Abstract
Coelenterazine (CTZ)-utilizing marine luciferases and their derivatives have attracted significant attention because of their ATP-independency, fast enzymatic turnover, and high bioluminescence brightness. However, marine luciferases typically emit blue photons and their substrates, including CTZ and the recently developed diphenylterazine (DTZ), have poor water solubility, hindering their in vivo applications. Herein, we report a family of pyridyl CTZ and DTZ analogs that exhibit spectrally shifted emission and improved water solubility. Through directed evolution, we engineered a LumiLuc luciferase with broad substrate specificity. In the presence of corresponding pyridyl substrates (i.e., pyCTZ, 6pyDTZ, or 8pyDTZ), LumiLuc generates highly bright blue, teal, or yellow bioluminescence. We compared our LumiLuc-8pyDTZ pair with several benchmark reporters in a tumor xenograft mouse model. Our new pair, which does not need organic cosolvents for in vivo administration, surpasses other reporters by detecting early tumors. We further fused LumiLuc to a red fluorescent protein, resulting in a LumiScarlet reporter with further red-shifted emission and enhanced tissue penetration. LumiScarlet-8pyDTZ was comparable to Akaluc-AkaLumine, the brightest ATP-dependent luciferase-luciferin pair, for detecting cells in deep tissues of mice. In summary, we have engineered a new family of ATP-independent bioluminescent reporters, which will have broad applications because of their ATP-independency, excellent biocompatibility, and superior in vivo sensitivity.
INTRODUCTION
In the past few decades, fluorescence imaging has evolved quickly and become a dominant visualization method for live-cell studies.1 However, fluorescence imaging has several limitations, such as photobleaching, phototoxicity, and poor tissue penetration, largely due to the need for light excitation. Unlike fluorescence, bioluminescence produces photons via enzyme-catalyzed biochemical reactions in which luciferases oxidize their corresponding small-molecule substrates (a.k.a., luciferins) to generate excited-state emitters. As a result, bioluminescence signals glow essentially on dark background, leading to excellent signal-to-background ratios. Moreover, even though the spatiotemporal resolution of bioluminescence imaging (BLI) is usually worse than that of fluorescence imaging, the emitted photons can escape through several centimeters of tissue.2 BLI is thus especially suited for diverse, noninvasive in vivo imaging applications.3-6
Photinus pyralis firefly luciferase (FLuc) and d-luciferin (λmax: 563 nm) constitute the most widely used luciferase-luciferin pair for in vivo BLI. Recently, research has been performed to develop FLuc and d-luciferin derivatives for brighter and more red-shifted emission. In particular, an Akaluc-AkaLumine luciferase-luciferin pair with near-infrared (NIR) emission (λmax: 650 nm) was reported for highly sensitive deep-tissue in vivo BLI.7 Despite the progress, AkaLumine has been shown to induce cytotoxicity.8–10 Moreover, FLuc, Akaluc, and other insect luciferases consume ATP for photon production; the bioluminescence reaction between FLuc and d-luciferin reduced the intracellular ATP-to-ADP ratio of live mammalian cells from > 40:1 to ~ 20:1,9 suggesting metabolic disruption by all ATP-dependent luciferases. Because ATP is required for the activation of the luciferin substrates, this metabolic disruption issue cannot be addressed by simply improving insect luciferases and the corresponding substrates.
In contrast to insect luciferases, a large family of marine luciferases and photoproteins, such as Renilla luciferase (RLuc), Gaussia luciferase (GLuc), Oplophorus luciferase (OLuc), and aequorin, are ATP-independent and use coelenterazine (CTZ, Figure 1) as their native substrate for bioluminescence production.11 The 19kDa catalytic domain of OLuc12 was recently engineered into NanoLuc, which has a fast enzyme turnover and produces intense blue bioluminescence (λmax: 456 nm) in the presence of a synthetic CTZ analog, furimazine (FRZ, Figure 1).13 To expand the color palette, NanoLuc was further engineered into teLuc, which emits red-shifted photons (λmax: 502 nm) when paired with a synthetic substrate diphenylterazine (DTZ, Figure 1).8 Since biological tissues significantly absorb and scatter short-wavelength photons,14 NanoLuc and teLuc have been fused to fluorescent proteins, resulting in Antares, Antares2, and enhanced Nano-Lanterns for further red-shifted emission via bioluminescence resonance energy transfer (BRET).8, 15, 16 The water solubility of CTZ, FRZ, and DTZ is adequate for protein- and cell-based assays, because their solubility is already higher than typical substrate concentrations in these in vitro assays. However, in vivo applications of existing ATP-independent bioluminescent reporters are greatly hindered by the low solubility of CTZ, FRZ, and DTZ. Small animals, such as mice, can only tolerate a small injection volume. To enhance bioluminescence brightness by delivering more luciferin substrate, hydroxypropyl-β-cyclodextrin, polyethylene glycols (PEGs), or other organic cosolvents are typically used to formulate CTZ, FRZ, or DTZ for in vivo administration.8, 15, 17, 18 These formulation ingredients are not biologically inert and can cause irritation or biotoxicity. It is also practically difficult to intravenously (i.v.) inject these highly viscous solutions into small animals. Furthermore, it is still of great interest to further red-shift marine luciferases for enhanced tissue penetration.
Figure 1.
