Abstract
Bacterial biofilms represent an essential part of Earth’s ecosystem that can cause multiple ecological, technological and health problems. The environmental resilience and sophisticated organization of biofilms are enabled by the extracellular matrix that creates a protective network of biomolecules around the bacterial community. Current anti-biofilm agents can interfere with extracellular matrix production but, being based on small molecules, are degraded by bacteria and rapidly diffuse away from biofilms. Both factors severely reduce their efficacy, while their toxicity to higher organisms create additional barriers to their practicality. In this paper we report on the ability of graphene quantum dots to effectively disperse mature amyloid-rich Staphylococcus aureus biofilms, interfering with the self-assembly of amyloid fibers - a key structural component of the extracellular matrix. Mimicking peptide-binding biomolecules, graphene quantum dots form supramolecular complexes with phenol soluble modulins, the peptide monomers of amyloid fibers. Experimental and computational results show that graphene quantum dots efficiently dock near the N-terminus of the peptide and change the secondary structure of phenol soluble modulins, which disrupts their fibrillation and represents a strategy for mitigation of bacterial communities.
Keywords: carbon nanomaterials, extracellular matrix, phenol-soluble modulins, nanoscale biomimetics, quorum sensing, functional amyloid, biofilm regulation
Most microorganisms survive in the form of biofilms - structurally and functionally sophisticated microbial communities. Biofilms are implicated in numerous technological problems from water membrane failures to airplane crashes1,2 as well as in various diseases from persistent infections to cancer.3,4 Biofilms also form a large component of the ecosphere on Earth and their dysregulation can also cause significant ecological consequences.5
Biofilms act like multicellular organisms and possess exceptional adaptability and resilience to environmental factors and pharmaceuticals.6,7 Both of these properties are afforded by the protective function of extracellular matrix (ECM), which is made from several types of biomolecules (i.e., polysaccharides, DNA, and peptides).8,9 To date only a few classes of compounds have been shown to be effective anti-biofilm agents.10,11 Peptides rich in amino acids such as proline, arginine, phenylalanine or tryptophan have shown broad-spectrum activity in killing bacteria within biofilms by targeting a universal stringent stress response in bacteria.12 However, they are currently being developed only as antibiotic adjuvants because they are subject to rapid proteolytic degradation and are relatively expensive to synthesize. Small molecules have been used to inhibit the quorum sensing (QS) system13 or interfere with the polymerization of amyloid proteins.14 While further exploration of these strategies of biofilm inhibition need to be explored, the rapid loss of the small molecules due to diffusion into environment reduce their efficacy and increase their environmental toxicity.
These problems with the design of effective anti-biofilm agents prompted us to search for other material platforms. Graphene quantum dots (GQDs) are a single layer graphene a few nanometers in diameter. They are often regarded as biocompatible alternatives of II-VI semiconductor NPs also known as quantum dots (QDs).15 GQDs have been investigated for antibacterial activity due to reactive oxygen species production and membrane disruption.16–18 While these functionalities highlight the significance of GQDs as a part of microbiology toolbox, we hypothesize that these nanoscale particles can interfere with the self-assembly processes of biomolecular components of the biofilms. Representing the common tendency of nanoscale particles to self-assemble and replicate some other properties of biological macromolecules,19 this hypothesis is grounded in the multiple observations of GQDs and other NPs to specifically interact and assemble with other nanoscale particles and biomolecules.20–28 As applied to biofilms, GQDs can form ‘decoy’ complexes with the key structural components of ECM, thereby inhibiting the biofilm formation. Recently this functionality was confirmed by observation of GQDs acting as inhibitors of fibrillation of the protein characteristic for Alzheimer’s and Parkinson’s disease29–32 Counteracting known problems of other anti-biofilm agents mentioned above, GQDs have high molecular weight (103 to 105 g/mol) that slows diffusion and are resistant to proteolytic degradation.
As an experimental model to study the anti-biofilm effects of GQDs, we used biofilms of Staphylococcus aureus -- a major cause of hospital- and community-associated bacterial infections in the U.S. and around the world.33 Formation of S. aureus biofilms on host tissues and implanted medical devices contributes to chronic infections, as biofilms are known to be exceptionally resistant to host immune response and tolerant to antibiotics.36 The formation and dispersion of staphylococcal biofilms is dependent on the secretion of the phenol-soluble modulins (PSMs)33–35 -- small α-helical amphipathic peptides that have previously been implicated in bacterial virulence.36,37 Assembly of PSMs into fibers36 promotes the maturation of ECM,33,38,39 whose integrity renders these bacterial communities resistant to dispersal by proteinase K, Dispersin B, DNase and sodium dodecyl sulfate.33
The self-assembly process of PSMs into amyloid-like fibers involves backbone hydrogen bonding and side-chain interaction (e.ghydrophobic interaction, π-stacking, and van der Waals attraction)40 We took advantage of the characteristics of GQDs including polarizability, amphiphilic character, ability to form hydrogen bonds and participation in π-π stacking.41 Based on microscopy and spectroscopy data, we found that the growth of fibers from PSM peptides was dramatically stunted in the presence of GQDs. Furthermore, GQDs disrupt the structure and morphology of mature fibers. These effects resulted in dispersal of mature S. aureus biofilms. Computer simulations indicate that GQD docks at the N-terminal of PSM peptides via carboxyl edge-group and alters the PSM conformation.
