Abstract
A plan to ‘genomicize’ epigenomics research and pave the way for breakthroughs in the prevention, diagnosis and treatment of human disease.
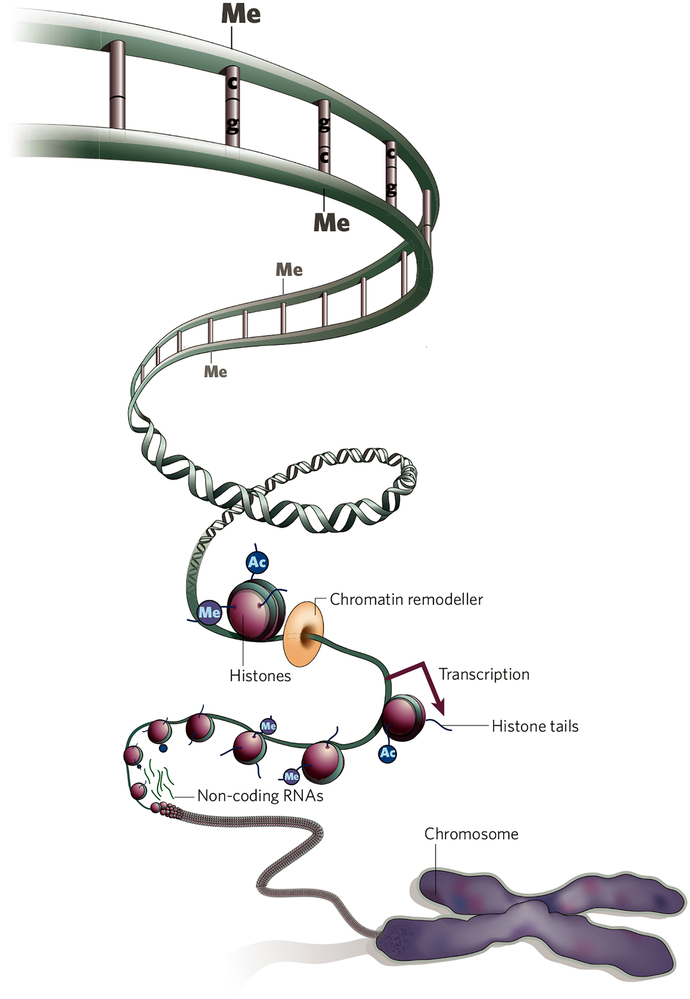
It is now possible to define whole epigenomes, representing the totality of epigenetic marks in a given cell type. Epigenetic processes are essential for packaging and interpreting the genome, are fundamental to normal development and are increasingly recognized as being involved in human disease. Epigenetic mechanisms include, among other things, histone modification, positioning of histone variants, nucleosome remodelling, DNA methylation, small and non-coding RNAs (Fig. 1). These mechanisms interact with transcription factors and other DNA-binding proteins to regulate gene-expression patterns inherited from cell to cell. The patterns underlie embryonic development, differentiation and cell identity, transitions from a stem cell to a committed cell and responses to environmental signals such as hormones, nutrients, stress and damage.
Figure 1 |. Epigenetic mechanisms.
The coding and structural information in the base sequence of DNA is organized in chromatin to form multiple epigenomes. DNA cytosine methylation and covalent modifications of the tails of histones and histone variants contribute information to nucleosomal remodelling machines that render genes and non-coding RNAs susceptible to transcription. Transcription factors (not shown) also play a major part in the competence and organization of the epigenome. The AHEAD project will map epigenetic marks in a defined set of epigenomes.
Although epigenomic changes are heritable in somatic cells, drug treatments could potentially reverse them. This has significant implications for the prevention, diagnosis and treatment of major human diseases and for ageing. Diseases to be targeted could include diabetes, cardio pulmonary diseases, Rett syndrome, other neurological disorders, imprinting disorders, autoimmune diseases and cancer, in which missteps in epigenetic programming have been directly implicated. Indeed, several inhibitors of chromatin-modifying enzymes including histone deacetylase (HDAC) inhibitors and DNA methyltransferase (DNMT) inhibitors have now been approved by the US Food and Drug Administration or are in clinical trials with good prognosis for tumour regression. Epigenetic therapy is now a reality, but to maximize the potential of such therapeutic approaches, it is crucial that there be a more comprehensive characterization of the epigenetic changes that occur during normal development, adult cell renewal and disease, and of the relation-ships between genetic and epigenetic variation and their impact on health.