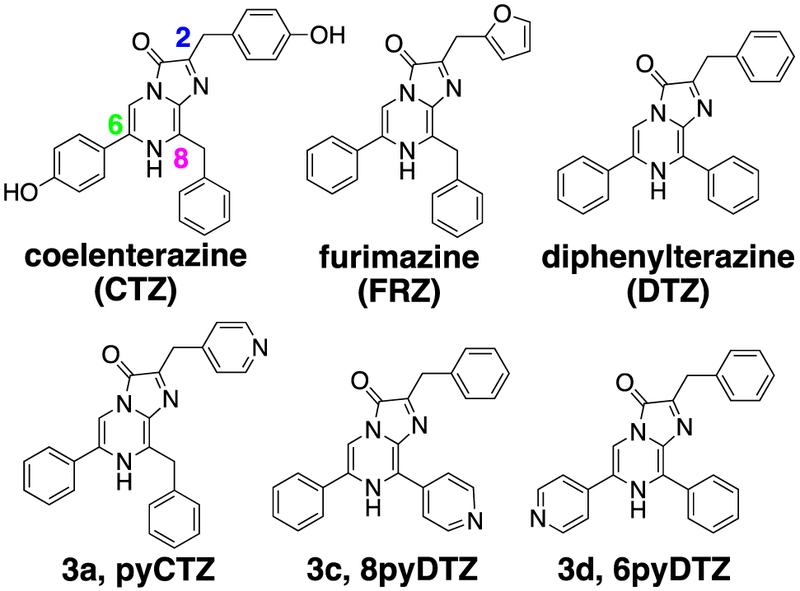
Chemical structures of coelenterazine (CTZ), furimazine (FRZ), diphenylterazine (DTZ), pyCTZ (3a), 8pyDTZ (3c) and 6pyDTZ (3d).
In this work, we chemically modified CTZ and DTZ for spectrally shifted emission and enhanced water solubility. Concurrently, we engineered teLuc into a LumiLuc luciferase, which is highly active toward the new substrates for intense blue, teal, and yellow emission. Moreover, by harnessing a recently reported high-quality red fluorescent protein, mScarlet-I, we developed a LumiScarlet reporter with significant emission longer than 600 nm. Our multipronged approach yielded a new family of ATP-independent bioluminescent reporters, which have improved biochemical and photophysical properties and are expected to have broad applications.
RESULTS
Design and synthesis of pyridyl CTZ and DTZ analogs with enhanced water solubility
Despite that recent studies have synthesized and tested a number of CTZ analogs with NanoLuc,19, 20 the luciferase has not yet been optimized to pair with these new substrates and the water solubility issue of the substrates has not yet been tackled systematically. We sought to develop CTZ and DTZ analogs with improved water solubility by using the concept of bioisostere replacements in medicinal chemistry. Pyridine is considered a biocompatible N-heterocycle substituent for benzene with enhanced water solubility, because pyridine-containing molecules can be readily turned into pyridinium salts. Therefore, we designed a convergent synthetic route to prepare a series of CTZ and DTZ analogs with pyridyl isomer substitutions at the C-2, C-6 and C-8 positions of the imidazopyrazinone core (Scheme 1). Briefly, we first used Suzuki or Negishi cross-coupling reactions to regioselectively functionalize 2-amino-3,5-dibromopyrazine with either pyridyl, phenyl, or benzyl functional groups to give monosubstituted products (1a-c), which were subsequently derivatized via Suzuki cross-coupling reactions to afford disubstituted intermediates (2a-f, see structures and synthetic methods in the Supporting Information). In the second cross-coupling step, the XPhos-Pd-G2 catalyst was used to enhance reaction yields and minimize the protodeboronation of pyridyl boronic acids.21 We further utilized an acid-catalyzed ring closing reaction22 in dioxane to derive various pyridyl CTZ and DTZ analogs (3a-f, Table 1) from the disubstituted intermediates and corresponding α-ketoacetals.
Scheme 1. Synthesis of pyridyl CTZ and DTZ analogs.
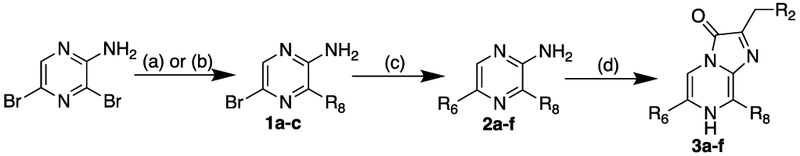
(a) Suzuki coupling: Pd(PPh3)4, Na2CO3, R8-B(OH)2, and EtOH; (b) Negishi coupling: PhCH2MgCl, ZnCl2, (PPh3)2PdCl2, and THF; (c) Suzuki coupling: XPhos-Pd-G2, Na2CO3, R6-B(OH)2, and EtOH; (d) Acid-catalyzed ring closing: corresponding α-ketoacetal, HCl, and dioxane.
Table 1.
Chemical and photoluminescence properties of synthetic pyridyl CTZ and DTZ analogs.