RESULTS AND DISCUSSION
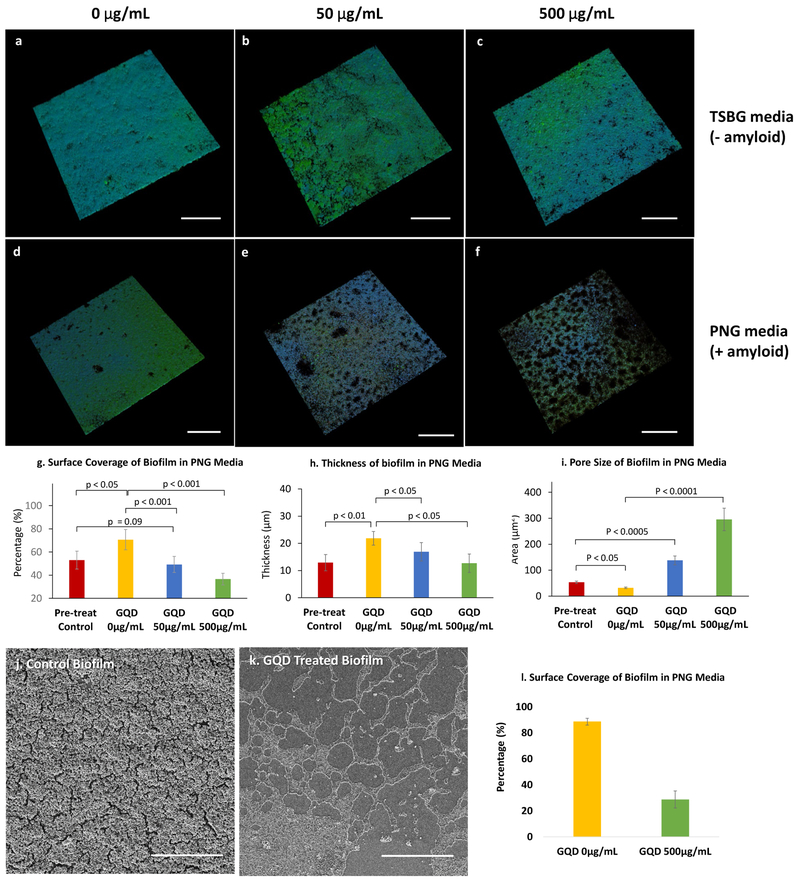
GQDs used in this study had a diameter of 2–8 nm15 and carried nine carboxyl edge-groups on average. (Figure S1) To investigate how GQDs affect amyloid-rich biofilms, S. aureus biofilms were grown in peptone-NaCl-glucose (PNG) media for four days in flow cells.33 S. aureus biofilms were also grown in tryptic soy broth supplemented with glucose (TSBG) media that inhibits amyloid synthesis as a negative control. GQD at concentrations up to 640 μg/mL have previously been shown to have minimal toxicity on mammalian cells.15,42–44 Hence, we applied 50 μg/mL and 500 μg/mL GQDs for the anti-biofilm investigations. Confocal images showed that S. aureus formed biofilm covering nearly the entire surface of the flow cell in both TSBG and PNG without GQDs treatment (Figure 1a, d). In the presence of 50 μg/mL GQDs, there is evidence of dispersion in the biofilm grown in PNG, whereas the biofilm on TSBG retained its structure (Figure 1b, e). A further dispersion of biofilm structure occurred in the presence of 500 μg/mL GQDs in PNG condition, while TSBG biofilm remaining fully present (Figure 1c, f). According to quantitative image analysis, surface coverage of the PNG biofilm (Figure 1g) decreased from 70.6 ± 8.8% to 49.3 ± 6.9% when cultured with 50 μg/mL GQDs; the surface coverage further decreased to 36.6 ± 5.0% for 500 μg/mL GQDs. The thickness of the PNG biofilm (Figure 1h) decreased from 21.9 ± 2.5 μm to 16.9 ± 3.4 μm when cultured with 50 μg/mL GQDs; the thickness further decreased to 12.7 ±3.4 μm with 500 μg/mL GQDs. The thickness for biofilms grown in TSBG media showed limited change with addition of any dose of GQDs (Figure S2). The decrease in surface coverage highlighted by the pores and heterogeneity of biofilm indicate areas of dispersion.45 Although the total surface coverage is quantitatively similar between the pre-treat controls and the 50 μg/mL GQDs (p = 0.09), the quantitative size of the voids (Figure 1i) in the 50 μg/mL GQDs is 2.6 times larger than the pre-treat controls, suggesting disruption of the architecture consistent with dispersal. This is confirmed by actual quantitative loss of coverage and further increase of void size at the higher dose (500 μg/mL). The structural changes of amyloid-rich biofilms before and after GQD (500 μg/mL) treatment was further verified by scanning electron microscopy (SEM) (Figure 1j, k). Here the change in surface coverage of the PNG biofilm is even more distinct than that in confocal microscopy, which decreased to 28.8 ± 6.6 % (Figure 1l). This is likely a result of dehydration process required for SEM sample preparation, which washed off weakly attached bacterial cells and loose ECM content. The phenomenon not only confirmed the decrease of biofilm surface coverage after GQD treatment but also indicated reduced integrity of remaining biomass.
Figure 1. Effects of GQDs on S. aureus biofilms.
(a-f) Confocal microscopy of S. aureus biofilm grown for 3 days and then treated for 1 day; stains - polysaccharide intercellular adhesin (PIA; green) and bacterial cells (blue). Biofilms grown in TSBG medium in presence of (a) 0 μg/mL (f) 50 μg/mL and (c) 500 μg/mL GQDs. Biofilms grown in PNG medium in presence of (d) 0 μg/mL (e) 50 μg/mL and (f) 500 μg/mL GQDs. Bar graphs of quantified (g) surface coverage, (h) thickness and (i) void size of biofilms analyzed from the z-stack images for each group in (a-f). (j, k) Representative SEM images and their (l) calculated surface coverage of S. aureus biofilms grown in PNG media. Scale bar: 200 μm (a-f), 50 μm (j, k).
Of note, the growth rate of planktonic S. aureus did not differ significantly with or without GQDs up to 200 μg/mL (Figure S3). This observation indicates that the GQDs are not toxic to the individual bacteria cells per se, and thus the biofilm dispersal is associated with change in ECM integrity only. Furthermore, the specificity for the GQD effect on PNG grown biofilm versus the TSBG biofilm suggests that amyloid is the specific target of GQDs among other components of the ECM.
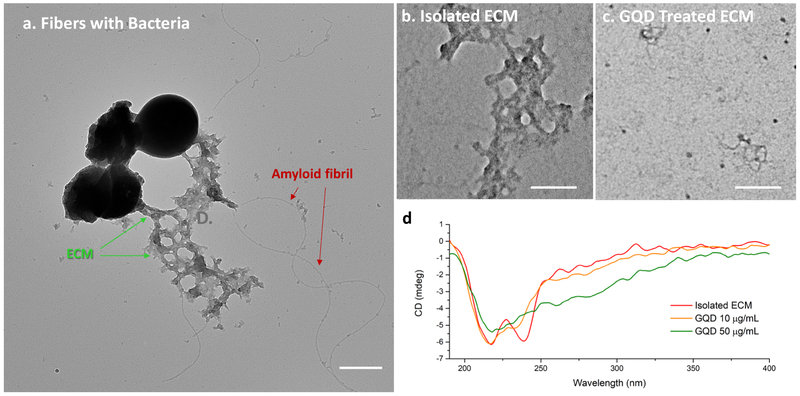
The formation of amyloid fibers in S. aureus biofilms grown in PNG were confirmed by transmission electron microscopy (TEM). The fibers, with a diameter of about 12 nm, were closely associated with bacterial cells and surrounding ECM; their dimensions are comparable to those observed in prior studies (Figure 2a). 33 Amyloid fibers and surrounding ECM were extracted from the biofilm cultures and further analyzed with respect to the effect of GQDs on matured amyloid fibers included in ECM. In general, various components from ECM may closely combine with the amyloid-fibril core (Figure 2a, b), which may contain PSM peptides, extracellular DNA and polysaccharide intercellular adhesion (PIA). The GQDs, with concentration of 50 μg/mL, dramatically altered the morphology and integrity (Figure 2c).
Figure 2. TEM and CD spectra of isolated ECM from S. aureus biofilm grown in PNG and exposed to GQDs.
(a) Amyloid fibrils in composition with ECM attached to bacteria; (b) Extracted composite ECM treated with (b) 0 μg/mL and (c) 50 μg/mL GQDs for 4 days, (d) CD spectra of isolated ECM treated with 0 μg/mL, 10 μg/mL, and 50 μg/mL GQD for 4 days. Scale bars: 500 nm (a), 100 nm (b, c). Only low concentrations of GQDs were evaluated in this test due to the low concentration of ECM obtained from the isolation procedure compared to that from intact biofilms.
Circular dichroism (CD) spectrometry was used to obtain complementary information about the secondary structure of the amyloid fibrils. A wide negative peak between 200 nm to 230 nm observed in the CD spectrum (Figure 2d) is characteristic of amyloid-like fibrils including typical β-sheet negative bands at 218 nm and α-helix negative bands at 208 nm and 222 nm. The negative CD peak at ≈245 nm is associated with extracellular DNA.46 All peaks between 200 nm to 250 nm were significantly decreased after two-hour incubation with GQDs suggesting that GQDs cause disruption of the fibril composite structure of extracted biofilm ECM within this timeframe.
