Abstract
Background:
Saliva has recently served as a primary investigative tool in forensics in the detection of crime, cases of sexual assaults, human and animal bite marks, poisoning, hormone identification, and alcohol and drug abuse.
Aim:
This scoping review aimed to comprehensively identify the role of saliva in comparative and reconstructive identification and propose the concept of salivary signature (SS) in forensics.
Methodology:
A literature search was performed on electronic databases, PubMed and Google Scholar with keywords, “salivary,” “microbiome,” and “forensics,” and relevant articles identified along with reference tracking.
Results:
SS model was based on salivary microbiome and biomarkers which together provide pertinent information about lifestyle, behavioral patterns, circadian rhythms, geolocation, cohabitation of individuals, postmortem intervals, systemic and oral ailments or cancers besides salivary flow and composition.
Conclusions:
This communication highlights the constituents of SS and their significance in forensics. It also enumerates factors altering SS, limitations owing to diversity in microbiome and biomarker status, and possible measures to improve its accuracy and robustness in forensics.
Keywords: Forensics, salivary biomarkers, salivary microbiome, salivary signature
Introduction
Saliva is the medium of choice among all body fluids in forensic and criminal investigations due to ease of availability and noninvasive cost-effective collection methods. Advents in DNA technology have undoubtedly improved the isolation of human DNA in dried saliva stains obtained from bite marks and lip prints and made it more accurate.[1] But in cases, where there is lack of DNA sample from suspect or unavailability of a suspect for comparative identification, saliva has also been used for biologic profiling to determine age, gender, and personal characteristics of individuals for reconstructive identification. This relatively new arena of salivary research is related to the study of human microbiome, primarily microbial DNA of microorganisms (bacteria, fungi, microeukarya, and viruses) in salivary secretion, which when combined with well-documented salivary biomarkers, can provide vital information about the suspect's lifestyle, cohabitation, and medical condition, thus aiding forensic analyses.[2,3]
Aim
This study aims to perform a scoping review of salivary constituents and their role in reconstructive profiling.
Objective
Based on the review, the authors propose the concept of salivary signature(SS) in forensics.
Methodology
A literature search was performed on electronic databases, PubMed (P) and Google Scholar (GS) with keywords, “salivary,” “microbiome,” and “forensics” in May 2018 which revealed three articles from (P) and 789 results from (GS). Relevant articles from databases and reference tracking were shortlisted on the basis of inclusion criteria pertaining to the role of saliva in reconstructive forensic identification.
Results and Discussion
The current scoping review listed the constituents of saliva related to its significance in forensics, specifically in biologic profiling of individual, based on which the authors proposed the concept of SS – [Figure 1], in reconstructive forensic identification.
Figure 1.
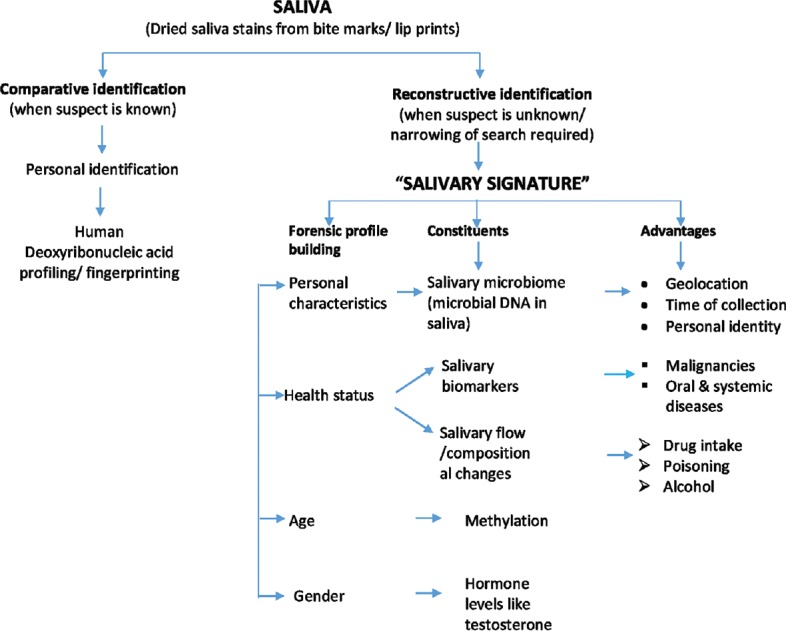
Figure depicts the role of saliva in forensic identification, both comparative and reconstructive. It explains further the role of salivary signature comprising of salivary microbiome, biomarkers, flow and composition changes in generation of biological profile of individual characteristics
Salivary constituents and their importance
Saliva comprises a scaffold-like structure housing electrolyte phase in water, proteins, and salivary micelles as well as lipoid material, epithelial, and bacterial cells. It constitutes of 99.5% water and remaining 0.5% enzymes, amylase and lipase, antibacterial compounds such as lysozyme and lactoferrin, electrolytes (sodium and potassium), growth factors, buffers, and also mucus.[3,4] The constituents of saliva coalesce with microorganisms of oral cavity, blood cells, airway secretions after entry into the oral cavity emanating from salivary ducts. Thus, the study of these elements provides pertinent information are about health status, quality of life, and lifestyle of individuals besides their age and gender.[5]
Concept of salivary signature and its role in forensics?