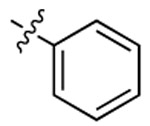
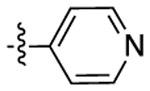
| Compound | R6 | R8 | R2 | Bioluminescence a λmax (nm) |
Chemiluminescence b λmax (nm) |
Water Solubility (μM) |
|---|---|---|---|---|---|---|
| 3a |  |
 |
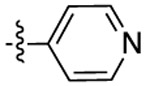 |
451 | 505 | 1416 |
| 3b | 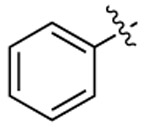 |
 |
 |
497 | 506 | 1813 |
| 3c | 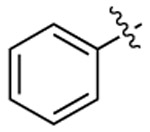 |
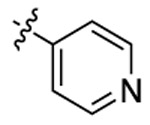 |
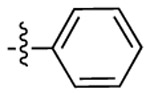 |
532 | 555 | 1711 |
| 3d |  |
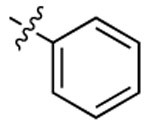 |
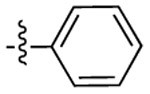 |
483 | 492 | 1736 |
| 3e | 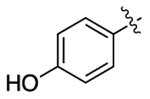 |
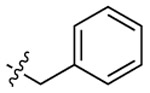 |
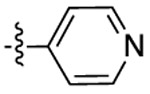 |
450 | 465 | 987 |
| 3f | 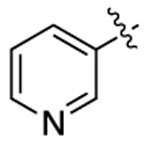 |
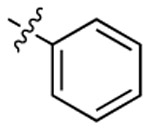 |
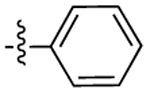 |
518 | 503 | 1562 |
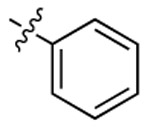
| CTZ | 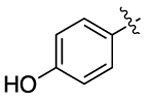 |
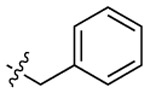 |
 |
455 | 461 | 256 |
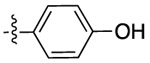
| DTZ | 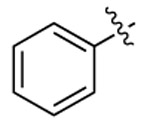 |
 |
 |
502 | 510 | 131 |
Note:
Determined with 1 nM teLuc in PBS;
Triggered by peroxymonocarbonate formed in situ.
We next used turbidimetric solubility assays23 to evaluate water solubility of these CTZ and DTZ analogs (Table 1 and Figure S1). Our newly synthesized pyridyl analogs enhanced the solubility by 4- to 14-fold from CTZ and DTZ. We also evaluated the autoluminescence and stability of these new analogs and they are comparable or even better than CTZ and FRZ (Figure S2). We further investigated their chemiluminescence, because the wavelength of bioluminescence is often related to the wavelength of substrate chemiluminescence, although the luciferase enzyme provides further electrostatic tuning which can reshape the emission (Figures S3). In addition, we evaluated the bioluminescence of these new substrates in the presence of several representative, ATP-independent luciferases such as RLuc8, NanoLuc, teLuc, and aequorin (Figures S4). Although each luciferase has different substrate preferences, the compound 3a (pyCTZ) generated strong blue bioluminescence in the presence of each of these tested luciferases. When paired with aequorin, the bioluminescence intensity of pyCTZ was comparable to native CTZ, suggesting that pyCTZ may be directly used to replace CTZ for aequorin-based calcium sensing.24 Furthermore, compared to DTZ, compounds 3c (8pyDTZ), and 3f were able to emit red-shifted chemiluminescence and/or bioluminescence, while 3b and 3d (6pyDTZ) caused hypsochromic shift (Figure S3 and Table 1). Molecular mechanisms governing the spectral shift properties of these synthetic substrates remain to be investigated. Because 3c showed the most red-shifted emission and red-shifted photons can penetrate through tissue better,25 we selected 3c (8pyDTZ) as our candidate substrate for further development of an optimized, red-shifted luciferase-luciferin pair.
Directed evolution of the teLuc luciferase for improved brightness
teLuc was previously optimized for DTZ, a substrate with conjugated disubstitutions on the imidazopyrazinone core. 8pyDTZ exhibits ~ 30 nm red-shift but the emission of teLuc-8pyDTZ has been greatly attenuated compared to teLuc-DTZ. We next engineered teLuc for increased photon flux in the presence of 8pyDTZ. On the basis of a published apo-nanoKAZ structure26 and our computational model,8 we first introduced random mutations to residues 18 and 19 close to a putative substrate-binding pocket (Figure 2). After screening for improved mutants, we further randomized residues 27, 28, and 29 located deeper in the putative catalytic site. From the first two rounds of protein engineering, a teLuc-L18Q/S19A/V27L/S28T mutant was identified, to which we further introduced random mutations using error-prone PCR. After eight additional rounds of mutagenesis and screening, we derived a LumiLuc luciferase with 12 total mutations and ~ 5-fold enhancement of 8pyDTZ bioluminescence from teLuc (Figures 2 and S5).
Figure 2. Engineering of a LumiLuc luciferase.

(a) Procedure to derive LumiLuc from teLuc. (b) Illustration of the putative substrate-binding site and LumiLuc mutations from teLuc. CTZ is shown as spheres and mutated residues are presented in sticks. (c) Bioluminescence of pyCTZ, 6pyDTZ, or 8pyDTZ in the presence of either teLuc or LumiLuc. (d) Normalized bioluminescence emission spectra of pyCTZ, 6pyDTZ, and 8pyDTZ in the presence of LumiLuc.
