Abstract
Background
Phlebotomine sand flies (Diptera: Psychodidae) are haematophagous insects that transmit the protozoan parasite Leishmania infantum (Kinetoplastida: Trypanosomatidae), the main causative agent of both zoonotic visceral leishmaniasis (VL) and canine leishmaniasis (CanL) in the Mediterranean basin. Eight species of sand flies have been previously recorded in Romania: Phlebotomus papatasi, Phlebotomus alexandri, Phlebotomus sergenti, Phlebotomus perfiliewi, Phlebotomus neglectus, Phlebotomus longiductus, Phlebotomus balcanicus and Sergentomyia minuta. Three of them (P. perfiliewi, P. neglectus and P. balcanicus) were incriminated as vectors of L. infantum. Recent reports of autochthonous CanL in Romania require updates on sand fly distribution and diversity in this country.
Methods
Between 2013–2014 and 2016–2018, CDC light traps and mouth aspirators were used to collect sand flies in 132 locations from Romania, indoors and around various animal species shelters. Species identification of collected specimens was done using morphological keys, genetic tools and MALDI-TOF protein profiling.
Results
Sand flies were present in seven localities (5.3%): Eibenthal, Baia Nouă, Gura Văii (south-western Romania, Mehedinţi County); Fundătura, Pâhneşti, Epureni (eastern Romania, Vaslui County); and Schitu (southern Romania, Giurgiu County). Of the total number of collected sand flies (n = 251), 209 (83.27%) were Phlebotomus neglectus, 39 (15.53%) P. perfiliewi, 1 (0.40%) P. papatasi, 1 (0.40%) P. balcanicus and 1 (0.40%) P. sergenti (sensu lato).
Conclusions
We confirmed the presence of five sand fly species previously recorded in Romania. However, their updated distribution differs from historical data. The diversity of sand fly species in Romania and their presence in areas with Mediterranean climatic influences constitutes a threat for the reemergence of vector-borne diseases. In the context of CanL and VL reemergence in Romania, but also due to imported cases of the diseases in both humans and dogs, updates on vector distribution are imperative.
Electronic supplementary material
The online version of this article (10.1186/s13071-019-3507-7) contains supplementary material, which is available to authorized users.
Keywords: Sand flies, Distribution, Diversity, Canine leishmaniasis, Romania
Background
Phlebotomine sand flies (Diptera: Psychodidae) are haematophagous insects demonstrated to transmit the protozoan parasite Leishmania infantum, the main causative agent of both zoonotic visceral leishmaniasis (VL) and canine leishmaniasis (CanL) in the Mediterranean region [1]. Sand flies are of high importance for both veterinary and public health because of their role as vectors for leishmaniases and other bacterial and viral diseases [2, 3].
Of approximately 25 sand fly species present in Europe, several species were proven or suspected of Leishmania transmission, mostly of the genus Larroussius (6 species) and Adlerius (1 species) [3]. Of these, three vectors of VL and CanL caused by L. infantum were recorded in Romania: Phlebotomus (Larroussius) perfiliewi Parrot, 1930; Phlebotomus (Larroussius) neglectus Tonnoir, 1921; and Phlebotomus (Adlerius) balcanicus Theodor, 1948. Other species were also recorded in the country: Phlebotomus (Phlebotomus) papatasi (Scopoli, 1786); Phlebotomus (Paraphlebotomus) alexandri Sinton, 1928; Phlebotomus (Paraphlebotomus) sergenti Parrot, 1917; Phlebotomus (Adlerius) longiductus Parrot, 1928; and Sergentomyia (Sergentomyia) minuta (Rondani, 1843). Their historical distribution and diversity in Romania were reported by various authors between 1910 and 1971 [4–20].
As no later update regarding the species composition of sand flies is available in Romania our study aimed to bring new data on their distribution and diversity. The need of updated knowledge of the vector distribution is more urgent after the recent reemergence of autochthonous VL and CanL [21, 22], followed by the demonstration of seropositivity in dogs [23]. Moreover, new cases of imported CanL cases in Romania were reported [24, 25] which raised the question of possible incrimination of locally available, vector competent species in the trigger of further local transmission.
Methods
Study area and design
Between 2013–2014 and 2016–2018, 267 CDC miniature light traps (Trappola per Monittoraggio Zanzare, IMT Original 2002, Italy and John W. Hock, model 512, Gainesville, FL, USA) were placed in 132 localities in 21 counties in Romania (see Additional file 1: Table S1), during the expected activity of sand flies (July-August). One to eight traps were set in each locality per night. In some localities, more than one trapping site was selected, according to the visual evaluation of the field conditions and the possible occurrence of sand flies, considering their environmental requirements. A standardized protocol was used [26]. Traps were placed over night (19:00–5:30 h), in and around animal shelters, close to the walls, at about 1.5 m height. The traps were placed in locations selected according to the known habitat preferences of sand flies: altitude below 750 m, old stables, with decomposing organic matter, abundant dejections, walls built on a wood structure with a composition of clay, dry grass or stables with large agglomerations of domestic animals, abandoned buildings of former stables and ruins, along with old fences built with rocks [27]. In general, nights with no wind and no precipitations were chosen for sampling. Geographical coordinates, animal-related and shelter-related data were recorded on site. In one location (Eibenthal, Mehedinţi County), sand flies were also collected directly from the walls by mouth aspirator (John W. Hock Company, USA).