The time is right for a major effort to decode the human epigenome, and we urge the community to join in a coordinated effort in support of the Alliance for the Human Epigenome and Disease (AHEAD) to help solve the problems of cancer and other intractable diseases. Just as the Human Genome Project provided a reference ‘normal’ sequence for studying human disease, the goal of the AHEAD project is to provide high-resolution reference epigenome maps. These maps would be of great use in basic and applied research, would have an immediate impact on under-standing many diseases, and would lead to the discovery of new means to control the diseases. Although the project should have a human focus, it will be essential to use model organisms to obtain mechanistic insights as to the functionality of epigenomic parameters or ‘codes’. Studying, genome-wide, the increasing number of interacting epigenetic mechanisms in a spectrum of cell types will involve data generation on a massive scale. An international project would provide the bioinformatics infra-structure needed to ensure that epigenomics research can be integrated with genomic data, and be utilized efficiently to advance the knowledge of human health and disease. Here, we discuss the benefits of the AHEAD framework to coordinate and plan an international Human Epigenome Project.
Early steps
Individual investigators have been studying epigenetics for several decades; however, concerted efforts to organize the epigenomics research community are quite recent, and none has sought to engage the community on a broad-based, international level.
Europe has a strong tradition for epigenetics research that has been recognized by European Union funding programmes and by individual national initiatives. More than €50 million (US$79 million) has been allocated to networks and consortia that focus on central epigenetic questions such as DNA methylation (HEP, Human Epigenome Project), chromatin profiling (HEROIC, High-Throughput Epigenetic Regulatory Organization In Chromatin), and treatment of neoplastic disease (EPITRON, EPIgenetic Treatment Of Neoplastic Disease). A special function is provided by the Epigenome Network of Excellence (NoE), created by the European Commission in 2004 (see Box 1).
Box 1 |. A European success story.
European researchers in the late 1990s were quick to take up the fast-emerging field of epigenetics, building on early discoveries in X-chromosome inactivation, Polycomb regulation, dosage compensation, imprinting, heterochromatin, DNA methylation, nucleosome modification and RNA interference (RNAi). The Epigenome Network of Excellence (NoE) was created by the European Commission in 2004 with the long-term aim of tackling major epigenetic questions in the post-genomic era. Under the direction of Thomas Jenuwein of the Research Institute of Molecular Pathology in Vienna, assisted by Phil Avner of the Pasteur Institute in Paris and Geneviève Almouzni of the Curie Institute in Paris, the intention was to create a durable structure for epigenetics research in Europe, promoting collaborations, sharing technologies and resources, and training and fostering the development of leaders in the field.
Now, after four of the five years planned, the NoE includes an original core of 25 members plus 35 associate members and 23 Newly Established Teams (NETs): a total of 83 laboratories from 12 European countries, as well as 350 laboratories worldwide that participate in ongoing activities through the NoE website. Expansion of the NoE resulted in the creation of Associate Members and, most importantly, the NETs. Through the NET programme, talented young European investigators in their first four years of independent research are selected to participate and provided with moderate financial support. The NET programme has been a resounding success and has given a powerful impetus to a cohort of young research talent.
The NoE programme of activities has promoted a common identity and generated numerous collaborations through training and mobility programmes, workshops, meetings and dissemination of technologies. In 2007 alone, the NoE collectively published some 300 research articles. A major effort has also been made to reach out to the broader community with a scientific website (www.epigenome-noe.net), and to the general public, through conferences, newspaper articles, and a public website (www.epigenome.eu), where news, information, explanations of the concepts, significance and applications of epigenetic research in lay terms are provided in seven languages. Several NoE members are involved in biotechnology companies and technology transfer has been the subject of NoE workshops.
