Abstract
Background
Phytohormones are key regulators of plant growth, development, and signalling networks involved in responses to diverse biotic and abiotic stresses. Transcriptional reference maps of hormone responses have been reported for several model plant species such as Arabidopsis thaliana, Oryza sativa, and Brachypodium distachyon. However, because of species differences and the complexity of the wheat genome, these transcriptome data are not appropriate reference material for wheat studies.
Results
We comprehensively analysed the transcriptomic responses in wheat spikes to seven phytohormones, including indole acetic acid (IAA), gibberellic acid (GA), abscisic acid (ABA), ethylene (ET), cytokinin (CK), salicylic acid (SA), and methyl jasmonic acid (MeJA). A total of 3386 genes were differentially expressed at 24 h after the hormone treatments. Furthermore, 22.7% of these genes exhibited overlapping transcriptional responses for at least two hormones, implying there is crosstalk among phytohormones. We subsequently identified genes with expression levels that were significantly and differentially induced by a specific phytohormone (i.e., hormone-specific responses). The data for these hormone-responsive genes were then compared with the transcriptome data for wheat spikes exposed to biotic (Fusarium head blight) and abiotic (water deficit) stresses.
Conclusion
Our data were used to develop a transcriptional reference map of hormone responses in wheat spikes.
Electronic supplementary material
The online version of this article (10.1186/s12864-019-5726-x) contains supplementary material, which is available to authorized users.
Keywords: Phytohormone, Transcriptome, Resistance, Fusarium head blight, Water deficit, Marker gene, Defence mechanism
Background
Common wheat (Triticum aestivum) is one of the most important cereal crops worldwide because of its production and use. Additionally, because of its unique processing quality, wheat is consumed in many specific forms, including as bread or steamed bread, pizza, noodles, cake, biscuits, and dumplings. Common wheat is a hexaploid species (2n = 6x = 42), with a large genome comprising 16 gigabases [1]. The complexity of the wheat genome has impeded the elucidation of the roles of phytohormones in wheat plants, in contrast to model plant species.
Phytohormones are key molecules for regulating plant growth, development, and signalling networks involved in responses to diverse biotic and abiotic stresses [2–4]. Moreover, they function as part of a complex network that finely regulates gene expression in response to environmental cues. The biosynthesis, catabolism, transport, and signalling pathways of the major hormones [i.e., auxin, gibberellin (GA), abscisic acid (ABA), cytokinin (CK), ethylene (ET), salicylic acid (SA), and jasmonic acid (JA)] have been widely investigated in model plant species such as Arabidopsis thaliana and rice [5–10]. Phytohormones affect wheat yield, pre-harvest sprouting, and Fusarium head blight (FHB) resistance, all of which are related to spikes. The global wheat yield has substantially increased since the 1960s largely because of the Green Revolution [11], with one of the wheat Green Revolution genes encoding a mutant GA-responsive protein, DELLA [12]. Auxins can increase the final wheat harvest [13]. Additionally, GA, ABA, and auxins (IAA) regulate pre-harvest sprouting and seed dormancy [14–16]. Both wheat yield and quality are negatively affected by FHB, which is a devastating disease worldwide [17]. Infections by Fusarium graminearum (i.e., the major causal agent of FHB) lead to altered endogenous phytohormone levels in the wheat spikes [18]. The effects of JA, ABA, IAA, ET, and SA on FHB have been reported [18–24]. Moreover, F. graminearum and JA and/or ABA treatments have a synergistic effect on the expression of ExpB6 (β-expansin 6), Pdf1.2 (plant defensin 1.2), and PR4 (pathogenesis-related protein 4). Furthermore, F. graminearum and JA treatments have an antagonistic effect on ATB2 (auxin-inducible oxidoreductase) expression [18].
The transcriptome refers to the total mRNA content in an organism or in a specific type of tissue or cell. A transcriptome analysis enables researchers to characterise the global transcriptional activity and to identify a subset of target genes relatively easily. Considering the importance of phytohormones, transcriptome analyses are essential for elucidating the key roles of phytohormones, and have been conducted for several model plant species such as A. thaliana (135 megabases) [10], rice (389 megabases) [25], and Brachypodium distachyon (272 megabases) [26]. The resulting comprehensive transcriptome data have been widely used in other studies regarding the largescale or gene-specific regulation of transcripts. However, these transcriptome data cannot be used as reference material for studies on wheat spike-related traits because of species differences and the complexity of the common wheat genome (16 gigabases). Nevertheless, transcriptome analyses are still widely used for the global and rapid identification of differentially expressed genes (DEGs) under various conditions. Therefore, the phytohormone-regulated transcriptomic changes in wheat spikes can and should be analysed.
In this study, we completed a comprehensive analysis of the transcriptomic changes in wheat spikes in response to seven phytohormones [IAA, GA (GA3), ABA, ET, CK (trans-zeatin), SA, and MeJA] to identify responsive genes, investigate the crosstalk among hormones, and develop quantitative real-time polymerase chain reaction (qRT-PCR) markers for hormone signalling. The resulting data were then used for combined analyses of the transcriptomic changes due to biotic (FHB) and abiotic (water deficit) stresses. The results presented herein may be useful for clarifying the effects of phytohormones on wheat spike-related traits.
Results
Identification of differentially expressed genes
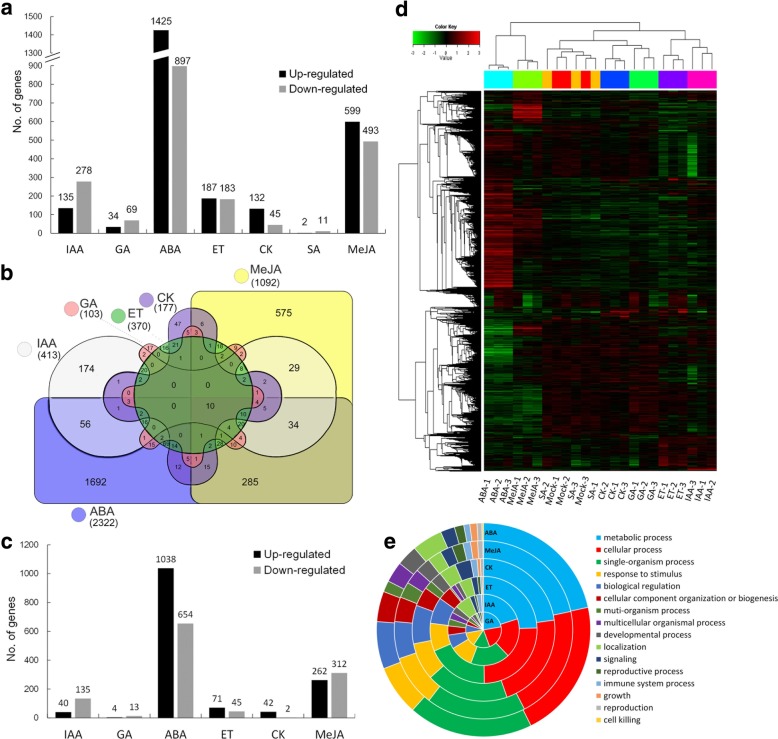
Microarray data revealed gene expression changes in wheat spikes in response to all seven tested hormones. A total of 3386 DEGs were identified (Additional file 1: Table S1). The application of exogenous phytohormones upregulated gene expression levels as follows: 135 genes for IAA, 34 genes for GA3, 1425 genes for ABA, 187 genes for ET, 132 genes for trans-zeatin, 2 genes for SA, and 599 genes for MeJA (Fig. 1a). The phytohormone treatments downregulated gene expression levels as follows: 278 genes for IAA, 69 genes for GA3, 897 genes for ABA, 183 genes for ET, 45 genes for trans-zeatin, 11 genes for SA, and 493 genes for MeJA (Fig. 1a). Moreover, ABA and SA exhibited the strongest and weakest effects on transcript abundance, respectively.
