Abstract
Heat stress has become a major threat to crop production due to global warming; however, the mechanisms underlying plant high-temperature sensing are not well known. In plants, the membrane-anchored receptor-like kinases (RLKs) relay environmental signals into the cytoplasm. In a previous study, we isolated a wall-associated RLK-like (WAKL) gene CaWAKL20 from pepper (Capsicum annuum L.). Here, the amino acid sequence of CaWAKL20 was characterized and found to consist of conserved domains of WAK/WAKL family, including an extracellular region containing a GUB-WAK binding domain and a degenerated EGF2-like domain; a transmembrane region; and an intercellular region with an STKc catalytic domain. Moreover, CaWAKL20 transcription was inhibited by heat stress, whereas it was induced by both ABA and H2O2 treatments. Silencing of CaWAKL20 enhanced pepper thermotolerance, while overexpression decreased Arabidopsis thermotolerance. Additionally, Arabidopsis lines overexpressing CaWAKL20 showed less sensitivity to ABA during seed germination and root growth. Finally, the survival rate of Arabidopsis seedlings under heat stress treatment was enhanced by ABA pre-treatment, while it was compromised by the overexpression of CaWAKL20. Furthermore, the heat-induced expression of several ABA-responsive genes and some key regulator genes for thermotolerance was decreased in Arabidopsis CaWAKL20-overexpression lines. These results suggest that CaWAKL20 negatively modulates plant thermotolerance by reducing the expression of ABA-responsive genes, laying a foundation for further investigation into the functional mechanisms of WAKs/WAKLs in plants undergoing environmental stresses.
Keywords: pepper, heat stress, wall-associated receptor-like protein kinase, abscisic acid, H2O2
Introduction
Plants frequently face adverse environmental conditions, including extreme temperature, drought, salinity, and flood that can negatively affect plant growth, development, production, and even survival. As global warming worsens, heat waves are occurring with increased frequency and longer duration, especially in lower latitude regions (IPCC, 2014), and the resultant heat stress is becoming an increasingly significant problem for crop production and world food security (Ohama et al., 2017).
Heat stress can cause protein denaturation and aggregation, reduce cellular functions, and even result in cell death (Driedonks et al., 2015). To defend against these negative effects, as sessile organisms, plants have developed a conserved heat stress response (HSR) system to induce the expression of stress-related genes (Mittler et al., 2012). However, relatively little is known about how plants sense high temperature, which then hinders the establishment of systems that protect against heat stress in crop breeding and/or cultivation.
Under heat stress, the HSR can be triggered by increased fluidity of the plasma membrane (Sangwan et al., 2002) or accumulation of unfolded proteins (Rütgers et al., 2017). The plasma membrane is suggested to be a primary heat sensor in plants (Hofmann, 2009), and changes in cytomembrane fluidity can be sensed by integral membrane proteins, such as ion channels and/or transporters, as well as membrane-anchored receptor-like kinases (RLKs) (Zhu, 2016). While the involvement of membrane calcium channels in plant HSR has been confirmed (Saidi et al., 2009; Finka et al., 2012), the contribution of RLKs to plant thermotolerance is poorly understood.
Plant RLKs are classed as transmembrane proteins containing an N-terminal extracellular domain and a C-terminal intracellular kinase domain. Receptor-like kinases are thought to participate in a diverse range of life processes including growth, development, hormone signalings, plant-microbe interactions, and responses to environmental stimuli (Shiu and Bleecker, 2001; Ye et al., 2017).
As an RLK subfamily, wall-associated receptor kinase (WAK) and WAK-like (WAKL) are characterized by the presence of an extracellular epidermal growth factor (EGF)-like domain. These WAKs/WAKLs physically link the cell wall to the plasma membrane, directly transmitting extracellular signals into the cytoplasm to regulate cell growth and stress responses (Anderson et al., 2001; Kohorn, 2016). Zhang et al. (2017) even suggested that “ZmWAK functions as a hub in the trade-off between maize growth and defense.”
