Abstract
This study investigated the influence of dietary fiber levels on the growth performance, digestion, metabolism, and cecal microbial community of rabbits with different diets at different age. The different levels of dietary natural detergent fiber (NDF) were formulated accordingly: 400(A), 350(B), 300(C), 250(D) g/kg original matter basis, respectively; the different ages were 52, 62, and 72 days. With NDF increasing, the average daily feed intake (ADFI) and feed conversion rate (FCR) were increased, whereas average daily gain (ADG) and mortality were decreased (p < 0.05). The stomach relative weight, stomach content relative weight, cecal relative weight, and cecal content weight increased with increasing NDF (p < 0.05). The NH 3‐N concentration of cecum dropped when the dietary NDF increased (p < 0.05). The diversity of the total microbiota increased significantly in Diets B, C (p = 0.011), and reached the lowest in 52 days for all diet groups. The richness index was decreased significantly in Diet A, D (p < 0.05) and in 62 days (p < 0.001), respectively. The phylum Firmicutes was higher (p < 0.01) in rabbits fed Diets B, C than Diets A, D and Bacteroidetes was highest in Diets C, D, and Proteobacteria was the highest in Diet A (p < 0.001). Among the classified genera, there were 14 that had levels of abundance of more than 1% and were commonly shared by all samples. Ruminococcus spp. that produced volatile fatty acid (VFA) abundance was highest from Diets B, C at 52 and 62 days. It is interesting to note that Bifidobacterium from Diet C was the most abundant genus during the entire experimental period (p < 0.01). The data from Venn diagrams, principal component analysis (PCA), and heat map plots of the bacterial communities showed that there were more groups of shared microbiota with aging. The above results indicate the cecal microbiota controlled by the 350 g/kg NDF diet can prevent gastrointestinal distress and exhibit good production performance.
Keywords: cecal microbiota, high‐throughput sequencing, Neutral detergent fiber, rabbits
1. INTRODUCTION
The hindgut of monogastric animals harbors many thousands of microbial species (Daly, Stewart, Flint, & Shirazi‐Beechey, 2001; Ivarsson, Roos, Liu, & Lindberg, 2014). The cecum occupies 40% of the whole‐tract content size, and ecosystem of rabbits have highly active microbiota, which lead to a important role in their digestive physiology (Monteils, Cauquil, Combes, Godon, & Gidenne, 2008). The dietary fiber hydrolyzed by cecal microbiota results in the release of soluble sugars, which are fermented to VFA, such as acetate, propionate, and butyrate, etc. The VFA absorbed across the epithelium of the large intestine provide 30–50% of the rabbit's body energy (Gidenne, Pinheiro, & e Cunha, 2000). A previous study suggested that sufficient dietary fiber supply can prevent intestinal diarrhea in the growing rabbit (Gidenne, Arveux, & Madec, 2001). The recommended dietary NDF levels of rabbits are about 20%–34% (Gidenne, Jehl, Segura, & Michalet‐Doreau, 2002). Because of the main energy substrate for the microbiota, dietary fiber levels significantly impact of the composition of the intestinal microbiota (Combes, Fortun‐Lamothe, Cauquil, & Gidenne, 2013; Sawicki et al., 2015), and most of their beneficial effects are associated with bacterial fermentation (Lattimer & Haub, 2010; Verbeke et al., 2015). The understanding of cecal microbiota structure is considered to have an important role in the nutrition and health of rabbits.
The objectives of the research were to investigate and discuss the effects of different dietary NDF levels on the cecal ecosystem of growing meat rabbits and to determine the appropriate dietary NDF level in growing meat rabbits.
2. EXPERIMENTAL PROCEDURES
2.1. Animals, housing, and sampling
Two hundred weanling New Zealand rabbits were assigned into four groups according to the mean body weight (1.46 ± 0.16 kg). They were fed each diet with 400 (Diet A), 350 (Diet B), 300 (Diet C), and 250 (Diet D) g/kg NDF (original matter basis), respectively (Table 1). Rabbits were provided with free access to feed and a water nipple.
Table 1.
Composition and nutrient levels of experimental diets (as fed basis) %
| Item | Experimental diets | |||
|---|---|---|---|---|
| A | B | C | D | |
| Corn | 5.0 | 15.0 | 25.0 | 30.0 |
| Wheat bran | 5.0 | 10.0 | 15.0 | 20.0 |
| Soybean meal | 15.0 | 15.0 | 15.0 | 15.0 |
| Corn germ meal | 5.0 | 5.0 | 5.0 | 5.0 |
| Medicago sativa | 16.5 | 16.5 | 16.5 | 16.5 |
| Soybean straw | 12.0 | 6.0 | 0.0 | 0 |
| Peanut vine | 39.0 | 30.0 | 21.0 | 11.0 |
| Bentonite | 1.0 | 1.0 | 1.0 | 1.0 |
| Salt | 0.5 | 0.5 | 0.5 | 0.5 |
| Vitamin‐mineral premixa | 1.0 | 1.0 | 1.0 | 1.0 |
| Total | 100.0 | 100.0 | 100.0 | 100.0 |
| Nutrient levelsb | ||||
| GE (MJ/kg) | 15.37 | 15.71 | 15.72 | 15.81 |
| Dry matter | 86.72 | 86.29 | 85.49 | 84.39 |
| Crude protein | 16.47 | 16.83 | 16.86 | 16.94 |
| Ether extract (EE) | 2.73 | 2.56 | 2.44 | 2.32 |
| Crude fiber | 22.37 | 18.46 | 14.32 | 10.03 |
| Neutral detergent fiber (NDF) | 41.84 | 37.36 | 31.72 | 25.24 |
| Acid detergent fiber (ADF) | 25.39 | 23.08 | 20.70 | 17.51 |
| Acid detergent lignin (ADL) | 7.88 | 6.90 | 6.10 | 5.19 |
| Ash | 10.28 | 9.35 | 8.20 | 7.30 |
| Ca | 0.94 | 0.95 | 0.84 | 0.81 |
| P | 0.75 | 0.80 | 0.71 | 0.73 |
Premix provided the following per kg of diets: VA 8000 IU; VD 31,000 IU; VE 50 mg; Lys 1.5 g; Met 1.5 g; Cu 50 mg; Fe 100 mg; Mn 30 mg; Mg 150 mg; I 0.1 mg; Se 0.1 mg.