These achievements have resulted from a conjunction of circumstance, hard work and the talent assembled by the NoE. Europe is in many respects preeminent in the field of epigenetics and the NoE could serve as a model for similar programmes worldwide. But, as its five-year mandate approaches its end, the future of the NoE remains uncertain. The organization, coordination and momentum achieved should be nurtured, supported and exploited to broaden connections with biotechnology, agriculture and particularly with the biochemical and pharmaceutical industries that are such key components of the European technological landscape. With the worldwide interest in epigenetics, this seems an inopportune moment for the European Commission to reduce support for this research and risk losing momentum and brainpower. On the contrary, the opportunity should be seized to forge close links with international efforts such as AHEAD.

In the past few years, there have been several efforts to organize the epigenetics research community in the United States and develop the support and structure for a human epigenome project. A 2004 National Cancer Institute (NCI)-sponsored Epigenetic Mechanisms in Cancer Think Tank concluded that the development of a US human epigenome project of analogous scope to the Human Genome Project should be of the highest priority (http://www.cancer. gov/think-tanks-cancer-biology/page7). This was followed by an NCI workshop in 2005 attended by programme staff from many National Institutes of Health (NIH) institutes. Key recommendations were: 1) comprehen-sive analysis of reference epigenomes, focused on stem cells and key differentiated lineages that can be modelled experimentally; 2) development of a standardized set of reagents, including monoclonal antibodies and common tissue substrates; and 3) development of a computational infrastructure for data sharing and display that puts epigenetic data in the hands of non-epigenetic investigators1.
The American Association for Cancer Research (AACR) organized a Human Epigenome Workshop in June 2005 and also a follow-up workshop in July 2006 that focused on planning an international project to map a defined number of human epigenomes (http://www.aacr.org/page9673.aspx)2. The consensus of workshop participants was that there are compelling reasons from both scientific and public-health perspectives to initiate a human epigenome project that could take full advantage of advances in several existing US and European initiatives.
On the heels of these workshops, the AACR Human Epigenome Task Force, a cross-disciplinary group of international investigators, was formed to design a strategy and develop a timetable for the implementation of an international Human Epigenome Project. As described below, the task force recommends the formation of AHEAD to coordinate a transdisciplinary, international project to map a defined subset of robust epigenetic markers in a limited number of human tissues at different stages of development and to develop a bioinformatics infrastructure to support the collection of epigenomic data. The efforts of this group will surely be bolstered by the recent and exciting decision of the NIH to fund epigenomics research as an interagency Roadmap initiative.
Asia, too, has been active in fostering epigenomics research, with a major emphasis placed on disease epigenomes, especially those in liver and gastric cancers. An international meeting, Genome-wide Epigenetics 2005, was held in Tokyo3. Scientists from Yonsei University (South Korea), the National Cancer Center (Japan), the Shanghai Cancer Institute (China) and the Genome Institute (Singapore) also organized meetings to facilitate the exchange of information in epigenomics and held their first meeting in Seoul in 2006, followed by another conference in Osaka in 2007. In December 2006, a Japanese Society for Epigenetics was formed. Clearly, Asia is now poised to contribute strongly to global epigenomics research.
National support for the Human Epigenome Project is also mounting in Australia with the formation of the Australian Alliance for Epigenetics to begin in late 2008. Australian meetings devoted to epigenetics were initiated in 1996 in Heron Island with Susan Clark of the Garvan Institute of Medical Research, Sydney, holding the first workshop on bisulphite sequencing and this has been followed by biannual meetings, the most recent being held in Perth in November 2007 to showcase Australia’s strengths in the global epigenetic research arena.
The scope of AHEAD
AHEAD would aim to provide reference epigenomes for key cellular states, such as a stem-cell phenotyope, proliferation, differentiation, senescence and stress, using common core specimens from different cell types, and to develop a bioinformatics infra-structure to support the collection and integration of epigenomic data. The precise objectives of such a project still need to be delineated. However, we would almost certainly expect AHEAD to 1) provide complete epigenome maps at very high resolution for important histone modifications across diverse cellular states in both human and mouse; 2) to complete and catalogue epigenome maps of model yeasts (Saccharomyces cerevisiae and S. pombe), plants (Arabidopsis and rice), and animals (Caenorhabditis elegans and Drosophila); 3) to deliver a high resolution DNA methylation map of the entire human genome in defined cell types and a landmark map for transcription start sites of all protein coding genes and a representative number of other features throughout the genome; 4) define non-coding and small RNAs; and 5) to establish a bioinformatics platform including a relational database, website and suite of analytical tools to organize, integrate and display whole epigenomic data on model organisms and humans. AHEAD will develop standardized procedures that assure data quality in the choice of reagents (antibodies), the experimental procedures (a minimal set of biological replicates) and data analysis (appropriate statistical procedures).