Since firstly reported in Arabidopsis (He et al., 1998), the contributions of WAKs/WAKLs to plant immunity have been widely studied, including AtWAKL22 in Arabidopsis resistance to Fusarium oxysporum (Diener and Ausubel, 2005), SlWAK1 in tomato resistance to Pst (Rosli et al., 2013), OsWAKs in rice resistance to fungal blast and bacterial blight disease (Li et al., 2009; Delteil et al., 2016; Harkenrider et al., 2016), ZmWAKs in maize resistance to northern corn leaf blight and head-smut disease (Hurni et al., 2015; Zhang et al., 2017; Yang et al., 2018), and TaWAKL4 in wheat resistance to Zymoseptoria tritici (Saintenac et al., 2018).
An increasing number of studies have reported on the involvement of WAKs/WAKLs in plant tolerance to abiotic stresses. For example, Sivaguru et al. (2003) reported that overexpression of AtWAK1 enhanced Arabidopsis tolerance to heavy metal stress, whereas Hou et al. (2005) found that T-DNA insertion in the AtWAKL4 promoter increased Arabidopsis hypersensitivity to salt stress. Furthermore, Xia et al. (2018) suggested that, when subjected to excess Cu, OsWAK11 can modify cell wall properties to retain Cu at the cell wall. However, it remains to be elucidated whether WAK/WAKL genes are involved in plant thermotolerance.
In our previous work, we isolated a heat-responsive gene Capana12g000852 from the pepper plant. Due to its high amino acid sequence homology with Arabidopsis AtWAKL20, Capana12g000852 was renamed CaWAKL20. In this study, we show that silencing the CaWAKL20 gene enhanced pepper tolerance to heat stress, whereas overexpressing CaWAKL20 weakened Arabidopsis thermotolerance. In addition, Arabidopsis lines overexpressing CaWAKL20 exhibited reduced sensitivity to abscisic acid (ABA) during seed germination and seedling growth. We also show that, under heat stress treatment, the induced expression of some ABA-responsive genes was reduced in CaWAKL20-overexpressing Arabidopsis lines. Our study may contribute to the understanding of the molecular mechanisms of plant tolerance against heat stress.
Materials and Methods
Plant Materials and Growth Conditions
The pepper R9 thermotolerant line (introduced from the World-Asia Vegetable Research and Development Center, PP0042-51) and Arabidopsis ecotype Col-0 were used in this study. Plant materials were grown in a chamber under 200 μmol⋅m–2⋅s–1 illumination intensity, a 16 h day/8 h night regime, and 70% relative humidity. The temperature was set to 26°C/20°C (day/night) for pepper and 22°C/18°C for Arabidopsis.
Analysis of the Deduced CaWAKL20 Amino Acid Sequence
The CaWAKL20 (Capana12g000852) amino acid sequence was downloaded from the PGD database1, and those of Arabidopsis WAKs/WAKLs were obtained from TAIR (The Arabidopsis Information Resource)2 (Verica and He, 2002). The conserved domains in the CaWAKL20 amino acid sequence were identified using the online SMART tool (Simple Modular Architecture Research Tool)3. Full-length CaWAKL20 and AtWAKs/WAKLs were aligned using the online Clustal Omega program4, and the phylogenetic tree was generated by MEGA 6 software using the neighbor-joining method, based on the p-distance substitution model, and with 1000 bootstrap replicates (Tamura et al., 2013).
Arabidopsis AtWAKL20 was used to predict the CaWAKL20 protein-protein interaction network in the STRING interaction database (Search Tool for the Retrieval of Interacting Genes/Proteins)5 with a 0.400 confidence score. Finally, the output was imported into Cytoscape_v3.4.0 (National Institute of General Medical Sciences, MD, United States) to generate the network map.
Subcellular Localization of CaWAKL20
The CaWAKL20 coding sequence (CDS) without a stop codon was amplified from the pepper line R9 using the GFP-CaWAKL20-F and GFP-CaWAKL20-R primer pair (Supplementary Table S1); the PCR product was then cloned into a GFP-tagged pBI221 transient expression vector. The empty vector was used as the control. Particle bombardment was used to transfect plasmid into onion epidermal cells, which were then incubated for 24 h at 28°C in the dark. The GFP signal was visualized using A1R confocal laser scanning microscopy (Nikon, Tokyo, Japan). For plasmolysis analysis, the transformed onion epidermal cells were treated in 0.8 M mannitol solution for 15 min before observation of GFP expression (Genovesi et al., 2008).