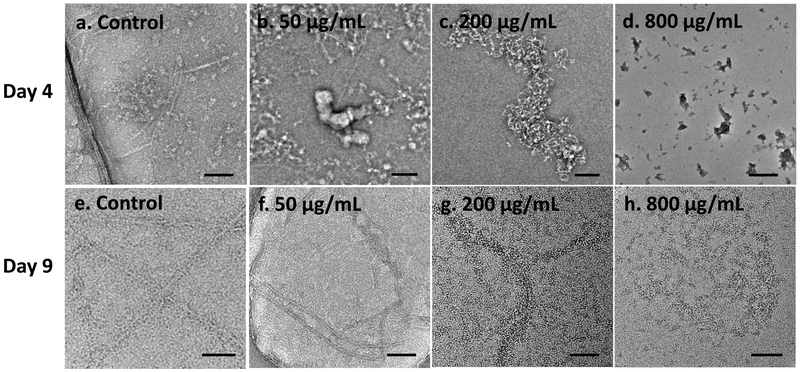
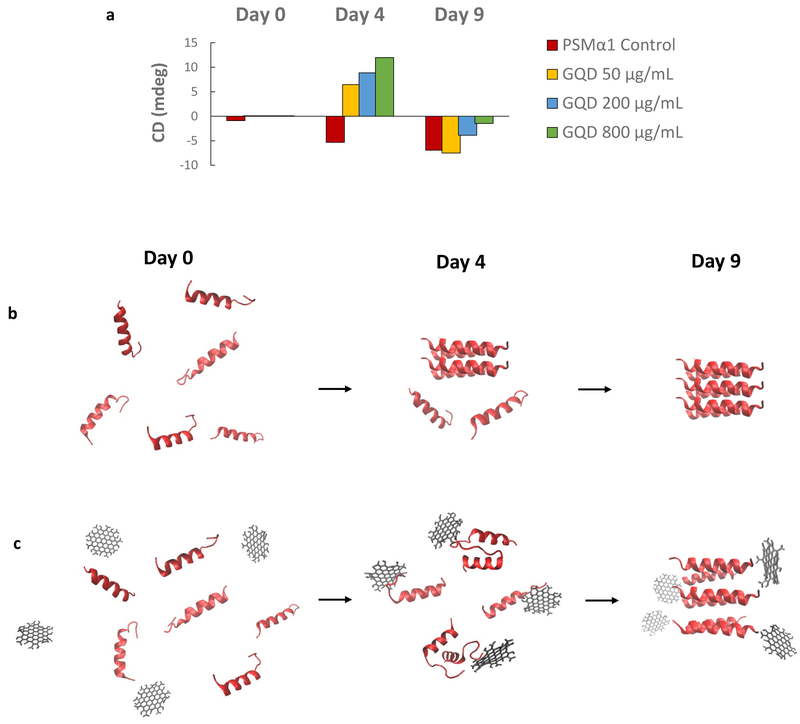
The main content of the isolated amyloid fibers from PNG biofilm consists of small α-helical amphipathic peptides, PSMs.33 PSMα1 is one of these PSM peptides with capacity to form amyloid-like fibrils. We used commercially available, synthetic PSMα1 to further investigate the effects of GQDs. Their fibrillation was monitored for 9 days with and without the addition of GQDs. The longer time of the fibrillation is caused by the different aggregation feature of PSMα1 in vitro from PSMs in ECM of biofilm.38 Control peptides start forming short fibers in diameter of 11.4 ± 1.79 nm and in length of 134.4 ± 59.5 nm on Day 4, and the fibers were continuously elongated till Day 9 with similar diameter of 12.4 ± 0.82 nm (Figure 3a, e). The length of the elongated fibers on Day 9 was not measured as the fiber length exceed the field of view and was confounded by nonlinear geometry. Meanwhile, to better understand the mechanism of their interactions, PSMα1 peptides were incubated with GQDs at the concentrations of 50, 200 and 800 μg/mL to obtain molar ratios of GQD to PSMα1 of 1:4, 1:1 and 4:1. After incubation with GQDs for four days, small aggregates in diameter of 96.1 ± 49.98 nm, 127.9 ± 47.58 nm and 102.4 ± 48.90 nm instead of short fibers were found respectively in the peptide solution incubated with GQDs at the concentrations of 50, 200 and 800 μg/mL (Figure 3b-d). While incubated with 50 μg/mL GQDs after nine days, a few fibers in diameter of 10.39 ± 2.06 nm were formed as well (Figure 3f), including short fibers with length of 201.59 ± 64.4 nm and elongated fibers coiled together. The density of the fibers was 13 ± 6 fibers/μm2 compared to 46 ± 12 fibers/μm2 in the control sample according to the average of 20 TEM images for each group. At the same time, there were few short fibers and limited elongated fibers formed in diameter of 12.02 ± 2.79 nm in the sample of 200 μg/mL GQDs (Figure 3g). However, there were only small aggregates in diameter of 32.28 ± 11.03 nm observed in the TEM images with 800 μg/mL GQDs (Figure 3h). After 9-day incubation of 800 μg/mL sample, large aggregates were sedimented at the bottom (Figure S4). Given the observed structures in TEM samples are from the suspension of incubation solution, the average diameter of measured aggregates decreased over time from day 4 to day 9. In all the images, the contrast of the agglomerates was enhanced with increasing GQD concentration, which is associated with the tendency of GQDs to co-assemble with amyloid peptides. The delayed fibrillation for 4 days suggested that the kinetics of fibril formation changed in presence of GQDs which matches the expectations of the formation of ‘decoy’ complexes. Similarly to NPs,47 these supramolecular structures affect the secondary structure of peptides that serve as nucleation centers for fibrillation. Thus, binding of GQDs to PSM monomers or small prefibrillar oligomers is likely to hinder the formation of critical nuclei and elongation of the fibrils.
Figure 3. PSMα1 fibril formation with and without addition of GQDs.
TEM images of PSMα1 peptides after addition of 0, 50, 200, 800 μg/mL GQDs (a-d) incubated for 4 days or (e-h) 9 days. Scale bars: 100 nm.
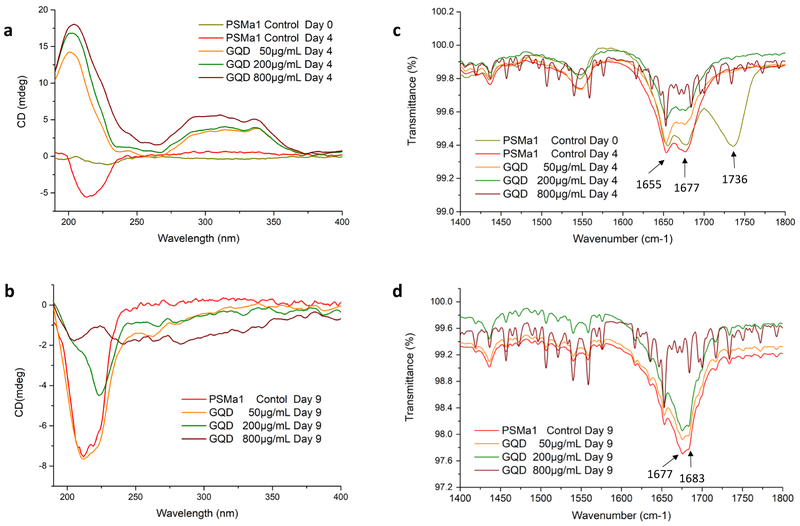
To verify this point, structural change in PSMα1 peptides interacting with GQDs were monitored for conformational conversion on Day 4 and Day 9 of incubation using CD spectrometry. As previously observed, there are two stages of conformational and morphological changes of peptide aggregation for PSM amyloid-like fibrils. Initially, the freshly dissolved PSMα1 peptides mainly contain α-helixes.48 The typical β-sheet signal with a negative band at ≈ 218 nm,38 appears after four days for PSMα1 without GQDs (Figure 4a). This suggested that formation of amyloid fibrils, which agreed with the result of ThT staining (Figure S5). Interestingly, secondary structures of PSMα1 incubated with GQDs showed enhancement of β-turns signal with a positive band at ≈208nm, in agreement with the theoretical peak of pure β-turns signal.49,50 Meanwhile, a broad peak from 250 nm and 375 nm was observed, corresponding to the absorption of GQDs onto peptide (Figure S6) and associated with the deformed (coiled) conformation of the graphene sheet.15 The combination of these two positive CD peaks of PSMα1 induced by GQDs in the solution are potentially contributed by the interaction between PSMα1 monomers and GQDs. Such interaction may change secondary structure of peptides incubated with GQDs, and/or enhance the CD signal of turns by GQD. Subsequently, the negative band at ≈ 218 nm of β-sheet was enhanced in PSMα1 self-assembling without GQDs after a nine-day incubation, which agrees with the significant increase in ThT fluorescence emission (Figure 4b, S5). The change of CD peak in PSMα1 control from Day 4 to Day 9 indicated the transition from lag phase of individual peptides and oligomer to fibril formation.47 Fibrillation of PSMα1 incubated with GQDs was inhibited when the molar ratio of peptides and GQDs are equal or more than 1:1 (GQD concentration at 200 μg/mL), However, the fibrillation CD signal of PSMα1 was further interrupted when incubated with excess amount of GQDs (800 μg/mL) on both Day 4 and 9 (Figure 4a, b).