Nutrient levels were measured values.
2.2. Production trial
All rabbits were weighed at the beginning (day 32) and the end (day 72) of the experimental period and the average daily gain (ADG), the average daily feed intake (ADFI), feed conversion rate (FCR), and mortality were recorded and calculated for the whole experimental period. The stomach weight, stomach content, cecal weight, cecal content weight, small intestine length, and rabbit body length were measured at 72 days, which were used for computing relative stomach weight (stomach weight/body weight), stomach content relative weight (stomach content weight/body weight), small intestine relative length (small intestine length/body length), cecal relative weight (cecal weight/body weight), cecal content relative weight (cecal content weight/body weight), mortality percentage (mortality number/rabbits total numbers per group). After slaughter, the cecum were removed immediately, cecal content (n = 8) used for measuring the pH value, NH3‐N concentration, VFA by the technique of Weatherburn (1967) and Tao and Li (2006).
2.3. Intestinal microbiota trial
Five healthy rabbits randomly selected from each diet treatment group were euthanized at 52, 62, and 72 days of age and harvested their cecal contents from the middle of the ccum, and stored at −80°C until microbial analyses.
Animal management and experimental procedures abided by the welfare guidelines of the Animal Care Committee, Shandong Agricultural University, People's Republic of China.
2.4. Chemical analysis of experimental diets
The diets were formulated according to the values of the National Research Council (NRC; 1977) and recommendations in “The Nutrition of the Rabbit” (De Blas et al., 1998), and the food was formed the as pellets of 4 mm diameter (Table 1), and food or drinking did not add antibiotics in the total experiment. The experimental diets were analyzed dietary ingredient according to the recommendations of the Association of Official Analytical Chemists (National Standards Recommend Method, China). The crude fiber was measured by the acid‐base method, and the levels of NDF and acid detergent fiber (ADF) were determined using the detergent method developed by Van Soest (1963).
2.5. DNA extraction and purification
For each sample, DNA was obtained using a QIAamp® DNA Stool Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. In order to avoid bias, DNA was extracted in duplicate (Michelland et al., 2010) for 16S rRNA sequencing. A260/280 measurement was used to assess the DNA quality by a DU640 Nucleic Acids and Protein Analyzer (Beckman Coulter, Brea, CA). DNA samples were sent to Hanyu Bio‐Tech (Shanghai, China) for V3–V4 region of the 16S rRNA gene high‐throughput sequencing with an Illumina MiSeq platform according to protocols described by previous studies (Caporaso et al., 2010).
2.6. Statistical and bioinformatic analyses
The data of production trial were subjected to one‐way analysis of variance (ANOVA) using the general linear models (GLM) procedure in SAS 9.1 (SAS institute Inc., Cary, NC). Intestinal microbiota richness and diversity indexes (Tables 5,6) were analyzed by one‐way or two‐way analysis of variance to examine the effects of diet, age, and diet×age. p < 0.05 and p < 0.01 indicates significant difference and highly significant difference among 16S sequences from different treatments, respectively. The valid sequences were filtered from the high‐throughput sequencing with standards previously reported (Hamady et al., 2008). Briefly, an average quality score<25 and min length <200 bp were considered as low‐quality sequences, and any mismatches to the primers, or a homopolymer longer than 8 bases were excluded. The passing sequences were assigned to the individual sample according to the 7‐bp barcodes, and the remaining sequences were clustered into operational taxonomic units (OTUs) with a cutoff of 97% identity. Rarefaction curves, abundance‐based coverage estimates (ACE), Chao1 (Chao, Chazdon, Colwell, & Shen, 2005), network analysis, heat map analysis, and beta diversity analysis indexes were generated at 97% sequence identity level using RDP classifier. In addition, a principal component analysis (PCA) was performed based on weighted UniFrac distance. The diversity index (Simpson/Shannon), Good's coverage, and classification for all sequences were determined with Mothur (Schloss et al., 2009).
3. RESULTS
3.1. Effects of different dietary NDF on growth performance and total tract apparent digestibility and nitrogen balance
ADFI, ADG, FCR, and mortality during the growth trial are summarized in Table 2. ADFI and FCR were increased when NDF increasing (p < 0.05). ADG trend is contrast with above. Mortality reached minimum levels in diet A and diet B (p < 0.01). The stomach relative weight, stomach content relative weight, cecal relative weight, and cecal content weight increased with increasing NDF (p < 0.05). The level of NDF did not affect small intestine relative length (Table 3).
Table 2.
Effects of NDF level on growth performance of growing meat rabbits (n = 50)
| Items | Diets | p‐value | |||
|---|---|---|---|---|---|
| A | B | C | D | ||
| Average daily feed intake (ADFI, g/d) | 156.91 ± 4.88a | 158.66 ± 4.64[Link] | 142.62 ± 3.39a | 138.45 ± 3.24[Link] | 0.022 |
| Average daily gain (ADG, g/d) | 18.53 ± 2.90B | 28.40 ± 2.33A | 32.20 ± 2.20A | 40.76 ± 2.29C | 0.001 |
| Feed conversion rate (FCR) | 8.47 ± 0.26A | 5.59 ± 0.16B | 4.43 ± 0.11C | 3.97 ± 0.21C | <0.001 |
| Mortality (%) | 1.7 ± 0.02A | 1.5 ± 0.05B | 4.8 ± 0.11C | 11.2 ± 0.31C | <0.00 1 |
Different small letter superscripts mean significant difference (p < 0.05).
A,B,C Different capital letter superscripts mean significant difference (p < 0.01); whereas with the same or no letter superscripts mean no significant difference (p > 0.05).
Table 3.