Virus-Induced Gene Silencing (VIGS) of CaWAKL20
To generate gene-silenced plants using VIGS, a conserved 346 bp fragment in the CaWAKL20 CDS was amplified from the pepper line R9 with the TRV2-CaWAKL20-F and TRV2-CaWAKL20-R primer pair (Supplementary Table S1), and the PCR product was inserted into a pMD19-T vector (Takara, Dalian, China). After being digested with EcoR I and BamH I, the resultant CaWAKL20 gene fragment was cloned into the pTRV2 virus expression vector to generate the TRV2:CaWAKL20 gene-silencing vector. The empty vector was used as the control and was referred to as TRV2:00, and the TRV2:CaPDS (phytoene desaturase gene) vector was used as the marker for successful gene silencing. The Agrobacterium tumefaciens strain GV3101 containing TRV2:CaWAKL20, TRV2:00, or TRV2:CaPDS was injected into the cotyledons of the pepper line for gene silencing. When the photo-bleaching phenotype was clearly observable in newly grown leaves of plants transformed with TRV2:CaPDS, the transcription of TRV1 and TRV2 in plants expressing TRV2:00 and TRV2:CaWAKL20 was assessed by semi-quantitative PCR using the TRV1-TL-F/TRV1-TL-R and TRV2-Coat P-F/TRV2-Coat P-R primer pairs, respectively (Supplementary Table S1, Tsaballa et al., 2011). The efficiency of silencing of CaWAKL20 expression in plants expressing TRV2:CaWAKL20 was assessed by qRT-PCR using the qCaWAKL20-F and qCaWAKL20-R primer pair (Supplementary Table S1).
Generation of Arabidopsis Lines Overexpressing CaWAKL20
The complete CaWAKL20 CDS was amplified from the R9 line using the CaWAKL20-F and CaWAKL20-R primer pair (Supplementary Table S1), and the amplification product was cloned into the pVBG2307 plant binary expression vector under the control of the CaMV35S promoter. Using A. tumefaciens strain GV3101, the 35S::CaWAKL20 vector was transformed into Arabidopsis ecotype Col-0 by the floral dip method (Clough and Bent, 1998). Transgenic plants were screened by adding kanamycin into the MS culture medium, and the T3 generation was then used for subsequent experiments.
Experimental Treatments and Collection of Samples
For tissue-specific expression analysis of CaWAKL20, samples of seeds, roots, stems, young leaves, flower buds, and young fruits were collected from pepper plants grown under normal conditions.
For analysis of CaWAKL20 expression under stress treatments, pepper seedlings at the six-leaf stage were either incubated at 45°C for heat stress treatment, sprayed evenly with a 0.1 mM ABA solution for ABA treatment, or immersed with roots in a 100 mM H2O2 solution for H2O2 treatment. The young leaves were collected at 0, 1, 3, 6, 12, and 24 h post-treatments, and all the samples were immediately frozen in liquid nitrogen and stored at −80°C for RNA extraction.
To assess the efficiency of silencing of CaWAKL20 expression, pepper seedlings containing TRV2:CaWAKL20 and TRV2:00 were incubated at 45°C for 1 h. To measure the thermotolerance of CaWAKL20-silenced plants, pepper seedlings containing TRV2:CaWAKL20 and TRV2:00 were incubated at 45°C for 10 h and then allowed to recover for 24 h. The pepper leaves were sampled at the end of both treatments and either stored at −80 °C for gene expression analysis or used immediately to determine malondialdehyde (MDA) content and the maximal photochemical efficiency of PSII (Fv/Fm).
For thermotolerance evaluation of plants overexpressing CaWAKL20, transgenic Arabidopsis lines overexpressing CaWAKL20 (CaWAKL20-OE) or containing the empty vector (EV) were used. Ten days old Arabidopsis seedlings on MS plates were immersed in a water bath at 45°C for 50 min and then allowed to recover for 2 days at 22°C. Three weeks old Arabidopsis seedlings in pots were incubated at 45°C for 12 h and allowed to recover for 7 days at 22°C. The survival rates of Arabidopsis seedlings were calculated at the end of both treatments, and leaves from seedlings in pot were sampled at 3 h post-heat stress treatment for gene expression analysis. Three weeks old Arabidopsis seedlings in pots were treated at 45°C for 12 h and then collected for the assessment of H2O2 accumulation.