Fig. 1.
Differentially expressed genes (DEGs) in hormone-treated wheat spikes. a Number of DEGs regulated by various plant hormones. b Overlapping and unique DEGs in response to various plant hormones (excluding SA). c Number of unique DEGs regulated by various plant hormones. d Heat map illustrating the hierarchical clustering results for the microarray data. Mock, water treatment. e Gene ontology classification of DEGs
Among the identified DEGs, only one (Ta.12812) was common to all seven phytohormones. We observed some overlapping gene expression, but the expression levels of many genes were uniquely upregulated or downregulated by individual hormones (Fig. 1b). Specifically, the expression levels of 40, 4, 1038, 71, 42, and 262 genes were uniquely upregulated by IAA, GA, ABA, ET, trans-zeatin, and MeJA, respectively, whereas the expression levels of 135, 13, 654, 45, 2, and 312 genes were uniquely downregulated by IAA, GA, ABA, ET, trans-zeatin, and MeJA, respectively (Fig. 1c). We did not detect any gene uniquely regulated by SA under our experimental conditions.
A hierarchical clustering analysis to compare global gene expression changes (Fig. 1d) indicated that the differences between the drought stress and hormone treatments (except SA) were significant. The global expression patterns induced by MeJA, GA, and ET were similar to those induced by ABA, CK, and IAA, respectively.
Gene ontology classification of DEGs affected by phytohormones
A gene ontology (GO) classification of DEGs was completed to identify biological processes affected by six of the seven hormone treatments (the exception was SA) (Fig. 1e). Genes associated with the GO terms ‘cellular processes’, ‘single-organism processes’, and ‘metabolic processes’ were generally the most affected by the hormone treatments. Notably, genes associated with the GO term ‘response to stimulus’ were strongly affected by the six hormone treatments. Significant GO terms were identified for each of the six hormone treatments (Additional file 1: Table S2). Additionally, the gene response patterns varied among the hormone treatments.
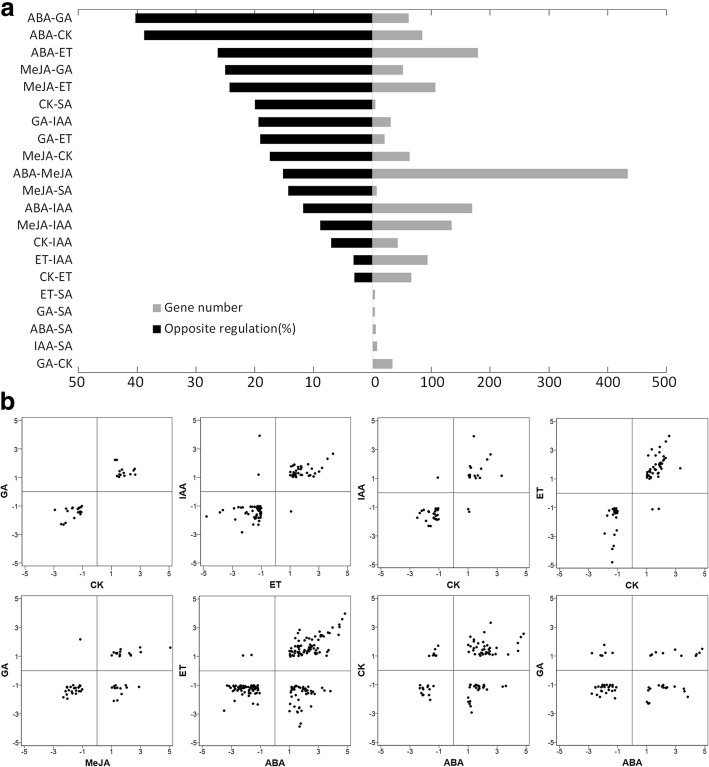
Antagonistic and synergistic interactions between phytohormones
In this study, 22.7% of the DEGs were regulated by two or more hormones. These overlapping genes might be important for the crosstalk among hormones. The percentage of antagonistically regulated genes obviously varied between different hormone pairs (Fig. 2a). In particular, almost no opposite responses were identified for the overlapping genes between GA and CK, IAA and ET, IAA and CK, and ET and CK (Fig. 2b), indicating extensive synergy between these hormones at the transcriptional level. In contrast, there were considerable opposite responses for the overlapping genes between GA and MeJA, ABA and ET, CK and ABA, and GA and ABA (Fig. 2b).
Fig. 2.
Antagonistic and synergistic interactions between phytohormones. a Percentage of antagonistically regulated genes between pairs of hormones. Gene number refers to the number of overlapping genes regulated by the two corresponding hormones. b Scatter plots of fold-changes in the overlapping genes between pairs of hormones. The vertical and horizontal ordinates indicate expression values presented in the log2-transformed form
Validation of gene expression by qRT-PCR
Microarray data were verified by the qRT-PCR analysis of 44 DEGs (Additional file 1: Table S1). The expression patterns of the 44 genes as determined by qRT-PCR were largely consistent with those obtained from the microarray analysis (Fig. 3).
Fig. 3.

Validation of the expression of 44 genes by qRT-PCR. Expression-level changes based on qRT-PCR and microarray data are indicated by RT and AR, respectively. Significant gene expression changes validated by qRT-PCR in response to hormones are listed on the right side. Two or more hormones in one line indicate overlapping results. Red and green refer to upregulated and downregulated expression, respectively
Some of the genes were identified as suitable hormone response markers in wheat spikes because they were significantly and differentially regulated by a specific phytohormone. A gene specifically induced by SA (Ta.5208.1.S1_x_at, which encodes a thionin-like protein) exhibited approximately 6.9-fold and 8.5-fold increases in expression levels in response to SA according to microarray and qRT-PCR analyses, respectively. Similarly, the expression levels of genes encoding an auxin-responsive protein (Ta.16912.1.S1_at), a seed maturation protein (Ta.9389.1.S1_x_at), a flavonol 3-sulfotransferase (Ta.12136.1.A1_at), and a chymotrypsin inhibitor (Ta.2632.1.S1_x_at) were upregulated by IAA, ABA, CK, and MeJA, respectively, according to the microarray and qRT-PCR data. Conversely, the expression of TaAffx.15441.1.A1_at, Ta.646.1.A1_at, Ta.8582.1.S1_at, and Ta.21556.1.S1_x_at was inhibited by GA, ABA, ET, and MeJA, respectively.
Expression of hormone-responsive genes during an F. graminearum infection
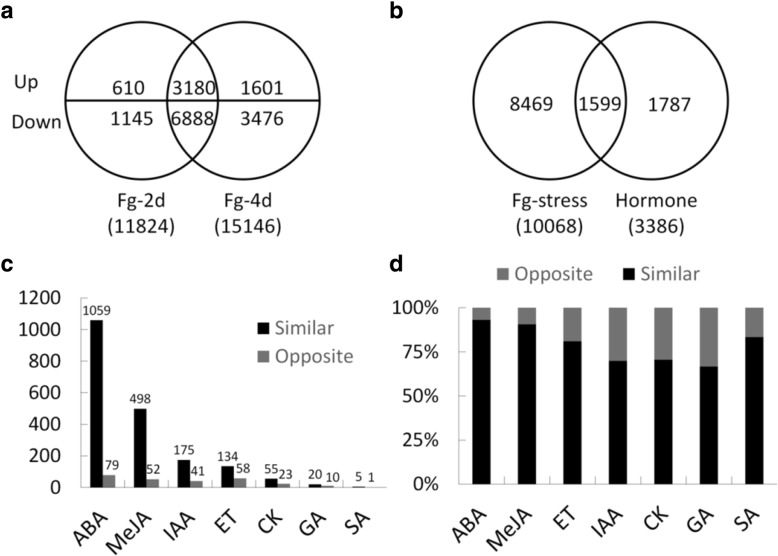
To better characterise the phytohormone functions related to wheat resistance against F. graminearum, the transcriptome data for T. aestivum cv. ‘Roblin’ plants infected with FHB (GEO record #GSE54554) were compared with our data. A total of 10,068 DEGs, including 3180 upregulated and 6888 downregulated genes, were identified at 2 and 4 days post-inoculation with F. graminearum (Fig. 4a). Moreover, 1599 of the 10,086 DEGs were responsive to both the F. graminearum infection and hormone treatments (Fig. 4b).