Figure 4. Secondary structure of synthetic PSMα1 peptides and fibrils with GQDs.
(a, b) CD spectra of PSMα1 with/without addition of 50, 200, 800 μg/mL GQDs for (a) 4 days and (b) 9 days. FTIR spectra of PSMα1 with/without addition of 50, 200, 800 μg/mL GQDs for (c) 4 days and (d) 9 days. The intense signals between 1735–1740 cm−1, for control PSMα1 on Day 0, are indicative of C=O stretch of carboxylic group, which disappeared after incubation for four days in all samples. This is contributed by the minimum remains of trifluoroacetic acid (TFA) and hexafluoroisopropanol (HFIP), which cleave synthesized peptides to mono-dispersed peptide (Figure S7). This peak disappeared after 4 hours at room temperature and the remains did not change the pH of the peptide solution.
To confirm the change of secondary structure in protein, we monitored PSMα1 fibrillation with or without GQDs using attenuated total reflectance - Fourier transform infrared spectroscopy (ATR-FTIR) (Figure 4c, d). Deconvolution of the absorbance spectra in the amide I region51 is indicative of the individual secondary structure components and their relative contribution to the main signal. PSMα1 peptides incubated with or without addition of GQDs for 4 days display common bands both at ≈ 1655 cm−1 and ≈ 1677 cm−1. The peak at 1655 cm−1 corresponds to α-helix which appears between 1650 cm−1 and 1657cm−1 and overlapped with turns between 1655 cm−1 and 1675 cm−1. The latter is attributed to the weak peak of antiparallel β-sheet structure 1670-1690 cm−1. With addition of GQDs, antiparallel β-sheet were significantly decreased and indicated the inhibition of amyloid fibrillation at early stage of fibril growth. Meanwhile, the band at ≈ 1655 cm−1 right-shifted and became narrower, which verified the slight changes from α-helix to β-turns in PSMα1 dispersion. After 9 days, the α-helix peak decreased in all samples. At the same time, the peaks at 1670-1690 cm−1 increased in the samples of control, 50 and 200μg/mL of GQDs, indicating the formation of β-sheet nucleation centers for fibrillation. The peak intensity of β-sheet at 1677 cm−1 and 1683 cm−1 became lower with presence of higher concentration of GQDs, which verifies the inhibitory effect of GQD on fibrillation.
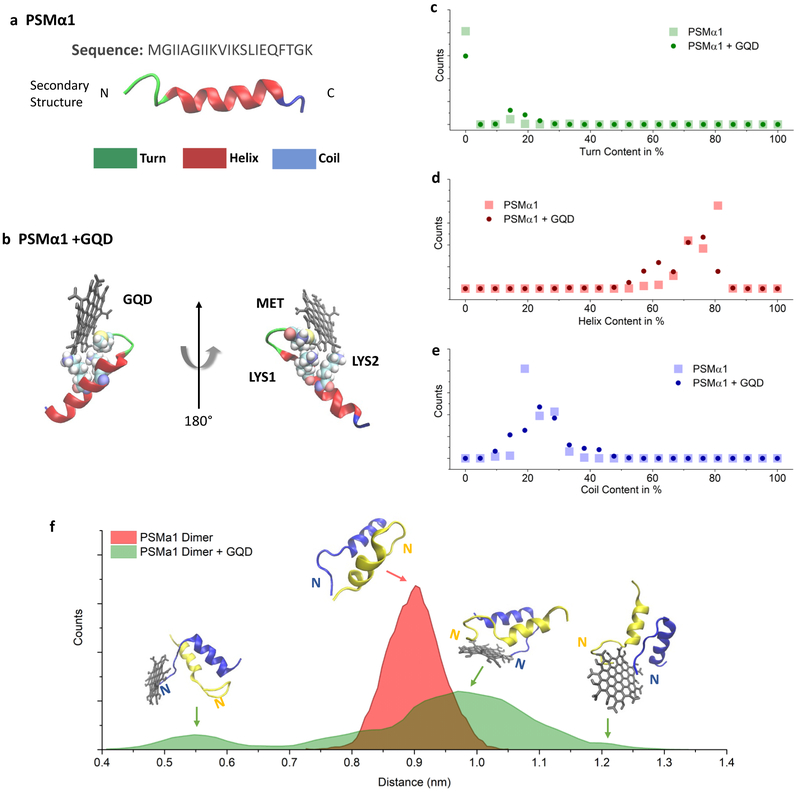
Molecular dynamics (MD) simulations were used to study the interactions between GQD and PSMα1 by monitoring the changes in secondary structure of the individual peptides. PSMα1 peptide contains 21 amino acids and consist of a single amphipathic helix with a slight bend near the N- and C-terminal (Figure 5a).52 Eight different simulations, with the GQD initially placed at the vertices of a cuboid encasing a single PSMα1, were tested to study the interactions between the two molecules. The results indicate that electrostatic interactions between the positive charged residues near the PSMα1 N-terminal, methionine (MET) and lysine (LYS), and –COO− groups present on the edges of GQD lead to the formation of stable conformational complexes (Figure 5b). The simulations show a large pool of such complexes, where the position and the distance of the carboxylic groups affect the structure of the aggregate. Therefore, due to the relative high number of stable configurations and the slow transition among them, we cannot make conclusive remarks about the details of the most stable complexes. However, the analysis of the PSMα1 secondary structure changes due to the GQD-peptide interactions is revealing. The PSMα1-GQD complex, shows an increase of 311.0 % in the number of turn motifs compared to the isolated peptide (Figure 5c), which is consistent with the enhanced CD signal of β-turns at ≈208 nm. At the same time, the number of peptide configurations containing both helix and coils decreases by 3.0 %, but the distributions are affected differently: the residues involved in helix secondary structure decrease in both number and frequency, while the variance of the number of residues in a coil configuration increase (Figure 5d, e). In other words, the peptide losses α-helix structure in favor of the formation of β-turns or coils. Overall, the formation of stable PSMα1/GQD complexes by electrostatic interactions leads to relevant changes in the secondary structure and peptide motifs, and therefore we expect the presence of GQDs to affect unfavorably the organization of PSMα1 peptides on a larger scale. Specifically, the increase in structural disorder alters the initial stage of PSMα1 fibrillation thus inhibit the formation of nucleation centers.
Figure 5. MD simulation of PSMα1 and GQDs.
Schemes of (a) PSMα152 and (b) PSMα1 and GQD complex, β-tiirns are shown in green, alpha helix in red and random coils in blue, (c-e) Histogram of secondary structure amounts in PSMα1 in the GQD/PSMα1 complex, (f) Histogram of the center of mass distance between two PSMα1 units showing the distance distribution with and without a GQD molecule. N: N-terminal of PSMα1; C: C-terminal of PSMα1.