Effects of NDF level on gastrointestinal development of growing meat rabbits (n = 8)
| Items | Diets | p‐value | |||
|---|---|---|---|---|---|
| A | B | C | D | ||
| Stomach relative weight/% | 6.85 ± 0.40a | 6.17 ± 0.37a | 5.30 ± 0.25a | 5.12 ± 0.15a | 0.012 |
| Stomach content relative weight/% | 4.87 ± 0.37a | 4.38 ± 0.33a,b | 3.64 ± 0.24a | 3.22 ± 0.11a | 0.031 |
| Small intestine relative length | 3.89 ± 0.28 | 3.81 ± 0.18 | 3.61 ± 0.19 | 3.38 ± 0.23 | 0.853 |
| Cecal relative weight/% | 8.84 ± 0.70a | 7.76 ± 0.33a,b | 6.93 ± 0.39a | 6.56 ± 0.6a | 0.022 |
| Cecal content relative weight/% | 4.87 ± 0.37a | 4.38 ± 0.33a,b | 3.64 ± 0.24a | 3.49 ± 0.24a | 0.031 |
Stomach relative weight = Stomach weight/body weight; Stomach content relative weight = Stomach content weight/body weight; Small intestine relative length = Small intestine length/body length; Cecal relative weight = Cecal weight/body weight; Cecal content relative weight = Cecal content weight/body weight.
a,b Different small letter superscripts mean significant difference (p < 0.05), whereas with the same or no letter superscripts mean no significant difference (p > 0.05).
The effect of different dietary NDF on cecal fermentation is shown in Table 4. The only NH3‐N concentration of cecum dropped when the dietary NDF increased (p < 0.05).
Table 4.
The cecal fermentation character of experimental rabbits (n = 8)
| Items | Diets | p‐value | |||
|---|---|---|---|---|---|
| A | B | C | D | ||
| Total volatile fatty acid (VFA, mmol/L) | 38.23 ± 3.13 | 41.97 ± 2.73 | 43.76 ± 3.69 | 44.07 ± 2.69 | 0.325 |
| Propionic acid proportion (100%) | 4.14 ± 0.63 | 3.85 ± 1.02 | 5.43 ± 2.02 | 5.32 ± 0.53 | 0.395 |
| Acetic acid proportion (100%) | 80.43 ± 0.88 | 78.98 ± 0.78 | 76.16 ± 0.58 | 74.76 ± 0.59 | 0.069 |
| Butyric acid proportion (100%) | 15.43 ± 1.68 | 16.18 ± 0.98 | 18.41 ± 1.68 | 20.12 ± 0.73 | 0.051 |
| NH3‐N (mmol/L) | 21.22 ± 1.13a | 22.36 ± 0.55a,b | 23.56 ± 0.48a,b | 26.18 ± 0.57a,b | 0.033 |
| pH value | 6.82 ± 0.03 | 6.73 ± 0.04 | 6.56 ± 0.07 | 6.53 ± 0.05 | 0.074 |
a,bDifferent small letter superscripts mean significant difference (p < 0.05), whereas with the same or no letter superscripts mean no significant difference (p > 0.05).
3.2. Illumina MiSeq derived metadata
The raw sequences with an average length of 402 bp were obtained from the Illumina Miseq platform. By quality filtering, the obtained valid sequences per sample (based on 97% sequence identity) were aligned in accordance with the RDP classifier alignment and clustered into OTUs. These OTUs were assigned to 10 different phyla (Figure 1).The rarefaction curves of all samples are shown in Supplementary Figure S1. Although the corresponding curve from each sample was not plateaued indicating that complete sampling in rabbits cecum had not yet been achieved, Good's coverage estimates pointed out a large part of the diversity in all samples captured with an average coverage of 96.2% (data not shown).
Figure 1.
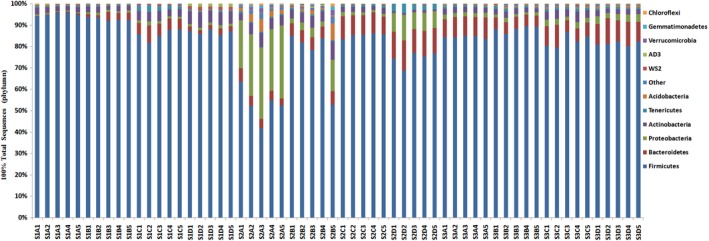
Bacterial composition of the different diet treatments at the phylum level. A, B, C, D represented the sequences of cecum from Diet A, B, C, D, respectively. A1, A2, A3, A4, A5……D5 represented five repeat per diet treatment, respectively; S represented sampling time at 52d (S1), 62d (S2), 72d (S3). The same as below
3.3. Microbial diversity and abundance
The Shannon–Weaver diversity index was calculated to assess the diversity of the microbial communities. Using two‐factor analysis, the DNA sequences from all five rabbits in each diet group were used individually for these calculations, and diversity indexes were compared over time among different diets (Table 5). The diversity of the total microbiota decreased significantly in diets A and D (p = 0.011), and the diversity levels reached the lowest in 52 days for all diet groups (Table 2, p < 0.001). The numbers of OTUs, the Chao 1, and ACE estimates at two cut‐off levels of 3% are provided in Supplementary Table S1, which showed OTUs number, Chao 1, and ACE, associated with richness significantly increased in Diets A, B (p = 0.03, p = 0.003, p = 0.002, respectively); the richness indexes for these diet groups significantly increased in 62 days (Table 2, p < 0.001).
Table 5.
Indexesa of microbial richness and diversity for all samples
| Diet | Age (days) | p level | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | 52 | 62 | 72 | RMSE | Diet | Age | Diet × Age | |
| Shannon index | 5.44a,b | 5.67a | 5.56a | 5.28b | 5.13B | 5.71A | 5.63A | 0.33 | 0.011 | <0.001 | <0.001 |
| ACE | 15300[Link] | 17890A | 2970B | 3290B | 2016B | 22729A | 4843B | 12409.43 | 0.002 | <0.001 | <0.001 |
| Chao 1 | 8561A | 10044A | 2651B | 2672B | 1817B | 12361A | 3769B | 6552.36 | 0.003 | <0.001 | <0.001 |
| OTUs | 3134.8ba | 3573.9a | 1665.1b | 1564.3b | 1250.1B | 4196.3A | 2007.3B | 2200.33 | 0.030 | <0.001 | 0.005 |
RMSE, Root mean square error; ACE, abundance‐based coverage estimates.