The resultant LumiLuc-8pyDTZ pair has an emission peak at 525 nm. Its in vitro maximal photon emission rate (Vmax) is ~ 60% and ~ 36% of NanoLuc-FRZ and teLuc-DTZ, respectively. The apparent Michaelis constant (KM) of LumiLuc-8pyDTZ was 4.6 μM, lower than that of teLuc-DTZ or NanoLuc-FRZ (Figure S6a). This reduced KM is practically beneficial, since LumiLuc-8pyDTZ would be relatively brighter when effective substrate concentrations are limited, such as in live cells (Figure S6b) and in vivo. Similar to NanoLuc and teLuc, the bioluminescence kinetics of LumiLuc is flash-type in phosphate buffer saline (PBS) and glow-type in a specially formulated assay buffer (Figure S6c).13
LumiLuc has broad substrate specificity. It improved the photon flux of 3a (pyCTZ) and 3d (6pyDTZ) from teLuc by ~ 120% and ~ 150%, respectively (Figure 2c). The directed evolution process to enhance photon flux of teLuc for 8pyDTZ did not preclude the luciferase from catalyzing other structurally relevant substrates. LumiLuc is capable of efficiently generating blue, teal, or yellow bioluminescence when paired with pyCTZ, 6pyDTZ or 8pyDTZ (λmax: 450, 476, and 525 nm, respectively; Figure 2d), thereby leading to a new family of ATP-independent bioluminescent reporters with water-soluble substrates.
LumiLuc-8pyDTZ in cultured mammalian cells
We next evaluated LumiLuc-8pyDTZ in human embryonic kidney (HEK) 293T cells transiently expressing the luciferase (Figure S7). The LumiLuc-8pyDTZ pair produced ~ 3- to 5-fold more bioluminescence than teLuc-8pyDTZ at all tested substrate concentrations (Figure S8). Moreover, despite that LumiLuc-8pyDTZ is less bright than teLuc-DTZ at saturated substrate concentrations, LumiLuc-8pyDTZ is notably brighter than teLuc-DTZ at low substrate concentrations (from 6.25 to 25 μM; Figure 3a). Far-red emission at wavelengths longer than 600 nm is more indicative of the in vivo performance of bioluminescent reporters, because mammalian tissue is more transparent in this spectral region.25 To compare far-red emission intensities of bioluminescent reporters, we imaged HEK 293T cells in the presence of a 600–700 nm bandpass filter. At substrate concentrations from 6.25 to 100 μM, LumiLuc-8pyDTZ consistently produces 1.6- to 3.9-fold higher photon flux than teLuc-DTZ (Figure 3b).
Figure 3. Bioluminescence of teLuc- and LumiLuc-expressing HEK 293T cells.
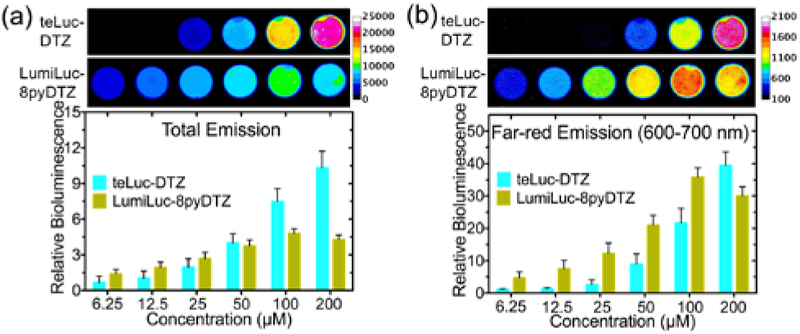
Images were acquired (a) without or (b) with a 600-700 nm bandpass filter. Values for relative brightness were normalized to teLuc in the presence of 6.25 μM DTZ.
ATP-dependent luciferases, such as FLuc and Akaluc consumes one ATP molecule in each catalytic cycle, leading to metabolic disruption.9 Instead, ATP-independent LumiLuc does not use ATP for catalysis. We monitored ATP/ADP ratios in live HEK 293T cells using PercevalHR, a previously reported fluorescent ATP/ADP biosensor.27 No ATP perturbation was observed from 8pyDTZ-treated, LumiLuc-expressing cells (Figure S9).
LumiLuc-8pyDTZ to track tumor growth in a mouse xenograft model
BLI has been a popular imaging modality for various animal models.14, 25 The recently reported Akaluc-AkaLumine and Antares2-DTZ pairs are two benchmark reporters for in vivo BLI.7, 8 We adapted a biologically-relevant tumor xenograft mouse model28 to compare these bioluminescent reporters. We first generated cervical cancer HeLa cell lines stably expressing individual luciferases, including teLuc, Antares2, LumiLuc, and Akaluc (Figure S10). Next, we injected 104 or 105 luciferase-expressing HeLa cells into the left or right dorsolateral trapezius and thoracolumbar regions of immunodeficient NU/J mice (day 0) and monitored tumor growth over 4 weeks. Bioluminescence was quantified in days 1, 3, 5, 7, 14, and 28 after tail vein injection of corresponding substrates. AkaLumine-HCl was delivered at a dose of 1.5 μmol per mouse. This dosage (~ 75 nmol/g), which is normalized against the body weights of mice, is identical to the previously reported dosage.7 Moreover, when 3 μmol of AkaLumine-HCl per mouse (~ 150 nmol/g) was used, we observed death for 2 out of 3 mice in our pilot experiment. 8pyDTZ was dissolved in normal saline to its saturation concentration and intravenously injected, resulting in a dose of 0.2 μmol per mouse (~ 10 nmol/g). The LumiLuc-8pyDTZ pair showed detectable bioluminescence on day 1 at sites inoculated with 104 cells, and kept exhibiting ~ 3-fold higher photon flux over Akaluc-AkaLumine up to day 7 (Figures 4a and 4b). The signals for Akaluc-AkaLumine at sites inoculated 104 cells were not consistently higher than background until day 3. Furthermore, the in vivo brightness of LumiLuc-8pyDTZ is comparable to, if not higher than, the Antares2-DTZ pair (Figures 4 and S11), despite the fact that the majority of emitted photons from LumiLuc-8pyDTZ has not yet exceeded 600 nm. These data collectively support that LumiLuc-8pyDTZ is a superior bioluminescent reporter system for high-sensitivity in vivo BLI.