To assess the effects of CaWAKL20 overexpression on Arabidopsis sensitivity to ABA, seeds of CaWAKL20-OE and EV lines were germinated on MS plates containing 0, 0.75, and 1 μM ABA. The germination rate, green cotyledon rate, and root length were determined at 4, 9, and 11 days after treatment, respectively. All experiments were performed with three biological replicates.
To evaluate the effects of exogenous ABA on the thermotolerance of plants overexpressing CaWAKL20, 10 days old seedlings of Arabidopsis lines of EV, OE2, and OE14 were transferred to MS medium with or without 5 μM ABA (Wang et al., 2017). After pre-treatment with ABA for 2 days, the plates were immersed in a water bath at 45°C for 50 min and then allowed to recover for 2 or 3 days at 22°C. The rate of green seedlings was calculated at different time points after treatments.
Determination of MDA Content, PSII Fv/Fm and H2O2 Accumulation
The MDA content of the pepper leaves was measured using the thiobarbituric acid assay (Dhindsa et al., 1981). The PSII Fv/Fm of pepper leaves was determined using the FluorCam7 fluorescence imaging system (EcoTech, China). The assessment of H2O2 accumulation in Arabidopsis seedlings was performed using the diaminobenzidine (DAB) staining method (Dang et al., 2013). All experiments were performed with three biological replicates.
Total RNA Extraction, cDNA Synthesis, and qRT-PCR Analysis
Total RNA was extracted from the leaves of pepper and Arabidopsis plants using the Trizol® kit (Invitrogen, Carlsbad, CA, United States), and the residual genomic DNA was digested with RNase-free DNase I (Promega, Madison, WI, United States). First-strand cDNA synthesis was performed using the PrimeScriptTM Kit (TaKaRa, Tokyo, Japan) according to the manufacturer’s instructions. Primer pairs were designed using Primer-BLAST in NCBI6 (Supplementary Table S1), and qRT-PCR was performed using SYBR® Premix Ex TaqTM II (TaKaRa). Relative gene expression levels were analyzed according to the 2–ΔΔCT method (Livak and Schmittgen, 2001), in which CaUBI3 and AtActin2 were used as internal controls for pepper and Arabidopsis, respectively. Significance tests for differences in gene expression between control and stress treatments were performed using the Student’s t-test method at the 0.05 and 0.01 significance levels.
Results
Analysis of the Deduced CaWAKL20 Amino Acid Sequence
Domains that are conserved in the WAK/WAKL protein family were identified in the amino acid sequence of Capana12g000852 (Supplementary Figures S1A,B) using the SMART online tool. The conserved domains included an extracellular region containing a signal peptide and a GUB-WAK binding domain (galacturonan-binding, pfam13947) at the N-terminus, a transmembrane region, and an intercellular region with a catalytic STKc domain (Serine/Threonine kinase, cd14066) at the C-terminus. The EGF domain, a marker for the WAK subfamily that distinguishes it from others in RLKs (Anderson et al., 2001), was absent in Capana12g000852 (Supplementary Figure S1A); however, a degenerated EGF2-like domain (Prosite: PS01187) (Verica et al., 2003) was present (Supplementary Figure S1B), which is also observed for AtWAKL20 (Verica and He, 2002). In addition, Capana12g000852 displayed a closer phylogenetic relationship with AtWAKL20 than with the other 25 WAK/WAKL members found in Arabidopsis (Verica and He, 2002; Supplementary Figure S1C), and was therefore renamed CaWAKL20.
To further understand its functional patterns, the CaWAKL20 protein-protein interaction network was predicted using the STRING online tool based on Arabidopsis interologs. Ten CaWAKL20 interaction partners were identified, including a WAK family member (WAKL7), a zinc ion binding protein, and eight protein phosphatase 2C (PP2C) family members, i.e., ABI2 (ABA Insensitive 2), AHG1 (ABA-Hypersensitive Germination 1), APD9 (Arabidopsis PP2C clade D9), EGR2 (E Growth-Regulating 2), HAI1 (Highly ABA-Induced PP2C gene 1), and PP2C74, 76, and 80 (Supplementary Figure S1D).