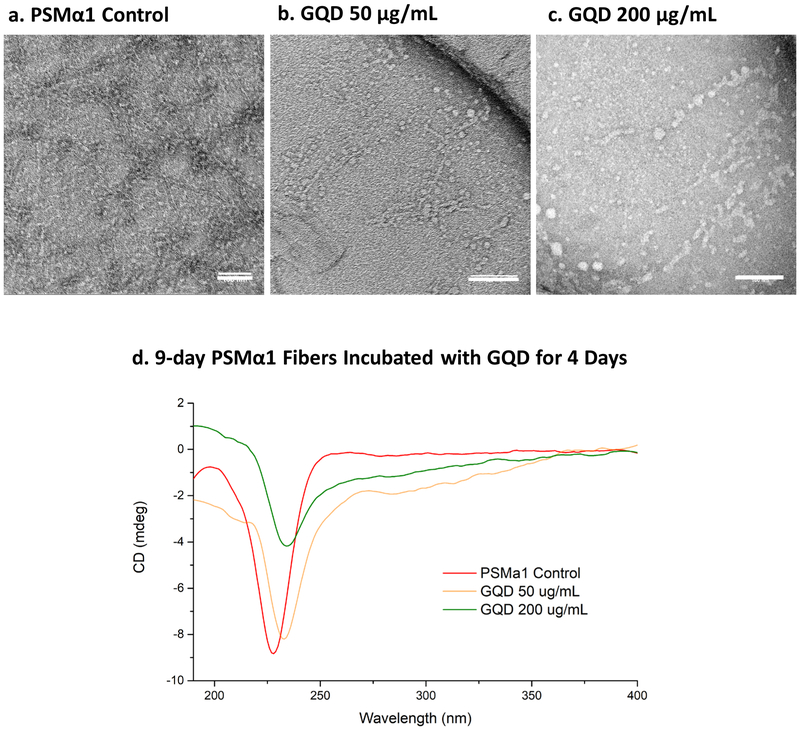
MD simulations were also used to elucidate the impact of GQDs on the assembly of PSMα1 monomers. Specifically, the statistics of the inter-peptide distance (center of mass) were collected and analyzed (Figure 5f). The results show that the average distance increased from 0.76 ± 0.062 nm to 1.03 ± 0.055 nm when a GQD is in close proximity. Moreover, the distribution changes from a relatively symmetric monomodal bell-shaped curve centered at 0.9 nm to a distribution with three peaks (at 0.57, 0.97 and 1.2 nm) with a wide shoulder at 0.8 nm. While each peak is composed by a variety of configurations (representative complexes are shown in correspondence of each peak in Figure 5f), the analysis of the trajectories suggests that the inter-peptide N-terminal/C-terminal interaction, responsible for the dimer stability is disrupted by the presence of the GQD. Additionally, as the GQD interacts with N-terminal of a peptide, it alters the contact angle of the peptide backbones and causes the groups close to the N-terminal to stretch. Such interaction can affect forming amyloid fibers observed experimentally in β-sheet signal in (Figure 6a), caused by the GQDs docking near the N-terminus of the peptides and by changes in the secondary structure (Figure 6b, c). Meanwhile, the interaction between GQD and PSM can also affect matured amyloid fibers by altering the distance between PSMs and secondary structures of PSMs in the outer layer of formed fibers. The effect of GQDs on matured fibers was experimentally confirmed by adding GQDs to 9-day incubated PSM fibers. The matured PSMα1 fibers incubated with 50 μg/mL and 200 μg/mL GQDs for an additional 4 days became collapsed and thickened (Figure 7a-c). This was correlated with a decrease and shift of the wide negative characteristic peak between 200 nm to 230 nm in the sample groups with addition of GQDs (Figure 7d). Overall, the interaction between GQD and PSM peptides strongly suggests a frustration of amyloid fibrillation.
Figure 6. Schematics of the effect of GQDs on PSM peptide fibrillation.
(a) The timeline of CD intensity at the typical β-sheet signal, a negative band at ≈ 218 nm indicates the process of PSM peptide fibrillation with/without addition of 50,200, 800 μg/mL GQDs. (b, c) GQDs interfere on N-terminal of PSM monomers or dimers and thereby frustrate fibrillation.
Figure 7. Effect pf GQDS on mature amyloid fibers.
PSMα1 fibers matured for 9 days were exposed to GQDs for 4 days. TEM images of PSMα1 fibers matured for 9 days (a) without GQD and with (b) 50 μg/mL and (c) 200 μg/mL GQDs were incubated for 4 more days. (d) CD spectra of PSMα1 fibers matured for 9 days with or without 50 μg/mL, and 200 μg/mL GQD for 4 more days. Scale bars: 100 nm (a-c).
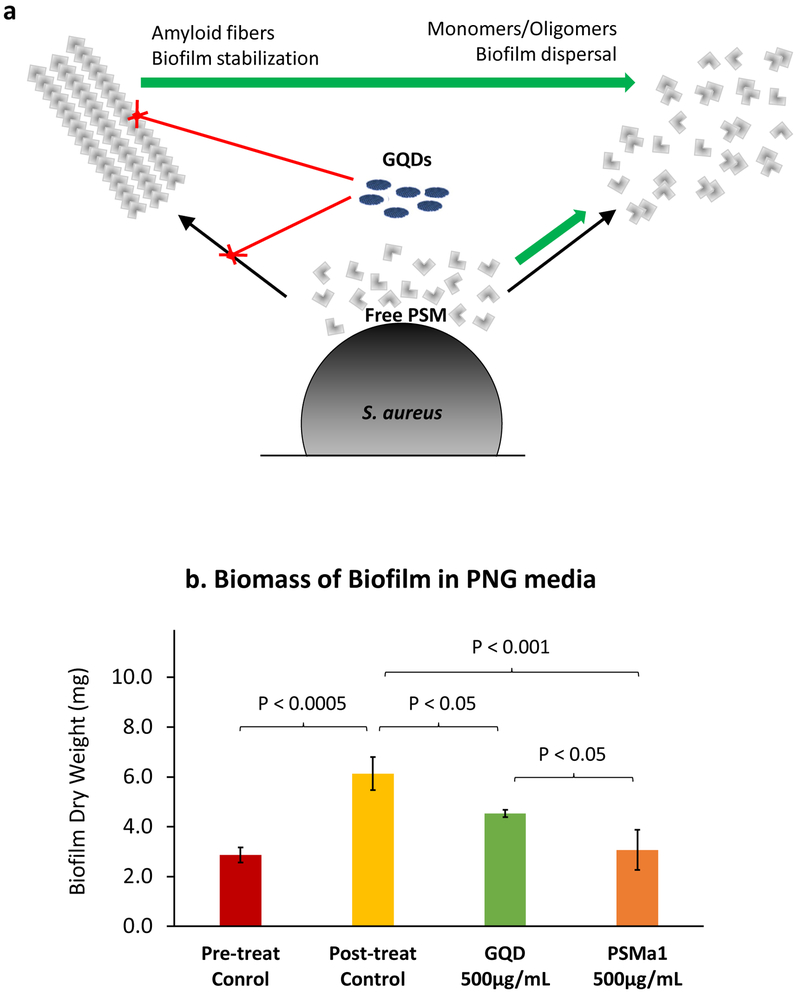
The results thus far suggest that GQDs disruption functional amyloids by inhibition of fibrillation and disruption of mature fibers. The mechanistic result is an increase in the concentration of free or oligomeric PSM peptides (Figure 8a). 33 To evaluate this hypothesis, we treated amyloid-rich biofilms with free PSMα1 monomers at the same concentration as GQDs and measured total biomass. The premise of this experiment is that free PSM should lead to a similar reduction in biomass as GQDs. Biomass of S. aureus amyloid-rich biofilms treated with GQDs was increased in relation to pre-treat controls (Figure 8b), suggesting inhibition with respect to total bacterial burden for this particular treatment regimen (single dose, single time point). Meanwhile, free PSMα1 monomers exhibit a similar if not more pronounced effect, supporting the hypothesis of increased free PSMs. The combination of reduced amyloid mediated biofilm stabilization and increased monomeric surfactant PSMs – which have been implicated in bacterial/ECM detachment36 – drives biofilm dispersion. Such supramolecular PSM-GQD complexes may also interfere with QS due to increased concentration of PSM monomers/oligomers that are tightly regulated by the Agr QS system.53,54
Figure 8. Schematics of GQD mediated staphylococcal biofilm dispersal.