Figure 4. Tracking of tumor growth in a xenograft mouse model with various luciferase-luciferin pairs.
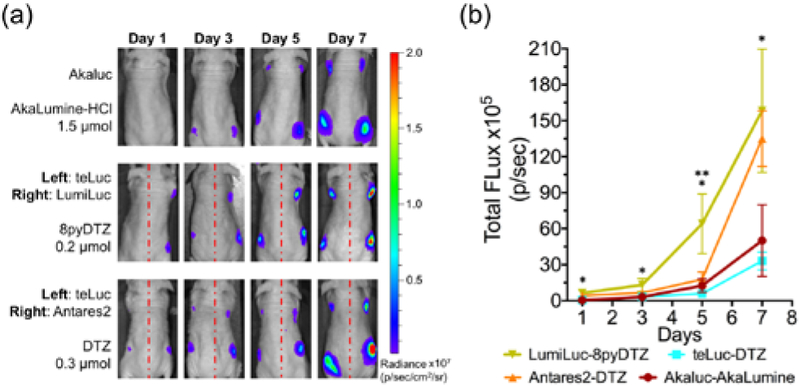
(a) BLI (n = 4) on day 1, 3, 5, and day 7. 104 luciferase-expressing HeLa cells were injected to the left and right dorsolateral trapezius regions and 105 cells were injected to the left and right dorsolateral thoracolumbar regions of NU/J mice. For i.v. administration of substrates, AkaLumine-HCl (1.5 μmol/mouse) and 8pyDTZ (0.2 μmol/mouse) were dissolved in normal saline, and DTZ (0.3 μmol/mouse) was formulated with a mixture of organic cosolvents. (b) Comparison of luciferase-luciferin pairs at tumor sites inoculated with 104 cells. (*p < 0.05 for LumiLuc-8pyDTZ and teLuc-DTZ, and for LumiLuc-8pyDTZ and Akaluc-AkaLumine; **p < 0.05 for LumiLuc-8pyDTZ and Antares2-DTZ).
The bioluminescence of Akaluc-AkaLumine eventually surpassed LumiLuc-8pyDTZ from day 14 (Figure S11). In addition to differences in biodistribution and pharmacokinetic properties of AkaLumine and 8pyDTZ, we interpret that 8pyDTZ may be a limiting reagent in large tumors because AkaLumine-HCl could be delivered into mice at a much higher dose than 8pyDTZ due to the higher solubility of AkaLumine-HCl. Presumably, we may further enhance the in vivo performance of marine luciferases and their derivatives by further increasing the water solubility and thus the administration dosage of CTZ and DTZ analogs.
Engineering of BRET-based LumiScarlet for deep-tissue BLI
mScarlet-I is a recently reported red fluorescent protein with high quantum yield and excellent performance as a Förster resonance energy transfer (FRET) acceptor.29 We thus hypothesized that LumiLuc could be genetically fused to mScarlet-I for BRET, thereby red-shifting the emission of LumiLuc. We explored several fusion strategies between LumiLuc and mScarlet-I, constructed libraries by randomizing the linkers, and screened for mutants with high BRET efficiency (Figures S12a and S12b). We identified a mutant, namely LumiScarlet (Figure 5a), which is a fusion protein of mScarlet-I (residues 1–225) linked to the N-terminus of LumiLuc (residues 2–169) through a single-residue “Lys” linker.
Figure 5. BRET-based LumiScarlet for deep tissue BLI.
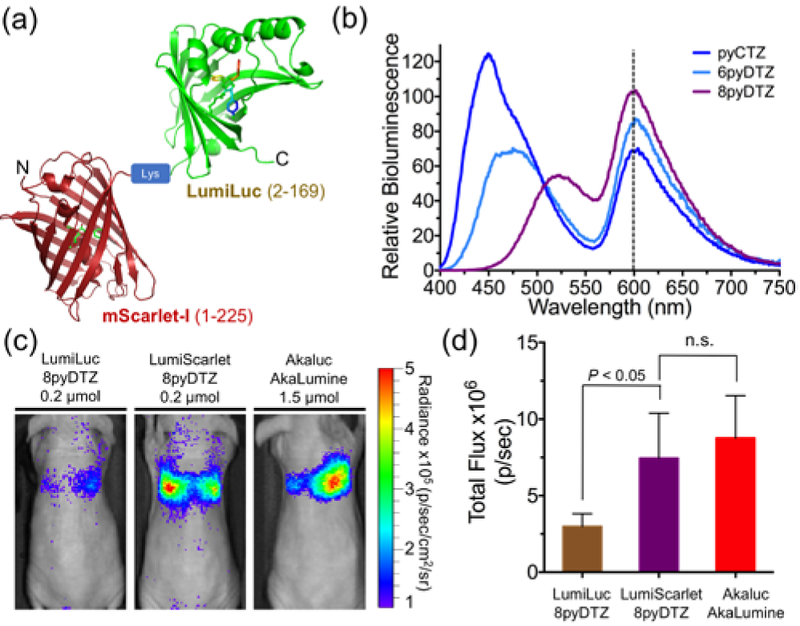
(a) Schematic diagram of LumiScarlet, a genetic fusion of mScarlet-I and LumiLuc. (b) Bioluminescence emission of LumiScarlet in the presence of pyCTZ, 6pyDTZ, or 8pyDTZ, showing significant emission longer than 600 nm. (c) Comparison of LumiLuc-8pyDTZ, LumiScarlet-8pyDTZ, and Akaluc-AkaLumine in NU/J mice (n = 4) at 4 h post i.v. injection of 106 luciferase-expressing HeLa cells. (d) Quantitative analysis of bioluminescence from the regions around the lungs in panel c (n.s.: not significant).