Species identification
After each trapping night, the sand flies were separated from other insects, and stored in 70% ethanol. For species identification, heads and genitalia of each specimen were dissected and slide-mounted in Swan solution, previously cleared with Marc-André solution (chloral hydrate/acetic acid). The sand flies were morphologically identified using entomological key [28] relying on specific morphological features of the pharynx and genitalia (spermathecae in females, external genitalia in males).
As the morphological identification of female specimens of species within the subgenera Larroussius and Adlerius may be challenging, molecular techniques were also applied for species confirmation. Seventeen specimens morphologically identified as different species (five P. perfiliewi, five P. neglectus, one P. papatasi, one P. sergenti sensu lato (s.l.) and specimens without a robust identification [four Phlebotomus (Larroussius) spp., one Phlebotomus (Adlerius) spp.] were randomly selected for DNA barcoding analysis. Additionally, other eight specimens belonging to P. perfiliewi morphospecies were subjected to MALDI-TOF protein profiling. The method was chosen as a new, time- and cost-effective molecular approach for species identification to help robustly identify P. perfiliewi females which can be challenging for morphological identification due to close resemblance with other Larroussius species, namely P. tobbi.
The DNA was extracted individually from the thorax of each specimen using the Qiagen DNeasy Blood and Tissue Kit (Qiagen, Austin, Texas, USA) following the manufacturer’s instructions and stored at − 20 °C. PCR amplifications of the cytochrome c oxidase subunit 1 (cox1) gene region (~ 660 bp) were performed in 50 µl reaction volume using LCO1490 and HCO2198 primers [29]. The amplification products were separated and visualized on 2% agarose gels, purified using the QIAquick PCR Purification Kit (Qiagen) and directly sequenced in both directions using the primers used for DNA amplification (ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit, Foster City, USA). Sequences were edited and aligned using BioEdit v.7.0.9.0 [30]. To construct taxon identity trees, obtained sequences and the similar sequences deposited in GenBank were subjected to Neighbor Joining (NJ) analysis using the Kimuraʼs 2- parameter (K2P) nucleotide substitution model in MEGA6.0 [31].
For MALDI-TOF protein profiling, thoraxes with wings and legs were manually homogenized, proteins were extracted from the homogenate by 25% formic acid and spotted on steel target plates with a matrix of an aqueous 60% acetonitrile/0.3% TFA solution of sinapinic acid (30 mg/ml; Sigma-Aldrich, St. Louis, USA) as previously described [32]. Protein mass spectra were measured on an Ultraflex III MALDI-TOF spectrometer (Bruker Daltonics, Bremen, Germany) within a mass range of 3–25 kDa and with external calibration using the Bruker Protein Calibration Standard I. Each spectrum represented an accumulation of 1000 laser shots (20 × 50 laser shots from different positions of the sample spot). For species identification, obtained protein profiles were processed using MALDI Biotyper 3.1 and searched against our in-house database which currently comprises reference spectra of 23 different sand fly species. Log score value (LSV) > 2.0 was accepted as the unambiguous assignment.
Mapping
The maps were generated using QGis 2.18.0 software (http://www.qgis.org). For historical data (presence/absence), georeferenced records were extracted from previously published data [23].
Results
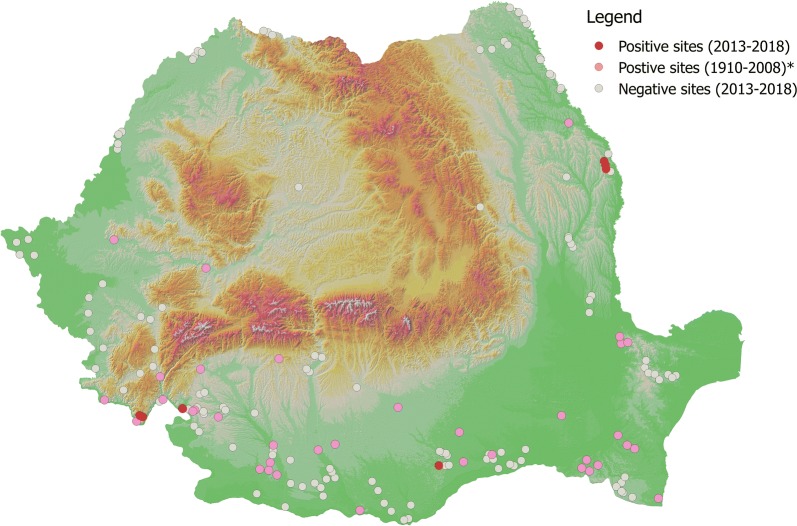
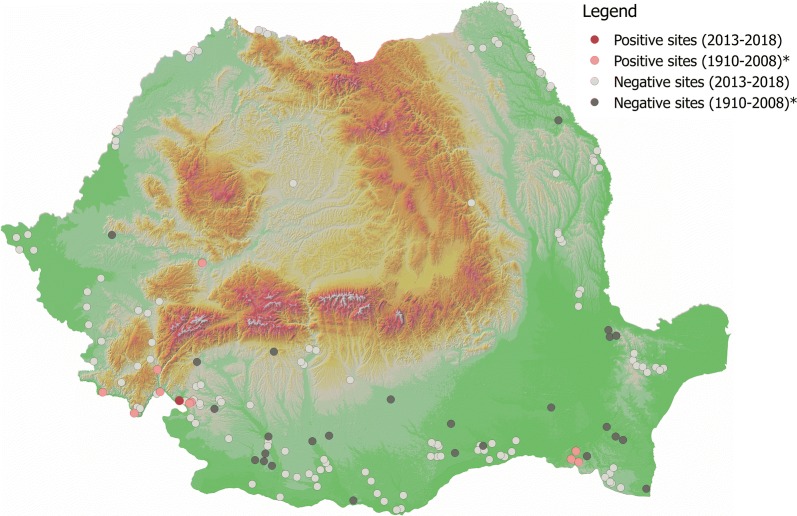
Sand flies were present in 7 out of the 132 sampled localities (5.3%). The positive and negative localities are shown in Fig. 1. A total of 251 sand flies (33 males and 218 females) were collected between 2013–2014 and 2016–2018. Out of 218 females collected, 42 (19.27%) were blood-fed and 5 (2.30%) were gravid. All specimens belonged to genus Phlebotomus (subgenera Phlebotomus, Paraphlebotomus, Adlerius, and Larroussius) (see Additional file 1: Table S1). The total number of positive localities for each species varied from one to five (Table 1).