Any trace of saliva left behind in bite marks or lip prints, which gives information about age, gender, personal characteristics, health status of individual may be used to create SS, shown diagrammatically in Figure 1. Following are the details of components of SS.
Salivary microbiome
It refers to the study of proportions of various bacteria present in saliva potentially useful for individual identification. Normally eight genera in the salivary microbiome constitutes >70% of the total bacterial population: Streptococcus, Neisseria, Prevotella, Haemophilus, Veillonella, Porphyromonas, Rothia, and Fusobacterium.[2] The composition of this salivary microbiome can be altered based on a person's age, circadian rhythms, lifestyle their eating habits, their cohabitants, the presence of pets, their habits of smoking, interactions such as kissing. and their overall health status.[6,7,8] Of these 700 bacterial species found in the mouth, it is inevitable that some of these species will be codependent in individuals residing in close association.[9,10] Hence, the constituents of saliva obtained from crime site can be matched to suspect's close associates. Another advantage of salivary bacterial DNA over human DNA recovered from sexual assault sites would be exclusivity of salivary microbiome profile over skin, whereas human DNA would be the same in both skin and saliva.[4] Literature also supports microbial analysis of bite marks to correlate specifically streptococcal DNA in saliva to that of suspect where human DNA availability is questionable.[11,12] Hence, proper collection and analysis of salivary sample for oral microbiome signatures are performed in the following steps:
Different stages of analysis of salivary microbiome
Stage 1: Collection of sample –salivary sample can be collected from the skin of victim in bite marks or objects such as cell phones or keyboards of the computer. They can also be matched to environmental microbiome DNA if required where samples can be collected from the surfaces of classrooms, air or soil. Microbial DNA signatures can be shared among cohabiting spouses or families[1]
Stage 2: Directed (16S rRNA sequences) and undirected (whole genome shotgun [WGS]) sequencing are done to identify the taxonomic distribution of bacterial species. Human Microbiome Project which studied microbiome on 15–18 body sites in 300 individuals led to the conclusion that bacterial compositions were more consistent in same body site in different individuals rather than the different site in the same individual.[1] Further studies proved that time-related consistency was found to be greater in gut and saliva than other body sites. Hence, strengthening the importance of spit signatures in forensic identification
Stage 3: Analysis of 16S rRNA sequences is made to match with the published dataset of salivary microbiome, while WGS is made to target all gene sequencing but is yet to be used as a forensic tool
Stage 4: Identify its role in forensics.