High BRET efficiency was achieved with LumiScarlet in the presence of either pyCTZ, or 6pyDTZ, or 8pyDTZ (Figure 5b). In particular, because the emission spectrum of LumiLuc-8pyDTZ overlaps well with the excitation spectrum of mScarlet-I (Figure S12c), ~ 51% of the total emission of LumiScarlet, when paired with 8pyDTZ, was longer than 600 nm (Table S1).
We next compared our newly engineered LumiLuc-8pyDTZ and LumiScarlet-8pyDTZ with Akaluc-AkaLumine for deep-tissue BLI. We injected a million HeLa cells stably expressing corresponding luciferases into each of NU/J mice via tail vein and performed BL imaging 4 h later. Immunodeficient mice were used here to minimize immune responses to HeLa cells, so that signals will be mostly from live cells trapped in the lungs. LumiScarlet gave ~ 3-fold higher detectable signals than LumiLuc under this condition (Figure 5c and 5d), even though the in vitro brightness of LumiScarlet is only ~ 70% of LumiLuc (Figure S12d). Moreover, the signals from LumiScarlet-8pyDTZ were comparable to the signals from Akaluc-AkaLumine.
We want to note that we observed some diffuse signals from areas other than the lungs. These signals were not caused by substrate background, as injection of 8pyDTZ into blank mice (Figure S13a) resulted in only weak background much lower than what we observed in Figure 5c. Luciferase activities were detectable in blood after tail vein injection of luciferase-expressing HeLa cells (Figure S13b), suggesting partial lysis of luciferase-labeled cells during cell injection. Also, in contrast to ATP-independent LumiLuc and LumiScarlet, ATP-dependent Akaluc is enzymatically inactive in serum because of relatively low ATP levels and further deactivation by serum components.9 Thus, diffuse signals from LumiScarlet in Figure 5c were likely from the LumiScarlet luciferase released into blood.
Collectively, our deep-tissue BLI results confirm that red-shifted BRET-based LumiScarlet has better mammalian tissue penetration than LumiLuc. Moreover, LumiScarlet-8pyDTZ is a novel, ATP-independent bioluminescent reporter with exceptional deep-tissue BLI performance comparable to ATP-dependent Akaluc-AkaLumine.
DISCUSSION
Conventionally, ATP-dependent bioluminescent reporters, such as FLuc and Akaluc, are considered to be more useful for in vivo BLI than ATP-independent marine luciferases, because the emission of ATP-dependent insect luciferases is often at the red end of the visible spectrum where the mammalian tissue is relatively transparent. However, these insect luciferases require ATP and Mg2+ for bioluminescence. The ATP- and Mg2+-dependency is sometimes problematic because ATP and Mg2+ levels may vary under different biological circumstances.30 In particular, ATP-dependent luciferases are inactive in extracellular space and common biological fluids such as blood and urine, where ATP accessibility is limited.9 Moreover, ATP-dependent luciferases consume ATP in bioluminescence reactions and may cause concerns such as metabolic disruption.9 In contrast, most ATP-independent marine luciferases are enzymatically active in extracellular space and common biological fluids; they do not consume ATP for bioluminescence. Furthermore, some marine luciferase derivatives have fast catalytic turnover and thus give high photon flux. It is therefore not surprising that marine luciferase and their derivatives, such as NanoLuc and Gaussia luciferase, have been widely used for in vitro bioluminescence assays. Currently, the in vivo applications of marine luciferases are hindered by their blue emission and poor substrate water solubility. In this study, we combined chemical synthesis and protein engineering approaches to enhance ATP-independent marine luciferases for in vivo BLI by developing red-shifted colors and water-soluble substrates.
First, we prepared a series of pyridyl CTZ and DTZ analogs with diverse emission profiles. The water solubility of these synthetic analogs generally increased by ~10-fold from their ancestors. These substrate analogs can not only be paired with the new luciferases engineered here, but also existing ATP-independent reporters, such as RLuc and aequorin.
We further engineered a luciferase for the 8pyDTZ substrate via directed protein evolution. The resultant LumiLuc-8pyDTZ bioluminescent reporter system exhibited reduced KM and red-shifted emission. These factors favored in vivo BLI. As a result, LumiLuc-8pyDTZ showed high sensitivity in a mouse xenograft model. In addition, LumiLuc-8pyDTZ did not perturb the intracellular ATP/ADP level, and 8pyDTZ could be dissolved up to ~ 2 mM in low-viscosity saline without using irritative and toxic organic cosolvent. Therefore, our effort enhanced not only the biocompatibility of bioluminescent reporters, but also reproducibility for intravenous injections.