Fig. 1.
All positive and negative sites for all sand fly species. Asterisk indicates historical data [23]
Table 1.
Trapped sand fly species by locality, sex, gravidity status and feeding status in Romania (2013–2014; 2016–2018)
| P. neglectus | P. perfiliewi | P. papatasi | P. balcanicus | P. sergenti s.l. | |
|---|---|---|---|---|---|
| Collected specimens | 209/251 (83.27%) | 39/251 (15.53%) | 1/251 (0.40%) | 1/251 (0.40%) | 1/251 (0.40%) |
| Positive localities | 3/132 (2.30%) | 5/132 (3.80%) | 1/132 (0.75%) | 1/132 (0.75%) | 1/132 (0.75%) |
| Collected males | 32/209 (15.31%) | 1/39 (2.60%) | 0/1 (0%) | 0/1 (0%) | 0/1 (0%) |
| Collected females | 177/209 (84.69%) | 38/39 (97.40%) | 1/1 (100%) | 1/1 (100%) | 1/1 (100%) |
| Blood-fed females | 42/177 (23.70%) | 0/38 (0%) | 0/1 (0%) | 0/1 (0%) | 0/1 (0%) |
| Un-fed females | 135/177 (76.30%) | 38/38 (100%) | 1/1 (100%) | 1/1 (100%) | 1/1 (100%) |
| Gravid females | 3/177 (1.70%) | 1/38 (2.64%) | 0/1 (0%) | 0/1 (0%) | 1/1 (100%) |
| Non-gravid females | 174/177 (98.30%) | 37/38 (97.36%) | 1/1 (100%) | 1/1 (100%) | 0/1 (0%) |
Two species, P. neglectus and P. perfiliewi were dominant (83.27% and 15.53%, respectively). Phlebotomus neglectus was found in Eibenthal, Baia Nouă and Gura Văii (south-western Romania, Mehedinţi County). Phlebotomus perfiliewi was found in Gura Văii (south-western Romania, Mehedinţi County), Fundătura, Pâhneşti, Epureni (eastern Romania, Vaslui County), and Schitu (southern Romania, Giurgiu County). Other three species, P. papatasi, P. balcanicus and P. sergenti s.l., were represented by a single specimen in a single locality (Gura Văii, Mehedinţi County, Romania). The present data (presence/absence) as well as historical (presence/absence) data for each of the five species identified in our study are shown in Figs. 2, 3, 4, 5 and 6.
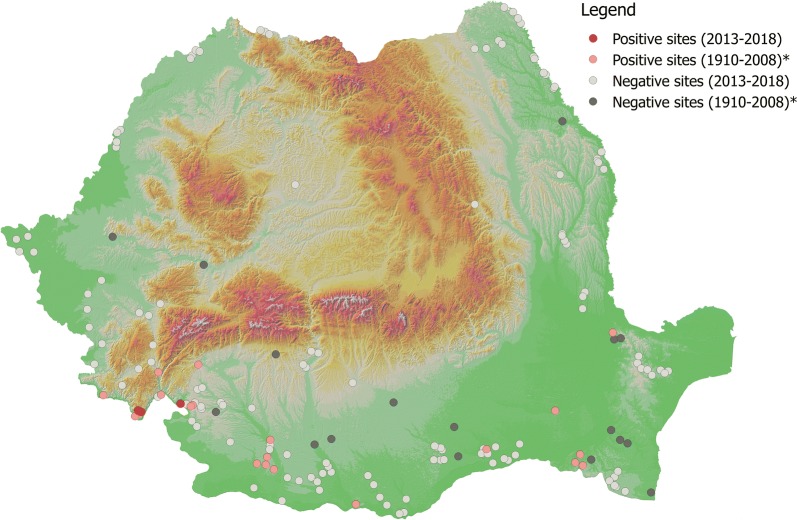
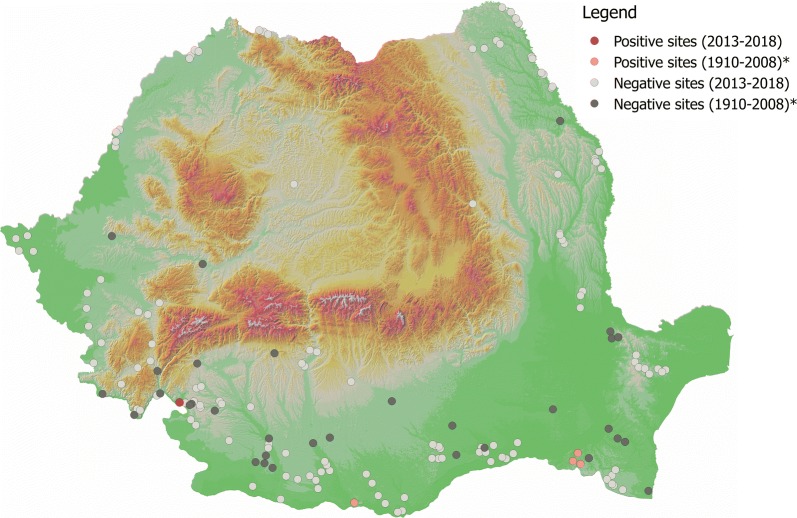
Fig. 2.