The operational taxonomic units (OTUs) were defined at 3% dissimilarity level. The richness estimators (ACE and Chao1) and diversity indexes (Shannon) were calculated using the Mothur analysis.
a,bDifferent small letter superscripts mean significant difference (p < 0.05);
A,B,C Different capital letter superscripts mean significant difference (p < 0.01); whereas with the same or no letter superscripts mean no significant difference (p > 0.05).
3.4. Taxon‐based analysis and Phylum‐level difference in microbial composition among group
To describe the bacterial composition of the different dietary treatment groups and how they changed during total experimental period, we conducted a taxon‐dependent analysis (Cole et al., 2013). Describing the distribution of DNA sequences at phylum level in Figure 1, the results showed that bacterial communities of all samples were composed primarily of Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria, which had overall majority of the total sequences. Significant fluctuations of Firmicutes, Bacterioidetes, Proteobacteria, and Actinobacteria were detected among the four diets and three ages (Table 6). The phylum Firmicutes was higher (p < 0.01) in rabbits fed Diets B and C when compared with the other two diets (Diets A and D). The phylum Bacteroidetes was higher when rabbits were fed with Diets C and D than the other diets (p < 0.001). The highest level of NDF (Diet A) increased the Firmicutes/Bacteroidetes ratio (p < 0.001), and a higher Firmicutes/Bacteroidetes ratio occurred in the 52‐d age group (p < 0.001). However, phylum Proteobacteria was the highest in Diet A (p < 0.001). Similarly, means from each age group among the four diets were compared by an analysis of diet × age (Table 6). For all of the above bacterial phyla, the interaction between diet and age was highly significant (p < 0.001).
Table 6.
Effect of different diets on Relative abundance of the main bacterial phylaa
| Diet | Age (days) | p level | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | 52 | 62 | 72 | RMSE | Diet | Age | Diet × Age | |
| Firmicutes | 77.43B | 85.77A | 84.49A | 80.80B | 90.02A | 72.22C | 84.12B | 4.8 | <0.001 | <0.001 | <0.001 |
| Bacteroidetes | 4.70B | 4.76B | 7.59A | 8.32A | 2.87B | 8.01A | 8.13A | 1.147 | <0.001 | <0.001 | <0.001 |
| Firmicutes/Bacteroidetesa | 126.89A | 25.62B | 12.04B | 18.09B | 115.57A | 10.1B | 11.31B | 80.35 | <0.001 | <0.001 | <0.001 |
| Proteobacteria | 10.71A | 2.63C | 1.76C | 4.44B | 1.00B | 11.36A | 2.30B | 2.12 | <0.001 | <0.001 | <0.001 |
| Actinobacteria | 3.37 | 3.44 | 3.50 | 3.17 | 3.78A | 3.50A | 2.82B | 0.75 | _ | <0.001 | <0.001 |
Relative abundance of a phylum in different libraries was calculated as percentage of the sequence of this phylum to all sequences in that sample.
Firmicutes/Bacteroidetes represented the their percentage of the sequence ratio.
Different capital letter superscripts mean significant difference (p < 0.01); whereas with the same or no letter superscripts mean no significant difference (p > 0.05).
3.5. Genus‐level differences in microbial composition among groups
As shown Figure 2, among the classified genera, there were 14 predominant genera (>1%) and were commonly shared by all samples (in any sample): Desulfovibrio, Arthrobacter, Ruminococcus, Adlercreutzia, Faecalibacterium, Dorea, Bifidobacterium, Eggerthella, Coprococcus, Rikenella, Clostridium, Oscillospira, Bibersteinia, and Bacteroides. Comparing the different diet treatments in rabbits of the same age shows that except for Bacteroides and Faecalibacterium at 52 days and Dorea at 72 days, the abundance levels of the above bacteria genera were significantly different among the four dietary treatments at 52, 62, and 72 days (p < 0.05). Ruminococcus was the most abundant genus among the four treatments, accounting for 22%–52% of the total number of high‐quality bacterial sequences. Moreover, the levels of Ruminococcus spp. differed significantly among the four treatments at the same sampling time (Figure 2). Except for the 72‐day sample, the Ruminococcus spp. abundance was highest in Diets B and C at 52 and 62 days. It is interesting to note that Bifidobacterium from Diet C was the most abundant genus during the entire experimental period (p < 0.01). However, the trends were similar to the abundance levels of Desulfovibrio in Diet C samples. Clostridium only occurred at very low levels (p < 0.01) in Diet C, except for the 62‐day samples; it was most abundant in rabbits fed Diet A at 52 days (p < 0.01) and Diet B at 72 days (p < 0.01). Oscillospira accounted for 6%–41% of the total bacterial effective sequences in all samples, whereas Oscillospira was more abundant in Diet A than the other diet groups in the 52‐ and 62‐day samples (p < 0.01) and was extremely highly abundant in rabbits fed Diets A and B in the 72‐day samples (p < 0.01). The distribution of some abundant genera also changed with aging. For instance, the genus Coprococcus was more abundant in rabbits fed Diets B and D at 52 days (p < 0.01), Diet C at 62 days (p < 0.01) and Diets A and C at 72 days (p < 0.01). Bibersteinia was highly abundant in rabbits fed Diet D (4.36%, p < 0.01) at 52 days, but was no longer abundant (0.09%, p < 0.01) at 62 and 72 days. The sequences assigned to Faecalibacterium were significantly affected by diets of different fiber levels (62 and 72 days, p < 0.01), whereas Diets B and C elicited higher percentages of total bacterial effective sequences at 62 days (4.5%). In addition, Diets A and C elicited a high abundance of Faecalibacterium at 62 days (3.52%; 3.07%) and Diet D (9.72%) at 72 days.
Figure 2.