Subcellular Localization of CaWAKL20
To confirm the WAK/WAKL classification of CaWAKL20, the GFP-tagged CaWAKL20 CDS was transiently expressed in onion epidermal cells under the control of the CaMV35S promoter. In the cells transformed with the empty control vector, the GFP signal was distributed throughout the entire cell. In contrast, green fluorescence was detected along the edge of the cells transformed with the CaWAKL20-GFP fusion vector (Figure 1A).
FIGURE 1.
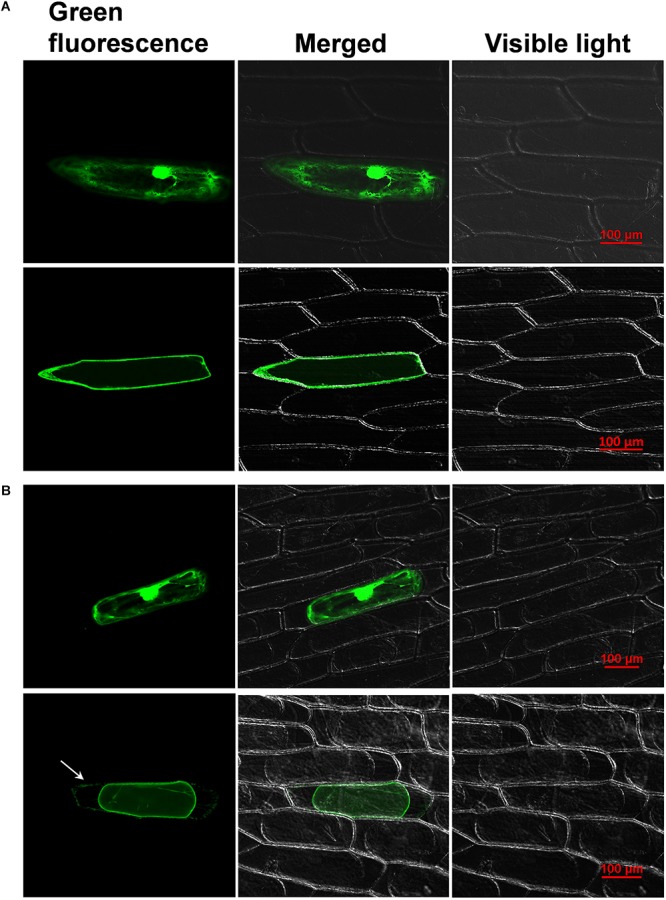
Fluorescence microscopy images of (A) normal and (B) plasmolyzed onion epidermal cells transiently expressing either free green fluorescent protein (GFP) (top) or 35S: CaWAKL20-GFP fusion proteins (bottom). The arrow shows the GFP signal at the cell wall. Scale bar = 100 μm.
To further test the membrane- and cell wall-binding properties of CaWAKL20, the onion epidermal cells transformed with CaWAKL20 were cultured in mannitol for plasmolysis. In plasmolyzed cells, the green fluorescence of the CaWAKL20-GFP protein was observed both in the plasma membrane and the cell wall (Figure 1B). This suggests that CaWAKL20 was localized at the plasma membrane and linked to the cell wall.
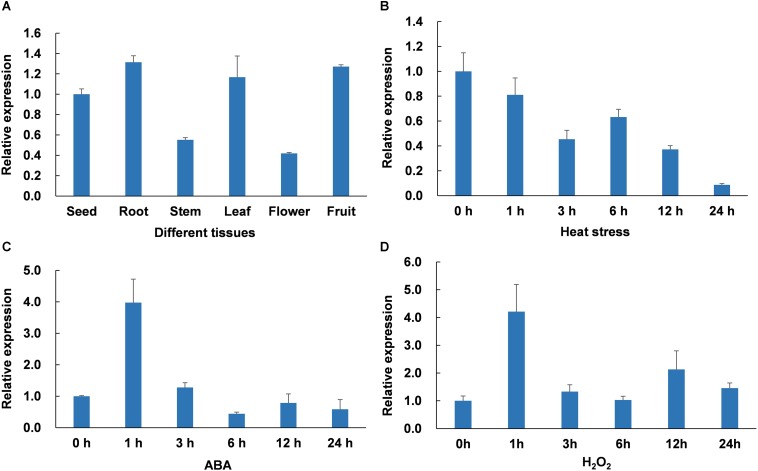
Expression of CaWAKL20 in Different Pepper Plant Tissues and Under Heat Stress, ABA, and H2O2 Treatments
To explore the expression patterns of CaWAKL20 in different pepper plant tissues, the roots, stems, leaves, flowers, and fruits of plants grown under normal conditions were sampled from the R9 thermotolerant line, and qRT-PCR was performed using a CaWAKL20-specific primer pair. The results indicated that CaWAKL20 was variably expressed in different tissues, and the expression levels were clearly lower in the stems and flowers than in the other tissues (Figure 2A).