Distribution of P. neglectus. Asterisk indicates historical data [23]
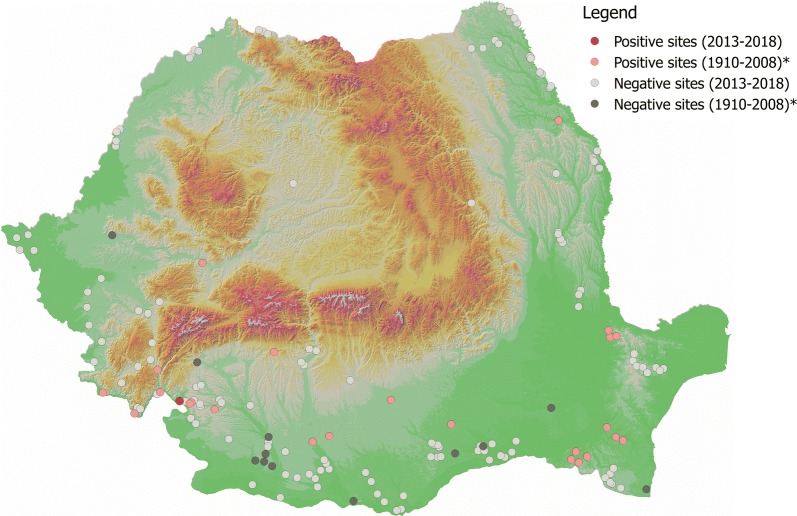
Fig. 3.
Distribution of P. perfiliewi. Asterisk indicates historical data [23]
Fig. 4.
Distribution of P. papatasi. Asterisk indicates historical data [23]
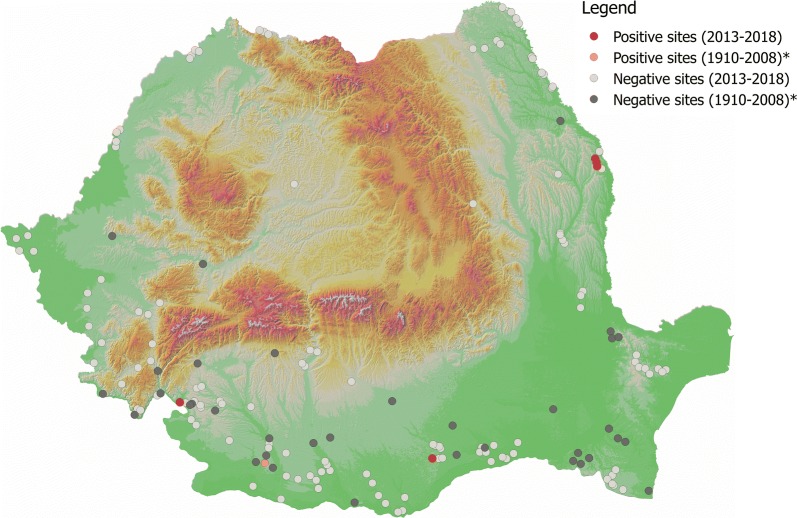
Fig. 5.
Distribution of P. balcanicus. Asterisk indicates historical data [23]
Fig. 6.
Distribution of P. sergenti s.l. Asterisk indicates historical data [23]
The aligned cox1 sequences of Romanian sand fly species were found to be 610 bp long without any deletions, insertions or stop codons. NJ analysis of these sequences with the ones available in GenBank and in our database showed that the specimens belonging to the same species clustered together. In addition, three Larroussius spp. females were assigned as P. perfiliewi and one as P. neglectus. The only specimen belonging to the subgenus Adlerius was grouped with P. balcanicus from Turkey (E. Kasap & B. Alten, unpublished data). Although the mean K2P distance between the Turkish and Romanian specimens was found to be high (5.3%), this difference could be attributed to the geographical origins of the individuals examined (Fig. 7). For the eight P. perfiliewi and five P. neglectus sequences analysed, only two and three unique haplotypes were detected, respectively. The sequence data of the haplotypes obtained for all the Romanian sand fly species are available in the GenBank database under the accession numbers MK425631–MK425638.
Fig. 7.
