) or NEDD4L-expressing vectors (b) for 48 h and then treated with TGF-β (1 ng/ml) for 4 h. Total cell lysates were prepared and subjected to immunoblotting analysis to determine the phosphorylation of SMAD2 and the expression of NEDD4L. GAPDH was detected as a loading control
miR-93 promotes TGF-β-induced EMT through downregulating NEDD4L expression
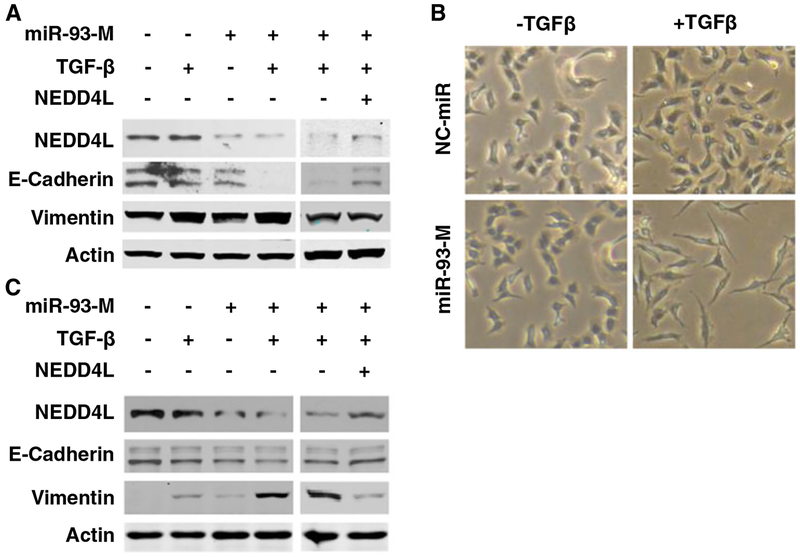
In terms of the finding that NEDD4L is a target of miR-93 and can inhibit TGF-β signaling, as well as the fact that TGF-β is able to induce EMT, we attempted to determine whether miR-93 can also enhance TGF-β-induced EMT in lung cancer cells by downregulating NEDD4L. To this end, we first transfected miR-93-M into A549 cells, treated with TGF-β, and then detected the expression of the epithelial marker E-cadherin and the mesenchymal marker vimentin [21, 22]. As expected, a low protein level of NEDD4L was consistently observed in cells overexpressing miR-93 (Fig. 4a). Although the extremely low dose of TGF-β (0.5 ng/ml) stimulation did not change the expression level of E-cadherin, vimentin expression was increased, indicating that an EMT occurred. Most importantly, A549 cells transfected with miR-93-M displayed a remarkably decreased expression of E-cadherin and an increased expression of vimentin (Fig. 4a, left panel), indicating that miR-93 promotes TGF-β-induced EMT. In addition, miR-93 overexpressing A549 cells exhibited an elongated mesenchymal-like morphology, which is more obvious than control miRNA-transfected cells, upon TGF-β treatment (Fig. 4b), further supporting the promoting role of miR-93 in TGF-β-induced EMT. Interestingly, cotransfection of A549 cells with both miR-93-M and NEDD4L-expressing vector counteracted miR-93-promoted, TGF-β-induced EMT, reflected by increased Ecadherin and decreased vimentin expression (Fig. 4a, right panel). We further repeated this study using another lung cancer cell line H1650. As shown in Fig. 4c, miR-93 overexpression significantly downregulated the expression of NEDD4L and enhanced TGF-β- induced vimentin expression, although their effect on Ecadherin in H1650 cells was not as clear as in A549 cells. Simultaneous transfection with miR-93 mimics and NEDD4L-expressing vectors hampered miR-93-promoted, TGF-β-induced vimentin overexpression as well as E-cadherin down expression. In summary, these data indicate that miR-93 promotes TGF-β-induced EMT by downregulating NEDD4L in lung cancer cells, and the EMT markers may respond differentially in different cancer cell lines.