FIGURE 2.
CaWAKL20 expression patterns (A) in different pepper tissues, under (B) heat stress induction at 45°C, (C) 0.1 mM ABA spraying, and (D) 100 mM H2O2 root-immersing. The expression of CaWAKL20 was normalized to the ubiquitin-conjugating protein-coding gene CaUBI3. The experiment was conducted with three biological replicates, and each replicate contained five pepper seedlings; error bars represent standard deviations for three biological replicates.
The CaWAKL20 protein was predicted to interact with several proteins related to ABA (Supplementary Figure S1D), a major phytohormone essential for plant responses to a wide range of stresses (Vishwakarma et al., 2017). In addition, reactive oxygen species (ROS) also play a key role in plants’ acclimation to abiotic stresses, including heat (Suzuki and Katano, 2018). Therefore, to elucidate the response of CaWAKL20 to heat stress, ABA and ROS treatments, CaWAKL20 expression was analyzed following incubation at 45°C and exposure to 0.1 mM ABA and 100 mM H2O2, respectively. The results showed that CaWAKL20 transcription responded to all treatments. For heat stress, CaWAKL20 expression decreased continuously during the whole treatment (Figure 2B), while with both ABA and H2O2 treatments, CaWAKL20 expression increased at 1 h, and then declined rapidly (Figures 2C,D).
Thermotolerance Was Enhanced in CaWAKL20-Silenced Pepper Seedlings
To understand the role of CaWAKL20 in pepper thermotolerance, CaWAKL20 was silenced in the thermotolerant R9 pepper line. The fragment of CaWAKL20 used for VIGS was predicted to have no off-target potential (by the VIGS tool in SGN7) and was therefore presumed to be specific for the CaWAKL20 gene. Approximately 1 month after injection for VIGS, the bleaching was clearly observed in the leaves of positive control pepper seedlings containing TRV2:CaPDS; however, no visible phenotype was observed in leaves with either TRV2:00 or TRV2:CaWAKL20 (Supplementary Figure S2A). Both the RNA1 segment in TRV1 (GenBank: AF406990) and the coat protein in TRV2 (GenBank: AF406991) were almost equally expressed in all virus-infected pepper plants (Supplementary Figure S2B). Compared to plants with TRV2:00, the expression of the CaWAKL20 gene was silenced to approximately 70% in the plants with TRV2:CaWAKL20, which became more evident after the heat stress of 45°C for 1 h (Supplementary Figure S2C).
After exposure to heat stress at 45°C for 10 h and recovery for 24 h under normal conditions, the leaves of the pepper plants containing TRV2:00 were severely parched, while those with TRV2:CaWAKL20 resumed growing (Figure 3A). Under heat stress treatment, the MDA content increased from 5.88 to 11.44 μmol*g–1 fresh weight (FW) in leaves of plants with TRV2:CaWAKL20, whereas for plants with TRV2:00 the MDA content in the leaves increased from 5.98 to 9.15 μmol*g–1FW. After heat treatment, the MDA content was significantly higher in the leaves of plants with TRV2:00 than those with TRV2:CaWAKL20 (p < 0.05) (Figure 3B). Furthermore, with prolonged heat stress, the PSII Fv/Fm declined continuously in the leaves of plants transformed with both TRV2:CaWAKL20 and TRV2:00; at the end of treatment, the Fv/Fm value was clearly higher in the former (0.60) than that in the latter (0.51) (p < 0.01) (Figures 3C,D).