Neighbor joining tree constructed using the mitochondrial cox1 sequences of sand fly species from Romania (tip labels in bold) and sequences obtained from GenBank. Bootstrap values higher than 70% are shown
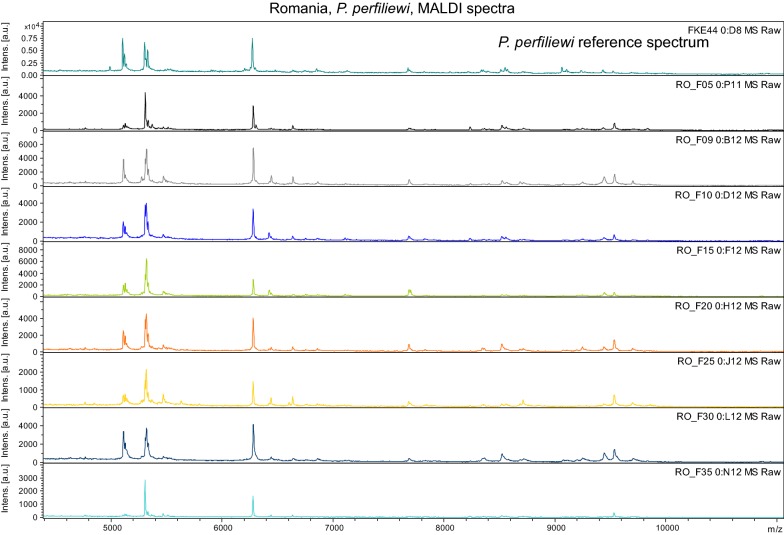
All specimens analysed by MALDI-TOF protein profiling were identified as P. perfiliewi (Fig. 8). Obtained spectra showed good quality and scored over a threshold of LSV 2, except for one specimen (RO_F35) which produced a spectrum of compromised quality and LSV < 2, nevertheless also for this specimen P. perfiliewi was the first suggested identification. Figure 8 shows overall protein spectra compared with a reference spectrum of P. perfiliewi from our in-house database.
Fig. 8.
MALDI spectra for P. perfiliewi (origin Romania)
Discussion
In the present study, five Phlebotomus species were identified out of the eight sand fly species previously described in Romania [33]. Three of them are of high importance in terms of L. infantum transmission: P. neglectus, P. perfiliewi and P. balcanicus [3]. Two of these three species represented 98.80% of all collected sand flies in the present study, highlighting the risk for local diseases transmission foci, as recent autochthonous and imported cases of VL and CanL were reported [21, 22, 24, 25].
Phlebotomus neglectus was the most abundant species in the present study (83.27%). It belongs to the P. major complex which currently comprises six species with different distribution: P. major in India, Nepal and Pakistan; P. wui in China; P. notus in Afghanistan; P. wenyoni in Iran and Iraq; P. syriacus in south-western part of Asia and Caucasia; and P. neglectus in south and south-eastern regions of Europe and Crimea) [34]. The species was reported for the first time in Romania in 1957 [13]. Its presence was historically recorded along the Danube valley and Bărăgan plain (south-western, southern and south-eastern Romania). It was mostly present outdoor, in arid to humid natural sites, at altitudes of 200–300 m, in valleys and at caves entrance, in crevices, but also peri-domestically, on building walls. The recorded temperature of its activity in Romania was between 24.9–29.8 °C, with average annual temperature of 11 °C [11, 15]. The current distribution of the species is limited to Mehedinţi Plateau (south-eastern Romania), at similar latitudes and environmental conditions as previously historically described [11, 15].
Phlebotomus neglectus has an important role as a vector for L. infantum in south-central, southern and eastern Europe and its presence and distribution was mostly correlated with CanL surveillance, in countries like Greece [35], Albania [36], Croatia [37], Montenegro [38] or Hungary [39]. In these countries, the species is also the most abundant and found at latitudes between 35.63°N (Karpathos Island, Greece), the southernmost, and 47.43°N, the northernmost (Törökbálint, Hungary). These geographical territories comprise regions with Mediterranean climate (Greece, Albania, Croatia and Montenegro), its transition to the temperate climate (northern Greece) and temperate climate with Mediterranean influences (southern Hungary and southern Romania).
The second most abundant species of the present study was P. perfiliewi. In Romania, the presence of P. perfiliewi was mentioned for the first time in 1956 [12]. The species was suspected for the first time to transmit VL and CanL in Romania in 1971 [11]. Its presence in Romania was described between the 10 °C and 11 °C isotherms [12]. In the present study, P. perfiliewi was present in five out of seven positive sites in south-western, southern and north-eastern Romania, the latter being a new locality, outside its previously known range. The species seems to have the widest geographical distribution in the country. It is also present in the neighbouring countries at similar average annual temperatures and latitudes [39–41].
Only one specimen of each P. papatasi, P. balcanicus and P. sergenti s.l. was collected from the same site, Gura Văii, Mehedinți County (south-western Romania). Phlebotomus papatasi was mentioned for the first time more than 100 years ago in Iaşi, north-eastern Romania [4]. In has also been found in parts of central, south-western, southern and south-eastern Romania [15, 16]. Phlebotomus papatasi is the vector for Leishmania major in North Africa and the Middle East and it is also known to transmit several phleboviruses [3]. The species is distributed from Portugal and Morocco in the West to India in the East [28]. In Serbia, a neighbouring country of Romania, P. papatasi was the predominant species in a recent study from 2017 [41]. In the present study, P. papatasi appears to have a more restricted distribution than previously described [15, 16] (Fig. 4).
Phlebotomus balcanicus was mentioned for the first time in Romania in 1958 [14]. The species was present in the central, south-western and southern regions of the country [11, 18]. Its current known distribution is limited to the Balkans, Turkey, Armenia and Georgia [41, 42]. Its current known distribution in Romania is limited to the south-eastern region of the country.
Phlebotomus sergenti s.l. was found in the southern and south-eastern parts of Romania [11]. The species is distributed in the semi-arid regions of the Mediterranean basin, North Africa, and Middle East, mostly at altitudes between 0–200 m [3, 43]. In the present study, its distribution in Romania is represented by south-eastern parts of the country.