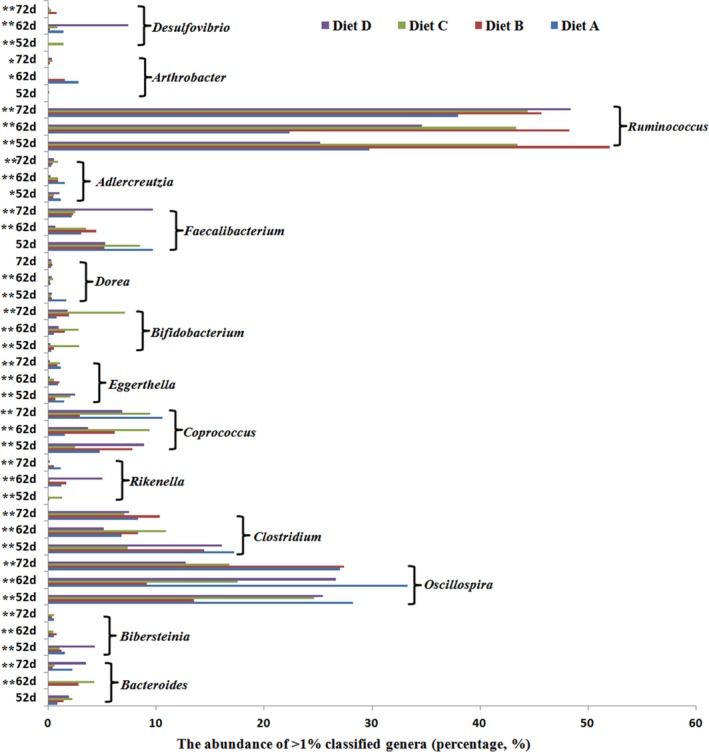
The histogram represented occurred at >1% abundance in at least one sample at genus level, and their comparing among four different NDF levels diets in 52, 62, and 72 days
3.6. Relationships of cecal bacterial communities feeding different dietary fiber levels
The principal component analysis of the weighted UniFrac distance matrix and heat map analysis were conducted to further evaluate the pyrosequencing data. The PCA score plot revealed that the majority of the samples harbored characteristic bacterial communities, and the samples from Diet C at 52 days (S1C1, S1C2, S1C3, S1C4, S1C5), Diets C and D at 62 days (S2C1, S2C2, S2C3, S2C4, S2C5), and Diets A, B, C and D at 72 days (S3A1, S3A2, S3A4, S3A5, S3B1, S3B2, S3B3, S3B4, S3B5, S3C1, S3C2, S3C3, S3C4, S3C5, S3D1, S3D2, S3D3, S3D4, S3D5) grouped to the right of the graph along PC1, which accounts for 56.86% of the total variations. Others were separate from the above samples along PC2, which represented 18.35% of the total variations (Figure 3). Overall, the two PCA axes explained 75.21% of the variation between the different communities. Analysis of the hierarchically clustered heat map based on the bacterial community profiles at the genus level revealed the same trend (Figure 4).
Figure 3.
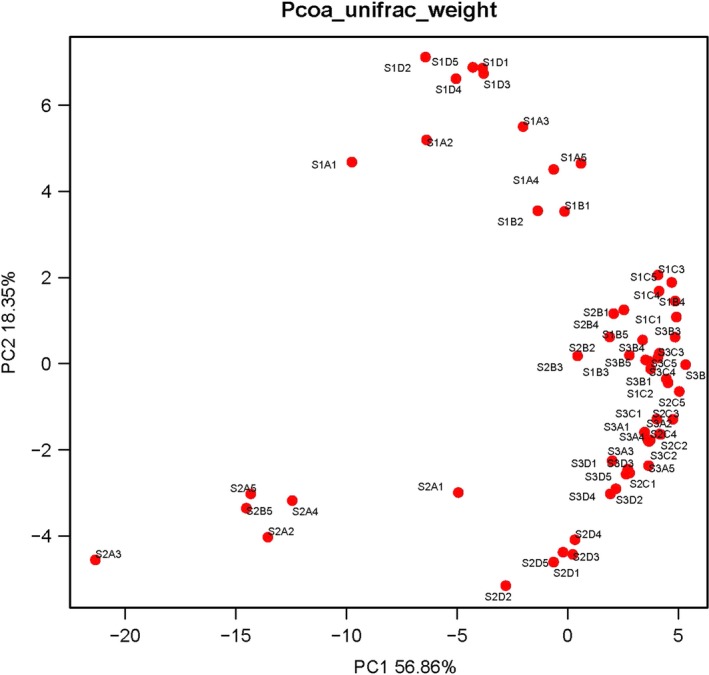
Sample sorting analysis
Figure 4.
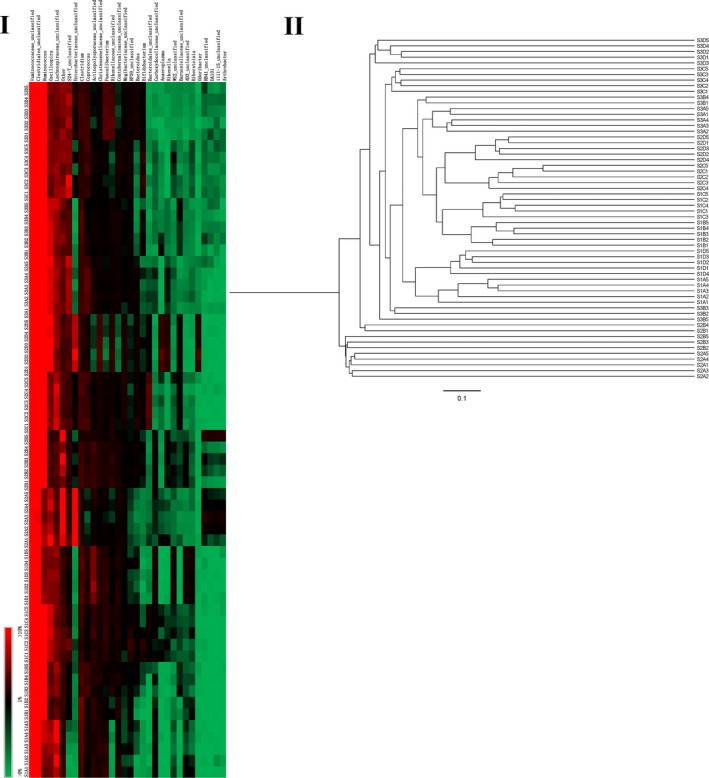
Bacterial distribution among all samples
Each Venn diagram represents the sum of genus‐level OTUs at each sampling time point of each sample (Figure 5). Examination of these genera captured the shared and unique genus‐level OTUs of all samples in the same age. Comparative analysis indicated 816, 1102 and 1162 OTUs shared by different dietary treatments at 52, 62, and 72 days, respectively. Therefore, the number of shared OTUs among different treatments tended to increase with age.