Fig. 4.
Expression of hormone-responsive genes during an F. graminearum infection. a Number of upregulated and downregulated genes in T. aestivum cv. ‘Roblin’ spikes at 2 (Fg-2d) and 4 days (Fg-4d) post-inoculation. Specific and common genes were categorised from the non-intersecting and intersecting areas, respectively. Up, upregulated genes; Down, downregulated genes. b Venn diagrams presenting the genes responsive to F. graminearum (Fg-stress) and hormone treatments. c Number of differentially expressed genes (DEGs) exhibiting similar or the opposite expression patterns in spike samples treated with F. graminearum or hormones. d Percentage of DEGs exhibiting similar or the opposite expression patterns in spike samples treated with F. graminearum or hormones
SA
Six DEGs were responsive to SA (Fig. 4c; Additional file 1: Table S3), and five of them, including Ta.5208.1.S1_x_at (i.e., SA-response marker gene) were similarly expressed following an F. graminearum infection (fungal stress) and SA treatment.
MeJA
A total of 550 DEGs were identified as MeJA-responsive genes, and 90.55% (498/550) of these DEGs exhibited similar expression patterns in response to fungal stress and MeJA (Fig. 4c and d; Additional file 1: Table S3). A GO enrichment analysis indicated that multiple defence-related processes were over-represented, including glutathione conjugation reactions and metabolic processes, sulfur metabolic processes, responses to oxidative stress, heterocycle biosynthetic processes, and peroxidase reactions (Additional file 1: Table S4). Excluding the interference of other hormones, 273 DEGs were specifically regulated by MeJA, 85 of which exhibited upregulated expression in response to both fungal stress and MeJA. An examination of the putative functions of these 85 genes revealed that they are involved in DON detoxification, phenylpropanoid pathways for secondary cell wall thickening, peroxidase reactions, GDSL-lipase reactions, cell wall defence, and defence signalling (Table 1).
Table 1.
Defence-related genes specifically upregulated by MeJA and F. graminearum
| Probe Set | Annotation | Flod change (log2) | ||
|---|---|---|---|---|
| Fg-2d | Fg-4d | MeJA | ||
| DON detoxification | ||||
| Ta.12808.1.S1_at | PDR-like ABC transporter | 18.06 | 16.20 | 2.39 |
| Ta.21281.2.A1_at | similar to PDR-like ABC transporter | 8.62 | 11.66 | 2.44 |
| TaAffx.1140.1.A1_at | glutathione S-transferase | 7.42 | 15.43 | 3.35 |
| Ta.13496.1.A1_at | UDP-glucosyltransferase | 3.24 | 4.13 | 2.21 |
| Ta.22589.1.S1_at | UDP-glucosyltransferase | 3.58 | 6.05 | 2.18 |
| Ta.18630.1.A1_at | cytochrome P450 | 10.84 | 13.91 | 2.25 |
| Ta.18630.1.A1_x_at | cytochrome P450 | 10.83 | 10.57 | 2.30 |
| Ta.1875.1.S1_at | cytochrome P450 | 10.18 | 51.10 | 2.40 |
| Ta.1875.2.S1_at | cytochrome P450 | 9.45 | 26.90 | 2.56 |
| Ta.1875.2.S1_x_at | cytochrome P450 | 10.05 | 34.93 | 2.37 |
| TaAffx.105598.1.S1_at | cytochrome P450 | 4.52 | 2.06 | 2.37 |
| Secondary cell wall | ||||
| Ta.8618.1.S1_at | shikimate kinase | 47.61 | 59.14 | 2.17 |
| Ta.9122.1.S1_at | arogenate dehydratase 1 | 13.86 | 28.33 | 2.72 |
| Ta.9122.1.S1_x_at | arogenate dehydratase 1 | 14.02 | 25.07 | 2.86 |
| Ta.9122.2.S1_at | arogenate dehydratase 1 | 5.91 | 4.88 | 2.31 |
| Ta.16968.1.A1_at | 4-coumarate--CoA ligase | 15.38 | 14.90 | 2.48 |
| Ta.8228.1.S1_at | agmatine coumaroyltransferase | 22.31 | 83.23 | 8.60 |
| TaAffx.109981.1.S1_x_at | agmatine coumaroyltransferase | 12.31 | 7.71 | 4.13 |
| TaAffx.29050.1.S1_s_at | agmatine coumaroyltransferase | 24.29 | 33.47 | 4.80 |
| Ta.14545.1.S1_at | O-methyltransferase | 267.12 | 205.33 | 2.99 |
| Peroxidase | ||||
| Ta.18497.1.S1_at | Peroxidase | 23.37 | 10.47 | 2.31 |
| Ta.21505.1.S1_at | peroxidase | 15.31 | 7.12 | 2.03 |
| Ta.24106.1.S1_x_at | peroxidase | 8.29 | 2.59 | 3.94 |
| Ta.24710.1.S1_at | Peroxidase | 6.01 | 2.46 | 4.13 |
| TaAffx.39568.2.S1_at | Peroxidase | 4.26 | 2.53 | 5.04 |
| Defense genes | ||||
| Ta.14766.1.S1_at | NBS-LRR type disease resistance protein RPG1-B | 17.96 | 5.14 | 2.28 |
| GDSL-lipases | ||||
| Ta.5520.1.S1_at | GDSL lipase | 3.40 | 2.36 | 2.06 |
| Cell wall defence | ||||
| Ta.21262.1.A1_at | xylanase inhibitor precursor | 16.40 | 23.60 | 2.45 |
| Ta.21262.1.A1_x_at | xylanase inhibitor precursor | 16.13 | 23.60 | 2.54 |
| Ta.19591.2.A1_a_at | Glucan 1,3-beta-glucosidase precursor | 2.88 | 3.78 | 2.38 |
| Transcription and signalling | ||||
| Ta.4678.1.S1_at | WRKY transcription factor | 9.67 | 31.06 | 2.31 |
| Ta.4678.1.S1_x_at | WRKY transcription factor | 8.87 | 29.85 | 2.28 |
| Ta.30507.2.S1_x_at | ZIM domain containing protein | 13.60 | 20.22 | 2.11 |
| Ta.30507.1.S1_a_at | ZIM domain containing protein | 12.70 | 18.32 | 2.30 |
| Ta.9507.2.S1_x_at | zinc-finger protein | 35.96 | 7.822 | 2.78 |
| TaAffx.120360.1.A1_at | similar to heat shock transcription factor | 15.24 | 33.62 | 3.34 |
| TaAffx.76510.1.S1_at | similar to MADS-box protein FDRMADS | 3.45 | 2.60 | 2.20 |
ABA
We identified 1138 DEGs as ABA-responsive genes, and 93.06% (1059/1138) of these genes were similarly expressed after fungal stress and ABA treatments (Fig. 4c and d; Additional file 1: Table S3). Among these DEGs, 777 were specifically regulated by ABA, and 92.41% (718/777) of these DEGs produced similar expression patterns in response to fungal stress and ABA treatments. We previously confirmed that JA and ABA differentially regulate wheat resistance against F. graminearum, with JA significantly enhancing resistance and ABA having the opposite effect [18]. Unexpectedly, multiple defence processes were over-represented among the DEGs upregulated by ABA, including glutathione conjugation reactions and metabolic processes, cell wall polysaccharide metabolic processes, xylan catabolic processes, and sulfur metabolic processes (Additional file 1: Table S4). In contrast, phenylpropanoid metabolic and biosynthetic processes were enriched among the DEGs downregulated by ABA. Indeed, the expression levels of multiple types of phenylpropanoid pathway genes involved in lignin and flavonoid biosynthesis were downregulated by ABA, including genes encoding caffeic acid-O-methyltransferase, dihydroflavonol-4-reductase, flavonoid 3′-monooxygenase, chalcone synthase, chitinase, and dirigent proteins (Table 2). Obviously, ABA can promote F. graminearum infections in wheat by inhibiting the biosynthetic processes related to the plant secondary cell wall, even though ABA also upregulates the expression of many genes associated with resistance against F. graminearum.