The highest diversity of sand fly species recorded in the present study comprising all five collected species was in Mehedinţi Plateau (south-eastern Romania). Compared to the other positive locations situated in southern and north-eastern regions, where only one species was present, Mehedinţi Plateau has some distinctive climatic characteristics: (i) it is the only geographical region in Romania that has Mediterranean climatic influences (rainy autumns and softer winters); (ii) the annual average temperatures are between 10–11 °C, with 2–3 °C higher compared to the rest of the country; (iii) the altitudes are between 400–600 m, with precipitations between 700–800 mm [44]. Significant correlations were described between such climatic parameters (latitude, altitude, annual average temperature), the first date of sand fly collection and the type of the abundance trend [42], or between a higher species diversity and altitudes of around 900 m [43].
Conclusions
The updates provided by the present study on the distribution and diversity of the sand fly species in Romania show changes in both parameters, compared to historical data. Most localities where sand flies were reported at the beginning of the 20th Century are nowadays negative. This could be explained by a series of factors like the widespread use of insecticides in Romania during the malaria eradication programmes (1958–1964) [33], or the alterations of the sand fly habitats as a consequence of environmental, demographic, human behavioural factors or climate changes in the last decades [3]. Also, three other species that appeared to be present in the country, P. alexandri, P. longiductus and S. minuta, were not present in the surveyed trapping sites in our study. The negative trapping sites do not exclude, though, their presence in the country as the study has its limitations (e.g. more than one team to work in the field, nonhomogeneous data collection). The diversity of sand fly species in Romania and their presence in areas with Mediterranean climatic influences constitutes a threat for the reemergence of vector-borne diseases and a continuous surveillance is recommended in order to monitor the multiannual dynamics and to understand the real sand fly diversity and distribution. In the context of CanL and VL re-emergence, but also due to imported cases of the diseases in both humans and dogs, updates on vector distribution are crucial.
Additional file
Additional file 1: Table S1. Sampling information, site codes and trapped sand flies in Romania (2013–2015; 2016–2018).
Abbreviations
- VL
visceral leishmaniasis
- CanL
canine leishmaniasis
- CDC
Centre of Disease Control and prevention
- MALDI-TOF
matrix-assisted laser desorption/ionization time of flight mass spectrometry
- LSV
log score value
- s.l.
sensu lato
- cox1
cytochrome c oxidase subunit 1 gene
- NJ
neighbor joining
Authorsʼ contributions
BA, ADM, OEK, VD and PV designed the study. PV, VD, CDC, IRP, AMI and GO participated in the field work. CDC and GO carried out the morphological speciation of sand flies. CDC, OEK and PH used genetic tools and MALDI-TOF protein profiling for species identification. CDC performed the GIS data management. BA, PV, VD, OEK, ADM and CDC performed the data analysis. CDC drafted the original manuscript. BA, ADM, VD, PV, OEK and MOD critically revised the manuscript for important intellectual content. All authors read and approved the final manuscript.
Funding
This study was supported from the following project: VectorNet, a European network for sharing data on the geographic distribution of arthropod vectors, transmitting human and animal disease agents (contract OC/EFSA/AHAW/2013/02-FWC1) funded by the European Food Safety Authority (EFSA) and the European Centre for Disease Prevention and Control (ECDC), PCCDI 57/2018 (UEFISCDI), TE299/2015 (UEFISCDI), Ministry of Research and Innovation through Program 1 - Development of the National Research and Development System, Subprogram 1.2 - Institutional Performance - Projects for Financing the Excellence in CDI, Contract no. 37PFE/06.11.2018, title of the project: “Increasing the institutional performance through consolidation and development of research directions within the USAMVCN”, Czech Science Foundation (15-04329S) and the Institute of Microbiology (RVO61388971). The training of CDC was done under EurNegVec COST TD1303 Action.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional files. The sequence data of the haplotypes obtained for all the Romanian sand fly species are available in GenBank database under the accession numbers MK425631–MK425638.
Ethics approval and consent to participate
The traps were placed in and around animal shelters with the consent of the owners.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Cristina Daniela Cazan, Email: cristina.cazan@usamvcluj.ro.
Ioana Raluca Păstrav, Email: ioana.pastrav@usamvcluj.ro.
Angela Monica Ionică, Email: ionica.angela@usamvcluj.ro.
Gizem Oguz, Email: gizemoguz90@gmail.com.
Ozge Erisoz Kasap, Email: erisoz@hacettepe.edu.tr.
Vit Dvorak, Email: icejumper@seznam.cz.
Petr Halada, Email: halada@biomed.cas.cz.
Mirabela Oana Dumitrache, Email: mirabela.dumitrache@usamvcluj.ro.
Petr Volf, Email: volf@cesnet.cz.
Bulent Alten, Email: kaynas@hacettepe.edu.tr.
Andrei Daniel Mihalca, Email: amihalca@usamvcluj.ro.
References
- 1.Killick-Kendrick R. The biology and control of phlebotomine sand flies. Clin Dermatol. 1999;17:279–289. doi: 10.1016/S0738-081X(99)00046-2. [DOI] [PubMed] [Google Scholar]
- 2.Depaquit J, Grandadam M, Fouque F, Andry P, Peyrefitte C. Arthropod-borne viruses transmitted by Phlebotomine sandflies in Europe: a review. Euro Surveill. 2010;15:195–207. [PubMed] [Google Scholar]
- 3.Maroli M, Feliciangeli MD, Bichaud L, Charrel RN, Grandoni L. Phlebotomine sandflies and the spreading of leishmaniases and other diseases of public health concern. Med Vet Entomol. 2013;27:123–147. doi: 10.1111/j.1365-2915.2012.01034.x. [DOI] [PubMed] [Google Scholar]
- 4.Leon N. Note sur les diptères buveurs de sang de Roumanie. In: Contribution à l’étude de parasites animaux de Roumanie (1924). Ed. Cultura Naţionalã, Bucarest. 1910;54:67–69.