(a) GQDs interact with PSM peptides in two ways. First, they inhibit fibrillation of free peptides. Second the disrupt already formed fibers. These events prevent robust stabilization of the biofilm. In addition, they lead to an increase in free monomeric and oligomeric PSM peptides which trigger dispersal events, (b) Dry weights of S. aureus biofilm in PNG media treated with 500 μg/mL GQD and PSMα1.
CONCLUSIONS
Amyloid-rich biofilms are exceptionally resistant to chemical disruption33,53 and the ability to disperse such biofilms as a potential therapeutic alone or in combination with other antimicrobials is worthy of consideration. Here we identify GQDs as a potential therapeutic for this purpose. The specificity for amyloid-rich biofilms and lack of direct bacterial toxicity implicate amyloid-forming PSM peptides in the ECM as the molecular target for GQDs. This mechanism was confirmed through experiments with isolated biofilm ECM and synthetic PSMα1 as well as MD simulations. We conclude that GQDs disperse S. aureus biofilms by competitive assembly with amyloid peptides. Given that amyloid-rich biofilms are resistant to most methods of dispersal33,53 this is a significant finding. Its merits for specific application will depend on the dose, frequency, and duration of therapy. Potential low therapeutic dosing and high in vivo biocompatibility of GQDs43 will propel broad practical applications on biofilms. Considering the broader implications of this work, additional studies on the interaction between GQDs and amyloid-forming peptides in other species (e.g., Escherichia coli and Pseudomonas spp) will provide different approaches and insights into the potential manipulation of microbial communities through their ECM. Further engineering of GQDs for enhanced association with amyloid peptides may lead to diverse effects on a wide range of biological processes, biomolecular functions, and signaling pathways.
METHODS
Synthesis and Characterization of GQDs.
GQDs were synthesized by a modified protocol through a top-down “oxidation-cutting” process.15 50 mg of carbon fibers was dispersed into a 4 mL mixture of sulfuric acid and nitric acid (3:1 v/v). The prepared solution was sonicated for 2 h and mechanically stirred for 24 h at 80 °C. Afterward, the mixture was cooled and diluted with deionized (DI) water to the concentration of 0.15 mg/mL. Sodium hydroxide was added into the solution to adjust the pH value to neutral. The mixture was further dialyzed in DI water for 3 days to obtain the final product. The size and shape of GQDs were characterized by TEM (JEOL 3011 HREM). The element analysis was performed by XPS (Kratos, Axis Ultra XPS). The light emission properties of GQDs and their assemblies were investigated by fluorescence spectroscopy (Horiba, Fluoromax-3).
Biofilm Culture.
S. aureus US A3 00 from −80 °C glycerol stock was plated on tryptic soy agar. Single colony inoculates were grown to mid-log, optical density 0.4 - 0.8 at 600 nm (OD600). Mid-log cultures were diluted to OD600 = 0.02 for seeding biofilms. For treating intact biofilms with GQDs 100 μl of this suspension had 106 S. aureus cells which were seeded into Stovall triple chamber flow cells. Biofilms were allowed to grow for 72 hours with either TSBG or PNG media flowing at 13 μl/min. Then 0, 50, or 500 μg/mL of GQDs were added and the flow was resumed for an additional 24 h. For isolating biofilm ECM, 100 μl suspensions containing 106 S. aureus were seeded on to polystyrene culture plates. Biofilms were then grown under static conditions for 5 days. All bacterial cultures were maintained at 37 °C.
Peptide Preparation.
PSMα1 was purchased from ChinaPeptides (Shanghai, China) with a purity >90%. They were dissolved to a final concentration of 0.5 mg/mL in a 1:1 mixture of trifluoroacetic acid (TFA) and hexafluoroisopropanol (HFIP). Peptides were then sonicated for 10 minutes and incubated for 1 h at room temperature. Stock solutions were divided into aliquots, solvent TFA/HFIP dried with a SpeedVac (Thermo Scientific, USA) at room temperature and stored at −80 °C. Samples were suspended in anhydrous dimethyl sulfoxide (5%) and sonicated for 10 minutes. This preparation yielded PSMs in monomeric form.
Thioflavin T (ThT) Assay.
Purified PSM solution was mixed with a ThT stock solution (prepared in 50 mM NaCl, 20 mM Tris, pH 8) to a final concentration of 2–4 μM peptides, 0.1-0.25 mM ThT. The final volume was 200 μl, and measurements were performed in 96-well plates with a clear bottom. Fixed excitation/emission filter of 430/480 nm (Ex/Em), respectively. Data was collected every hour for 15 hours.
Circular dichroism (CD) Spectrometry.
CD spectra were measured in a Jasco-815 spectropolarimeter (Jasco, Japan) thermostated at 25 °C. Aggregated peptides were prepared at 20 μM and measured immediately. Spectra were recorded from 190 to 400 nm, at 0.5 nm intervals, 1 nm bandwidth, and a scan speed of 100 nm/min.
Confocal Imaging.
For biofilm visualization by confocal laser scanning microscopy (Nikon A1 plus), stains were added to TSBG or PNG within the flow cells at the conclusion of the biofilm culture experiment. The stains were incubated under static conditions in the flow cells for 20 minutes. Wheat Germ Agglutinin (WGA) was used to visualize PIA while cells were stained with DAPI (4',6-diamidino-2-phenylindole). Three sets of samples were prepared at different time periods. Nine to fifteen images were taken in each sample for image analysis.
Image Analysis.
Digital images captured from confocal fluorescence microscopy and SEM were processed in ImageJ in the following manner: (1) Using process subtract background, background corrections were made. (2) Using image adjust threshold, a constant threshold level was set across a set of conditions to ensure that enough area of signal was present to identify the target including PIA, bacterial cells. (3) A constant area was applied to measure the area and integrated fluorescent density of the measured content. (4) Particle size analysis tool was used for quantification of void sizes in PNG biofilms and void area between 4 to 4000 μm22 was counted.
Biofilm ECM Isolation.
Biofilm ECM was harvested by disrupting biofilms using vortex and sonication as previously described.33 Briefly fibers were collected after 5-day static growth by scraping biofilms into 3 mL of potassium phosphate buffer (50 mM, pH 7.4). The biofilm suspensions were homogenized using a tissue homogenizer (Power 4, 3 minutes) to shear fibers free from the cell walls. Supernatants were clarified by repeated (×2) centrifugation at 4,000 g for 15 minutes to remove cells. The cell-free supernatant was incubated in 200 mM NaCl for 24 hours at room temperature. The fibers were isolated using Millipore Amicon Ultra Centrifugal Filter Units with a pore size of 30 kDa.
Biomass Test.
Biofilms grown in flow cells with PNG media for 72 hours were treated with 0 or 500 μg/mL of GQDs or PSMα1 for an additional 24 h. For biomass measurement, the tubing and the convertible top from the flow cell were removed, followed by aspirating off all media. The flow cells containing biofilms were dried in a vacuum oven at −30 in of Hg (−762 mmHg) and 50 C° for 15-20 min. The dried flow cells were weighed before and after removing the biofilms.
Scanning Electron Microscopy (SEM).
Biofilms were cultured on sample holders as the standard procedure. GQDs (500 μg/mL) were added incubated for 24 hours at 37 °C. Similarly, isolated ECM was placed on sample holders and treated with GQDs. All samples were fixed in glutaraldehyde, serially dehydrated in ethanol, air dried at room temp, sputter-coated with gold and visualized using AMRAY 1910 Field Emission Scanning Electron Microscope.
Transmission Electron Microscopy (TEM).
The TEM employed in the characterization is JEOL 3011 HREM. Carbon-coated 400 mesh copper TEM grids were placed coated-side-down for 60 s onto sample drops (10 μl) of PSMα1 solution with/without addition of 50, 200, 800 μg/mL GQDs for 4 and 9 days. The experiment was repeated three times and in average of 30 fibers and/or aggregates were measured in each sample group for image analysis. The grids were then retrieved, washed with deionized water (two droplets). The sample was stained with 1% (w/v) uranyl acetate for 40 s. Grids were then blotted and air-dried before imaging. For imaging cells with amyloid containing ECM, biofilms were scrapped directly on to copper TEM grids, stained with uranyl acetate, dried and imaged.