AHEAD would differ from and complement other ongoing projects such as ENCODE (ENCyclopedia Of DNA Elements). The ENCODE project is focused on defining the functional sequences in the genome, whereas AHEAD would define the patterns of epigenetic regulation occurring at those sequences in different cell states. The potential synergies between these two projects in terms of development of innovative molecular and computational techniques would enhance each of them. It would be valuable to coordinate such individual projects in the interests of efficiency and potential scientific insights. Some of these objectives are exemplified by the current efforts on model organisms carried out by the modENCODE (Model Organism ENCyclopedia Of DNA Elements) programme.
Reference epigenomes
In a multicellular organism, there are many potential epigenomes that define each cell type and reflect the current and past environments of individual cells. Epigenetic programming may even precede lineage choice and amplify signals from the environment. A major goal of AHEAD would be to map the epigenome in normal tissues and differentiating cells so that they could be compared with disease states, including but not restricted to cancer, that arise in these tissues. Therefore, the selection of which cell types to use and which epigenetic modifications to examine deserves careful consideration. Many environmental factors, such as ageing, hormonal milieu, diet and infection must be taken into account. Emphasis should be placed on comparing embryonic stem cells, adult stem and precursor cells, and related differentiated cells of primary human tissues, with the diseased state.
The reference systems selected should meet several criteria: 1) cells should be easy to sample in a reproducible fashion (renewable if possible); 2) cell numbers should be sufficient for analyses of DNA methylation and chromatin modifications; 3) cell progenitors should be identified that can be suitably manipulated and harvested in a pure state; 4) where possible, cells should be amenable to tissue reconstruction and three-dimensional model systems; and, of course, 5) systems must provide insight into key differentiation and related disease states. Some examples of suitable systems are given in Table 1.
Table 1 |.
Potential reference cell systems for epigenomic analysis
| Organ system | Tissue progenitor | Differentiated cell | Altered disease state |
|---|---|---|---|
| Embryo | Embryonic stem cells | ||
| Haematopoietic | CD34+ | Polymorphonuclear leukocytes, lymphoid, etc. | Acute myelogenous leukemia, Acute lymphoblastic leukemia, Chronic myelogenous leukemia, Myelodysplastic syndrome |
| Breast | CD24+and/or CD44+ | Myoepithelial, luminal, fibroblasts | Breast cancer |
| Liver | Oval cell | Hepatocytes, stromal cells | Cirrhosis, viral disease, Hepatocellular carcinoma |
Specific modifications to histones often correlate with gene activation or repression — for example lysine acetylation and trimethylation of lysine 4 of histone H3 (H3K4me3) are permissive for gene activation whereas H3K9me2 and H3K27me3 correlate with transcriptional silencing. Often, activating and repressive histone marks co-exist at gene start sites, reflecting perhaps epigenetic heterogeneity among otherwise similar cells, and it is the balance between these marks that determines gene expression states4–6. Because there are so many chromatin modifications, it may prove challenging to examine all of them in every cell type. We recommend that the initial set of reference epigenomes focus on several of the better understood covalent markers, such as those listed in Table 2, that could be mapped in a ‘first pass’ project defining reference epigenomes.
Table 2 |.
Recommended histone markers with known function to define reference epigenomes
| Histone modification | Function |
|---|---|
| H3K9ac, H4K16ac and H3K4me | Transcriptional ‘ON’ states |
| H3K9me and H3K27me | Transcriptional ‘OFF’ states |
| H3K36me | Elongation; primary transcripts |
| H3K9me and HP1 | Chromosome organization |
| H4K20me and H2BS14ph | Stress/damage response |
Several high-profile studies, including the human ENCODE maps7 and genome-wide profiling for histone modifications in mouse and human cells8,9, have been published recently. Although they present valuable first-generation epigenetic maps, these efforts largely focused on transcriptional regulation and functional sequences. We still need to know more about epigenetic control and plasticity in intergenic regions, mechanisms of heritability (Box 2), the role of repetitive elements, non-coding and small RNAs, and finally how epigenetic marking contributes to chromosome segregation, stress response and transducing signals from the environment.
