Abstract
Aims
The aim of this study was to identify and compare the gut microbial community of wild and captive Tor tambroides through 16S rDNA metagenetic sequencing followed by functions prediction.
Methods and results
The library of 16S rDNA V3‐V4 hypervariable regions of gut microbiota was amplified and sequenced using Illumina MiSeq. The sequencing data were analyzed using Quantitative Insights into Microbial Ecology (QIIME) pipeline and Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt). The most abundant bacterial phyla in both wild and captive T. tambroides were Firmicutes, Proteobacteria, Fusobacteria and Bacteroidetes. Cetobacterium spp., Peptostreptococcaceae family, Bacteroides spp., Phosphate solubilizing bacteria PSB‐M‐3, and Vibrio spp. were five most abundant OTU in wild T. tambroides as compared to Cetobacterium spp., Citrobacter spp., Aeromonadaceae family, Peptostreptococcaceae family and Turicibacter spp. in captive T. tambroides.
Conclusion
In this study, the specimens of the wild T. tambroides contain more diverse gut microbiota than of the captive ones. The results suggested that Cetobacterium spp. is one of the core microbiota in guts of T. tambroides. Besides, high abundant Bacteroides spp., Citrobacter spp., Turicibacter spp., and Bacillus spp. may provide important functions in T. tambroides guts.
Significance and impact of the study
The results of this study provide significant information of T. tambroides gut microbiota for further understanding of their physiological functions including growth and disease resistance.
Keywords: 16S rDNA, gut microbiota, Illumina MiSeq, metagenetic sequencing, next‐generation sequencing, Tor tambroides
1. INTRODUCTION
Fish of the genus Tor, commonly known as mahseers, are important to most nations in the Asian region due to its biodiversity and high‐value (Ng, 2004). Tor tambroides also known as “empurau” in Sarawak or “kelah merah” in Peninsular Malaysia is the most valued freshwater fish species in Malaysia (Ingram, Sungan, Tinggi, Sim, & De Silva, 2007). The T. tambroides has generated much interest in its artificial propagation for both conservation and aquaculture production due to its high market demand, high flesh quality and high commercial value (Ng, Abdullah, & De Silva, 2008).
One of the major problems with T. tambroides captive breeding is the slow growth of T. tambroides (Lee et al., 2014). There were many studies on the different feed formulations and feed additives that improved the growth rate of T. tambroides (Ishak, Kamarudin, Ramezani‐Fard, & Yusof, 2016; Kamarudin, Ramezani‐Fard, Saad, & Harmin, 2011; Misieng, Kamarudin, & Musa, 2011; Ng & Andin, 2011; Ng et al., 2008; Ramezani‐Fard, Kamarudin, Saad, Harmin, & Goh, 2012). There were also studies on effects of host gut‐derived probiotic bacteria to T. tambroides where the probiotic improved nutrients utilization and metabolism, adjusting gut microbiota balance and enhanced growth by promoting muscle fiber hypertrophy (Asaduzzaman, Iehata, et al., 2018; Asaduzzaman, Sofia, et al., 2018). Nevertheless, there is no report on the phylogenetic and functional characterization of gut microbiota of T. tambroides.
Gut microbiota can be considered as an “extra organ” due to its crucial role in intestinal development, homeostasis and immunological protection, growth and health (O'Hara & Shanahan, 2006). The gut microbiota in vertebrate is complex and contains diverse and abundant bacteria, archaea, viruses, and fungi (Liu et al., 2015; Neuman & Koren, 2015). Gut microbiota of aquatic animals is transient and has higher fluidity than terrestrial animals; thus, changes in environmental factors such as temperature, salinity, trophic level, and host phylogeny may affect the gut microbial community (Denev, Staykov, Moutafchieva, & Beev, 2009; Guerreiro et al., 2016; Ringø et al., 2016; Sullam et al., 2012). More than 99% of environmental prokaryotes including the gut microbiota of animals are unculturable in laboratory that limits our understanding of microbial physiology, genetics, and community ecology (Schloss & Handelsman, 2005). The development of next‐generation sequencing (NGS) technology allows the recognition of discrete populations (culturable and unculturable) based on DNA sequences in the environmental samples (Konstantinidis & Rosselló‐Móra, 2015; Tarnecki, Burgos, Ray, & Arias, 2017). Esposito and Kirschberg (2014) clarified that the metagenomic study means the whole genome sequencing and analysis of each member of the microbial community in an environmental sample by 16S rDNA‐based sequencing should be called as metagenetic sequencing.
Illumina MiSeq (Illumina, USA) has been widely used for 16S rRNA gene sequencing of gastrointestinal tract microbiota of freshwater fishes such as blunt snout, grass carp, mandarin fish, topmouth cutler, common carp, crucian carp, silver carp, bighead carp, and Prussian carp (Kashinskaya et al., 2015; Liu et al., 2015) and marine fishes such as emerald rockcod, crocodile icefish, ploughfish, bald rockcod, yellowtail scad, brown‐marbled grouper, spotted coral grouper and Atlantic salmon (Dehler, Secombes, & Martin, 2017; Hennersdorf et al., 2016; Song et al., 2016).
The objectives of this study were to identify and compare gut microbiota in wild and captive T. tambroides. Determination of core bacteria and prediction of their functions in gut microflora lead to identification of potential bacteria that could be used as probiotics to improve growth performance and disease resistance of T. tambroides in captivity.
2. MATERIALS AND METHODS
2.1. Fish sampling and species verification
Three captive adult T. tambroides (standard length 35.77 ± 1.39 cm, weight 960.57 ± 58.29 g) were obtained from hatchery at Agro‐Biotechnology Institute (ABI) on 6 April 2015. The captive fish were obtained from the wild and reared in hatchery for 3 years. They were fed twice daily (8.00 a.m. and 4.00 p.m.) with commercial floating pellet containing 42% crude protein and 6% lipid. Fish were reared in rectangular fiberglass tank with 1,500 L of dechlorinated tap water with continuous aeration. Each tank was attached to a recirculating aquaculture system (RAS) with 30% water changes fortnightly. Three wild adult T. tambroides (standard length 31.73 ± 0.78 cm, weight 630.27 ± 56.32 g) were obtained by angling from Kenyir Lake, Terengganu, Malaysia (GPS Coordinates: 5°0′14″N, 102°38′19″E) on 13 April 2015. These fish were packed in Kenyir Lake water and transported alive to ABI, Serdang, Selangor, Malaysia (GPS Coordinates: 2°59′18″N, 101°41′52″E) (approx. 5 hr). These fish were processed upon arrived at the destination.
DNA of the fish was extracted from dorsal fin samples using Phenol‐Chloroform‐Isoamyl‐Alcohol (PCI) DNA extraction method (Tan et al., 2008). The cytochrome b gene was amplified using GluDG‐L (5′‐TGACTTGAARAACCAYCGTTG‐3′) and CB2‐H (5′‐CCCTCAGAATGATATTTGTCCTCA‐3′) primers (Palumbi et al., 2002). Reaction mixture (25 μl) included HotStarTaq Plus Master Mix (10 μl) (Qiagen, Germany), forward and reverse primers (1 μM and 5 μl of each) and template DNA (10 ng). Amplification conditions were the following: initial denaturation at 95°C for 5 min; denaturation at 95°C for 45 s, annealing at 47°C for 45 s, elongation at 72°C for 45 s (25 cycles); final elongation at 72°C for 7 min. The PCR products were purified using QIAquick PCR Purification Kit (Qiagen, Germany). DNA sequencing was outsourced to First BASE Laboratories Sdn. Bhd. (Malaysia). The results were analyzed using NCBI BLASTn (NCBI, 1988).
2.2. Fish dissection and DNA extraction
Tor tambroides were anesthetized using 30 ppm clove oil (Neiffer & Stamper, 2009) and euthanized by pithing (Leary et al., 2013). Fish skin was disinfected with 70% ethanol prior to autopsy. The abdomen of fish was dissected using sterile instruments in laminar flow cabinet. The gut samples were removed and separated from other internal organs. The gut parts from esophagus to anus were then cut into small pieces and placed in sterile phosphate‐buffered saline (PBS) (Nie, Zhou, Qiao, & Chen, 2017) followed by mixing with vortex and kept at −80°C. Gut microbiota DNA in these gut samples was extracted using PCI DNA extraction method (Tan et al., 2008).