- 5.Nitzulescu V. Contributions à l’étude des phlébotomes de Roumanie. Ann Parasitol Hum Comp. 1929;7:430–437. doi: 10.1051/parasite/1929075430. [DOI] [Google Scholar]
- 6.Nitzulescu V. Sur le Phlebotomus chinensis. Ann Parasitol Hum Comp. 1930;8:362–375. doi: 10.1051/parasite/1930083362. [DOI] [Google Scholar]
- 7.Nitzulescu V. Essai de classification des phlébotomes. Ann Parasitol Hum Comp. 1931;9:271–275. doi: 10.1051/parasite/1931093271. [DOI] [Google Scholar]
- 8.Nitzulescu G, Nitzulescu V. Essai de table dichotomique pour la détermination des phlébotomes européens. Ann Parasitol Hum Comp. 1931;9:122–133. doi: 10.1051/parasite/1931092122. [DOI] [Google Scholar]
- 9.Nitzulescu V, Nitzulescu G. Sur la présence à Jassy de Phlebotomus chinensis var longiductus Parrot 1928. Bull Acad Méd Roumaine. 1938;5:166–170. [Google Scholar]
- 10.Duport M, Teodorescu AM. Contribuţiuni la studiul flebotomilor în România. Rev Stiint Med. 1946;35:46–55. [Google Scholar]
- 11.Duport M, Lupaşcu GH, Cristescu A. Contribution à l’étude des phlébotomes des biotopes naturels de Roumanie. Arch Roum Pathol Exp Microbiol. 1971;30:387–398. [PubMed] [Google Scholar]
- 12.Lupaşcu G, Agvriloaei P, Costin ME, Zelig M, Radvov G, Feodorovici S, et al. Contribuţii la studiul febrei papatace. Rev Stiint Med. 1956;8:265–269. [PubMed] [Google Scholar]
- 13.Lupaşcu G, Duport M, Cristescu A, Manoiu C, Raicu V. Présence de Phlebotomus perfiliewi dans la Republique Populaire Roumaine. Stud Cercet Inframicrobiol. 1957;8:121–126. [PubMed] [Google Scholar]
- 14.Lupaşcu GH, Dancescu P, Costin P, Stefanescu J. Contribuţii la studiul speciilor de flebotomi din R. P. R. Nota II. Prezenţa în ţara noastrã a speciilor de Phlebotomus major şi Phlebotomus chinensis var. simici. Stud Cercet Inframicrobiol. 1958;9:515–529. [Google Scholar]
- 15.Lupaşcu G, Dancesco P, Cheles N. Contribution à l’étude des espèces de phlébotomes (Diptera, Psychodidae) existant en Roumanie III, Présence de l’espèce Phlebotomus (Larroussius) major Annandale 1910, dans la région de Dobroudja. Observations sur la morphologie et l’écologie de l’espèce. Arch Roum Pathol Exp Microbiol. 1965;24:187–194. [PubMed] [Google Scholar]
- 16.Lupaşcu GH, Dancesco P, Cristesco A. Recherches sur les espèces de phlébotomes sauvages de Roumanie IV Présence de l’espèce Phlebotomus alexandri Sinton, 1928. Arch Roum Pathol Exp Microbiol. 1965;24:741–746. [PubMed] [Google Scholar]
- 17.Dancesco P. Recherches sur les espèces de phlébotomes de Roumanie. Académie Roumaine: Thèse docteur ès sciences; 1959. [Google Scholar]
- 18.Dancesco P. Donnés sur la sous-espèce Phlebotomus chinensis balcanicus Theodor, 1958 (Diptera, Psychodidae) en Roumanie. Arch Inst Pasteur Tunis. 1968;45:185–194. [Google Scholar]
- 19.Dancesco P, Cristesco A, Costin P, Stefanesco I, Mazilu V, Schirer A, et al. Nouvelles stations de capture de Phlebotomus longiductus Parrot, 1928 en Roumanie. Arch Inst Pasteur Tunis. 1970;47:57–63. [Google Scholar]
- 20.Cristesco A, Dancesco P. Contributions à la différentiation morphologiques des femelles appartenant aux espèces Phlebotomus major et Phlebotomes perfiliewi (Diptera, Psychodidae) de Roumanie. Arch Roum Pathol Exp Microbiol. 1967;26:319–326. [PubMed] [Google Scholar]
- 21.Gogoaşe MG, Teodorescu I, Preda C, Ionescu SC. Two case reports on visceral leishmaniasis diagnosed in Romania. Roum Arch Microbiol Immunol. 2013;72:49–62. [PubMed] [Google Scholar]
- 22.Mircean V, Dumitrache MO, Mircean M, Bolfa P, Györke A, Mihalca AD. Autochthonous canine leishmaniasis in Romania: neglected or (re)emerging? Parasit Vectors. 2014;7:135. doi: 10.1186/1756-3305-7-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dumitrache MO, Nachum-Biala Y, Gilad M, Mircean V, Cazan CD, Mihalca AD, Baneth G. The quest for canine leishmaniasis in Romania: the presence of an autochthonous focus with subclinical infections in an area where disease occurred. Parasit Vectors. 2016;9:297. doi: 10.1186/s13071-016-1583-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pavel G, Timofte D, Mocanu D, Malancus R, Solcan C. Imported leishmaniasis in a dog in a sandfly-populated area in northeastern Romania. J Vet Diagn Invest. 2017;29:683–685. doi: 10.1177/1040638717708391. [DOI] [PubMed] [Google Scholar]
- 25.Tanase OI, Daraban C, Velescu E, Boghean D, Bocaneti-Daraban F. Symptomatic leishmaniasis in an Italian Segugio dog from northeastern Romania: a case report. Iran J Parasitol. 2018;13:673–678. [PMC free article] [PubMed] [Google Scholar]
- 26.European Centre for Disease Prevention and Control; European Food Safety Authority. Field sampling methods for mosquitoes, sandflies, biting midges and ticks—VectorNet project 2014–2018. Stockholm and Parma: ECDC and EFSA. 2018.