Box 2 |. Heritability.
Epigenetics is often defined as somatically heritable changes in gene expression that do not involve changes in base sequence. DNA methylation patterns are known to be heritable through S phase of the cell cycle, however it has been more difficult to demonstrate mechanisms for the heritability of other chromatin modifications that will be mapped. Also, the abilities of transcription factors to reprogram the epigenome in normal and experimental situations need to be considered. The AHEAD project will help illuminate processes of heritability and reprogramming by providing genome-wide reference maps.
Advances in technology
Our understanding of the epigenome has been transformed in recent years by a succession of technological innovations. Approaches involving microarrays and, most recently, ultra-high-throughput sequencing technology have been applied to map chromatin modifications, cytosine methylation and non-coding RNAs across chromosomes and even entire genomes.
Genome-scale studies of histone modifications and other aspects of chromatin structure have typically relied on an immunological procedure, chromatin immunoprecipitation, in which specific antibodies are used to enrich chromatin. For example, an antibody against histone H3 acetylated at lysine 9 can be used to isolate genomic regions associated with this activating modification. Isolated DNA is then interrogated by microarray hybridization or by deep sequencing. Microarrays are a well-established readout, particularly suited to studies that require high coverage of a small subset of a given genome (for example, annotated gene promoters). Resolution on the order of the nucleosome (a few hundred bases) can be achieved with sufficiently dense tiling arrays. Recent studies — several dozen whole mammalian genome data sets have already been reported8,9 — have leveraged emerging ultra-high-throughput sequencing technology to ‘deep-sequence’ chromatin-immunoprecipitated DNA to generate genome-wide chromatin state maps. This sequencing approach offers potential advantages over arrays in precision, throughput and genome coverage. Notably, sequencing requires orders-of-magnitude less DNA than microarrays, and this enhanced sensitivity should enable studies of primary tissues, disease samples, and other limited cell populations of high biological importance.
The quality of the data derived from studies relying on chromatin immunoprecipitation depends mainly on the quality of the antibodies used, and there is an essential need for high-titre polyclonal antibodies with high specificity. Standardization of monoclonal antibodies has proven difficult because of low avidity and only limited epitope recognition. Reference maps defined by AHEAD, as well as those already developed from studies in human cells and model organisms, would provide a platform to assess the quality and robustness of modification-specific antibodies.
Rapid progress has also been made towards characterizing the global distribution of cytosine methylation. Genome-scale assays for detecting this epigenetic modification fall into two general categories. The gold-standard technique for reading out the methylation state of individual cytosines is bisulphite sequencing developed by Clark and Marianne Frommer at the University of Sydney10, in which unmethylated cytosines are converted to uracils and read as thymine, while methylated ones are protected from conversion. Although this method yields precise nucleotide-resolution data, bisulphite sequencing is challenging to scale — regions of interest must be sequenced several times in order for methylation patterns in a given cell type to be appreciated. A recent study showed the potential of combining bisulphite methodology with ultra-high-throughput sequencing by deciphering the entire Arabidopsis ‘DNA methylome’ at nucleotide resolution11. Still, bisulphite studies of the much larger human methylome have thus far been limited to relatively small subsets of genome12. For more comprehensive coverage, many investigators have turned to alternative approaches that involve the isolation of methylated (or unmethylated) fractions of genome by methylation-sensitive restriction or immunoprecipitation with a methylcytosine-specific antibody. The isolated fractions are then interrogated on microarrays. Several successes have been described recently using this genome fractionation/microarray approach, including a profile of promoter methylation in human fibroblasts13 and drafts of the complete Arabidopsis methylome14–16.
Model organisms
Model organisms have led the way in understanding epigenetic mechanisms of gene regulation. Position effect variegation, imprinting, transposon silencing, DNA methylation and histone modification were all discovered in model organisms and later found in humans. RNA interference and its role in epigenetics were also first described in model organisms (plants and worms).