2.3. Library preparation and MiSeq sequencing
V3–V4 hypervariable regions of 16S rRNA genes of gut microbiota were amplified by polymerase chain reaction (PCR) using primers (Forward primer: 5′‐TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG‐3′ and Reverse primer: 5′GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC‐3′) according to the manufacturer's instructions. Reaction mixture (25 μl) included 2X KAPA HiFi HotStart ReadyMix (12.5 μl) (Kapa Biosystems, USA), forward and reverse primers (1 μM and 5 μl of each), and template DNA (7.5 ng). Amplification conditions were as the following: initial denaturation at 95°C for 3 min; denaturation at 95°C for 30 s, annealing at 55°C for 30 s, elongation at 72°C for 30 s (25 cycles); final elongation at 72°C for 5 min.
Amplicons were cleaned up followed by PCR to attach unique index adapter pairs to the amplicons using Nextera XT Index kit (Illumina). These indexed DNA libraries were cleaned up with Agencourt AMPure XP (Beckman Coulter, USA) followed by concentration quantification using Qubit dsDNA HS Assay Kit and Qubit 2.0 Fluorometer (Thermo Fisher Scientific, USA) and size validation using Agilent DNA 1000 Kit and Agilent 2100 Bioanalyzer (Agilent, USA).
The libraries were serial diluted and quantified by quantitative real‐time PCR (qRT‐PCR) through Eppendorf Mastercycler RealPlex2 (Eppendorf, Germany) followed by normalization to 4 nM and pooled into one tube. Pooled DNA libraries were denatured and spiked with 15% denatured PhiX as quality control. A mixture of 600 μl of denatured pooled DNA libraries with denatured PhiX loaded into the sample well in MiSeq Reagent Kit v2 (2 × 250 cycles) (Illumina) and sequenced using Illumina MiSeq (Illumina) at Malaysia Genome Institute (MGI), Kajang, Selangor, Malaysia. All sequences were also submitted to NCBI Sequence Read Archive (SRA).
2.4. Data analysis using Quantitative Insights into Microbial Ecology (QIIME)
The analysis of MiSeq sequencing results was done using Quantitative Insights into Microbial Ecology (QIIME ver. 1.9.0) pipeline (Caporaso, Kuczynski, et al., 2010). Adapter sequences were trimmed from the paired‐end forward and reverse reads and merged. Merged reads were quality filtered at Phred Quality Score of 20 (Q20) (Cock, Fields, Goto, Heuer, & Rice, 2010). Length filter was used to remove reads shorter than 100 bp (below 20% of the library length) to avoid unspecific match that will disturb the accuracy of the calling (Edgar, 2010). Chimeric sequences were removed using RDP Gold databases as reference (Edgar, Haas, Clemente, Quince, & Knight, 2011). De novo OTU picking strategy was used as it did not cause any information lost although may be time‐consuming for large datasets (Edgar, 2010; Edgar et al., 2011).
The OTUs in generated OTU BIOM file were summarized into different taxonomic levels. Taxa summary plots were plotted to show the differences in taxonomic levels of the samples. Alpha rarefactions curves were plotted to determine the adequacy of sequencing depth. Alpha diversity indexes (Chao1, Shannon and Simpson) were calculated to explain the species richness and diversity in each sample (Udayangani et al., 2017). Good's Coverage estimator was used to estimate the percentage of the total species that are represented in a sample. In beta diversity analysis, the number of sequences per sample had been rarified to equal number based on the sample which had the lowest sequences number. Principle coordinates analysis (PCoA) was used to visualize similarities or dissimilarities of data based on phylogenetic or count‐based distance metrics. Weighted UniFrac was used in the PCoA analysis of this study because it accounted for differences in relative abundances of each taxon within the communities (Lozupone, Hamady, Kelley, & Knight, 2007). Mann–Whitney U test was used to determine the differences of the gut microbial communities in the wild and captive T. tambroides (Jonsson, Österlund, Nerman, & Kristiansson, 2016).
2.5. Functions prediction using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt)
PICRUSt (ver. 1.1.0) was used to predict the metabolic functions of the microbial communities in each sample (Langille et al., 2013). Closed reference OTU picking strategy was used with the GreenGenes database (version 13.5) as reference at 97% identity threshold (Caporaso, Bittinger, et al., 2010; DeSantis et al., 2006). The OTU table was then normalized, and the microbiota functions were predicted with referenced to Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology (KO) database (Kanehisa & Goto, 2000).
3. RESULTS
3.1. Dissection and species verification
Autopsy of T. tambroides revealed that the gut digesta color of wild T. tambroides obtained from Kenyir Lake was green while it was brown in captive T. tambroides obtained from hatchery which was fed with commercial floating feed pellets. The cytochrome b gene of both wild and captive T. tambroides was analyzed using NCBI BLASTn and found to be 98%–99% similar to cytochrome b gene in complete mitochondrial genome of T. tambroides (GenBank Accession Number: JX444718.1) (National Center for Biotechnology Information (NCBI), 1988; Norfatimah, Teh, Salleh, Mat Isa, & SitiAzizah, 2014).
3.2. Metagenetic sequencing of wild and captive T. tambroides gut microbiota with QIIME analysis
The de novo OTU picking generated 7,749 and 9,468 OTUs for wild and captive T. tambroides gut microbiota, respectively (Table 1). Nevertheless, the number of species found in wild T. tambroides was 501 as compared to 442 in captive ones. 304 genera were shared between wild and captive T. tambroides (Supporting Information Table SA). All sequencing data (three wild and three captive T. tambroides gut microbiome) were submitted to NCBI Sequence Read Archive (SRA) under accession number of SRP094031.
Table 1.
Summary of gut microbiome in wild and captive Tor tambroides
| Sample | Sequences after merging of forward and reverse reads | Sequences after QC quality filter | Sequences after length filter (>100 bp) | Sequences after chimera filter | Operational taxonomic units (OTUs) after OTU picking |
|---|---|---|---|---|---|
| Wild | 785,128 | 751,324 | 751,318 | 558,171 | 7,749 |
| Captive | 1,024,668 | 983,240 | 983,010 | 702,194 | 9,468 |
Each value was a mean value calculated from the raw data.
3.3. Alpha diversity analysis of T. tambroides gut microbiota
The OTUs found in both wild and captive T. tambroides were reduced as the number of sequences increased at 50,000 sequences per sample (Figure 1). In Table 2, Good's Coverage confirmed that the sequencing covered up to 99% of all gut microbiota in wild and captive T. tambroides. Chao1 index showed that captive T. tambroides gut microbiota had higher species richness than wild T. tambroides. Nevertheless, both Shannon and Simpson indexes for wild T. tambroides gut microbiota were higher than captive T. tambroides, indicated higher species diversity in fish that live in natural environment.
Figure 1.
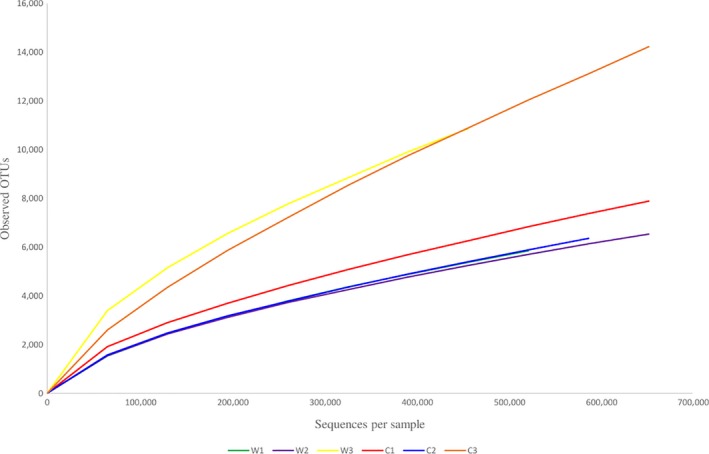
Alpha rarefaction curves of wild and captive Tor tambroides gut microbiota (C1–C3: biological replicates of captive T. tambroides; W1–W3: biological replicates of wild T. tambroides)
Table 2.