Geolocation
Different geographical locations may exhibit a difference in composition and function of microbial communities existing in saliva owing to different eating patterns, oral hygiene measures, humidity, climate, temperature, or even disease outbreaks.[13] Various studies have identified an ethnicity signature to oral microbiome reflected in saliva, of which one study particularly found a significant association of 16S rRNA sequencing of salivary microbiome variation in a study of 120 individuals with the distance of locations of the subjects from equator.[1,14]
Time of collection
Bacterial taxonomic data vary with different time points, hence may be linked to time of death based on postmortem interval estimation. However, more than salivary microbiome signature, gut, bones, and skin are majorly researched during decomposition.[1,15]
Personal identity
Metagenomic tags causing microbial fingerprints in saliva on bite mark left behind on the body of victim in a sexual assault case may link the suspect to the crime. The microbial DNA also relate categorically to the ancestral genomic background identifying the ethnicity of the suspect.[1] Literature holds the prospect of fingerprinting bacteria, specifically Streptococcus salivarius of viridans group present in saliva in association with suspect's saliva collected from bite marks in sexual assault cases.[1,12]
Variation with age
Colonization of bacteria begins 6 h after birth, a stage where the oral cavity is completely sterile. The early colonization is marked with the presence of Staphylococcus, Streptococcus, Enterococcus, Micrococcus, and Veillonella. This slowly transforms into S. salivarius in infants found in the tongue. Diverse Streptococci species and Actinomyces dominate the bacterial flora with the eruption of teeth and formation of dental plaque. The flora becomes comparable to adult from the age of 5 years with the absence of Spirochaetes and Prevotella melaninogenica as exceptions. However, loss of teeth can cause a marked decrease in Spirochaetes, Streptococcus sanguinis, Lactobacillus, and Streptococcus mutans. Although some research has been directed toward establishing the stability of microbiome,[16] further studies are required specific to each age group to establish its role in forensics.[2]
Persistence
Bacterial DNA can persist for a greater amount of time than human DNA making them valuable for forensics for a longer time than human DNA. This is because the prokaryotic bacterial cells are protected by a cell wall and have a circular DNA compared to a cell membrane in eukaryotic cells with a linear DNA, making it easier to be destroyed.[2]
Salivary biomarkers
The presence of proteomic and genomic biomarkers in saliva may indicate conditions of underlying systemic illnesses, oral diseases, detection of oral cancers, and malignant tumors. For example, increase in levels of salivary transcriptase as compared to controls indicates pancreatic cancers. c-erbB-2 is another proteome in saliva which is a prognostic marker for malignancies in females. Salivary traces of vitamin B12 or cobalamin indicate pernicious anemia, oral candida suggests leukemia/iron-deficiency anemia, amyloid material suggests multiple myeloma/amyloidosis, thickened saliva with purulent discharge, and candida suggests Sjogren's syndrome, mast cells indicate mastocytosis, and candida with glucose and crevicular fluid suggests diabetes mellitus. Hence, detection of these markers in saliva maybe suggestive of systemic conditions associated to suspects under consideration.[4,5]
Salivary composition and flow rate
Drugs-reduced salivary flow while intake of anticholinergic drugs such as antihistamines, antidepressants, antihypertensives, or anxiolytics may alter the composition of saliva, which may give clue about drug history of suspects[4]
Alcohol consumption: salivary flow gets affected in excessive alcohol intake due to overdose of ethanol, altered output of proteins, amylase, and decrease in electrolytes
Salivary flow index: intensity, duration, and type of stimulus results in elevation in sodium, calcium, proteins, bicarbonates and chloride levels, and reduced phosphates and magnesium. Knowledge of such compositional changes helps in forensic investigations.
Age
Methylation of DNA from saliva can yield the age of a person within 5 years approximately.[4]
Gender
Unstimulated whole saliva flow rates as well as salivary constituents of testosterone, etc., are different in males and females.[4]
Shortcomings of salivary signature
The statistical power of salivary microbiomes owing to their vast diversity is still questionable in ascertaining individual identification in forensic science
Majority of systemic and oral diseases are still unexplored for their salivary constituents
Biomarkers may show some overlapping in various diagnostic conditions and are not exclusive
Microbial taxa database for comparative geological or personal identification is limited across multiple geographical locations
Microbial sample storage, time and temperature of storage such as repeated thawing and freezing of samples at crime site may influence the microbiome consistency
Standardized sampling techniques to prevent the contamination, as well as the accuracy of microbial signatures, form trace quantity of samples is yet to be accomplished
Microbial taxa is itself very vast with over 700 different species of bacteria located in the oral cavity, and many of these species are codependent. Newer species are also identified over time and variations arising from various environmental influences, making the task of identification challenging
Maintaining a taxa dataset similar to DNA fingerprinting database has legal implications as it may reveal personal information compromising privacy rights of individual.
Measures proposed to make salivary signature more robust
Routine oral microbiome analysis in dentistry, which can be maintained in microbial databanks
Population-based data can be collected to aid in geological identification or ethnicity of individuals
Microbiome clusters can be formulated for various dietary and lifestyle conditions such as smoking
Latest sequencing techniques and high throughput sequencing machines should be used
Multiple systemic and oral diseases should be explored for their salivary biomarkers.