Fig. 4.
miR-93 promotes TGF-β- induced EMT through downregulating NEDD4L expression. A549 (a, b) and H1650 (c) cells were transfected with miR-93 mimic or transfected with mR-93 mimic and NEDD4L expression construct simultaneously for 24 h, treated with TGF-β (0.5 and 5 ng/ml for A549 and H1650 cells, respectively) for 3 days. Whole cell lysates were prepared, and western blot analysis was carried out to detect the protein level of NEDD4L, E-cadherin, and vimentin. Actin was also detected as a loading control (a, c). Representative phase contrast images of A549 cells after TGF-β treatment were shown (b)
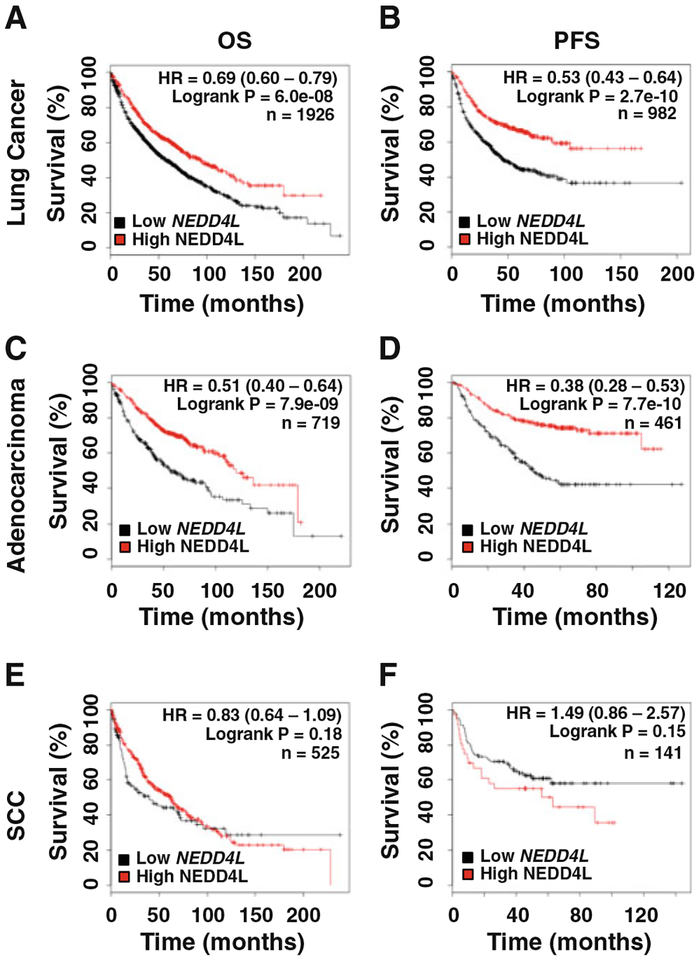
NEDD4L expression levels positively correlate with survival of lung adenocarcinoma patients
A previous study of 79 cases of lung cancer showed that NEDD4L mRNA expression was significantly lower in tumors than in normal lung tissues [13]. To further determine the important role of NEDD4L in lung cancer progression, we analyzed the relationship between the NEDD4L mRNA expression level and the survival rate of different types of lung cancers using publicly available datasets (http://kmplot.com/analysis/index.php?p=service&cancer=lung). Kaplan-Meier analyses demonstrated that lower NEDD4L mRNA expression in lung cancer patients is correlated with a poor patient survival rate in the overall survival (OS), as well as progression-free survival (PFS) of lung cancer patients (Fig. 5a, b). These correlations are more pronounced in patients with adenocarcinoma but not squamous cell carcinoma (Fig. 5c–f). These data further confirmed the tumor suppressor role of NEDD4L in lung adenocarcinoma and supported the negative correlation between the miR-93 expression level and the outcomes of lung cancer patients.
Fig. 5.
The NEDD4L expression level is positively correlated with the survival of lung cancer patients. The effect of NEDD4L mRNA expression level on the overall survival (OS) (a, c, e) and progressionfree survival (PFS) (b, d, f) in all lung cancer patients (a, b), adenocarcinoma (c, d), and squamous cell carcinoma lung cancer patients (e, f) was analyzed, and the Kaplan-Meier plots were generated by Kaplan-Meier Plotter (http://www.kmplot.com)
Discussion
miR-93, belonging to the miR-106b-25 cluster, is a novel oncogenic miRNA. The aberrant expression and dysfunction of miR-93 have been reported in many types of human tumors [23], and the overexpression of miR-93 has been associated with tumor progression, metastasis, and poor prognosis in breast cancer [23], colorectal cancer [24], head and neck squamous cell carcinoma [25], and lung cancer [8]. Through downregulation of multiple tumor suppressor genes, such as FUS1, TP53INP1, EBTB4, and DAB2, miR-93 is able to promote cancer cell growth and invasion [7, 8, 26]. In this study, we demonstrated that miR-93 is able to enhance TGF-β-induced EMT by downregulating NEDD4L, providing a new explanation for the negative correlation between miR-93 expression level and survival of lung cancer patients.