Furthermore, we developed a BRET-based LumiScarlet reporter for further red-shifted emission. The emission of LumiLuc-8pyDTZ overlaps well with the excitation of mScarlet-I, an excellent red-emitting resonance energy transfer acceptor. LumiScarlet-8pyDTZ exhibited high brightness, significant emission longer than 600 nm, and excellent tissue penetration. LumiScarlet-8pyDTZ was comparable to NIR-emitting Akaluc-AkaLumine in a mouse model for deep-tissue BLI. Moreover, because LumiScarlet is enzymatically active in blood, we envision that it would be an excellent reporter for monitoring targets of interest in the blood of in vivo models.
LumiLuc is a luciferase with broad substrate specificity. When it was paired with different substrates, intense blue, teal, and yellow bioluminescence was generated. Subsequently, we gained different emission profiles from LumiScarlet in the presence of different substrates. We demonstrated the use of LumiScarlet-8pyDTZ for deep-tissue imaging. In addition, because the two emission peaks of LumiScarlet-pyCTZ or LumiScarlet-6pyDTZ are more separated than LumiScarlet-8pyDTZ, we envision that LumiScarlet-pyCTZ and LumiScarlet-6pyDTZ would be useful for studying protein-protein interactions or constructing BRET-based biosensors.
In summary, we have developed several engineered luciferase-luciferin pairs that emit photons spanning an appreciable range in the visible spectrum. Our effort has greatly enhanced the biocompatibility and sensitivity of ATP-independent bioluminescent reporters for in vivo BLI. We expect that future studies will continuously increase the water-solubility of CTZ and DTZ analogs and red-shift the emission of marine luciferases. Subsequently, we expect that a large array of bioluminescent biosensors will be developed on the basis of these bright, ATP-independent bioluminescent reporters.31 The new reporters and biosensors will further ease non-invasive imaging of freely moving animals, leading to new biological insights.
METHODS
Experimental details are given in the Supporting Information.
Supplementary Material
ACKNOWLEDGMENT
Research reported in this publication was supported in part by the University of Virginia, and the National Institute of General Medical Sciences of the National Institutes of Health under Awards R01GM118675 and R01GM129291.
Footnotes
ASSOCIATED CONTENT
The Supporting Information is available free of charge via the Internet.
Experimental details, synthetic methods, compound characterization, and supporting figures, tables, and results (PDF).
NOTES
The University of Virginia filed a provisional patent application that is partially based on results described in the manuscript. H.-w.A. and H.-W.Y. are listed as coinventors.
REFERENCES
- 1.Rodriguez EA, Campbell RE, Lin JY, Lin MZ, Miyawaki A, Palmer AE, Shu X, Zhang J, and Tsien RY (2017) The Growing and Glowing Toolbox of Fluorescent and Photoactive Proteins, Trends Biochem. Sci. 42, 111–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sadikot RT, and Blackwell TS (2005) Bioluminescence imaging, Proc. Am. Thorac. Soc. 2, 537–540, 511–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mezzanotte L, van ‘t Root M, Karatas H, Goun EA, and Lowik C (2017) In Vivo Molecular Bioluminescence Imaging: New Tools and Applications, Trends Biotechnol. 35, 640–652. [DOI] [PubMed] [Google Scholar]
- 4.Yao Z, Zhang BS, and Prescher JA (2018) Advances in bioluminescence imaging: new probes from old recipes, Curr. Opin. Chem. Biol. 45, 148–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams ST Jr., and Miller SC (2014) Beyond D-luciferin: expanding the scope of bioluminescence imaging in vivo, Curr. Opin. Chem. Biol. 21c, 112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeh HW, and Ai HW (2019) Development and Applications of Bioluminescent and Chemiluminescent Reporters and Biosensors, Annu. Rev. Anal. Chem. 12, DOI: 10.1146/annurev-anchem-061318-115027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwano S, Sugiyama M, Hama H, Watakabe A, Hasegawa N, Kuchimaru T, Tanaka KZ, Takahashi M, Ishida Y, Hata J, Shimozono S, Namiki K, Fukano T, Kiyama M, Okano H, Kizaka-Kondoh S, McHugh TJ, Yamamori T, Hioki H, Maki S, and Miyawaki A (2018) Single-cell bioluminescence imaging of deep tissue in freely moving animals, Science 359, 935–939. [DOI] [PubMed] [Google Scholar]
- 8.Yeh HW, Karmach O, Ji A, Carter D, Martins-Green MM, and Ai HW (2017) Red-shifted luciferase-luciferin pairs for enhanced bioluminescence imaging, Nat. Methods 14, 971–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeh HW, Wu T, Chen M, and Ai HW (2019) Identification of Factors Complicating Bioluminescence Imaging, Biochemistry 58, 1689–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saito R, Kuchimaru T, Higashi S, Lu SW, Kiyama M, Iwano S, Obata R, Hirano T, Kizaka-Kondoh S, and Maki SA (2019) Synthesis and luminescence properties of near-infrared N-heterocyclic luciferin analogues for in vivo optical imaging, B Chem. Soc. Jpn. 92, 608–618. [Google Scholar]