Summary of alpha diversity of wild and captive Tor tambroides gut microbiota
| Sample | Chao1 | Shannon | Simpson | Good's coverage |
|---|---|---|---|---|
| Wild | 19,280.31 | 4.83 | 0.87 | 0.99 |
| Captive | 26,814.36 | 4.44 | 0.81 | 0.99 |
3.4. Gut taxonomy of wild and captive T. tambroides
The gut microbiota of wild T. tambroides was dominated mainly by Firmicutes followed by Fusobacteria, Proteobacteria, and Bacteroidetes which together accounted for 85.7% of total population. Gut of captive T. tambroides was dominated mainly by Proteobacteria followed by Fusobacteria, Firmicutes and Bacteroidetes which together accounted for 91.67% of total population (Figure 2). The most abundant genus in wild T. tambroides gut microbiota was Cetobacterium (23.48%), followed by genera in Peptostreptococcaceae family (11.87%), Bacteroides (9.60%), PSB‐M‐3 from Erysipelotrichaceae family (7.70%), Vibrio (4.94%), and others (42.41%). Cetobacterium (29.07%) also was the most abundant genus in captive T. tambroides gut microbiota, followed by Citrobacter (9.35%), genera in Aeromonadaceae family (8.63%), genera in Peptostreptococcaceae family (7.66%), Turicibacter (6.47%), and others (38.82%) (Supporting Information Table SA). The 10 most abundant unique species in either wild or captive samples were listed in Table 3. These unique species only existed in small percentages (<0.31%).
Figure 2.
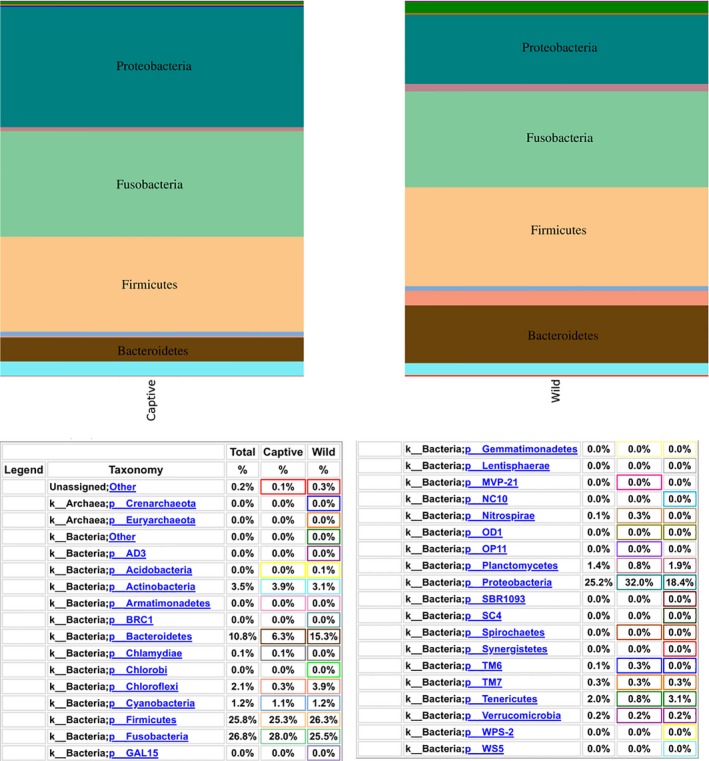
Relative abundance of phyla found in wild and captive Tor tambroides gut microbiome
Table 3.
Ten most abundant unique OTUs in either wild or captive Tor tambroides gut
| Wild T. tambroides | Percentage (%) | Captive T. tambroides | Percentage (%) |
|---|---|---|---|
| g__Synechococcus | 0.306 | g__Virgibacillus | 0.200 |
| o__SHA‐20;f__;g__ | 0.185 | g__Geobacillus | 0.049 |
| c__Betaproteobacteria; Other; Other; Other | 0.107 | c__OP11‐4;o__;f__;g__ | 0.014 |
| g__Methylocaldum | 0.105 | g__Salinivibrio | 0.013 |
| o__HOC36;f__;g__ | 0.099 | g__Pseudoxanthomonas | 0.012 |
| c__Betaproteobacteria;o__;f__;g__ | 0.080 | g__Jeotgalicoccus | 0.011 |
| o__ASSO‐13;f__;g__ | 0.077 | g__Cloacibacterium | 0.008 |
| f__A4b;g__ | 0.075 | g__Ureibacillus | 0.007 |
| f__Methylocystaceae; Other | 0.061 | f__[Tissierellaceae];Other | 0.005 |
| f__Pseudanabaenaceae; Other | 0.045 | g__Tepidimicrobium | 0.004 |
3.5. Beta diversity analysis of wild and captive T. tambroides gut microbiota
The PCoA plots in Figure 3 showed clusters based on wild and captive samples were observed at Principal Coordinate 1 versus Principal Coordinate 2 (PC1 vs PC2) and PC3 vs PC2. Cetobacterium spp. was the highest OTU found in both wild and captive T. tambroides gut microbiota but there was no any significant difference between both samples (Figure 4). Unclassified species from genus PSB‐M‐3, unclassified order from class CK‐1C4‐19, Caldilinea spp., and Clostridium spp. were significantly higher (p < 0.05) in wild T. tambroides. On the other hand, Turicibacter spp., unclassified genus from Rhodospirillaceae family, unclassified genus from Microbacteriaceae family, Bacillus spp., Citrobacter spp., and unclassified genus from Xanthomonadaceae family were significantly higher (p < 0.05) in captive T. tambroides.
Figure 3.
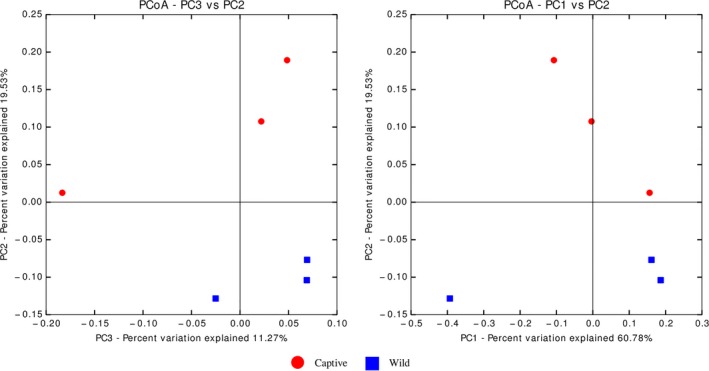
Principal Coordinates Analysis (PCoA) plots of beta diversity analysis based on weighted UniFrac distance metric
Figure 4.
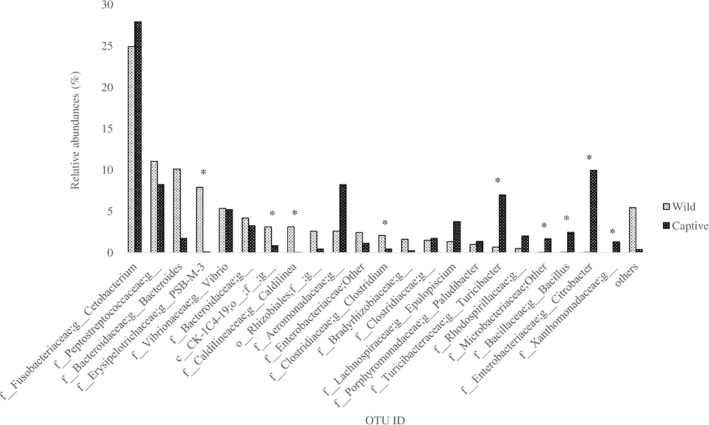
Comparison of top 15 most abundant observed bacteria in wild and captive Tor tambroides guts (Bars with * indicated significant differences between wild and captive T. tambroides samples)
3.6. Predicted metabolic functions using PICRUSt
PICRUSt analyses revealed a total of 293 predicted functions where 277 functions existed in both samples (Supporting Information Table SB). 10 unique functions were found only in wild T. tambroides gut microbiota while six unique functions were only found in captive T. tambroides gut microbiota. Bile secretion and lysine biosynthesis were significantly higher (p < 0.05) in wild T. tambroides gut microbiota while carbohydrate metabolism was significantly higher (p < 0.05) in captive T. tambroides (Figure 5).
Figure 5.