TGF-β is secreted by many cell types including macrophages in the tumor microenvironment. TGF-β is a multifunctional cytokine that regulates cell proliferation, differentiation, and tissue homeostasis [27]. TGF-β acts as a potent driver of cancer progression through the induction of EMT [28], which plays a key role in the early process of metastasis of cancer cells [29], and allowing migration, extravasation, and metastasis dissemination of cancer cells. Thus, restricting the TGF-β signal transduction in cancer cells should prevent tumor metastasis, while enhancing the TGF-β response would promote tumor progression via EMT. Our finding that miR-93 enhances TGF-β-induced EMT in lung cancer cells further supports the oncogenic role of miR-93 in tumors.
miR-93 has been suggested to downregulate TGF-β signaling because miR-93 can reduce the expression of the mRNA for TGF-βR2 in breast cancer cells [30] and nasopharyngeal carcinoma cells [31]. However, we observed an upregulated TGF-β signaling by miR-93 in lung cancer cells, and this can be attributed to the downregulated NEDD4L. Given that miR-93 has multiple targets, which act in different directions in regulating the TGF-β signaling, it is likely that miR-93 may selectively repress certain genes in different tumors.
NEDD4L is identified as a target of miR-93 in this study. Besides its function as a regulator of ion receptors and regulators, NEDD4L has also been demonstrated to downregulate the TGF-β signaling by facilitating the turnover of activated SMAD2/SMAD3 [16]. The NEDD4L-mediated attenuation of the TGF-β signaling could be one of the contributors to the positive correlation between NEDD4L expression and survival of lung cancer patients. In addition, the association of both NEDD4L and miR-93 levels with lung patient survival further demonstrates the relevance of the miR-93/ NEDD4L pathway in lung cancer progression.
By using the luciferase reporter assay, we revealed that the NEDD4L gene is a direct target of miR-93. We have shown that miR-93 bound to the 3′-UTR of the NEDD4L mRNA directly and decreased NEDD4L expression at protein level but had no influence on its mRNA level. Given that miRNAs inhibit protein synthesis either by repressing translation and/or by facilitating degradation of target mRNA [32, 33], our results suggest that miR-93 downregulates the expression of NEDD4L by interfering its translation.
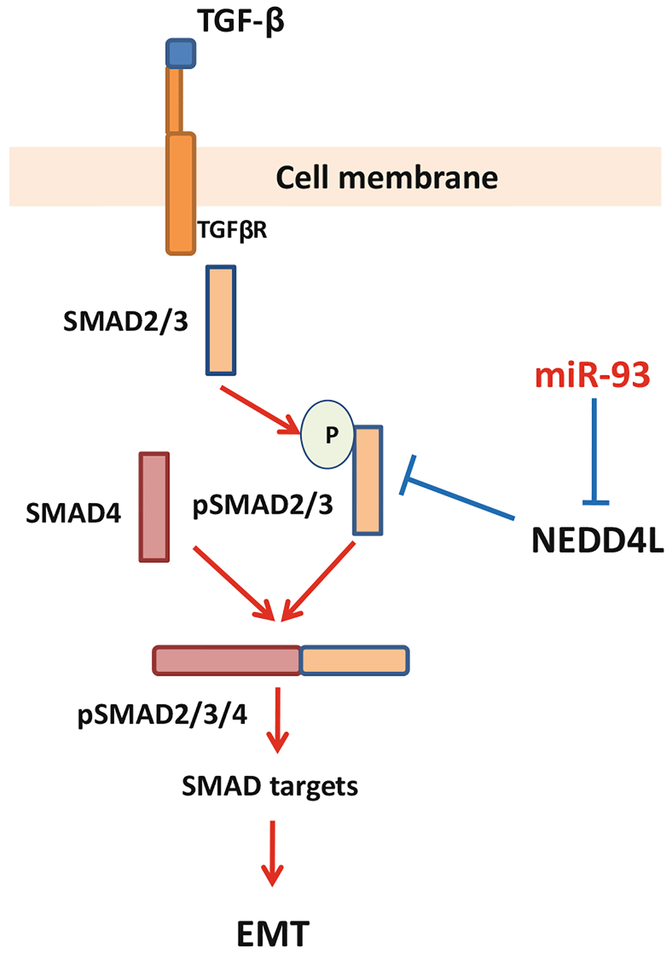
In conclusion, our results demonstrated that miR-93 may exert its oncogenic function to increase lung cancer aggressiveness by downregulating the expression of NEDD4L. Reduced NEDD4L alleviates the degradation of the activated SMAD2/SMAD3, enhancing the TGF-β signal transduction and promoting TGF-β-induced EMT (Fig. 6). Thus, targeting overexpressed miR-93 may serve as a promising therapeutic strategy for treating lung cancer patients.