Table 2.
Phenylpropanoid pathway genes downregulated by ABA and F. graminearum
| Probe set | Category | Annotation | Fold change (log2) | ||
|---|---|---|---|---|---|
| ABA | Fg-2d | Fg-4d | |||
| TaAffx.115378.1.S1_at | Phenylpropanoid biosynthesis | caffeic acid 3-O-methyltransferase | 0.42 | 0.07 | 0.12 |
| Ta.9172.1.S1_x_at | flavonoid biosynthetic process | chalcone synthase | 0.25 | 0.07 | 0.13 |
| Ta.9172.2.S1_x_at | flavonoid biosynthetic process | chalcone synthase | 0.41 | 0.28 | 0.21 |
| Ta.9172.3.S1_x_at | flavonoid biosynthetic process | chalcone synthase | 0.32 | 0.29 | 0.18 |
| Ta.12690.2.S1_x_at | flavonoid biosynthetic process | dihydroflavonol-4-reductase | 0.44 | 0.30 | 0.28 |
| TaAffx.37978.1.A1_at | flavonoid biosynthetic process | Flavonoid 3′-monooxygenase | 0.31 | 0.39 | 0.32 |
| Ta.4385.2.A1_at | Lignin Biosynthesis | chitinase-like protein 2 | 0.43 | 0.22 | 0.15 |
| Ta.4455.1.A1_at | lignin Biosynthesis | laccase - like protein | 0.36 | 0.17 | 0.14 |
| Ta.25384.1.S1_at | Lignin Biosynthesis | dirigent-like protein | 0.38 | 0.45 | 0.31 |
| TaAffx.132123.1.A1_x_at | Lignin Biosynthesis | dirigent protein | 0.22 | 0.28 | 0.19 |
IAA
A total of 216 IAA-responsive DEGs were detected, of which 81.02% (175/216) were similarly expressed following fungal stress and IAA treatments (Fig. 4c and d). Similar to the effects of MeJA and ABA, the expression levels of many defence-related genes were upregulated by IAA, but 75.79% (72/95) of the upregulated genes were not specifically regulated by IAA.
ET
We determined that 192 DEGs were responsive to ET, and 69.79% (134/192) of these DEGs exhibited similar expression patterns in response to fungal stress and ET (Fig. 4c and d). Only 40 DEGs were specifically regulated by ET, of which 23 were similarly expressed during exposures to fungal stress and exogenous ET.
CK and GA
Among the detected DEGs, 78 and 30 were responsive to CK and GA, respectively (Fig. 4c; Additional file 1: Table S3). Additionally, 70.51% (55/78) and 66.67% (20/30) of these DEGs produced similar expression patterns in response to fungal stress and the corresponding hormone, respectively (Fig. 4d). Only 8 and 5 of these DEGs were specifically regulated by CK and GA, respectively.
Expression of hormone-responsive genes in the wheat glume under water deficit conditions
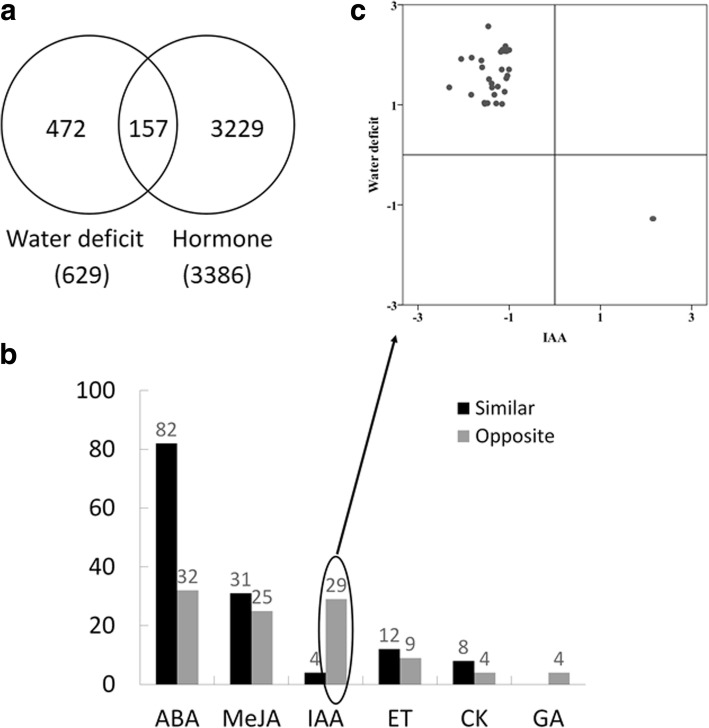
Wheat glume, which is the main green tissue of spikes, facilitates photosynthesis and delays aging under drought conditions [27]. To clarify the effects of the major hormones in the wheat glume in response to drought stress, we compared the available data for 629 DEGs under water deficit conditions at 6 days after anthesis [28] with the data generated in the current study. Of these 629 DEGs, 157 were identified as hormone-responsive genes (Fig. 5a; Additional file 1: Table S5).
Fig. 5.
Expression of hormone-responsive genes in the wheat glume in response to water deficit stress. a Venn diagrams presenting the genes responsive to water deficit stress and hormone treatments. b Number of differentially expressed genes exhibiting similar and the opposite expression patterns in spike samples treated with water deficit stress or hormones. c Scatter plots of the fold-changes for the 29 genes that exhibited the opposite expression patterns in response to water deficit stress and an IAA treatment. The vertical and horizontal ordinates indicate the expression values presented in the log2-transformed form
We determined that 71.92% (82/114) of the ABA-responsive genes (Fig. 5b) were similarly expressed in response to ABA and drought stress, whereas 87.88% (29/33) of the IAA-responsive genes (Fig. 5b; Additional file 1: Table S6) exhibited the opposite expression patterns following IAA and water deficit treatments. Moreover, the expression levels of almost all of these IAA-responsive genes (28/29) were downregulated by IAA, but were upregulated by drought stress (Fig. 5c). These results suggested that the drought resistance of the wheat glume may be improved by inhibiting IAA signalling. Functional annotations further implied that these 29 genes contribute to the drought resistance of the wheat glume by encoding the vesicle-associated membrane protein, serine/threonine protein kinase, receptor-like protein kinase, cytochrome P450, xyloglucan endotransglucosylase/hydrolase protein, lipoxygenase, and the cold acclimation protein (Additional file 1: Table S6).