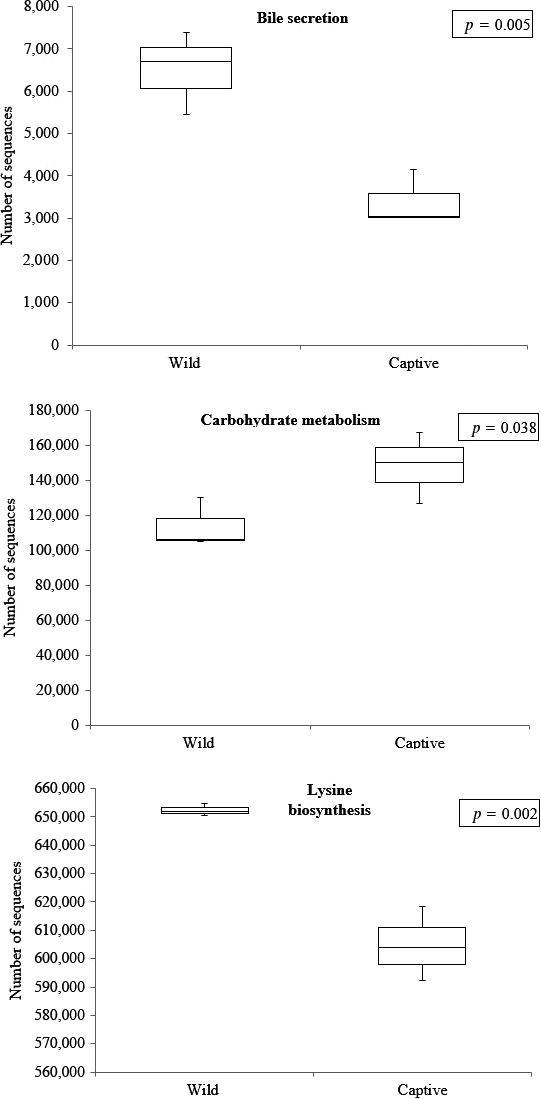
Box‐plots showed three significant predictive functions (bile secretion, carbohydrate metabolism and lysine biosynthesis) of gut microbial communities in wild and captive Tor tambroides
4. DISCUSSION
Although previous studies (Esa et al., 2008; Sati et al., 2013) mostly used cytochrome c oxidase subunit I (COI) for mahseer species identification, all six T. tambroides used in this study for metagenetic analysis were identified using mitochondrial Cytochrome b (CytB) gene. Comparison of CytB gene and COI gene showed that CytB gene is more accurate to construct phylogeny trees and reveal evolutionary relationships, and it gave better resolution during separating species based on sequence data (Tobe, Kitchener, & Linacre, 2011). Although Hampala showed considerable geographical variation in coloration and morphological characteristics, the mitochondrial cytochrome b gene sequencing was able to resolve phylogenetic relationship of Hampala fishes (Ryan & Esa, 2006). It was necessary to accurately verify the species of the mahseer fish used in this study prior to MiSeq sequencing since other fishes such as Tor spp. and Neolissochilus spp. are morphological similar to T. tambroides (Laskar et al., 2013). Identification of fish species based on morphological appearances is also subjective and can lead to misidentification.
Due to the different types of the feeds, eating habits and habitats of the wild and captive T. tambroides, it is anticipated that their gut microbiota community will be different (Li et al., 2014; Ringø et al., 2016). The wild T. tambroides in Kenyir Lake lives in natural environment that contains various types of algae belonging to cyanophytes, bacillariophytes and chlorophytes (Rouf, Phang, & Ambak, 2010). Digesta of all three wild T. tambroides were green in color indicating these fish may be fed in various kinds of algae or plant as food in Kenyir Lake. In contrast, the gut digesta of captive T. tambroides was brown due to the formulated pellet diet which consists of complete nutrients from animal and plant sources. Therefore, T. tambroides in captivity would grow faster than wild T. tambroides at the same age. Temperature fluctuation had effect to the composition of the gut microbiota in farmed Atlantic salmon (Neuman et al., 2016). Various habitats with different environmental factors formed the gut microbiota composition of Atlantic salmon parr (Dehler et al., 2017). Higher species richness of gut microbiota was found in Atlantic salmon parr exposed to open water natural environment than in captive reared ones (Dehler et al., 2017). Although the weight and sizes of captive T. tambroides were higher compared to wild T. tambroides used in this study, it was anticipated that their gut microbiota will be highly influenced by the living environment, feed, and feeding habits.
Operational taxonomic units (OTUs) are groups of sequences that clustered together based on percent similarity threshold (typically 97%) assuming they delineate a species (Nguyen, Warnow, Pop, & White, 2016). Number of OTUs found in wild T. tambroides guts were lower than captive T. tambroides. However, one OTU does not represent one species because OTUs were clustered based on similarity of the other sequences in the bacterial community regardless of whether the sequence is represented by references within a taxonomy outline (Schloss & Westcott, 2011). Thus, a few OTUs may refer to the same species. In alpha diversity analysis, high Chao1 value indicated high species richness (Hughes, Hellmann, Ricketts, & Bohannan, 2001). In our study, the Shannon and Simpson indexes of the wild T. tambroides gut microbiota were higher indicating higher bacterial diversity compared to captive T. tambroides gut microbiota. Shannon and Simpson indexes are calculated based on both species richness and species evenness of the microbial community (Gihring, Green, & Schadt, 2011; Spellerberg & Fedor, 2003).
Firmicutes and Bacteroidetes are able to degrade wide range of polysaccharides (Cockburn & Koropatkin, 2016). This may explain the higher percentages of Firmicutes and Bacteroidetes in wild T. tambroides gut microbiota as their habitat in Kenyir Lake contains huge amount of periphyton algae (Rouf et al., 2010). Cell wall of green algae contains various polysaccharides such as cellulose, pectins, hemicelluloses, lignin, and others (Domozych et al., 2012). Digesta in the wild T. tambroides guts used for this study appeared to be green in color indicated these fish consumed a lot of algae or plant as food which had cell walls made of polysaccharides. In PCoA plots, sample data points cluster together indicated high similarity of gut microbial population among the samples. One of the data of captive T. tambroides sample was not cluster close to the rest as captive T. tambroides samples that were obtained from separated tanks in hatchery where the different microbial community in the tank water could contribute to these dissimilarities. Bacterial communities in water affected Nile tilapia larvae gut microbial communities (Giatsis et al., 2015).
Cetobacterium spp. was the most abundant species without any significant differences in both wild and captive samples suggesting that it is a core species in T. tambroides guts. The colonization of this species in captive T. tambroides even after 3 years of rearing in hatchery condition may indicate their roles and functions in the fish gut. Anaerobic Cetobacterium spp. promotes decomposition of consumed organic debris, phytoplankton, or zooplankton (Borsodi et al., 2017). This species was also common in intestinal tracts of goldfish, common carp, grass carp, ayu, tilapia, zebrafish, rainbow trout, channel catfish, largemouth bass, and bluegill (Adeoye et al., 2016; Etyemez & Balcázar, 2015; Larsen, Mohammed, & Arias, 2014; Roeselers et al., 2011; Tsuchiya, Sakata, & Sugita, 2007; Van Kessel et al., 2011). However, the effects of this species have never been tested in fish. This may be due to the fact that Cetobacterium spp. is obligate anaerobe that will die under normal atmospheric condition thus hinder the possibility of using this species as probiotics in aquaculture production. There was a report stated that bacteria‐mediated cobalamin biosynthesis was supported by the presence of cobalamin synthesizers such as Bacteroides, Lactobacillus, and Cetobacterium (Koo et al., 2017). Besides, C. somerae was reported to produce vitamin B12 which also known as cobalamin (Tsuchiya et al., 2007).
The number of Bacillus spp. was higher in captive T. tambroides gut samples. The Bacillus spp. may originate from probiotics capsules that were added into the tanks few years ago. Bacillus species have been widely used as probiotics in aquaculture industry. Bacillus spp. was discovered to possess anti‐pathogenic properties such as antibacterial and anti‐quorum sensing properties (Chu, Zhou, Zhu, & Zhuang, 2014). Bacillus licheniformis and Bacillus pumilus showed antibacterial activity against Aeromonas hydrophila infection (Ramesh, Vinothkanna, Rai, & Vignesh, 2015; Shobharani, Padmaja, & Halami, 2015). Bacillus subtilis in diets increased growth rate of T. tambroides, upregulated immune‐related genes, and improved stress tolerance toward temperature changes (Nguyen, 2015). Spore production ability of B. subtilis has the potential to be used as delivery system for vaccine or recombinant spores that expressed surface enzyme which induced innate and adaptive immunity, systemic and local mucosal immunity (Jiang et al., 2017).