Fig. 6.
A schematic model of miR-93-enhanced TGF-β signal transduction. TGF-β binds to TGF-βR and activates SMAD2/3 by phosphorylation. The phosphorylated SMAD2/3 can be targeted by NEDD4L for proteolytic degradation, and the TGF-β signal transduction is restricted. miR-93 is able to repress the expression of NEDD4L, thus preventing the degradation of the activated SMAD2/3, leading to an enhanced TGF-β signaling transduction and the subsequent EMT
Acknowledgments
This work was supported by the National Institute of Health (CA151248 to Q.E.W.). This research also received grant support from Shandong Provincial Education Association for International Exchanges (M. H. Q.) and the Shandong Provincial Natural Science Foundation, China, ZR2011HM043 (Z. G.).
Footnotes
Conflicts of interest None
References
- 1.Siegel RL, Miller KD, Jemal A: Cancer statistics, 2015. CA Cancer J Clin 2015 [DOI] [PubMed] [Google Scholar]
- 2.Robinson KW, Sandler AB. The role of MET receptor tyrosine kinase in non-small cell lung cancer and clinical development of targeted anti-MET agents. Oncologist. 2013;18:115–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng Y, Thiagarajan PS, Ma PC. MET signaling: novel targeted inhibition and its clinical development in lung cancer. J Thorac Oncol. 2012;7:459–67. [DOI] [PubMed] [Google Scholar]
- 4.Nichols L, Saunders R, Knollmann FD. Causes of death of patients with lung cancer. Arch Pathol Lab Med. 2012;136:1552–7. [DOI] [PubMed] [Google Scholar]
- 5.Petrocca F, Vecchione A, Croce CM. Emerging role of miR-106b- 25/miR-17–92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 2008;68:8191–4. [DOI] [PubMed] [Google Scholar]
- 6.Zhu W, He J, Chen D, Zhang B, Xu L, Ma H, et al. Expression of miR-29c, miR-93, and miR-429 as potential biomarkers for detection of early stage non-small lung cancer. PLoS One. 2014;9, e87780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du L, Schageman JJ, Subauste MC, Saber B, Hammond SM, Prudkin L, et al. Pertsemlidis A: miR-93, miR-98, and miR-197 regulate expression of tumor suppressor gene FUS1. Mol Cancer Res. 2009;7:1234–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du L, Zhao Z, Ma X, Hsiao TH, Chen Y, Young E, et al. miR-93 directed downregulation of DAB2 defines a novel oncogenic pathway in lung cancer. Oncogene. 2014;33:4307–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savita U, Karunagaran D. MicroRNA-106b-25 cluster targets beta- TRCP2, increases the expression of Snail and enhances cell migration and invasion in H1299 (non small cell lung cancer) cells. Biochem Biophys Res Commun. 2013;434:841–7. [DOI] [PubMed] [Google Scholar]
- 10.Goel P, Manning JA, Kumar S. NEDD4–2 (NEDD4L): the ubiquitin ligase for multiple membrane proteins. Gene. 2015;557:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dahlberg J, Nilsson LO, von WF, Melander O. Polymorphism in NEDD4L is associated with increased salt sensitivity, reduced levels of P-renin and increased levels of Nt-proANP. PLoS One. 2007;2, e432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russo CJ, Melista E, Cui J, DeStefano AL, Bakris GL, Manolis AJ, et al. Association of NEDD4L ubiquitin ligase with essential hypertension. Hypertension. 2005;46:488–91. [DOI] [PubMed] [Google Scholar]
- 13.Sakashita H, Inoue H, Akamine S, Ishida T, Inase N, Shirao K, et al. Identification of the NEDD4L gene as a prognostic marker by integrated microarray analysis of copy number and gene expression profiling in non-small cell lung cancer. Ann Surg Oncol. 2013;20 Suppl 3:S590–8. [DOI] [PubMed] [Google Scholar]