Molecular Dynamics (MD) Simulation.
All the simulations were performed with the NAMD in explicit water using the classical all-atom force field CHARMM. A time step of 1 fs was employed to integrate the equations of motion. A cutoff of 1.2 nm was used in conjunction with the Particle Mesh Ewald method to evaluate long-range columbic forces. The systems were minimized for 1000 steps before the production runs. Single peptide simulations for a total or more than 650 ns, were performed in a canonical ensemble (300 K, with a time constant of 1 ps) in a cubic simulation box with side 7.5 nm. Production runs of the PSMα1 dimers were performed in a canonical ensemble in a cubic simulation box with a side of 10 nm.
Supplementary Material
ACKNOWLEDGEMENTS
This project was sponsored by the University of Michigan Mcubed Award and Defense Advanced Research Projects Agency (DARPA) HR00111720067 “Electromagnetic Processes and Normal Modes in Bacterial Biofilms”. Parts of the work were also supported by NIH under grant K08 AI128006 and NSF project “Energy- and Cost-Efficient Manufacturing Employing Nanoparticles” NSF 1463474. We are acknowledging the financial and programmatic support from the University of Michigan College of Engineering's Blue Sky Initiative. We are also acknowledging the support from UM electron microscopy facility MC2 for its assistance with electron microscopy, and for the NSF grant #DMR-9871177 for funding of the JEOL 2010F analytical electron microscope used in this work. The authors also thank Yang Zhao and John Soukar for assistance with experiments.
Footnotes
SUPPORTING INFORMATION
Supporting information file is available online and includes Figures S1-S7 as described below. Characterization of as synthesized GQDs; quantitative image analysis derived from confocal microscopy of TSBG grown S. aureus biofilms; Planktonic growth curve for S. aureus in the presence of GQDs; the photo of PSMα1 with/without addition of GQDs; ThT staining of PSMα1; Absorption spectra of PSMα1 with or without GQDs; FTIR of control solution.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- (1).Hill EC; Hill GC Microbial Contamination and Associated Corrosion in Fuels, during Storage, Distribution and Use. Adv. Mater. Res. 2008, 38, 257–268. [Google Scholar]
- (2).Porcelli N; Judd S Chemical Cleaning of Potable Water Membranes: A Review. Sep. Purif. Technol. 2010, 77, 137–143. [DOI] [PubMed] [Google Scholar]
- (3).Costerton JW Bacterial Bofilms: A Common Cause of Persistent Infections. Science (80-.). 1999, 284, 1318–1322. [DOI] [PubMed] [Google Scholar]
- (4).Castaño-Rodríguez N; Goh K-L; Fock KM; Mitchell HM; Kaakoush NO Dysbiosis of the Microbiome in Gastric Carcinogenesis. Sci. Rep. 2017, 7, 15957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Davey ME; O'toole GA Microbial Biofilms: From Ecology to Molecular Genetics. Microbiol. Mol. Biol. Rev. 2000, 64, 847–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Neethirajan S; Clond MA; Vogt A Medical Biofilms - Nanotechnology Approaches. J. Biomed. Nanotechnol. 2014, 10, 2806–2827. [DOI] [PubMed] [Google Scholar]
- (7).Hall-Stoodley L; Costerton JW; Stoodley P Bacterial Biofilms: From the Natural Environment to Infectious Diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [DOI] [PubMed] [Google Scholar]
- (8).Branda SS; Vik Å; Friedman L; Kolter R Biofilms: The Matrix Revisited. Trends Microbiol. 2005, 13, 20–26. [DOI] [PubMed] [Google Scholar]
- (9).Tursi SA; Tükel ç. Curli-Containing Enteric Biofilms Inside and Out: Matrix Composition, Immune Recognition, and Disease Implications. Microbiol. Mol. Biol. Rev. 2018, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Olsen I Biofilm-Specific Antibiotic Tolerance and Resistance. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 877–886. [DOI] [PubMed] [Google Scholar]
- (11).Arciola CR; Campoccia D; Speziale P; Montanaro L; Costerton JW Biofilm Formation in Staphylococcus Implant Infections. A Review of Molecular Mechanisms and Implications for Biofilm-Resistant Materials. Biomaterials 2012, 33, 5967–5982. [DOI] [PubMed] [Google Scholar]
- (12).Brogden KA Antimicrobial Peptides: Pore Formers or Metabolic Inhibitors in Bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [DOI] [PubMed] [Google Scholar]
- (13).Wu H; Moser C; Wang H-Z; HØiby N; Song Z-J Strategies for Combating Bacterial Biofilm Infections. Int. J. Oral Sci. 2014, 7, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Romero D; Sanabria-Valentín E; Vlamakis H; Kolter R Biofilm Inhibitors That Target Amyloid Proteins. Chem. Biol. 2013, 20, 102–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Suzuki N; Wang Y; Elvati P; Qu Z-B; Kim K; Jiang S; Baumeister E; Lee J; Yeom B; Bahng JH; Lee J; Violi A; Kotov N A Chiral Graphene Quantum Dots. ACS Nano 2016, 10, 1744–1755. [DOI] [PubMed] [Google Scholar]
- (16).Sun H; Gao N; Dong K; Ren J; Qu X Graphene Quantum Dots-Band-Aids Used for Wound Disinfection. ACS Nano 2014, 8, 6202–6210. [DOI] [PubMed] [Google Scholar]
- (17).Zeng Z; Yu D; He Z; Liu J; Xiao F-X; Zhang Y; Wang R; Bhattacharyya D; Tan TTY Graphene Oxide Quantum Dots Covalently Functionalized PVDF Membrane with Significantly-Enhanced Bactericidal and Antibiofouling Performances. Sci. Rep. 2016, 6, 20142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Hui L; Huang J; Chen G; Zhu Y; Yang L Antibacterial Property of Graphene Quantum Dots (Both Source Material and Bacterial Shape Matter). ACS Appl. Mater. Interfaces 2016, 8, 20–25. [DOI] [PubMed] [Google Scholar]
- (19).Kotov NA Inorganic Nanoparticles as Protein Mimics. Science (80-. ). 2010, 330, 188–189. [DOI] [PubMed] [Google Scholar]
- (20).Tang Z; Kotov NA; Giersig M Spontaneous Organization of Single CdTe Nanoparticles into Luminescent Nanowires. Science (80-. ). 2002, 297, 237–240. [DOI] [PubMed] [Google Scholar]
- (21).Zhou Y; Marson RL; van Anders G; Zhu J; Ma G; Ercius P; Sun K; Yeom B; Glotzer SC; Kotov NA Biomimetic Hierarchical Assembly of Helical Supraparticles from Chiral Nanoparticles. ACS Nano 2016, 10, 3248–3256 [DOI] [PubMed] [Google Scholar]
- (22).Park J I1; Nguyen TD; de Queirós Silveira G; Bahng JH; Srivastava S; Zhao G; Sun K; Zhang P; Glotzer SC; Kotov NA Terminal Supraparticle Assemblies from Similarly Charged Protein Molecules and Nanoparticles. Nat. Commun. 2014, 5, 3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Huang R; Carney RP; Stellacci F; Lau BLT Protein-Nanoparticle Interactions: The Effects of Surface Compositional and Structural Heterogeneity Are Scale Dependent. Nanoscale 2013, 5, 6928–6935. [DOI] [PubMed] [Google Scholar]