The development of infrastructure to study model-organism epigenomes is under way and is having a major impact on human epigenome research, with hundreds of whole-genome or whole-chromosome profiles of histone modifications and DNA methylation already published in yeast, Arabidopsis, Drosophila and the mouse. Importantly, mutants in epigenetic modification, gene silencing, development and metabolism, as well as disease models, can be readily obtained and manipulated, and will allow the epigenetic pathways that are responsible for controlling genomic output to be determined first in model organisms. Other tools such as lineage-specific green fluorescent protein reporters and tagged histones have been engineered in mouse and Arabidopsis, allowing cell-type specific epigenomic profiles to be determined through cell sorting or immunoprecipitation. It is anticipated that new technologies such as massively parallel sequencing will make the generation of these profiles so straightforward that the limitations will be in the data analysis and the ability to take advantage of such genetic and cell biological tools. Reference epigenomes for several model organisms including S. cerevisiae, Arabidopsis, Drosophila, C. elegans, mouse and others will form an integral part of AHEAD. Clearly, AHEAD would enable coordination and expansion of these projects and rapid translation into human studies.
Computational challenges
As a comprehensive human epigenome project will require a strong bioinformatics platform, an early priority of AHEAD would be to establish a central relational database and a web interface to present data to the scientific community. This website would include analytical and statistical tools that could be used dynamically to process data for visualization. It would have tremendous utility for investigators, allowing immediate access to detailed maps for a locus of interest in the same way that the Human Genome Project provides sequence data. By initially focusing on establishing data interoperability, it is hoped that the database, or later versions of it, would allow for the simultaneous display of integrated epigenomic parameters on the entire human genome. In addition, not yet addressed is how the complex system of epigenomic regulation should be treated as a whole: the data could be a powerful resource for the emerging field of systems biology, allowing insights into the stability of the epigenomic networks and how they can be perturbed in disease states.
The development of these resources would be coordinated with parallel efforts already under way as part of the caBIG (the NIH Cancer Biomedical Informatics Grid) and Cancer Genome Atlas initiatives, among others. These resources could be distributed through the MGED (Microarray Gene Expression Data Society) and the BioConductor (open-source software) organizations. In addition, one could build on the sophisticated approaches of existing genome browsers used for data access and visualization, such as those at University of California, Santa Cruz, and ENSEMBL, to develop innovative means of data representation and web-based data analysis that would be of immense value to and foster collaborations among the scientific community.
Summary and perspectives
Dramatic changes in technologies have made AHEAD eminently feasible. Successful coordination of this multifaceted project through an AHEAD International Steering Committee would be essential if rapid progress is to be made, and it will require leadership from many stakeholders at all levels. The major challenges of AHEAD include the initial selection of the most important epigenomic markers, the identification of appropriate, pure cell populations, and the handling and presentation of the data. None of these challenges is insurmountable, and there are enormous potential scientific and public health benefits.
AHEAD would effectively integrate epigenetics research that is currently being conducted on a piecemeal basis around the world and would pave the way for breakthroughs in understanding normal and pathological processes. This research would offer significant scientific opportunities to maximize translational research to prevent and cure human diseases. The selection of epigenetics as a new NIH Roadmap initiative in the United States and the European Union’s Network of Excellence are important in helping establish the infra-structure needed to advance the field. We must now move AHEAD in earnest to achieve the goals of an international Human Epigenome Project.
Acknowledgements
We gratefully acknowledge Kimberly Sabelko from the AACR for her assistance in editing this manuscript. We also thank the AACR and Margaret Foti for supporting and funding the establishment of the Human Epigenome Task Force.