Discussion
Comprehensive transcriptome analyses following hormone treatments have been completed for model plant species, including A. thaliana [10], rice [25], and B. distachyon [26]; however, these studies focused on the seedling stage. Gene expression patterns differ dramatically between organs and tissues because of a temporally and spatially regulated process involving the selective expression of specific parts of the genome [29, 30]. Common wheat, which is an allohexaploid species comprising three genomes (A, B, and D), has multiple orthologous genes. The expression of these genes for coordinated responses to diverse stimuli involves a very complex mechanism [31, 32]. Additionally, the complexity of the wheat genome contributes to the difficulties associated with analyses using transcriptional data from diploid species. We previously determined that caution should be exercised when using traditional A. thaliana marker genes to investigate wheat [20]. In the current study, we comprehensively analysed the transcriptomic changes in wheat spikes induced by seven phytohormones. A comparison between the transcriptome data for B. distachyon and rice [25, 26] and the data generated in this study indicated that only a few genes exhibited the same expression pattern in wheat spikes (data not shown), highlighting the importance of the transcriptional reference map of hormone responses in wheat spikes.
Satisfying the growing demand for wheat worldwide has been challenging [33]. Research on the effects of phytohormones may contribute to increased productivity to narrow the gap between the demand and supply. Since the 1960s, the global wheat yield has substantially increased as a result of the the manipulation of GA signaling [11, 12]. Other GA-responsive dwarfism genes, such as Rht4, Rht5, Rht8, Rht12, and Rht13, have the potential to increase bread wheat yield without compromising aerial biomass or coleoptile length. The use of uniconazole, a plant growth regulator that inhibits GA biosynthesis, also enhances wheat production [34]. Additionally, an earlier investigation proved that increasing the auxin level positively influences the final wheat yield [13]. In rice, OsCKX2 encodes a cytokinin oxidase/dehydrogenase, which degrades CK. Thus, downregulated OsCKX2 expression results in the accumulation of CK in rice inflorescence meristems and increases the number of reproductive organs, ultimately resulting in increased grain yield [35]. Identifying hormone-responsive genes in the wheat spike may provide important insights for the cloning of specific genes encoding regulators of wheat production.
Pre-harvest sprouting in wheat refers to the germination of seeds in the spikes after physiological maturity, but before harvest, which leads to decreased grain yield and end-use quality as well as considerable economic losses [36]. Pre-harvest sprouting in wheat mainly results from the breaking or lack of seed dormancy under humid conditions [37]. Seed dormancy is primarily regulated by the balance between ABA and GA [38]. However, in A. thaliana, IAA also controls seed dormancy via its stimulatory effects on ABA signalling [15]. Accordingly, the application of exogenous IAA enhances the accumulation of ABA in wheat spikes [18]. There is little information available on the effects of endogenous hormones in wheat spikes on pre-harvest sprouting. Related research may benefit from the gene expression data generated in this study.
Drought stress adversely impacts many aspects of plant physiology, especially the photosynthetic capacity, thereby diminishing crop growth and productivity. Drought tolerance is a complex trait controlled by various genes, transcription factors, microRNAs, hormones, proteins, co-factors, ions, and metabolites [39]. In addition to quantitative trait locus mapping, transcriptomic, proteomic, and metabolomic techniques have been used to identify drought signalling pathways in wheat [39–44]. However, there has been relatively little research focused on wheat spikes, which are an important photosynthate source for the grain-filling period [45]. Consequently, the molecular mechanism underlying drought tolerance in wheat spikes remains relatively uncharacterised [28]. Hormone responses and homeostasis are key physiological mechanisms associated with drought stress tolerance [39, 43, 46]. Reddy et al. [47] reported that auxin, ABA, brassinosteroid, CK, ET, GA, and JA are involved in the drought stress response of wheat leaves, and that ABA, auxin, and ET are particularly important for this response. In the current study, we determined that ABA, JA, and auxin (IAA) play a major role in the drought stress response of wheat spikes (Fig. 5b). The accumulated ABA is thought to activate the acclimation and adaptation response that allows longer term survival under drought stress conditions [48, 49]. Additionally, decreases in the auxin content to inhibit growth may also mediate drought stress responses [50]. Our results suggest that ABA signalling is a key factor for improving the drought resistance of wheat spikes, whereas IAA decreases the drought tolerance of wheat spikes. Our data may be useful for clarifying the key molecular mechanism regulating drought responses in wheat spikes during the grain-filling period.
The contribution of phytohormones to the wheat defence mechanisms against FHB remains relatively unknown, although studies have been completed to elucidate the roles of phytohormones related to wheat FHB resistance [19, 20, 23, 51–53]. The infection of wheat heads by F. graminearum leads to a significant increase in the accumulation of SA, JA, ABA, and IAA [18]. Additionally, SA signalling is reportedly important for A. thaliana and wheat defences against F. graminearum [22–24, 54]. Previous studies confirmed that JA signalling is a key factor for improving wheat resistance to FHB [18, 19, 52]. A comparison of the transcriptomic data of FHB-susceptible and -resistant wheat lines revealed a strong correlation between the upregulation of JA signalling and wheat FHB resistance. The application of exogenous JA and the inhibition of JA biosynthesis by BSMV (barley stripe mosaic virus)-VIGS (virus induced gene silencing) can increase and decrease FHB resistance, respectively [18]. In the current study, we confirmed that exogenous MeJA specifically upregulates the expression of numerous genes, many of which are closely related to defence response processes (Table 1). These genes may be important for JA-induced FHB resistance. Exogenous ABA reportedly increases the susceptibility of wheat to F. graminearum infections [18, 53]. However, we observed that ABA induced the expression of the highest number of FHB-responsive genes among the seven tested phytohormones, including many detoxification-related genes. A possible mechanism underlying the negative effect of ABA on FHB tolerance involves the suppressed expression of phenylalanine pathway genes due to ABA. Previous studies of A. thaliana [55, 56] demonstrated that ABA may suppress lignin production by regulating phenylpropanoid biosynthesis. Secondary cell wall thickening is one of the main mechanisms that prevents the spread of F. graminearum in wheat, and is due to the deposition of hydroxycinnamic acid amides, flavonoids, and lignin, which are synthesised via a phenylpropanoid metabolic shunt [57]. Suppression of the phenylalanine pathway involved in flavonoid and lignin biosynthesis may decrease FHB resistance by weakening the physical barriers to the fungus. Additionally, IAA may be crucial for the interaction between wheat and F. graminearum. An earlier investigation indicated that F. graminearum can produce IAA, thereby contributing to dramatic changes in the IAA contents of infected wheat heads [18]. However, the mechanism by which fungal-derived IAA affects wheat FHB resistance remains unclear. Interestingly, IAA induces the accumulation of ABA [18], which represents a possible explanation. Our transcriptome analysis described herein may promote future research into the role of IAA during F. graminearum infections. Moreover, our transcriptional data may provide new insights into the contribution of phytohormones to wheat FHB resistance.
Conclusions
In this study, we comprehensively analysed the transcriptomic changes in wheat spikes induced by seven phytohormones (IAA, GA, ABA, ET, CK, SA, and MeJA), ultimately resulting in a transcriptional reference map of hormone responses in wheat spikes. We applied this map to investigate the role of hormone signalling pathways in wheat responses to biotic (FHB) and abiotic (water deficit) stresses. The data presented herein may be valuable for elucidating the contribution of phytohormones to wheat spike-related traits.
Methods
Plant material and growth conditions
Triticum aestivum cv. ‘Roblin’ (provided by Agriculture Canada) plants were grown in climate-controlled chambers under a 16-h day (25 °C):8-h night (20 °C) cycle. The plants were watered as needed and 15–15-15 (N-P-K) fertiliser was applied weekly.