- 27.Lawyer PG, Perkins PV. Leishmaniasis and trypanosomiasis. In: Eldridge BF, Edman JD, editors. Medical entomology—a textbook on public health and veterinary problems caused by arthropods. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2004. pp. 231–293. [Google Scholar]
- 28.Lewis DJ. A taxonomic review of the genus Phlebotomus (Diptera: Psychodidae) Bull Br Mus Nat Hist. 1982;45:171–209. [Google Scholar]
- 29.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 30.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic acids symposium series. 1999;41:95–98. [Google Scholar]
- 31.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dvorak V, Halada P, Hlavackova K, Dokianakis E, Antoniou M, Volf P. Identification of phlebotomine sand flies (Diptera: Psychodidae) by matrix-assisted laser desorption/ionization time of flight mass spectrometry. Parasit Vectors. 2014;7:21. doi: 10.1186/1756-3305-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dancesco P. Species of sandflies (Diptera: Psychodidae) in Romania, some aspects of their ecology and new capture stations. Trav Mus Nat Hist Grigore Antipa. 2008;51:185–199. [Google Scholar]
- 34.Léger N, Depaquit J. Systématique et biogéographie des Phlébotomes (Diptera: Psychodidae) Ann Soc Entomol France. 2002;38:163–175. [Google Scholar]
- 35.Tsirigotakis N, Pavlou C, Christodoulou V, Dokianakis E, Kourouniotis C, Alten B, Antoniou M. Phlebotomine sand flies (Diptera: Psychodidae) in the Greek Aegean Islands: ecological approaches. Parasit Vectors. 2018;11:1. doi: 10.1186/s13071-018-2680-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Velo E, Bongiorno G, Kadriaj P, Myrseli T, Crilly J, Lika A, et al. The current status of phlebotomine sand flies in Albania and incrimination of Phlebotomus neglectus (Diptera, Psychodidae) as the main vector of Leishmania infantum. Schallig HDFH. PLoS ONE. 2017;12:6. doi: 10.1371/journal.pone.0179118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bosnić S, Gradoni L, Khoury C, Maroli M. A review of leishmaniasis in Dalmatia (Croatia) and results from recent surveys on phlebotomine sandflies in three southern counties. Acta Trop. 2006;99:42–49. doi: 10.1016/j.actatropica.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 38.Ivović V, Ivović M, Miščević Z. Sandflies (Diptera: Psychodidae) in the Bar area of Montenegro (Yugoslavia) Ann Trop Med Parasitol. 2003;97:193–197. doi: 10.1179/000349803235001543. [DOI] [PubMed] [Google Scholar]
- 39.Farkas R, Tánczos B, Bongiorno G, Maroli M, Dereure J, Ready PD. First surveys to investigate the presence of canine leishmaniasis and its phlebotomine vectors in Hungary. Vector Borne Zoonotic Dis. 2011;11:823–834. doi: 10.1089/vbz.2010.0186. [DOI] [PubMed] [Google Scholar]
- 40.Melaun C, Krüger A, Werblow A, Klimpel S. New record of the suspected leishmaniasis vector Phlebotomus (Transphlebotomus) mascittii Grassi, 1908 (Diptera: Psychodidae: Phlebotominae)—the northernmost phlebotomine sandfly occurrence in the Palearctic region. Parasitol Res. 2014;113:2295–2301. doi: 10.1007/s00436-014-3884-y. [DOI] [PubMed] [Google Scholar]
- 41.Vaselek S, Ayhan N, Oguz G, Kasap O, Savić S, Di Muccio T, et al. Sand fly and Leishmania spp. survey in Vojvodina (Serbia): first detection of Leishmania infantum DNA in sand flies and the first record of Phlebotomus (Transphlebotomus) mascittii Grassi, 1908. Parasit Vectors. 2017;10:1. doi: 10.1186/s13071-017-2386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alten B, Maia C, Afonso MO, Campino L, Jimenez M, Gonzalez E, et al. Seasonal dynamics of phlebotomine sand fly species proven vectors of Mediterranean leishmaniasis caused by Leishmania infantum. PLoS Negl Trop Dis. 2016;10:2. doi: 10.1371/journal.pntd.0004458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simsek FM, Alten B, Caglar SS, Ozbel Y, Aytekin AM, Kaynas S, et al. Distribution and altitudinal structuring of phlebotomine sand flies (Diptera: Psychodidae) in southern Anatolia, Turkey: their relation to human cutaneous leishmaniasis. J Vector Ecol. 2007;32:269–279. doi: 10.3376/1081-1710(2007)32[269:DAASOP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 44.Ielenicz M, Pătru I. Physical geography of Romania. Bucharest: Academic; 2005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Sampling information, site codes and trapped sand flies in Romania (2013–2015; 2016–2018).
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional files. The sequence data of the haplotypes obtained for all the Romanian sand fly species are available in GenBank database under the accession numbers MK425631–MK425638.