- 11.Haddock SH, Moline MA, and Case JF (2010) Bioluminescence in the sea, Ann. Rev. Mar. Sci. 2, 443–493. [DOI] [PubMed] [Google Scholar]
- 12.Inouye S, and Sasaki S (2007) Overexpression, purification and characterization of the catalytic component of Oplophorus luciferase in the deep-sea shrimp, Oplophorus gracilirostris, Protein Expr. Purif. 56, 261–268. [DOI] [PubMed] [Google Scholar]
- 13.Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T, Robers MB, Benink HA, Eggers CT, Slater MR, Meisenheimer PL, Klaubert DH, Fan F, Encell LP, and Wood KV (2012) Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate, ACS Chem. Biol. 7, 1848–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weissleder R, and Ntziachristos V (2003) Shedding light onto live molecular targets, Nat. Med. 9, 123–128. [DOI] [PubMed] [Google Scholar]
- 15.Chu J, Oh Y, Sens A, Ataie N, Dana H, Macklin JJ, Laviv T, Welf ES, Dean KM, Zhang F, Kim BB, Tang CT, Hu M, Baird MA, Davidson MW, Kay MA, Fiolka R, Yasuda R, Kim DS, Ng HL, and Lin MZ (2016) A bright cyan-excitable orange fluorescent protein facilitates dual-emission microscopy and enhances bioluminescence imaging in vivo, Nat. Biotechnol. 34, 760–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki K, Kimura T, Shinoda H, Bai G, Daniels MJ, Arai Y, Nakano M, and Nagai T (2016) Five colour variants of bright luminescent protein for real-time multicolour bioimaging, Nat. Commun. 7, 13718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teranishi K, and Shimomura O (1997) Solubilizing coelenterazine in water with hydroxypropyl-beta-cyclodextrin, Biosci. Biotech. Bioch. 61, 1219–1220. [Google Scholar]
- 18.Morse D, and Tannous BA (2012) A water-soluble coelenterazine for sensitive in vivo imaging of coelenterate luciferases, Mol. Ther. 20, 692–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shakhmin A, Hall MP, Machleidt T, Walker JR, Wood KV, and Kirkland TA (2017) Coelenterazine analogues emit red-shifted bioluminescence with NanoLuc, Org. Biomol. Chem. 15, 8559–8567. [DOI] [PubMed] [Google Scholar]
- 20.Shakhmin A, Hall MP, Walker JR, Machleidt T, Binkowski BF, Wood KV, and Kirkland TA (2016) Three Efficient Methods for Preparation of Coelenterazine Analogues, Chem-Eur. J. 22, 10369–10375. [DOI] [PubMed] [Google Scholar]
- 21.Jedinak L, Zatopkova R, Zemankova H, Sustkova A, and Cankar P (2017) The Suzuki-Miyaura Cross-Coupling Reaction of Halogenated Aminopyrazoles: Method Development, Scope, and Mechanism of Dehalogenation Side Reaction, J. Org. Chem. 82, 157–169. [DOI] [PubMed] [Google Scholar]
- 22.Adamczyk M, Akireddy SR, Johnson DD, Mattingly PG, Pan Y, and Reddy RE (2003) Synthesis of 3,7-dihydroimidazo[1,2a]pyrazine-3-ones and their chemiluminescent properties, Tetrahedron 59, 8129–8142. [Google Scholar]
- 23.Kerns EH, Di L, and Carter GT (2008) In vitro solubility assays in drug discovery, Curr. Drug Metab. 9, 879–885. [DOI] [PubMed] [Google Scholar]
- 24.Rogers KL, Picaud S, Roncali E, Boisgard R, Colasante C, Stinnakre J, Tavitian B, and Brulet P (2007) Non-invasive in vivo imaging of calcium signaling in mice, Plos One 2, e974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zinn KR, Chaudhuri TR, Szafran AA, O’Quinn D, Weaver C, Dugger K, Lamar D, Kesterson RA, Wang X, and Frank SJ (2008) Noninvasive bioluminescence imaging in small animals, ILAR J. 49, 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inouye S, Sato J, Sahara-Miura Y, Yoshida S, and Hosoya T (2014) Luminescence enhancement of the catalytic 19 kDa protein (KAZ) of Oplophorus luciferase by three amino acid substitutions, Biochem. Biophys. Res. Commun. 445, 157–162. [DOI] [PubMed] [Google Scholar]
- 27.Tantama M, Martinez-Francois JR, Mongeon R, and Yellen G (2013) Imaging energy status in live cells with a fluorescent biosensor of the intracellular ATP-to-ADP ratio, Nat. Commun. 4, 2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morton CL, and Houghton PJ (2007) Establishment of human tumor xenografts in immunodeficient mice, Nat. Protoc. 2, 247–250. [DOI] [PubMed] [Google Scholar]
- 29.Bindels DS, Haarbosch L, van Weeren L, Postma M, Wiese KE, Mastop M, Aumonier S, Gotthard G, Royant A, Hink MA, and Gadella TW Jr. (2017) mScarlet: a bright monomeric red fluorescent protein for cellular imaging, Nat. Methods 14, 53–56. [DOI] [PubMed] [Google Scholar]
- 30.Salin K, Auer SK, Rey B, Selman C, and Metcalfe NB (2015) Variation in the link between oxygen consumption and ATP production, and its relevance for animal performance, P. Roy. Soc. B-Biol. Sci. 282, 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greenwald EC, Mehta S, and Zhang J (2018) Genetically Encoded Fluorescent Biosensors Illuminate the Spatiotemporal Regulation of Signaling Networks, Chem. Rev. 118, 11707–11794. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