Hormone treatments
Only flowering heads were used for hormone treatments. Two florets of each fully developed spikelet from a whole spike at the mid-anthesis stage were treated with 10 μl 10% methanol:water solution with or without (control) 2 mM IAA, 1 mM SA, 1 mM MeJA, 0.38 mM ABA, 0.5 mM GA3, or 0.5 mM trans-zeatin (a type of cytokinin). Regarding the ET treatment, each head was sealed in a plastic bag with 2 ml freshly prepared 25 mM ethephon (pH = 11). All of the hormone solutions were used to treat florets within 1 h of being prepared. The heads were collected at 24 h after the hormone treatments, and then ground to a fine powder in liquid nitrogen. Each treatment was completed with three biological replicates, each of which comprised at least five heads. No unusual morphological changes were observed in the treated heads. The hormone concentrations of the treatments were based on previous studies [18, 20, 58].
RNA isolation and microarray and qRT-PCR analyses
For the microarray analysis, total RNA was extracted with the TRIzol reagent (Invitrogen, Shanghai, China). The quality of the extracted RNA was monitored with the ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) and by agarose gel electrophoresis. The Affymetrix wheat genome array, with 61,127 probe sets representing up to 55,052 transcripts, was used for gene expression profiling at CapitalBio Corporation (Beijing, China). Normalised values are herein presented in the log2-transformed form. The normalised data were analysed with the R package SAM (significance analysis of microarrays) [59] to identify candidate genes that were significantly and differentially expressed according to the following criteria: q-value < 0.05, fold-change ≥2 in the expression ratio (i.e. log2 ratio ≥ 1.0 or ≤ − 1.0; hormone treatment vs control), and signal intensity > 1000 for at least one of the probes for a given gene. A hierarchical clustering analysis of the RNA data involving the average linkage method was used to estimate the global changes between biological replicates and treatments.
Regarding the qRT-PCR analyses, RNA samples were extracted with the TRIzol reagent (Invitrogen). The RNA quality was monitored with the ND-1000 spectrophotometer and by agarose gel electrophoresis before and after a DNase I treatment (Takara, Dalian, China). The RNA was purified with the RNeasy kit (Tiangen, Beijing, China), after which cDNA was synthesised with the PrimeScript RT reagent kit (Takara) and 1 μg total RNA as the template. Primers were designed and qRT-PCR analyses were completed as previously described [60]. Details regarding the qRT-PCR primers are listed in Additional file 1: Table S7. The primers were designed based on the consensus sequences in the NCBI unigene database (http://www.ncbi.nlm.nih.gov). Three housekeeping genes encoding aldehyde oxidase (AOx, NCBI UniGene Ta.6172), glyceraldehyde-3-phosphate dehydrogenase (w-GAPDH, Ta.66461), and heterogeneous nuclear ribonucleoprotein Q (hn-RNPQ, Ta.10105) were amplified as reference genes for the normalisation of the data [20].
Gene annotation and GO term enrichment analysis
Blast2GO (version 2.8) was used to annotate the gene transcripts with GO terms. For each treatment, all of the enriched GO terms (biological process, level 2) were identified based on a singular enrichment analysis, which was completed with the agriGO tool (http://systemsbiology.cau.edu.cn/agriGOv2/) [61].
Additional file
Table S1. List of genes differentially expressed in wheat spikes in response to seven hormones. Table S2. Gene enrichment analysis of DEGs in response to various hormones. Table S3. List of 1599 DEGs responsive to both F. graminearum and hormone treatments. Table S4. Gene enrichment analysis of DEGs commonly regulated by hormones (ABA, MeJA, or IAA) and F. graminearum. Table. S5. List of 157 DEGs responsive to water deficit stress in the glume and hormone treatments. Table S6. List of 29 genes that exhibited the opposite expression patterns in response to water deficit stress and IAA. Table S7. Details regarding the qRT-PCR primers. (XLSX 583 kb)
Acknowledgements
We thank Robbie Lewis, MSc, from Liwen Bianji, Edanz Group China (https://www.liwenbianji.cn/), for editing the English text of a draft of this manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (31230053 and 31570335), the National Basic Research Program of China (2014CB147200), and the National Key Research and Development Program of China (2016YFD0102000). The funding agencies were not involved in designing the study or in collecting, analysing, and interpreting the data or in writing the manuscript.
Availability of data and materials
The datasets generated and/or analysed during the current study are available in the Gene Expression Omnibus (GEO) repository (accession code GSE103430). The other supporting data were included as additional files.
Abbreviations
- ABA
Abscisic acid
- AOx
Aldehyde oxidase
- BSMV-VIGS
Barley stripe mosaic virus virus induced gene silencing
- CK
Cytokinin
- DEG
Differentially expressed gene
- ET
Ethylene
- FHB
Fusarium head blight
- GA
Gibberellic acid
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- GO
Gene ontology
- hn-RNPQ
Heterogeneous nuclear ribonucleoprotein Q
- IAA
Indole acetic acid
- MeJA
Methyl jasmonic acid
- SA
Salicylic acid
- SAM
Significance analysis of microarrays
Authors’ contributions
PFQ and YLZ designed the experiments. PFQ, YFJ, and QC wrote the manuscript and analysed the data. PFQ and YFJ prepared the figures. PFQ, ZRG, and YFJ revised the manuscript. PFQ, YFJ, QC, LJZ, ZZW, YW, ZRG, YZZ, BJX, KL, MD, JRW, GYC, QTJ, XJL, WL, and YMW performed the experiments. TO provided key reagents and advice. All authors reviewed the results and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Peng-Fei Qi, Phone: +86-28-82650337, Email: pengfeiqi@hotmail.com.
Yun-Feng Jiang, Email: jiangyunfeng2018@163.com.
Zhen-Ru Guo, Email: guozr1993@163.com.
Qing Chen, Email: qingchen83@sicau.edu.cn.
Thérèse Ouellet, Email: therese.ouellet@agr.gc.ca.
Lu-Juan Zong, Email: zonglujuan@163.com.
Zhen-Zhen Wei, Email: zhenzhen-wei@hotmail.com.
Yan Wang, Email: wyan810@163.com.
Ya-Zhou Zhang, Email: yazhou1012@gmail.com.
Bin-Jie Xu, Email: binjiexu@outlook.com.
Li Kong, Email: kongli088@163.com.
Mei Deng, Email: 183433899@qq.com.
Ji-Rui Wang, Email: jirui.wang@gmail.com.
Guo-Yue Chen, Email: guoyuech74@hotmail.com.
Qian-Tao Jiang, Email: qiantaojiang@sicau.edu.cn.
Xiu-Jin Lan, Email: lanxiujin@163.com.
Wei Li, Email: liw03@163.com.
Yu-Ming Wei, Email: ymwei@sicau.edu.cn.
You-Liang Zheng, Phone: +86-28-82650337, Email: ylzheng@sicau.edu.cn.