The American Association for Cancer Research Human Epigenome Task Force
Peter A. Jones1, Trevor K. Archer2, Stephen B. Baylin3, Stephan Beck4, Shelley Berger5, Bradley E. Bernstein6, John D. Carpten7, Susan J. Clark8, Joseph F. Costello9, Rebecca W. Doerge10, Manel Esteller11, Andrew P. Feinberg12, Thomas R. Gingeras13, John M. Greally14, Steven Henikoff15, James G. Herman3, Laurie Jackson-Grusby16, Thomas Jenuwein17, Randy L. Jirtle18, Young-Joon Kim19, Peter W. Laird1, Bing Lim20, Robert Martienssen21, Kornelia Polyak22, Henk Stunnenberg23, Thea Dorothy Tlsty24, Benjamin Tycko25, Toshikazu Ushijima26, and Jingde Zhu27
The European Union, Network of Excellence, Scientific Advisory Board
Vincenzo Pirrotta28, C. David Allis29, Sarah C. Elgin30, Peter A. Jones1, Robert Martienssen21, Jasper Rine31, and Carl Wu32
1USC/Norris Comprehensive Cancer Center, Los Angeles, California 90089, USA. 2NIH–NIEHS, Research Triangle Park, North Carolina 27709, USA. 3Johns Hopkins University, Baltimore, Maryland 21231, USA. 4Wellcome Trust Sanger Institute, Hinxton, Cambridge CB10 1SA, UK. 5The Wistar Institute, Philadelphia, Pennsylvania 19104, USA. 6Massachusetts General Hospital, Charlestown, Massachusetts 02129, USA. 7TGen, Phoenix, Arizona 85004, USA. 8Garvan Institute of Medical Research, Sydney, NSW 2010, Australia. 9Department of Neurological Surgery, University of California, San Francisco, San Francisco, California 94143, USA. 10Purdue University, West Lafayette, Indiana 47907, USA. 11Spanish National Cancer Center, Madrid, Spain. 12Departments of Medicine, Oncology and Molecular Biology & Genetics, Johns Hopkins University School of Medicine, Baltimore, Maryland 21205, USA. 13Affymetrix, Santa Clara, California, 95051, USA. 14Albert Einstein College of Medicine, Bronx, New York 10461, USA. 15Fred Hutchinson Cancer Research Center, Seattle, Washington 98104, USA. 16Harvard Medical School, Boston, Massachusetts 02115, USA. 17Research Institute of Molecular Pathology, Vienna 1030, Austria. 18Duke University Medical Center, Durham, North Carolina 27710, USA. 19Yonsei University, Seoul 120-719, Korea. 20Genome Institute of Singapore, Singapore 138672. 21Cold Spring Harbor Laboratory, Cold Spring Harbor, New York 11724, USA. 22Dana-Farber Cancer Institute, Boston, Massachusetts 02115, USA. 23Radboud University, Nijmegen 6525, the Netherlands. 24Department of Pathology, University of California, San Francisco, San Francisco, California, 94143, USA. 25Columbia University, New York, New York 10032, USA. 26National Cancer Center Research Institute, Tokyo 104-0045, Japan. 27Shanghai Cancer Institute, Shanghai 200032, China. 28Rutgers University, Piscataway, New Jersey 08854, USA. 29Rockefeller University, New York, New York 10021, USA. 30Washington University, Saint Louis, Missouri 63130, USA. 31University of California Berkeley, Berkeley, California 94720-3202, USA. 32National Cancer Institute, Bethesda, Maryland 20892, USA.
References
- 1.Feinberg AP, Jones PA & Ault G National Cancer Institute (NCI), Division of Cancer Biology (DCB) Workshop on Defining the Epigenome: Addressing the Value and Scope of a Human Epigenome Project. http://dcb.nci.nih.gov/Workshoprpt.cfm.
- 2.Jones PA & Martienssen R Cancer Res. 65, 11241–11246 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Lieb JD et al. Cytogenet. Genome Res. 114, 1–15 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azuara V et al. Nature Cell Biol. 8, 532–538 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Bernstein BE et al. Cell 125, 315–326 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Guenther MG, Levine SS, Boyer LA, Jaenisch R & Young RA Cell 130, 77–88 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birney E et al. Nature 447, 799–816 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barski A et al. Cell 129, 823–837 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Mikkelsen TS et al. Nature 448, 553–560 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark SJ, Harrison J, Paul CL & Frommer MF Nucleic Acids Res. 22, 2990–2997 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cokus SJ et al. Nature 452, 215–219 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckhardt F et al. Nature Genet. 38, 1378–1385 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber M et al. Nature Genet. 39, 457–466 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Zhang X et al. Cell 126, 1189–1201 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Zilberman D, Gehring M, Tran RK, Ballinger T & Henikoff S Nature Genet. 39, 61–69 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Vaughn MW et al. PLoS Biol. 5, e174 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]