Dominance of Citrobacter genus was observed in captive T. tambroides gut microbiome. Citrobacter freundii had inhibitory effects against A. hydrophila (Aly, Ahmed, Ghareeb, & Mohamed, 2008). In contrast, C. freundii isolated from intestinal tract of farmed grass carp showed pathogenicity to mice and zebrafish (Lü et al., 2011). Clostridium spp. was higher in wild T. tambroides gut. Clostridium butyricum was reported as a potential probiotic that has strong adhesion and antagonistic activity against A. hydrophila and Vibrio anguillarum (Pan et al., 2008).
Bile is essential for digestion and absorption of fats and removal of excess cholesterol, bilirubin, drugs, and toxic compounds (Kanehisa, Tanabe, Sato, & Morishima, 2017). Gut microbiota is capable to convert bile acids into secondary bile acids which modulate its signaling properties that regulate diverse metabolic pathways in the host (Ramírez‐Pérez, Cruz‐Ramón, Chinchilla‐López, & Méndez‐Sánchez, 2017). Eubacterium lentum and Clostridium perfringens were reported to possess the capability to produce iso‐bile acids (Hirano & Masuda, 1981; Hirano, Masuda, Oda, & Mukai, 1981). Enzymes from gut microbiota may contribute significantly to bile acid metabolism and essential for bile acid homeostasis in the host and contributed to host health (Long, Gahan, & Joyce, 2017). Higher bile secretion functions of gut microbiota in wild T. tambroides may offer protection to the fishes that exposed to natural environment. In this study, the Clostridium spp. was 2.09% in wild samples as compared to 0.42% in captive ones.
Carbohydrate needed to be digested to monosaccharides prior to absorption in the small intestine (Kanehisa et al., 2017). Some fishes may able to digest mono‐, di‐, and oligosaccharides but not for indigestible complex carbohydrates such as hemicellulose and cellulose which usually plenty in plants (Krogdahl, Hemre, & Mommsen, 2005). High carbohydrate and high lipid diets have been widely used in aquaculture to reduce cost, but they also caused excessive lipid accumulation in the fish liver (Xie et al., 2017). In contrast, wild T. tambroides may consume algae, fruits, small fishes, and crustaceans. Thus, the gut microbiota in captive T. tambroides showed higher carbohydrate metabolism function. Many Bacteroides spp. such as Bacteroides thetaiotaomicron are capable of metabolize polysaccharides in gut (Ravcheev, Godzik, Osterman, & Rodionov, 2013). Besides, these bacteria also can provide energy from indigestible polysaccharides comprising part of the host diet (Schwalm & Groisman, 2017).
Lysine biosynthesis evolved separately into two pathways which are diaminopimelic acid (DAP) and aminoadipic acid (AAA) pathways (Liu, White, & Whitman, 2010). Lysine is an essential amino acid for living organism especially those consume vegetarian or low animal protein diet. Lysine biosynthesis functions were higher in wild T. tambroides gut microbiota, and this could be due to consumption of microalgae that was abundant in Kenyir Lake. PICRUSt predictions were made based on available genome sequences of bacteria thus some OTUs that lack of closely related genomes may be underpredicted (Salinas & Magadán, 2017). Lysine produced by gut microbiota was reported to be absorbed at the host's small intestines (Metges, 2000). Bacillus sphaericus and Bacillus megaterium were reported to have the capability to synthesize lysine (Bartlett & White, 1986; Ekwealor & Obeta, 2005).
In conclusion, species diversity was higher in wild T. tambroides gut microbiota as compared to captive T. tambroides. The samples of wild and captive T. tambroides gut microbiota could be clustered in the PCoA plots based on origin of the samples where the gut microbial composition of wild T. tambroides was different compared to captive T. tambroides. The results suggested that Cetobacterium spp. is one of the core microbiota in guts of T. tambroides. The other bacteria may be important in T. tambroides guts included those presented in high abundance, like Bacteroides spp., Citrobacter spp., Turicibacter spp., and Bacillus spp. Metagenetic sequencing in this study revealed much bacteria existed in the guts of wild and captive T. tambroides, and future research could be focused on isolate those bacteria that may be use as potential probiotics. This requires the development of specific media and analysis of growing conditions.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interests.
AUTHORS CONTRIBUTION
Anjas and Tan conceived of the study conception and experimental design. Tan, Marilyn, and Nazrien were responsible for the acquisition of data. Anjas, Tan, Natrah, and Iswan analyzed and interpreted the data. Anjas and Tan drafted the manuscript. All authors discussed the results and contributed to the final manuscript.
ETHICAL STATEMENT
This study was approved by Malaysia Institute Pharmaceuticals and Nutraceuticals Animal Ethic Committee (IPAEC) with approval number of NIBM/IPharm/PTR (S) 100‐7/7‐ 4 (2017‐004).
Supporting information
ACKNOWLEDGMENTS
The author truthfully appreciates the funding for this study from Ministry of Science, Technology and Innovation (MOSTI) Malaysia. The author gratefully thanking Malaysia Genome Institute (MGI) for the laboratory facility for MiSeq sequencing and computer facility for bioinformatics data analysis.
Tan CK, Natrah I, Suyub IB, Edward MJ, Kaman N, Samsudin AA. Comparative study of gut microbiota in wild and captive Malaysian Mahseer (Tor tambroides). MicrobiologyOpen. 2019;8:e734 10.1002/mbo3.734
DATA ACCESSIBILITY
All sequences were also submitted to NCBI Sequence Read Archive (SRA) under accession number of https://www.ncbi.nlm.nih.gov/bioproject/PRJNA355218/.
REFERENCES
- Adeoye, A. A. , Yomla, R. , Jaramillo‐Torres, A. , Rodiles, A. , Merrifield, D. L. , & Davies, S. J. (2016). Combined effects of exogenous enzymes and probiotic on Nile tilapia (Oreochromis niloticus) growth, intestinal morphology and microbiome. Aquaculture, 463, 61–70. 10.1016/j.aquaculture.2016.05.028 [DOI] [Google Scholar]
- Aly, S. M. , Ahmed, Y. A. G. , Ghareeb, A. A. A. , & Mohamed, M. F. (2008). Studies on Bacillus subtilis and Lactobacillus acidophilus, as potential probiotics, on the immune response and resistance of Tilapia nilotica (Oreschromis niloticus) to challenge infections. Fish and Shellfish Immunology, 25(1–2), 128–136. 10.1016/j.fsi.2008.03.013 [DOI] [PubMed] [Google Scholar]
- Asaduzzaman, M. , Iehata, S. , Akter, S. , Kader, M. A. , Ghosh, S. K. , Khan, M. N. A. , & Abol‐Munafi, A. B. (2018). Effects of host gut‐derived probiotic bacteria on gut morphology, microbiota composition and volatile short chain fatty acids production of Malaysian Mahseer Tor tambroides . Aquaculture Reports, 9, 53–61. 10.1016/j.aqrep.2017.12.003 [DOI] [Google Scholar]
- Asaduzzaman, M. , Sofia, E. , Shakil, A. , Haque, N. F. , Khan, M. N. A. , Ikeda, D. , … Abol‐Munafi, A. B. (2018). Host gut‐derived probiotic bacteria promote hypertrophic muscle progression and upregulate growth‐related gene expression of slow‐growing Malaysian Mahseer Tor tambroides . Aquaculture Reports, 9, 37–45. 10.1016/j.aqrep.2017.12.001 [DOI] [Google Scholar]
- Bartlett, A. T. M. , & White, P. J. (1986). Regulation of the enzymes of lysine biosynthesis in Bacillus sphaericus NCTC 9602 during vegetative growth. Journal of General Microbiology, 132, 3169–3177. [Google Scholar]
- Borsodi, A. K. , Szabó, A. , Krett, G. , Felföldi, T. , Specziár, A. , & Boros, G. (2017). Gut content microbiota of introduced bigheaded carps (Hypophthalmichthys spp.) inhabiting the largest shallow lake in Central Europe. Microbiological Research, 195, 40–50. 10.1016/j.micres.2016.11.001 [DOI] [PubMed] [Google Scholar]
- Caporaso, J. G. , Bittinger, K. , Bushman, F. D. , DeSantis, T. Z. , Andersen, G. L. , & Knight, R. (2010). PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics, 26(2), 266–267. 10.1093/bioinformatics/btp636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso, J. G. , Kuczynski, J. , Stombaugh, J. , Bittinger, K. , Bushman, F. D. , Costello, E. K. , … Knight, R. (2010). QIIME allows analysis of high‐throughput community sequencing data. Nature Methods, 7(5), 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, W. , Zhou, S. , Zhu, W. , & Zhuang, X. (2014). Quorum quenching bacteria Bacillus sp. QSI‐1 protect zebrafish (Danio rerio) from Aeromonas hydrophila infection. Nature Scientific Reports, 4, 5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock, P. J. A. , Fields, C. J. , Goto, N. , Heuer, M. L. , & Rice, P. M. (2010). The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Research, 38(6), 1767–1771. 10.1093/nar/gkp1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockburn, D. W. , & Koropatkin, N. M. (2016). Polysaccharide degradation by the intestinal microbiota and its influence on human health and disease. Journal of Molecular Biology, 428(16), 3230–3252. 10.1016/j.jmb.2016.06.021 [DOI] [PubMed] [Google Scholar]