Figure 5.
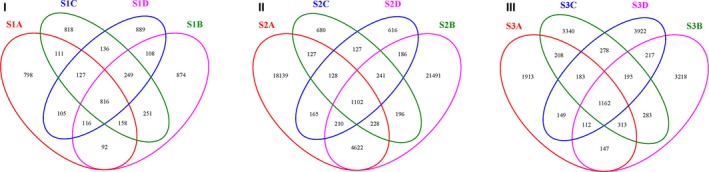
Shared OTUs analysis of the different libraries. Venn diagram showing the unique and shared OTUs (3% distance level) among the different libraries in 52 days (I), 62 days (II), and 72 days (III)
4. DISCUSSION
Although the rabbit relies on the intake of large quantities of fiber that can be fermented by the microbiota found predominantly within the cecum, the change in NDF level does not affect the cecal pH and VFA concentration, which indicates no relation between quantity of NDF and concentration of VFA in the cecum (Bellier & Gidenne, 1996). Previous studies focused particularly on the analysis of the effects of the dietary fiber level on the whole cecal bacterial community in rabbits using molecular profiles (Michelland et al., 2010, 2011; Monteils et al., 2008; Rodríguez‐Romero, Abecia, & Fondevila, 2013). Our study showed that the cecal microbiota abundance increased significantly when NDF levels reached 350–400 g/kg (original matter basis) (p < 0.001). However, 400 g/kg and 250 g/kg NDF groups decreased the diversity of the cecal microbiota (p = 0.01). Therefore, feeding rabbits with different dietary fiber levels resulted in alteration of the structure of the cecal bacteria community (diversity and relative abundance) (Crowley et al., 2017). We also noticed that the cecal bacterial diversity increases with age, whereas bacterial abundance maintains a dynamic balance (in this case, increased first and then decreased).
We found that more than 90% of DNA sequences of the cecal contents from all samples in the total experiment period belong to the Firmicutes, Bacteroides, Actinobacteria, and Proteobacteria, which is consistent with the findings of Cauquil and Gidenne (2012). In this study, rabbits feeding on high‐fiber diets (Diet A) required high cecal ratio of Firmicutes/Bacteroidetes (p < 0.001). Combes et al. (2011) reported ratio Firmicutes/Bacteroidetes was proposed as an indicator for microbiota maturity. Thus, our results indicated that the higher NDF level diet can accelerate the maturation of intestinal microbiota. In addition, the mice and human studies suggest that a higher Firmicutes/Bacteroidetes impels the gut microbiota to extract efficiently from the diet, which reveal one cause of adiposity (Marcobal et al., 2011; Ley, Lozupone, Hamady, Knight, & Gordon, 2008; Yasuda et al., 2015). However, it is clearly established that an increase in NDF level results in a reduction in the ADG in our study (p < 0.001), which apparently contradicts previous results. Here the result seems to be more related with physiological conditions, such as the rabbit cecum is different significantly from mice and human.
Ruminococcus, Coprococcus, Oscillospira, Bifidobacteria, and Clostridium were known as VFA‐producing genera, interestingly, our study also suggests that Only Coprococcus and Oscillospira richness was positively correlated with NDF levels. In fact, the quantity of all kinds of VFA among different treatments had no difference. But the effect of NDF levels on ADFI significantly increased with NDF reducing (p < 0.01). More research will be needed to understand the relation between production and various microbial groups in the cecum. Bifidobacterium species are frequently associated with health‐promoting effects in the human and animal intestinal tracts (Arboleya, Watkins, Stanton, & Ross, 2016; O'Callaghan & van Sinderen, 2016; Combes et al., 2013). Notably, the 300 g/kg NDF group (Diet C) led to significantly increased Bifidobacterium. Additionally, we observed what appears to be a substrate‐related stimulatory effect of Diet C on Desulfovibrio spp. (producers of toxic sulfides). Inness, McCartney, Khoo, Gross, and Gibson (2007) reported, also, modulation of by increasing Bifidobacteria and decreasing Desulfovibrio spp. may be beneficial to cats with IBD. Moreover, in our study, the diarrhea index was found to be significantly higher in Diets C and D than in Diets A and B (p < 0.001, data not shown), which indicated a positive relation with higher Desulfovibrio spp. (Diet C and Diet D, respectively, in 52 and 62 days, p < 0.001, Figure 2). In additional, our study showed that rabbit production performance was elevated, that is, the ADG reached the highest levels in Diets C and D (p = 0.001, Table 2). This result indicated that abundance of Desulfuricans spp. may reflect elevated production (Luo et al., 2018). On the other hand, Bibersteinia is an important pathogen that is associated with serious infection (Bleich, Sargeant, & Wiedmann, 2018; Parr, Smith, Jenks, & Thompson, 2018; Heinse, Hardesty, & Harris, 2016) and is implicated in the high incidence of diarrhea in Diet D at 52 days.
Venn diagrams, PCA, and heat map plots of the bacterial communities derived from rabbit cecum showed that a shift of dietary fiber can lead to quick changes in the composition of the microbiota, which is a reflection of a rapid adaptation to reach a new equilibrium in response to a nutritional disturbance (Michelland et al., 2011; Zhu, Sun, Wang, & Li, 2017). Meanwhile, we found that in the cecum, the structure of the bacterial community has no difference between rabbits of the same age or fed the same diet treatment (Michelland et al., 2010).
5. CONCLUSION
We provide clear evidence that the influence of dietary NDF levels on growth performance, digestibility and metabolism, the cecal bacterial community differed between rabbits of different ages. Indeed, better production performance, increasing the cecal microbiota diversity and abundance were all benefited from suitable dietary NDF level in Diet B (350 g/kg NDF, original matter basis).