- (24).Lundqvist M; Stigler J; Elia G; Lynch I; Cedervall T; Dawson KA Nanoparticle Size and Surface Properties Determine the Protein Corona with Possible Implications for Biological Impacts. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 14265–14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Harkness KM; Turner BN; Agrawal AC; Zhang Y; McLean JA; Cliffel DE Biomimetic Monolayer-Protected Gold Nanoparticles for Immunorecognition. Nanoscale 2012, 4, 3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Sun J; DuFort C; Daniel M-C; Murali A; Chen C; Gopinath K; Stein B; De M; Rotello VM; Holzenburg A; Kao CC; Dragnea B Core-Controlled Polymorphism in Virus-like Particles. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 1354–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Yoo S I1; Yang M; Brender JR; Subramanian V; Sun K; Joo NE; Jeong S-H; Ramamoorthy A; Kotov NA Inhibition of Amyloid Peptide Fibrillation by Inorganic Nanoparticles: Functional Similarities with Proteins. Angew. Chemie Int. Ed. 2011, 50, 5110–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Carrillo-Carrion C; Atabakhshi-Kashi M; Carril M; Khajeh K; Parak WJ Taking Advantage of Hydrophobic Fluorine Interactions for Self-Assembled Quantum Dots as a Delivery Platform for Enzymes. Angew. Chemie Int. Ed. 2018, 57, 5033–5036. [DOI] [PubMed] [Google Scholar]
- (29).Mahmoudi M; Akhavan O; Ghavami M; Rezaee F; Ghiasi SM A. Graphene Oxide Strongly Inhibits Amyloid Beta Fibrillation. Nanoscale 2012, 4, 7322. [DOI] [PubMed] [Google Scholar]
- (30).Liu Y; Xu L-P; Dai W; Dong H; Wen Y; Zhang X Graphene Quantum Dots for the Inhibition of β Amyloid Aggregation. Nanoscale 2015, 7, 19060–19065. [DOI] [PubMed] [Google Scholar]
- (31).Wang J; Cao Y; Li Q; Liu L; Dong M Size Effect of Graphene Oxide on Modulating Amyloid Peptide Assembly. Chem. - A Eur. J. 2015, 21, 9632–9637. [DOI] [PubMed] [Google Scholar]
- (32).Kim D; Yoo JM; Hwang H; Lee J; Lee SH; Yun SP; Park MJ; Lee M; Choi S; Kwon SH; Lee S ; Kwon S-H; Kim S; Park YJ; Kinoshita M; Lee Y-H; Shin S; Paik SR; Lee SJ; Lee S; et al. Graphene Quantum Dots Prevent α-Synucleinopathy in Parkinson’s Disease. Nat. Nanotechnol. 2018, 13, 812–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Schwartz K; Syed AK; Stephenson RE; Rickard AH; Boles BR Functional Amyloids Composed of Phenol Soluble Modulins Stabilize Staphylococcus Aureus Biofilms. PLoSPathog. 2012, 8, el002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Periasamy S; Chatterjee SS; Cheung GYC; Otto M Phenol-Soluble Modulins in Staphylococci. Commun. Integr. Biol. 2012, 5, 275–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Bleem A; Francisco R; Bryers JD; Daggett V Designed α-Sheet Peptides Suppress Amyloid Formation in Staphylococcus Aureus Biofilms. npj Biofilms Microbiomes 2017, 3, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Cheung GYC; Joo H-S; Chatteijee SS; Otto M Phenol-Soluble Modulins – Critical Determinants of Staphylococcal Virulence. FEMS Microbiol. Rev. 2014, 38, 698–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Tayeb-Fligelman E; Tabachnikov O; Moshe A; Goldshmidt-Tran O; Sawaya MR; Coquelle N; Colletier J-P; Landau M The Cytotoxic Staphylococcus Aureus PSMα3 Reveals a Cross-α Amyloid-like Fibril. Science. 2017, 355, 831–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Marinelli P; Pallares I; Navarro S; Ventura S Dissecting the Contribution of Staphylococcus Aureus α-Phenol-Soluble Modulins to Biofilm Amyloid Structure. Sci. Rep. 2016, 6, 34552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Pollitt EJG; Crusz SA; Diggle SP Staphylococcus Aureus Forms Spreading Dendrites That Have Characteristics of Active Motility. Sci. Rep. 2015, 5, 17698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Pauling L; Corey RB Configurations of Polypeptide Chains With Favored Orientations Around Single Bonds: Two New Pleated Sheets. Proc. Natl. Acad. Sci. U. S. A. 1951, 37, 729–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Elvati P; Baumeister E; Violi A Graphene Quantum Dots: Effect of Size, Composition and Curvature on Their Assembly. RSC Adv. 2017, 7, 17704–17710. [Google Scholar]
- (42).Nurunnabi M; Khatun Z; Huh KM; Park SY; Lee DY; Cho KJ; Lee Y-K In Vivo Biodistribution and Toxicology of Carboxylated Graphene Quantum Dots. ACS Nano 2013, 7, 6858–6867. [DOI] [PubMed] [Google Scholar]
- (43).Chong Y; Ma Y; Shen H; Tu X; Zhou X; Xu J; Dai J; Fan S; Zhang Z The in Vitro and in Vivo Toxicity of Graphene Quantum Dots. Biomaterials 2014, 35, 5041–5048. [DOI] [PubMed] [Google Scholar]
- (44).Wang S; Cole IS; Li Q The Toxicity of Graphene Quantum Dots. RSC Adv. 2016, 6, 89867–89878. [Google Scholar]
- (45).Boles BR; Horswill AR Staphylococcal Biofilm Disassembly. Trends Microbiol. 2011, 19, 449–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Das T; Kutty SK; Tavallaie R; Ibugo AI; Panchompoo J; Sehar S; Aldous L; Yeung AWS; Thomas SR; Kumar N; Gooding JJ; Manefield M Phenazine Virulence Factor Binding to Extracellular DNA Is Important for Pseudomonas Aeruginosa Biofilm Formation. Sci. Rep. 2015, 5, 8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Arosio P; Knowles TPJ; Linse S On the Lag Phase in Amyloid Fibril Formation. Rhys. Chem. Chem. Rhys. 2015, 77, 7606–7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Sreerama N; Woody RW Estimation of Protein Secondary Structure from Circular Dichroism Spectra: Comparison of CONTIN, SELCON, and CDSSTR Methods with an Expanded Reference Set. Anal. Biochem. 2000, 287, 252–260. [DOI] [PubMed] [Google Scholar]
- (49).Bush CA; Sarkar SK; Kopple KD Circular Dichroism of β Turns in Peptides and Proteins. Biochemistry 1978, 77, 4951–4954. [DOI] [PubMed] [Google Scholar]
- (50).Greenfield N Using Circular Dichroism Spectra to Estimate Protein Secondary Structure. Nat. Protoc. 2007, 1, 2876–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Spectroscopic Methods for Analysis of Protein Secondary Structure. Anal. Biochem. 2000, 277, 167–176. [DOI] [PubMed] [Google Scholar]
- (52).Towle KM; Lohans CT; Miskolzie M; Acedo JZ; van Belkum MJ; Vederas JC Solution Structures of Phenol-Soluble Modulins A1, A3, and B2, Virulence Factors from Staphylococcus Aureus. Biochemistry 2016, 55, 4798–4806. [DOI] [PubMed] [Google Scholar]
- (53).Rice KC; Murphy E; Yang S-J; Bayles KW; Smeltzer MS; Projan SJ; Beenken KE; Cassat J; Palm KJ; Dunman PM Transcriptional Profiling of a Staphylococcus Aureus Clinical Isolate and Its Isogenic Agr and SarA Mutants Reveals Global Differences in Comparison to the Laboratory Strain RN6390. Microbiology 2006, 152, 3075–3090. [DOI] [PubMed] [Google Scholar]
- (54).Xu T; Wang X-Y; Cui P; Zhang Y-M; Zhang W-H; Zhang Y The Agr Quorum Sensing System Represses Persister Formation through Regulation of Phenol Soluble Modulins in Staphylococcus Aureus. Front. Microbiol. 2017, 8, 2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.