- Dehler, C. E. , Secombes, C. J. , & Martin, S. A. M. (2017). Environmental and physiological factors shape the gut microbiota of Atlantic salmon parr (Salmo salar L.). Aquaculture, 467, 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denev, S. , Staykov, Y. , Moutafchieva, R. , & Beev, G. (2009). Microbial ecology of the gastrointestinal tract of fish and the potential application of probiotics and prebiotics in finfish aquaculture. International Aquatic Research, 1, 1–29. [Google Scholar]
- DeSantis, T. Z. , Hugenholtz, P. , Larsen, N. , Rojas, M. , Brodie, E. L. , Keller, K. , … Andersen, G. L. (2006). Greengenes, a chimera‐checked 16S rRNA gene database and workbench compatible with ARB. Applied and Environmental Microbiology, 72(7), 5069–5072. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domozych, D. S. , Ciancia, M. , Fangel, J. U. , Mikkelsen, M. D. , Ulvskov, P. , & Willats, W. G. T. (2012). The cell walls of green algae: A journey through evolution and diversity. Frontiers in Plant Science, 3, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 26(19), 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Edgar, R. C. , Haas, B. J. , Clemente, J. C. , Quince, C. , & Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics, 27(16), 2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwealor, I. A. , & Obeta, J. A. N. (2005). Studies on lysine production by Bacillus megaterium . African Journal of Biotechnology, 4(7), 633–638. [Google Scholar]
- Esa, Y. , Siraj, S. S. , Daud, S. K. , Ryan, J. J. R. , Rahim, K. A. A. , & Tan, S. G. (2008). Molecular systematics of Mahseers (Cyprinidae) in Malaysia inferred from sequencing of a mitochondrial cytochrome C Oxidase I (COI) gene. Pertanika Journal of Tropical Agricultural Science, 31(2), 263–269. [Google Scholar]
- Esposito, A. , & Kirschberg, M. (2014). How many 16S‐based studies should be included in a metagenomic conference? It may be a matter of etymology. FEMS Microbiology Letters, 351(2), 145–146. 10.1111/1574-6968.12375 [DOI] [PubMed] [Google Scholar]
- Etyemez, M. , & Balcázar, J. L. (2015). Bacterial community structure in the intestinal ecosystem of rainbow trout (Oncorhynchus mykiss) as revealed by pyrosequencing‐based analysis of 16S rRNA genes. Research in Veterinary Science, 100, 8–11. 10.1016/j.rvsc.2015.03.026 [DOI] [PubMed] [Google Scholar]
- Giatsis, C. , Sipkema, D. , Smidt, H. , Heilig, H. , Benvenuti, G. , Verreth, J. , & Verdegem, M. (2015). The impact of rearing environment on the development of gut microbiota in tilapia larvae. Scientific Reports, 5, 18206 10.1038/srep18206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gihring, T. M. , Green, S. J. , & Schadt, C. W. (2011). Massively parallel rRNA gene sequencing exacerbates the potential for biased community diversity comparisons due to variable library sizes. Environmental Microbiology, 14(2), 285–290. [DOI] [PubMed] [Google Scholar]
- Guerreiro, I. , Serra, C. R. , Enes, P. , Couto, A. , Salvador, A. , Costas, B. , & Oliva‐Teles, A. (2016). Effect of short chain fructooligosaccharides (scFOS) on immunological status and gut microbiota of gilthead sea bream (Sparus aurata) reared at two temperatures. Fish and Shellfish Immunology, 49, 122–131. 10.1016/j.fsi.2015.12.032 [DOI] [PubMed] [Google Scholar]
- Hennersdorf, P. , Kleinertz, S. , Theisen, S. , Abdul‐Aziz, M. A. , Mrotzek, G. , Palm, H. W. , & Saluz, H. P. (2016). Microbial diversity and parasitic load in tropical fish of different environmental conditions. PLoS ONE, 11(3), e0151594 10.1371/journal.pone.0151594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano, S. , & Masuda, N. (1981). Transformation of bile acids by Eubacterium lentum . Applied and Environmental Microbiology, 42(5), 912–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano, S. , Masuda, N. , Oda, H. , & Mukai, H. (1981). Transformation of bile acids by Clostridium perfringens . Applied and Environmental Microbiology, 42(3), 394–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, J. B. , Hellmann, J. J. , Ricketts, T. H. , & Bohannan, B. J. M. (2001). Counting the uncountable: Statistical approaches to estimating microbial diversity. Applied Environmental Microbiology, 67(10), 4399–4406. 10.1128/AEM.67.10.4399-4406.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram, B. , Sungan, S. , Tinggi, D. , Sim, S. Y. , & De Silva, S. S. (2007). Breeding performance of Malaysian mahseer, Tor tambroides and T. duoronensis broodfish in captivity. Aquaculture Research, 38, 809–818. 10.1111/j.1365-2109.2007.01716.x [DOI] [Google Scholar]
- Ishak, S. D. , Kamarudin, M. S. , Ramezani‐Fard, E. , & Yusof, Y. A. (2016). Effects of varying dietary carbohydrate levels on growth performance, body composition and liver histology of Malaysian mahseer fingerlings (Tor tambroides). Journal of Environmental Biology, 37(4), 756–764. [PubMed] [Google Scholar]
- Jiang, H. , Chen, T. , Sun, H. , Tang, Z. , Yu, J. , Lin, Z. , … Yu, X. (2017). Immune response induced by oral delivery of Bacillus subtilis spores expressing enolase of Clonorchis sinensis in grass carps (Ctenopharyngodon idellus). Fish and Shellfish Immunology, 60, 318–325. 10.1016/j.fsi.2016.10.011 [DOI] [PubMed] [Google Scholar]
- Jonsson, V. , Österlund, T. , Nerman, O. , & Kristiansson, E. (2016). Statistical evaluation of methods for identification of differentially abundant genes in comparative metagenomics. BMC Genomics, 17, 78 10.1186/s12864-016-2386-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamarudin, M. S. , Ramezani‐Fard, E. , Saad, C. R. , & Harmin, S. A. (2011). Effects of dietary fish oil replacement by various vegetable oils on growth performance, body composition and fatty acid profile of juvenile Malaysian mahseer, Tor tambroides . Aquaculture Nutrition, 18(5), 532–543. [Google Scholar]
- Kanehisa, M. , & Goto, S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Research, 28(1), 27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa, M. , Tanabe, M. , Sato, Y. , & Morishima, K. (2017). KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Research, 45(D1), D353–D361. 10.1093/nar/gkw1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashinskaya, E. N. , Belkova, N. L. , Izvekova, G. I. , Simonov, E. P. , Andree, K. B. , Glupov, V. V. , … Solovyev, M. M. (2015). A comparative study on microbiota from the intestine of Prussian carp (Carassius gibelio) and their aquatic environmental compartments, using different molecular methods. Journal of Applied Microbiology, 119(4), 948–961. 10.1111/jam.12904 [DOI] [PubMed] [Google Scholar]
- Konstantinidis, K. T. , & Rosselló‐Móra, R. (2015). Classifying the uncultivated microbial majority: A place for metagenomic data in the Candidatus proposal. Systematic and Applied Microbiology, 38(4), 223–230. 10.1016/j.syapm.2015.01.001 [DOI] [PubMed] [Google Scholar]
- Koo, H. , Hakim, J. A. , Powell, M. L. , Kumar, R. , Eipers, P. G. , Morrow, C. D. , … Bej, A. K. (2017). Metagenomics approach to the study of the gut microbiome structure and function in zebrafish Danio rerio fed with gluten formulated diet. Journal of Microbiological Methods, 135, 69–76. 10.1016/j.mimet.2017.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogdahl, A. , Hemre, G. I. , & Mommsen, T. P. (2005). Carbohydrates in fish nutrition: Digestion and absorption in postlarval stages. Aquaculture Nutrition, 11(2), 103–122. 10.1111/j.1365-2095.2004.00327.x [DOI] [Google Scholar]
- Langille, M. G. I. , Zaneveld, J. , Caporaso, J. G. , McDonald, D. , Knights, D. , Reyes, J. , … Huttenhower, C. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnology, 31(9), 814–821. 10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen, A. M. , Mohammed, H. H. , & Arias, C. R. (2014). Characterization of the gut microbiota of three commercially valuable warmwater fish species. Journal of Applied Microbiology, 116(6), 1396–1404. 10.1111/jam.12475 [DOI] [PubMed] [Google Scholar]
- Laskar, B. A. , Bhattacharjee, M. J. , Dhar, B. , Mahadani, P. , Kundu, S. , & Ghosh, S. K. (2013). The species dilemma of Northeast Indian Mahseer (Actinopterygii: Cyprinidae): DNA barcoding in clarifying the riddle. PLoS ONE, 8(1), e53704 10.1371/journal.pone.0053704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary, S. , Underwood, W. , Anthony, R. , Cartner, S. , Corey, D. , Grandin, T. , … Yanong, R. (2013). AVMA Guidelines for the Euthanasia of animals. 2013 ed. American Veterinary Medical Association.