CONFLICT OF INTEREST
All authors declare that there are no conflicts of interest.
Supporting information
ACKNOWLEDGMENTS
This work was supported by grants from the Natural Science Foundation of Shandong Province of China (Grant No: ZR2014CM017); the Science and Technology Plan Project of Shandong Province of China (G17KA135); the Funds of Shandong “Double Tops” Program (SYL2017YSTD11), and the Earmarked Fund for Modern Agro‐industry Technology Research System (Grant No: CARS‐43‐B‐1).
Wu Z, Zhou H, Li F, Zhang N, Zhu Y. Effect of dietary fiber levels on bacterial composition with age in the cecum of meat rabbits. MicrobiologyOpen. 2019;8:e708 10.1002/mbo3.708
*Zhenyu Wu and Hailiang Zhou contributed equally to this paper.
DATA ACCESSIBILITY
MiSeq Illumina sequencing raw sequence reads data: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA291670, which associated with the “Illumina MiSeq derived metadata” section in the article.
REFERENCES
- Arboleya, S. , Watkins, C. , Stanton, C. , & Ross, R. P. (2016). Gut bifidobacteria populations in human health and aging. Frontiers in microbiology, 7, 1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellier, R. , & Gidenne, T. (1996). Consequences of reduced fibre intake on digestion, rate of passage and caecal microbial activity in the young rabbit. British Journal of Nutrition, 75, 353–363. 10.1079/BJN19960139 [DOI] [PubMed] [Google Scholar]
- Bleich, V. C. , Sargeant, G. A. , & Wiedmann, B. P. (2018). Ecotypic variation in population dynamics of reintroduced bighorn sheep. The Journal of Wildlife Management, 82, 8–18. 10.1002/jwmg.21381 [DOI] [Google Scholar]
- Caporaso, J. G. , Kuczynski, J. , Stombaugh, J. , Bittinger, K. , Bushman, F. D. , Costello, E. K. , … Knight, R. (2010). QIIME allows analysis of high‐throughput community sequencing data. Nature Methods, 7, 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauquil, K. M. S. C. L. , & Gidenne, O. Z. T . (2012). High throughput 16S‐DNA sequencing for phylogenetic affiliation of the caecal bacterial community in the rabbit: Impact of the hygiene of housing and of the intake level In Symposium on Gut Microbiology (Vol. 18, p. 21th). Conference: INRA‐Rowett Symposium on Gut Microbiology: At Clermont‐Ferrand, France. [Google Scholar]
- Chao, A. , Chazdon, R. L. , Colwell, R. K. , & Shen, T. J. (2005). A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecology Letters, 8, 148–159. [Google Scholar]
- Cole, J. R. , Wang, Q. , Fish, J. A. , Chai, B. , McGarrell, D. M. , Sun, Y. , … Tiedje, J. M. (2013). Ribosomal database project: Data and tools for high throughput rRNA analysis. Nucleic Acids Research, 42, D633–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combes, S. , Fortun‐Lamothe, L. , Cauquil, L. , & Gidenne, T. (2013). Engineering the rabbit digestive ecosystem to improve digestive health and efficacy. Animal, 7, 1429–1439. 10.1017/S1751731113001079 [DOI] [PubMed] [Google Scholar]
- Combes, S. , Michelland, R. J. , Monteils, V. , Cauquil, L. , Soulié, V. , Tran, N. U. , … Fortun‐Lamothe, L. (2011). Postnatal development of the rabbit caecal microbiota composition and activity. FEMS microbiology ecology, 77, 680–689. 10.1111/j.1574-6941.2011.01148.x [DOI] [PubMed] [Google Scholar]
- Crowley, E. J. , King, J. M. , Wilkinson, T. , Worgan, H. J. , Huson, K. M. , Rose, M. T. , & McEwan, N. R. (2017). Comparison of the microbial population in rabbits and guinea pigs by next generation sequencing. PLoS ONE, 12, e0165779 10.1371/journal.pone.0165779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly, K. , Stewart, C. S. , Flint, H. J. , & Shirazi‐Beechey, S. P. (2001). Bacterial diversity within the equine large intestine as revealed by molecular analysis of cloned 16S rRNA genes. FEMS Microbiology Ecology, 38, 141–151. 10.1111/j.1574-6941.2001.tb00892.x [DOI] [Google Scholar]
- De Blas, C. , & Wiseman, J. (1998). Nutrition of the Rabbit, first edn. Oxon, Wallingford, CABI North American Office, U.K. PP. 241–245. [Google Scholar]
- Gidenne, T. , Arveux, P. , & Madec, O. (2001). The effect of the quality of dietary lignocellulose on digestion, zootechnical performance and health of the growing rabbit. Animal Science, 73, 97–104. 10.1017/S1357729800058094 [DOI] [Google Scholar]
- Gidenne, T. , Jehl, N. , Segura, B. , & Michalet‐Doreau, B. (2002). Microbial activity in the caecum of the rabbit around weaning: Impact of a dietary fiber deficiency and of intake level. Animal Feed science and Technology, 99, 107–118. 10.1016/S0377-8401(02)00138-4 [DOI] [Google Scholar]
- Gidenne, T. , Pinheiro, V. & e Cunha, L. F . (2000). A comprehensive approach of the rabbit digestion: Consequences of a reduction in dietary fibre supply. Livestock Science, 64, 225–237. 10.1016/S0301-6226(99)00141-4 [DOI] [Google Scholar]
- Hamady, M. , Walker, J. J. , Harris, J. K. , Gold, N. J. , & Knight, R. (2008). Error‐correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nature Methods, 5(3), 235–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinse, L. M. , Hardesty, L. H. , & Harris, R. B. (2016). Risk of pathogen spillover to bighorn sheep from domestic sheep and goat flocks on private land. Wildlife Society Bulletin, 40, 625–633. 10.1002/wsb.718 [DOI] [Google Scholar]
- Inness, V. L. , McCartney, A. L. , Khoo, C. , Gross, K. L. , & Gibson, G. R. (2007). Molecular characterisation of the gut microflora of healthy and inflammatory bowel disease cats using fluorescence in situ hybridisation with special reference to Desulfovibrio spp. Journal of Animal Physiology and Animal Nutrition, 91, 48–53. 10.1111/j.1439-0396.2006.00640.x [DOI] [PubMed] [Google Scholar]