Potential scope of salivary signature in forensic dentistry
Forensic dentistry inevitably uses teeth for reconstructive profiling of individuals by age and gender assessment. This can be further strengthened by analysis of oral fluids like saliva for its constituents and hence, SS to ascertain the complete biologic profile of the individual. The forensic dentist can prove instrumental in initiating oral microbial databanks which are geographical location and population specific. Further, multiple systemic and oral conditions can be explored for specific salivary genomes or biomarkers which invariably holds great research potential in forensic dentistry.
Conclusions
The proposed SS can be useful in forensics once the techniques of collection and storage of saliva are made more robust. Furthermore, oral microbiome and salivary biomarker databanks for different populations and locations should be created to provide the scope for comparative geological and personal identification.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Clarke TH, Gomez A, Singh H, Nelson KE, Brinkac LM. Integrating the microbiome as a resource in the forensics toolkit. Forensic Sci Int Genet. 2017;30:141–7. doi: 10.1016/j.fsigen.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Leake SL, Pagni M, Falquet L, Taroni F, Greub G. The salivary microbiome for differentiating individuals: Proof of principle. Microbes Infect. 2016;18:399–405. doi: 10.1016/j.micinf.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Leake SL. Is human DNA enough? – Potential for bacterial DNA. Front Genet. 2013;4:282. doi: 10.3389/fgene.2013.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajshekar M, Tennant M, Thejaswini B. Salivary biomarkers and their applicability in forensic identification. Sri Lanka J Forensic Med Sci Law. 2013;4:10–5. [Google Scholar]
- 5.Fuentes AM, Sanchez CN. New Trends in Biomarkers and Diseases Research: An Overview. 1st ed. UAE: Bentham Science Publishers; 2017. Biomarkers in saliva as a tool for health diagnosis; pp. 436–68. [Google Scholar]
- 6.Takayasu L, Suda W, Takanashi K, Iioka E, Kurokawa R, Shindo C, et al. Circadian oscillations of microbial and functional composition in the human salivary microbiome. DNA Res. 2017;24:261–70. doi: 10.1093/dnares/dsx001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song SJ, Lauber C, Costello EK, Lozupone CA, Humphrey G, Berg-Lyons D, et al. Cohabiting family members share microbiota with one another and with their dogs. Elife. 2013;2:e00458. doi: 10.7554/eLife.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alan G, Sarah JP. Microbes as forensic indicators. Trop Biomed. 2012;29:311–30. [PubMed] [Google Scholar]
- 9.Lamont RJ, Jenkinson HF. Oxford, UK: Wiley-Blackwell; 2010. Oral Microbiology at a Glance. [Google Scholar]
- 10.Parahitiyawa NB, Scully C, Leung WK, Yam WC, Jin LJ, Samaranayake LP. Exploring the oral bacterial flora: Current status and future directions. Oral Dis. 2010;16:136–45. doi: 10.1111/j.1601-0825.2009.01607.x. [DOI] [PubMed] [Google Scholar]
- 11.Kennedy DM, Stanton JA, García JA, Mason C, Rand CJ, Kieser JA, et al. Microbial analysis of bite marks by sequence comparison of streptococcal DNA. PLoS One. 2012;7:e51757. doi: 10.1371/journal.pone.0051757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown KA, Elliot TR, Rogers AH, Thonard JC. The survival of oral streptococci on human skin and its implication in bite-mark investigation. Forensic Sci Int. 1984;26:193–7. doi: 10.1016/0379-0738(84)90217-2. [DOI] [PubMed] [Google Scholar]
- 13.Li J, Quinque D, Horz HP, Li M, Rzhetskaya M, Raff JA, et al. Comparative analysis of the human saliva microbiome from different climate zones: Alaska, Germany, and Africa. BMC Microbiol. 2014;14:316. doi: 10.1186/s12866-014-0316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nasidze I, Li J, Quinque D, Tang K, Stoneking M. Global diversity in the human salivary microbiome. Genome Res. 2009;19:636–43. doi: 10.1101/gr.084616.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damann FE, Williams DE, Layton AC. Potential use of bacterial community succession in decaying human bone for estimating postmortem interval. J Forensic Sci. 2015;60:844–50. doi: 10.1111/1556-4029.12744. [DOI] [PubMed] [Google Scholar]
- 16.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–7. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]