- Lee, K. S. , Lihan, S. , Dasthagir, F. F. G. , Mikal, K. M. , Collick, F. , & Ng, K. H. (2014). Microbiological and physicochemical analysis of water from empurau fish (Tor tambroides) farm in Kuching, Sarawak, Malaysian Borneo. International Journal of Scientific and Technology Research, 3(6), 285–292. [Google Scholar]
- Li, J. , Ni, J. , Li, J. , Wang, C. , Li, X. , Wu, S. , … Yan, Q. (2014). Comparative study on gastrointestinal microbiota of eight fish species with different feeding habits. Journal of Applied Microbiology, 117(6), 1750–1760. 10.1111/jam.12663 [DOI] [PubMed] [Google Scholar]
- Liu, H. , Guo, X. W. , Gooneratne, R. , Lai, R. F. , Zeng, C. , Zhan, F. B. , & Wang, W. M. (2015). The gut microbiome and degradation enzyme activity of wild freshwater fishes influenced by their trophic levels. Scientific Reports, 6, 24340 10.1038/srep24340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , White, R. H. , & Whitman, W. B. (2010). Methanococci use the diaminopimelate aminotransferase (DapL) pathway for lysine biosynthesis. Journal of Bacteriology, 192(13), 3303–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, S. L. , Gahan, C. G. M. , & Joyce, S. A. (2017). Interactions between gut bacteria and bile in health and disease. Molecular Aspects of Medicine, 56, 54–65. 10.1016/j.mam.2017.06.002 [DOI] [PubMed] [Google Scholar]
- Lozupone, C. A. , Hamady, M. , Kelley, S. T. , & Knight, R. (2007). Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Applied and Environmental Microbiology, 73(5), 1576–1585. 10.1128/AEM.01996-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü, A. , Hu, X. , Zheng, L. , Zhu, A. , Cao, C. , & Jiang, J. (2011). Isolation and characterization of Citrobacter spp. from the intestine of grass carp Ctenopharyngodon idellus . Aquaculture, 313(1–4), 156–160. [Google Scholar]
- Metges, C. C. (2000). Contribution of microbial amino acids to amino acid homeostasis of the host. Journal of Nutrition, 130(7), 1857S–1864S. 10.1093/jn/130.7.1857S [DOI] [PubMed] [Google Scholar]
- Misieng, J. D. , Kamarudin, M. S. , & Musa, M. (2011). Optimum dietary protein requirement of Malaysian mahseer (Tor tambroides) fingerling. Pakistan Journal of Biological Sciences, 14(3), 232–235. [DOI] [PubMed] [Google Scholar]
- National Center for Biotechnology Information (NCBI) (1988). Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information. Retrieved from https://www.ncbi.nlm.nih.gov/
- Neiffer, D. L. , & Stamper, M. A. (2009). Fish sedation, anesthesia, analgesia, and euthanasia: Considerations, methods, and types of drugs. Institute for Laboratory Animal Research Journal, 50(4), 343–360. 10.1093/ilar.50.4.343 [DOI] [PubMed] [Google Scholar]
- Neuman, C. , Hatje, E. , Zarkasi, K. Z. , Smullen, R. , Bowman, J. P. , & Katouli, M. (2016). The effect of diet and environmental temperature on the faecal microbiota of farmed Tasmanian Atlantic Salmon (Salmo salar L.). Aquaculture . Research, 47(2), 660–672. [Google Scholar]
- Neuman, H. , & Koren, O. (2015). The gut microbiome. Encyclopedia of Cell Biology, 2, 799–808. [Google Scholar]
- Ng, C. K. (2004). King of the rivers: Mahseer in Malaysia and the region. Inter Sea Fishery (M). Kuala Lumpur, Malaysia: Sdn Bhd. [Google Scholar]
- Ng, W. K. , Abdullah, N. , & De Silva, S. S. (2008). The dietary protein requirement of the Malaysian mahseer, Tor tambroides (Bleeker), and the lack of protein‐sparing action by dietary lipid. Aquaculture, 284(1–4), 201–206. 10.1016/j.aquaculture.2008.07.051 [DOI] [Google Scholar]
- Ng, W. K. , & Andin, V. C. (2011). The Malaysian mahseer, Tor tambroides (Bleeker), requires low dietary lipid levels with a preference for lipid sources with high omega‐6 and low omega‐3 polyunsaturated fatty acids. Aquaculture, 322–323, 82–90. 10.1016/j.aquaculture.2011.09.021 [DOI] [Google Scholar]
- Nguyen, T. M. (2015). Effects of dietary probiotics and temperatures stress on growth and immunity related genes expression in Malaysian Mahseer (Tor tambroides). Unpublished master's thesis, Universiti Malaysia Terengganu. Retrieved from http://umt-ir.umt.edu.my/xmlui/handle/123456789/5279
- Nguyen, N. P. , Warnow, T. , Pop, M. , & White, B. (2016). A perspective on 16S rRNA operational taxonomic unit clustering using sequence similarity. Nature Partner Journal Biofilms and Microbiomes, 2, 16004. Retrieved from https://www.nature.com/articles/npjbiofilms20164.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie, L. , Zhou, Q. J. , Qiao, Y. , & Chen, J. (2017). Interplay between the gut microbiota and immune responses of ayu (Plecoglossus altivelis) during Vibrio anguillarum infection. Fish and Shellfish Immunology, 68, 479–487. 10.1016/j.fsi.2017.07.054 [DOI] [PubMed] [Google Scholar]
- Norfatimah, M. Y. , Teh, L. K. , Salleh, M. Z. , Mat Isa, M. N. , & SitiAzizah, M. N. (2014). Complete mitochondrial genome of Malaysian Mahseer (Tor tambroides). Gene, 548(2), 263–269. 10.1016/j.gene.2014.07.044 [DOI] [PubMed] [Google Scholar]
- O'Hara, A. M. , & Shanahan, F. (2006). The gut flora as a forgotten organ. EMBO Reports, 7(7), 688–693. 10.1038/sj.embor.7400731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbi, S. , Martin, A. , Romano, S. , McMillan, W. O. , Stice, L. , & Grabowski, G. (2002). The simple fool's guide to PCR version 2.0. Department of Zoology and Kewalo Marine Laboratory, University of Hawaii, Honolulu. Retrieved from http://palumbi.stanford.edu/SimpleFoolsMaster.pdf
- Pan, X. , Wu, T. , Zhang, L. , Song, Z. , Tang, H. , & Zhao, Z. (2008). In vitro evaluation on adherence and antimicrobial properties of a candidate probiotic Clostridium butyricum CB2 for farmed fish. Journal of Applied Microbiology, 105(5), 1623–1629. 10.1111/j.1365-2672.2008.03885.x [DOI] [PubMed] [Google Scholar]
- Ramesh, D. , Vinothkanna, A. , Rai, A. K. , & Vignesh, V. S. (2015). Isolation of potential probiotic Bacillus spp. and assessment of their subcellular components to induce immune responses in Labeo rohita against Aeromonas hydrophila . Fish and Shellfish Immunology, 45(2), 268–276. 10.1016/j.fsi.2015.04.018 [DOI] [PubMed] [Google Scholar]
- Ramezani‐Fard, E. , Kamarudin, M. S. , Saad, C. R. , Harmin, S. A. , & Goh, Y. M. (2012). Dietary lipid levels affect growth and fatty acid profiles of Malaysian Mahseer Tor tambroides . North American Journal of Aquaculture, 74(4), 530–536. 10.1080/15222055.2012.690829 [DOI] [Google Scholar]