- Ivarsson, E. , Roos, S. , Liu, H. Y. , & Lindberg, J. E. (2014). Fermentable non‐starch polysaccharides increases the abundance of Bacteroides–Prevotella–Porphyromonas in ileal microbial community of growing pigs. Animal, 8, 1777–1787. 10.1017/S1751731114001827 [DOI] [PubMed] [Google Scholar]
- Lattimer, J. M. , & Haub, M. D. (2010). Effects of dietary fibre and its components on metabolic health. Nutrients, 2, 1266–1289. 10.3390/nu2121266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley, R. E. , Lozupone, C. A. , Hamady, M. , Knight, R. , & Gordon, J. I. (2008). Worlds within worlds: Evolution of the vertebrate gut microbiota. Nature Reviews Microbiology, 6, 776–788. 10.1038/nrmicro1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, Y. , Chen, H. , Yu, B. , He, J. , Zheng, P. , Mao, X. , … Chen, D. (2018). Dietary pea fibre alters the microbial community and fermentation with increase in fibre degradation‐associated bacterial groups in the colon of pigs. Journal of Animal Physiology and Animal Nutrition, 102, e254–e261. 10.1111/jpn.12736 [DOI] [PubMed] [Google Scholar]
- Marcobal, A. , Barboza, M. , Sonnenburg, E. D. , Pudlo, N. , Martens, E. C. , Desai, P. , … Sonnenburg, J. L. (2011). Bacteroides in the infant gut consume milk oligosaccharides via mucus‐utilization pathways. Cell Host & Microbe, 10, 507–514. 10.1016/j.chom.2011.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelland, R. J. , Combes, S. , Monteils, V. , Cauquil, L. , Gidenne, T. , & Fortun‐Lamothe, L. (2010). Molecular analysis of the bacterial community in digestive tract of rabbit. Anaerobe, 16, 61–65. 10.1016/j.anaerobe.2009.05.002 [DOI] [PubMed] [Google Scholar]
- Michelland, R. , Combes, S. , Monteils, V. , Cauquil, L. , Gidenne, T. , & Fortun‐Lamothe, L. (2011). Rapid adaptation of the bacterial community in the growing rabbit cæcum after a change of dietary fibre supply. Animal, 5, 1761–1768. 10.1017/S1751731111001005 [DOI] [PubMed] [Google Scholar]
- Monteils, V. , Cauquil, L. , Combes, S. , Godon, J. J. , & Gidenne, T. (2008). Potential core species and satellite species in the bacterial community within the rabbit caecum. FEMS Microbiology Ecology, 66, 620–629. 10.1111/j.1574-6941.2008.00611.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Callaghan, A. , & van Sinderen, D. (2016). Bifidobacteria and their role as members of the human gut microbiota. Frontiers in Microbiology, 7, 925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr, B. L. , Smith, J. B. , Jenks, J. A. , & Thompson, D. J. (2018). Population dynamics of a bighorn sheep (Ovis canadensis) herd in the Southern Black Hills of South Dakota and Wyoming. The American Midland Naturalist, 179, 1–14. 10.1674/0003-0031-179.1.1 [DOI] [Google Scholar]
- Rodríguez‐Romero, N. , Abecia, L. , & Fondevila, M. (2013). Microbial ecosystem and fermentation traits in the caecum of growing rabbits given diets varying in neutral detergent soluble and insoluble fibre levels. Anaerobe, 20, 50–57. 10.1016/j.anaerobe.2013.02.001 [DOI] [PubMed] [Google Scholar]
- Sawicki, C. , Livingston, K. , Obin, M. , Roberts, S. , Chung, M. , & McKeown, N. (2015). Dietary fiber and the human gut microbiome: Application of evidence mapping methodology. The FASEB Journal, 29(736), 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss, P. D. , Westcott, S. L. , Ryabin, T. , Hall, J. R. , Hartmann, M. , Hollister, E. B. , … Weber, C. F. (2009). Introducing mothur: Open‐source, platform‐independent, community‐supported software for describing and comparing microbial communities. Applied and environmental microbiology, 75, 7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Z. Y. , & Li, F. C. (2006). Effects of dietary neutral detergent fibre on production performance, nutrient utilization, caecum fermentation and fibrolytic activity in 2‐to 3‐month‐old New Zealand rabbits. Journal of Animal Physiology and Animal Nutrition, 90, 467–473. 10.1111/j.1439-0396.2006.00628.x [DOI] [PubMed] [Google Scholar]
- Van Soest, P. J. (1963). Use of detergents in the analysis of fibrous feeds. II. A rapid method for the determination of fiber and lignin. Journal of the Association of Official Agricultural Chemists, 46, 829–835. [Google Scholar]
- Verbeke, K. A. , Boobis, A. R. , Chiodini, A. , Edwards, C. A. , Franck, A. , Kleerebezem, M. , … Tuohy, K. M. (2015). Towards microbial fermenta‐tion metabolites as markers for health benefits of prebiotics. Nutrition research reviews, 2, 42–66. 10.1017/S0954422415000037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherburn, M. W. (1967). Phenol‐hypochlorite reaction for determination of ammonia. Analytical Chemistry, 39, 971–974. 10.1021/ac60252a045 [DOI] [Google Scholar]
- Yasuda, K. , Oh, K. , Ren, B. , Tickle, T. L. , Franzosa, E. A. , Wachtman, L. M. , … Morgan, X. C. (2015). Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host & Microbe, 17, 385–391. 10.1016/j.chom.2015.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y. , Sun, Y. , Wang, C. , & Li, F. (2017). Impact of dietary fibre: Starch ratio in shaping caecal archaea revealed in rabbits. Journal of Animal Physiology and Animal Nutrition, 101, 635–640 10.1111/jpn.12585 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MiSeq Illumina sequencing raw sequence reads data: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA291670, which associated with the “Illumina MiSeq derived metadata” section in the article.