- 14.Kovacevic Z, Chikhani S, Lui GY, Sivagurunathan S, Richardson DR. The iron-regulated metastasis suppressor NDRG1 targets NEDD4L, PTEN, and SMAD4 and inhibits the PI3K and Ras signaling pathways. Antioxid Redox Signal. 2013;18:874–87. [DOI] [PubMed] [Google Scholar]
- 15.Tanksley JP, Chen X, Coffey RJ. NEDD4L is downregulated in colorectal cancer and inhibits canonical WNT signaling. PLoS One. 2013;8, e81514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao S, Alarcon C, Sapkota G, Rahman S, Chen PY, Goerner N, et al. Ubiquitin ligase Nedd4L targets activated SMAD2/3 to limit TGF-beta signaling. Mol Cell. 2009;36:457–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui T, Srivastava AK, Han C, Yang L, Zhao R, Zou N, et al. XPC inhibits NSCLC cell proliferation and migration by enhancing ECadherin expression. Oncotarget. 2015;6:10060–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang YK, Zhu WY, He JY, Chen DD, Huang YY, Le HB, et al. miRNAs expression profiling to distinguish lung squamous-cell carcinoma from adenocarcinoma subtypes. J Cancer Res Clin Oncol. 2012;138:1641–50. [DOI] [PubMed] [Google Scholar]
- 19.He S, Deng J, Li G, Wang B, Cao Y, Tu Y. Down-regulation of Nedd4L is associated with the aggressive progression and worse prognosis of malignant glioma. Jpn J Clin Oncol. 2012;42: 196–201. [DOI] [PubMed] [Google Scholar]
- 20.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–79. [DOI] [PubMed] [Google Scholar]
- 21.Thomson S, Buck E, Petti F, Griffin G, Brown E, Ramnarine N, et al. Epithelial to mesenchymal transition is a determinant of sensitivity of non-small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer Res. 2005;65: 9455–62. [DOI] [PubMed] [Google Scholar]
- 22.Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith AL, Iwanaga R, Drasin DJ, Micalizzi DS, Vartuli RL, Tan AC, et al. The miR-106b-25 cluster targets SMAD7, activates TGF-beta signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer. Oncogene. 2012;31:5162–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang Q, Zou Z, Zou C, Zhang Q, Huang R, Guan X, et al. MicroRNA-93 suppress colorectal cancer development via Wnt/ beta-catenin pathway downregulating. Tumour Biol. 2015;36: 1701–10. [DOI] [PubMed] [Google Scholar]
- 25.Li G, Ren S, Su Z, Liu C, Deng T, Huang D, et al. Increased expression of miR-93 is associated with poor prognosis in head and neck squamous cell carcinoma. Tumour Biol. 2015;36: 3949–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeung ML, Yasunaga J, Bennasser Y, Dusetti N, Harris D, Ahmad N, et al. Roles for microRNAs, miR-93 and miR-130b, and tumor protein 53-induced nuclear protein 1 tumor suppressor in cell growth dysregulation by human T-cell lymphotrophic virus 1. Cancer Res. 2008;68:8976–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Massague J TGFbeta in cancer. Cell. 2008;134:215–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–58. [DOI] [PubMed] [Google Scholar]
- 29.Gavert N, Ben-Ze’ev A. Epithelial-mesenchymal transition and the invasive potential of tumors. Trends Mol Med. 2008;14:199–209. [DOI] [PubMed] [Google Scholar]
- 30.Liu S, Patel SH, Ginestier C, Ibarra I, Martin-Trevino R, Bai S, et al. MicroRNA93 regulates proliferation and differentiation of normal and malignant breast stem cells. PLoS Genet. 2012;8, e1002751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyu X, Fang W, Cai L, Zheng H, Ye Y, Zhang L, et al. TGFbetaR2 is a major target of miR-93 in nasopharyngeal carcinoma aggressiveness. Mol Cancer. 2014;13:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14. [DOI] [PubMed] [Google Scholar]
- 33.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. [DOI] [PubMed] [Google Scholar]