- Ramírez‐Pérez, O. , Cruz‐Ramón, V. , Chinchilla‐López, P. , & Méndez‐Sánchez, N. (2017). The role of the gut microbiota in bile acid metabolism. Annals of Hepatology, 16(1), s21–s26. [DOI] [PubMed] [Google Scholar]
- Ravcheev, D. A. , Godzik, A. , Osterman, A. L. , & Rodionov, D. A. (2013). Polysaccharides utilization in human gut bacterium Bacteroides thetaiotaomicron: Comparative genomics reconstruction of metabolic and regulatory networks. BMC Genomics, 14, 873 10.1186/1471-2164-14-873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringø, E. , Zhou, Z. , Vecino, J. L. , Wadsworth, S. , Romero, J. , Krogdahl, Å. , … Merrifield, D. L. (2016). Effect of dietary components on the gut microbiota of aquatic animals. A never‐ending story? Aquaculture Nutrition, 22, 219–282. 10.1111/anu.12346 [DOI] [Google Scholar]
- Roeselers, G. , Mittge, E. K. , Stephens, W. Z. , Parichy, D. M. , Cavanaugh, C. M. , Guillemin, K. , & Rawls, J. F. (2011). Evidence for a core gut microbiota in the zebrafish. ISME Journal, 5(10), 1595–1608. 10.1038/ismej.2011.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouf, A. J. M. A. , Phang, S. M. , & Ambak, M. A. (2010). Depth distribution and ecological preferences of periphytic algae in Kenyir Lake, the largest tropical reservoir of Malaysia. Chinese Journal of Oceanology and Limnology, 28(4), 856–867. 10.1007/s00343-010-9088-0 [DOI] [Google Scholar]
- Ryan, J. R. J. , & Esa, Y. B. (2006). Phylogenetic analysis of Hampala fishes (Subfamily Cyprininae) in Malaysia inferred from partial mitochondrial cytochrome b DNA sequences. Zoological Science, 23(10), 893–901. 10.2108/zsj.23.893 [DOI] [PubMed] [Google Scholar]
- Salinas, I. , & Magadán, S. (2017). Omics in fish mucosal immunity. Developmental and Comparative Immunology, 75, 99–108. 10.1016/j.dci.2017.02.010 [DOI] [PubMed] [Google Scholar]
- Sati, J. , Sah, S. , Pandey, H. , Ali, S. , Sahoo, P. K. , Pande, V. , & Barat, A. (2013). Phylogenetic relationship and molecular identification of five Indian Mahseer species using COI sequence. Journal of Environmental Biology, 34(5), 933–939. [PubMed] [Google Scholar]
- Schloss, P. D. , & Handelsman, J. (2005). Metagenomics for studying unculturable microorganisms: Cutting the Gordian knot. Genome Biology, 6, 229 10.1186/gb-2005-6-8-229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss, P. D. , & Westcott, S. L. (2011). Assessing and improving methods used in operational taxonomic unit‐based approaches for 16S rRNA gene sequence analysis. Applied and Environmental Microbiology, 77(10), 3219–3226. 10.1128/AEM.02810-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalm, N. D. , & Groisman, E. A. (2017). Navigating the gut buffet: Control of polysaccharide utilization in Bacteroides spp. Trends in Microbiology, 25(12), 1005–1015. 10.1016/j.tim.2017.06.009 [DOI] [PubMed] [Google Scholar]
- Shobharani, P. , Padmaja, R. J. , & Halami, P. M. (2015). Diversity in the antibacterial potential of probiotic cultures Bacillus licheniformis MCC2514 and Bacillus licheniformis MCC2512. Research in Microbiology, 166(6), 546–554. 10.1016/j.resmic.2015.06.003 [DOI] [PubMed] [Google Scholar]
- Song, W. , Li, L. , Huang, H. , Jiang, K. , Zhang, F. , Chen, X. , … Ma, L. (2016). The gut microbial community of antarctic fish detected by 16S rRNA gene sequence analysis. BioMed Research International, 2016, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellerberg, I. F. , & Fedor, P. J. (2003). A tribute to Claude Shannon (1916–2001) and a plea for more rigorous use of species richness, species diversity and the ‘Shannon–Wiener’ Index. Global Ecology and Biogeography, 12(3), 177–179. 10.1046/j.1466-822X.2003.00015.x [DOI] [Google Scholar]
- Sullam, K. E. , Essinger, S. D. , Lozupone, C. A. , O'Connor, M. P. , Rosen, G. L. , Knight, R. , … Russel, J. A. (2012). Environmental and ecological factors that shape the gut bacterial communities of fish: A meta‐analysis. Molecular Ecology, 21(13), 3363–3378. 10.1111/j.1365-294X.2012.05552.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, W. S. , Abdullah, J. O. , Sieo, C. C. , Shafee, N. , Mustafa, S. , Leow, T. C. A. , … Abdullah, N. (2008). A laboratory manual on molecular techniques for identification of bacteria. Serdang, Selangor: Universiti Putra Malaysia. [Google Scholar]
- Tarnecki, A. M. , Burgos, F. A. , Ray, C. L. , & Arias, C. R. (2017). Fish intestinal microbiome: Diversity and symbiosis unravelled by metagenomics. Journal of Applied Microbiology, 123, 2–17. 10.1111/jam.13415 [DOI] [PubMed] [Google Scholar]
- Tobe, S. S. , Kitchener, A. C. , & Linacre, A. (2011). Assigning confidence to sequence comparisons for species identification: A detailed comparison of the cytochrome b and cytochrome oxidase subunit I mitochondrial genes. Forensic Science International: Genetics Supplement Series, 3(1), e246–e247. [Google Scholar]
- Tsuchiya, C. , Sakata, T. , & Sugita, H. (2007). Novel ecological niche of Cetobacterium somerae, an anaerobic bacterium in the intestinal tracts of freshwater fish. Letters in Applied Microbiology, 46(1), 43–48. [DOI] [PubMed] [Google Scholar]
- Udayangani, R. M. C. , Dananjaya, S. H. S. , Nikapitiya, C. , Heo, G. J. , Lee, J. , & Zoysa, M. D. (2017). Metagenomics analysis of gut microbiota and immune modulation in zebrafish (Danio rerio) fed chitosan silver nanocomposites. Fish and Shellfish Immunology, 66, 173–184. 10.1016/j.fsi.2017.05.018 [DOI] [PubMed] [Google Scholar]
- Van Kessel, M. A. H. J. , Dutilh, B. E. , Neveling, K. , Kwint, M. P. , Veltman, J. A. , Flik, G. , … Camp, J. M. O. D. (2011). Pyrosequencing of 16S rRNA gene amplicons to study the microbiota in the gastrointestinal tract of carp (Cyprinus carpio L.). AMBI Express, 1, 41 10.1186/2191-0855-1-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, D. , Yang, L. , Yu, R. , Chen, F. , Lu, R. , Qin, C. , & Nie, G. (2017). Effects of dietary carbohydrate and lipid levels on growth and hepatic lipid deposition of juvenile tilapia, Oreochromis niloticus . Aquaculture, 479, 696–703. 10.1016/j.aquaculture.2017.07.013 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequences were also submitted to NCBI Sequence Read Archive (SRA) under accession number of https://www.ncbi.nlm.nih.gov/bioproject/PRJNA355218/.