FIGURE 3.
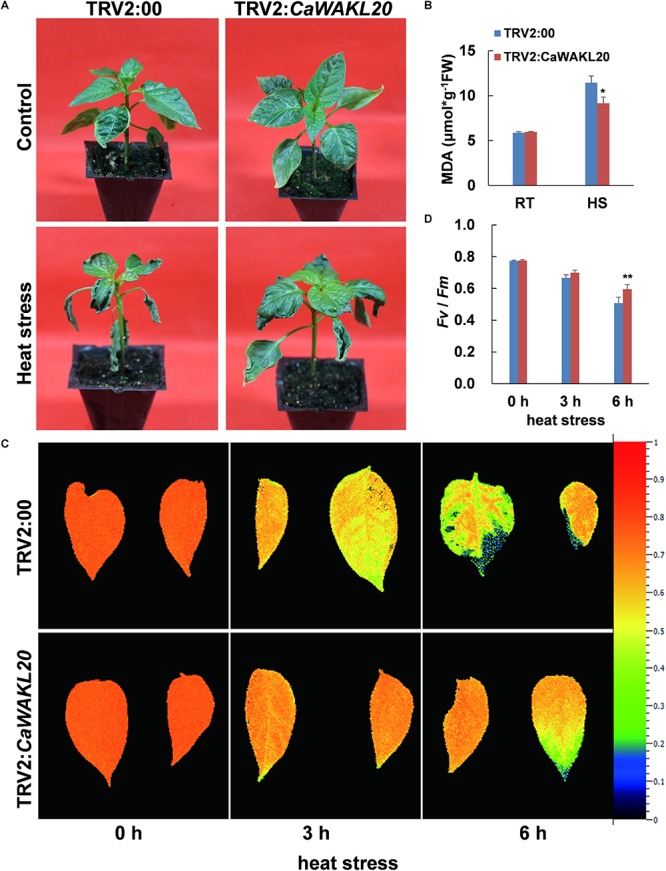
Thermotolerance of CaWAKL20-silenced pepper seedlings. (A) Performance of intact plants, (B) MDA contents in leaves, and (C,D) the maximal photochemical efficiency of PSII (Fv/Fm) after heat stress. RT, room temperature; HS, heat stress incubation at 45°C for 10 h, and then recovery for 24 h. Error bars represent standard deviations for three biological replicates; * and ** indicate significant differences at the 0.05 and 0.01 levels, respectively.
Thermotolerance Was Reduced in CaWAKL20-Overexpressing Arabidopsis Lines
To further understand the role of CaWAKL20 in plant thermotolerance, Arabidopsis lines overexpressing CaWAKL20 (OE) were generated, with OE2 and OE14 being selected and used for further studies (Supplementary Figures S2D,E).
For 10 days old Arabidopsis seedlings on MS plates, after induction of heat stress at 45°C for 50 min followed by recovery for 2 days, the survival rate was higher than 80% in the EV line compared to only approximately 12% for seedlings in either CaWAKL20-OE line (Figures 4A,B). For 3 weeks old Arabidopsis in pots, after induction of heat stress at 45°C for 12 h and recovery for 7 days, the survival rate was higher than 80% in the EV line compared to only 31% in the OE2 line and 34% in the OE14 line (Figures 4C,D). Meanwhile, the seedlings from the CaWAKL20-OE lines that survived had fewer green leaves than those from the EV line. After heat stress treatment at 45°C for 12 h, the level of DAB staining increased in all Arabidopsis seedlings. Compared with that of the EV line, the staining intensity in the seedlings of both OE2 and OE14 were obviously raised (Figure 4E), suggesting the higher levels of H2O2 accumulation in CaWAKL20-OE lines.
FIGURE 4.

Thermotolerance of CaWAKL20-overexpressing Arabidopsis lines. (A,C) Phenotypes, (B,D) survival rates, and (E) H2O2 accumulation of Arabidopsis seedlings grown (A,B) on plates or (C–E) in pots under heat stress (45°C in a water bath) (A,B) for 50 min, and then recovery for 2 days or for (C–E) 12 h and then (C,D) recovery for 7 days. OE2 and OE14, transgenic Arabidopsis lines overexpressing CaWAKL20; EV, control trangenic Arabidopsis line with the empty vector. Error bars represent standard deviations for three replicates, and each replicate contained five plates (A,B) or 12 Arabidopsis seedlings (C–E); ** indicates a significant difference at the 0.01 level.