References
- 1.International Wheat Genome Sequencing Consortium Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. 2018;361(6403):eaar7191. doi: 10.1126/science.aar7191. [DOI] [PubMed] [Google Scholar]
- 2.Robert-Seilaniantz A, Navarro L, Bari R, Jones JD. Pathological hormone imbalances. Curr Opin Plant Biol. 2007;10(4):372–379. doi: 10.1016/j.pbi.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Bari R, Jones JD. Role of plant hormones in plant defence responses. Plant Mol Biol. 2009;69(4):473–488. doi: 10.1007/s11103-008-9435-0. [DOI] [PubMed] [Google Scholar]
- 4.Durbak A, Yao H, McSteen P. Hormone signaling in plant development. Curr Opin Plant Biol. 2012;15(1):92–96. doi: 10.1016/j.pbi.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, et al. The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci U S A. 2011;108(45):18512–18517. doi: 10.1073/pnas.1108434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin Z, Zhong S, Grierson D. Recent advances in ethylene research. J Exp Bot. 2009;60(12):3311–3336. doi: 10.1093/jxb/erp204. [DOI] [PubMed] [Google Scholar]
- 7.Vicente RS, Plasencia J. Salicylic acid beyond defence: its role in plant growth and development. J Exp Bot. 2011;62(10):3321–3338. doi: 10.1093/jxb/err031. [DOI] [PubMed] [Google Scholar]
- 8.Hirano K, Aya K, Hobo T, Sakakibara H, Kojima M, Shim RA, et al. Comprehensive transcriptome analysis of phytohormone biosynthesis and signaling genes in microspore/pollen and tapetum of rice. Plant Cell Physiol. 2008;49(10):1429–1450. doi: 10.1093/pcp/pcn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang DL, Yang Y, He Z. Roles of plant hormones and their interplay in Rice immunity. Mol Plant. 2013;6(3):675–685. doi: 10.1093/mp/sst056. [DOI] [PubMed] [Google Scholar]
- 10.Goda H. The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J. 2008;55(3):526–542. doi: 10.1111/j.1365-313X.2008.03510.x. [DOI] [PubMed] [Google Scholar]
- 11.Grassini P, Eskridge KM, Cassman KG. Distinguishing between yield advances and yield plateaus in historical crop production trends. Nat Commun. 2013;4:2918. doi: 10.1038/ncomms3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature. 1999;400(6741):256–261. doi: 10.1038/22307. [DOI] [PubMed] [Google Scholar]
- 13.Ali A, Ahmad T, Abbasi NA, Hafiz IA. Effect of different concentrations of auxins on in vitro rooting of olive cultivar ‘Moraiolo. Pak J Bot. 2009;41(3):1223–1231. [Google Scholar]
- 14.Yang J, Liu Y, Pu Z, Zhang L, Yuan Z, Chen G, et al. Molecular characterization of high pI α-amylase and its expression QTL analysis in synthetic wheat RILs. Mol Breed. 2014;34(3):1075–1085. doi: 10.1007/s11032-014-0098-z. [DOI] [Google Scholar]
- 15.Liu X, Zhang H, Zhao Y, Feng Z, Li Q, Yang HQ, et al. Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc Natl Acad Sci U S A. 2013;110(38):15485–15490. doi: 10.1073/pnas.1304651110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shu K, Liu XD, Xie Q, He ZH. Two faces of one seed: hormonal regulation of dormancy and germination. Mol Plant. 2016;9(1):34–45. doi: 10.1016/j.molp.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 17.Goswami RS, Kistler HC. Heading for disaster: Fusarium graminearum on cereal crops. Mol Plant Pathol. 2004;5(6):515–525. doi: 10.1111/j.1364-3703.2004.00252.x. [DOI] [PubMed] [Google Scholar]
- 18.Qi PF, Balcerzak M, Rocheleau H, Leung W, Wei YM, Zheng YL, et al. Jasmonic acid and abscisic acid play important roles in host-pathogen interaction between Fusarium graminearum and wheat during the early stages of fusarium head blight. Physiol Mol Plant Pathol. 2016;93:39–48. doi: 10.1016/j.pmpp.2015.12.004. [DOI] [Google Scholar]
- 19.Li G, Yen Y. Jasmonate and ethylene signaling pathway may mediate Fusarium head blight resistance in wheat. Crop Sci. 2008;48(5):1888–1896. doi: 10.2135/cropsci2008.02.0097. [DOI] [Google Scholar]
- 20.Qi PF, Johnston A, Balcerzak M, Rocheleau H, Harris LJ, Long XY, et al. Effect of salicylic acid on Fusarium graminearum , the major causal agent of fusarium head blight in wheat. Fungal Biol. 2012;116(3):413–426. doi: 10.1016/j.funbio.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Ding L, Xu H, Yi H, Yang L, Kong Z, Zhang L, et al. Resistance to hemi-biotrophic F. graminearum infection is associated with coordinated and ordered expression of diverse defense signaling pathways. PLoS One. 2012;6(4):e19008. doi: 10.1371/journal.pone.0019008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makandar R, Nalam V, Chaturvedi R, Jeannotte R, Sparks AA, Shah J. Involvement of salicylate and jasmonate signaling pathways in Arabidopsis interaction with Fusarium graminearum. Mol Plant Mocrobe Interact. 2010;23(7):861–870. doi: 10.1094/MPMI-23-7-0861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makandar R, Nalam VJ, Lee H, Trick HN, Dong Y, Shah J. Salicylic acid regulates basal resistance to Fusarium head blight in wheat. Mol Plant Mocrobe Interact. 2012;25(3):431–439. doi: 10.1094/MPMI-09-11-0232. [DOI] [PubMed] [Google Scholar]
- 24.Makandar R, Essig JS, Schapaugh MA, Trick HN, Shah J. Genetically engineered resistance to Fusarium head blight in wheat by expression of Arabidopsis NPR1. Mol Plant Mocrobe Interact. 2006;19(2):123–129. doi: 10.1094/MPMI-19-0123. [DOI] [PubMed] [Google Scholar]
- 25.Garg R, Tyagi AK, Jain M. Microarray analysis reveals overlapping and specific transcriptional responses to different plant hormones in rice. Plant Signal Behav. 2012;7(8):951–956. doi: 10.4161/psb.20910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kakei Y, Mochida K, Sakurai T, Yoshida T, Shinozaki K, Shimada Y. Transcriptome analysis of hormone-induced gene expression in Brachypodium distachyon. Sci Rep. 2015;5:14476. doi: 10.1038/srep14476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia S, Lv J, Jiang S, Liang T, Liu C, Jing Z. Response of wheat ear photosynthesis and photosynthate carbon distribution to water deficit. Photosynthetica. 2015;53(1):95–109. doi: 10.1007/s11099-015-0087-4. [DOI] [Google Scholar]
- 28.Liu C, Mi H, Liu H, Xie S, Wu Y, Ning P, et al. Response to water deficit in glume of wheat: expression profiling by microarray analysis. Euphytica. 2017;213:26. doi: 10.1007/s10681-016-1827-7. [DOI] [Google Scholar]
- 29.Meyerowitz EM. Plants compared to animals: the broadest comparative study of development. Science. 2002;295(5559):1482–1485. doi: 10.1126/science.1066609. [DOI] [PubMed] [Google Scholar]
- 30.Ma L, Sun N, Liu X, Jiao Y, Zhao H, Deng XW. Organ-specific expression of Arabidopsis genome during development. Plant Physiol. 2005;138(1):80–91. doi: 10.1104/pp.104.054783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfeifer M, Kugler KG, Sandve SR, Zhan B, Rudi H, Hvidsten TR, et al. Genome interplay in the grain transcriptome of hexaploid bread wheat. Science. 2014;345(6194):1250091. doi: 10.1126/science.1250091. [DOI] [PubMed] [Google Scholar]
- 32.Feldman M, Levy AA, Fahima T, Korol A. Genomic asymmetry in allopolyploid plants: wheat as a model. J Exp Bot. 2012;63(14):5045–5059. doi: 10.1093/jxb/ers192. [DOI] [PubMed] [Google Scholar]
- 33.Reynolds M, Bonnett D, Chapman SC, Furbank RT, Manès Y, Mather DE, et al. Raising yield potential of wheat. I. Overview of a consortium approach and breeding strategies. J Exp Bot. 2010;62(2):439–452. doi: 10.1093/jxb/erq311. [DOI] [PubMed] [Google Scholar]
- 34.Yang W, Yu Z, Yu S, Fan G, Han H, Dong Z, et al. Effects of uniconazole waterless-dressing seed on yield of wheat. Acta Agron Sin. 2004;30(5):502–506. [Google Scholar]
- 35.Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, et al. Cytokinin oxidase regulates rice grain production. Science. 2005;309(5735):741–745. doi: 10.1126/science.1113373. [DOI] [PubMed] [Google Scholar]
- 36.Buchanan A, Nicholas E. Sprouting, alpha-amylase, and breadmaking quality. Cereal Res Commun. 1980;8:23–28. [Google Scholar]
- 37.Derera NF. The effect of preharvest rain. In: Derera NF, editor. Preharvest field sprouting in cereals. Boca Raton: CRC Press; 1989. pp. 1–25. [Google Scholar]
- 38.Shu K, Meng Y, Shuai H, Liu W, Du J, Liu J, et al. Dormancy and germination: how does the crop seed decide? Plant Biol. 2015;17(6):1104–1112. doi: 10.1111/plb.12356. [DOI] [PubMed] [Google Scholar]
- 39.Budak H, Hussain B, Khan Z, Ozturk NZ, Ullah N. From genetics to functional genomics: improvement in drought signaling and tolerance in wheat. Front Plant Sci. 2015;6:1012. doi: 10.3389/fpls.2015.01012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hussain B, Lucas SJ, Ozturk L, Budak H. Mapping QTLs conferring salt tolerance and micronutrient concentrations at seedling stagein wheat. Sci Rep. 2017;7(1):15662. doi: 10.1038/s41598-017-15726-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lucas SJ, Salantur A, Yazar S, Budak H. High-throughput SNP genotyping of modern and wild emmer wheat for yield and root morphology using a combined association and linkage analysis. Funct Integr Genomics. 2017;17(6):667–685. doi: 10.1007/s10142-017-0563-y. [DOI] [PubMed] [Google Scholar]
- 42.Galvez S, Meridagarcia R, Camino C, Borrill P, Abrouk M, Ramirezgonzalez RH, et al. Hotspots in the genomic architecture of field drought responses in wheat as breeding targets. Funct Integr Genomics. 2019;19(2):295–309. [DOI] [PMC free article] [PubMed]
- 43.Budak H, Akpinar BA, Unver T, Turktas M. Proteome changes in wild and modern wheat leaves upon drought stress by two-dimensional electrophoresis and nanoLC-ESI–MS/MS. Plant Mol Biol. 2013;83:89–103. doi: 10.1007/s11103-013-0024-5. [DOI] [PubMed] [Google Scholar]
- 44.Ullah N, Yuce M, Gokce ZN, Budak H. Comparative metabolite profiling of drought stress in roots and leaves of seven Triticeae species. BMC Genomics. 2017;18(1):969. doi: 10.1186/s12864-017-4321-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aranjuelo I, Cabrera-Bosquet L, Morcuende R, Avice JC, Nogues S, Araus JL, et al. Does ear C sink strength contribute to overcoming photosynthetic acclimation of wheat plants exposed to elevated CO2? J Exp Bot. 2011;62:3957–3969. doi: 10.1093/jxb/err095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peleg Z, Blumwald E. Hormone balance and abiotic stress tolerance in crop plants. Curr Opin Plant Biol. 2011;14(3):290–5. [DOI] [PubMed]
- 47.Reddy SK, Liu S, Rudd JC, Xue Q, Payton P, Finlayson SA, et al. Physiology and transcriptomics of water-deficit stress responses in wheat cultivars TAM 111 and TAM 112. J Plant Physiol. 2014;171(14):1289–1298. doi: 10.1016/j.jplph.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 48.Kuromori T, Seo M, Shinozaki K. ABA transport and plant water stress responses. Trends Plant Sci. 2018;23(6):513–522. doi: 10.1016/j.tplants.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 49.Yoshida T, Mogami J, Yamaguchi-Shinozaki K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr Opin Plant Biol. 2014;21:133–139. doi: 10.1016/j.pbi.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 50.Naser V, Shani E. Auxin response under osmotic stress. Plant Mol Biol. 2016;91(6):661–672. doi: 10.1007/s11103-016-0476-5. [DOI] [PubMed] [Google Scholar]
- 51.Gottwald S, Samans B, Lück S, Friedt W. Jasmonate and ethylene dependent defence gene expression and suppression of fungal virulence factors: two essential mechanisms of Fusarium head blight resistance in wheat? BMC Genomics. 2012;13(1):369. doi: 10.1186/1471-2164-13-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun Y, Xiao J, Jia X, Ke P, He L, Cao A, et al. The role of wheat jasmonic acid and ethylene pathways in response to Fusarium graminearum infection. Plant Growth Regul. 2016;80(1):69–77. doi: 10.1007/s10725-016-0147-1. [DOI] [Google Scholar]
- 53.Buhrow LM, Cram D, Tulpan D, Foroud NA, Loewen MC. Exogenous abscisic acid and gibberellic acid elicit opposing effects on Fusarium graminearum infection in wheat. Phytopathology. 2016;106(9):986–996. doi: 10.1094/PHYTO-01-16-0033-R. [DOI] [PubMed] [Google Scholar]
- 54.Qi PF, Zhang YZ, Liu CH, Zhu J, Chen Q, Guo ZR, et al. Fusarium graminearum ATP-binding cassette transporter gene FgABCC9 is required for its transportation of salicylic acid, fungicide resistance, mycelial growth and pathogenicity towards wheat. Int J Mol Sci. 2018;19(8):2351. doi: 10.3390/ijms19082351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mohr PG, Cahill DM. Suppression by ABA of salicylic acid and lignin accumulation and the expression of multiple genes, in Arabidopsis infected with Pseudomonas syringae pv. tomato. Funct Integr Genomics. 2007;7(3):181–191. doi: 10.1007/s10142-006-0041-4. [DOI] [PubMed] [Google Scholar]
- 56.Asselbergh B, De Vleesschauwer D, Höfte M. Global switches and fine-tuning—ABA modulates plant pathogen defense. Mol Plant-Microbe Interact. 2008;21(6):709–719. doi: 10.1094/MPMI-21-6-0709. [DOI] [PubMed] [Google Scholar]
- 57.Gunnaiah R, Kushalappa AC, Duggavathi R, Fox S, Somers DJ. Integrated metabolo-proteomic approach to decipher the mechanisms by which wheat QTL (Fhb1) contributes to resistance against Fusarium graminearum. PLoS One. 2012;7(7):e40695. doi: 10.1371/journal.pone.0040695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen X, Steed A, Travella S, Keller B, Nicholson P. Fusarium graminearum exploits ethylene signalling to colonize dicotyledonous and monocotyledonous plants. New Phytol. 2009;182(4):975–983. doi: 10.1111/j.1469-8137.2009.02821.x. [DOI] [PubMed] [Google Scholar]
- 59.Irizarry RA, Hobbs B, Collin F, Beazerbarclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 60.Wang JR, Wang L, Gulden S, Rocheleau H, Balcerzak M, Hattori J, et al. RNA profiling of fusarium head blight-resistant wheat addition lines containing the Thinopyrum elongatum chromosome 7E. Can J Plant Pathol. 2010;32(2):188–214. doi: 10.1080/07060661003740512. [DOI] [Google Scholar]
- 61.Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, et al. agriGO v2. 0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017;45:122–129. doi: 10.1093/nar/gkx382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of genes differentially expressed in wheat spikes in response to seven hormones. Table S2. Gene enrichment analysis of DEGs in response to various hormones. Table S3. List of 1599 DEGs responsive to both F. graminearum and hormone treatments. Table S4. Gene enrichment analysis of DEGs commonly regulated by hormones (ABA, MeJA, or IAA) and F. graminearum. Table. S5. List of 157 DEGs responsive to water deficit stress in the glume and hormone treatments. Table S6. List of 29 genes that exhibited the opposite expression patterns in response to water deficit stress and IAA. Table S7. Details regarding the qRT-PCR primers. (XLSX 583 kb)
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the Gene Expression Omnibus (GEO) repository (accession code GSE103430). The other supporting data were included as additional files.