ABA Sensitivity Decreased in Arabidopsis CaWAKL20-OE Lines
No difference was observed in the seed germination rate between CaWAKL20-OE and EV Arabidopsis lines under normal growth conditions. Under ABA treatments, the seed germination rate was reduced, but more so in the EV line than in the CaWAKL20-OE lines (Figure 5A). When the treatments were extended to 9 days, the growth of CaWAKL20-OE seedlings was clearly better than those of the EV line (Figure 5B). Under 0.75 and 1 μM ABA treatments, the percentage of green cotyledons decreased to 47 and 35%, respectively, in the EV line, but to 77 and 65%, respectively, in the CaWAKL20-OE lines (Figure 5C). Furthermore, root growth was suppressed by ABA treatments in both the CaWAKL20-OE and EV lines; however, root length was clearly greater in the former than in the latter (Figure 5D). For example, under 1 μM ABA treatment, the root length of the CaWAKL20-OE seedlings was approximately twofold greater than that of EV seedlings (Figure 5E).
FIGURE 5.
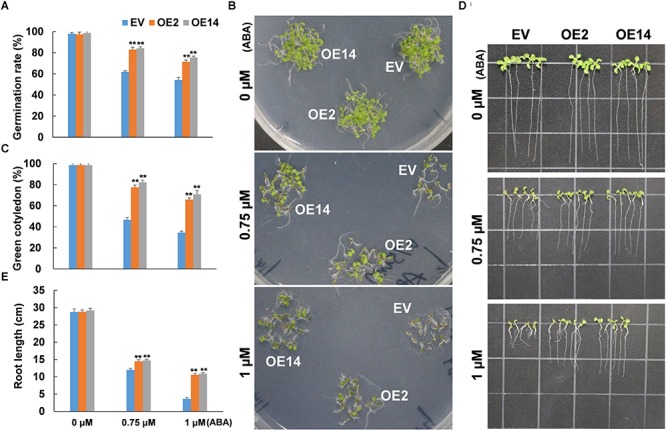
The sensitivity to ABA in terms of seed germination and seedling growth in CaWAKL20-overexpressing Arabidopsis lines. (A) Seed germination rate, (B,C) green cotyledon rate, and (D,E) root length of Arabidopsis at the (A) 4, (B,C) 9, and (D,E) 11 days after treatments with 0, 0.75, and 1 μM ABA, respectively. OE2 and OE14, Arabidopsis transgenic lines overexpressing CaWAKL20; EV, control Arabidopsis transgenic line containing the empty vector. Error bars represent standard deviations for three replicates, and each replicate contained (A–C) 5 plates or (D,E) 12 Arabidopsis seedlings. ** indicates a significant difference at the 0.01 level.
ABA-Enhanced Thermotolerance Was Compromised by Overexpression of CaWAKL20 in Arabidopsis
To clarify the relationship between ABA treatment and CaWAKL20 expression in the development of plant thermotolerance, the seedlings of Arabidopsis CaWAKL20-OE were pretreated with ABA for 2 days and then exposed to heat stress. The results showed that the heat stress decreased the survival rate of Arabidopsis seedlings of all three lines in both with and without ABA pre-treatment, and the survival rates in OE2 and OE14 were significantly lower than those in EV line in both the tested time points (Figure 6). Compared to those without ABA pre-treatment (ABA-), however, the survival rate of Arabidopsis seedlings under heat stress was enhanced by ABA pre-treatment (ABA+) in all three Arabidopsis lines of EV, OE2, and OE14 (Figure 6A). Furthermore, under the treatment of ABA-, in OE2 and OE14, the survival rate was lower than in EV line by about 35 and 31% after 2 days of heat stress, and about 34 and 35% after 3 days of heat stress, respectively. Under the ABA+ treatment, the survival rate was lower in OE2 and OE14 than in EV line by about 15 and 13% after 2 days of heat stress, and about 44 and 42% after 3 days of heat stress, respectively (Figure 6B).
FIGURE 6.
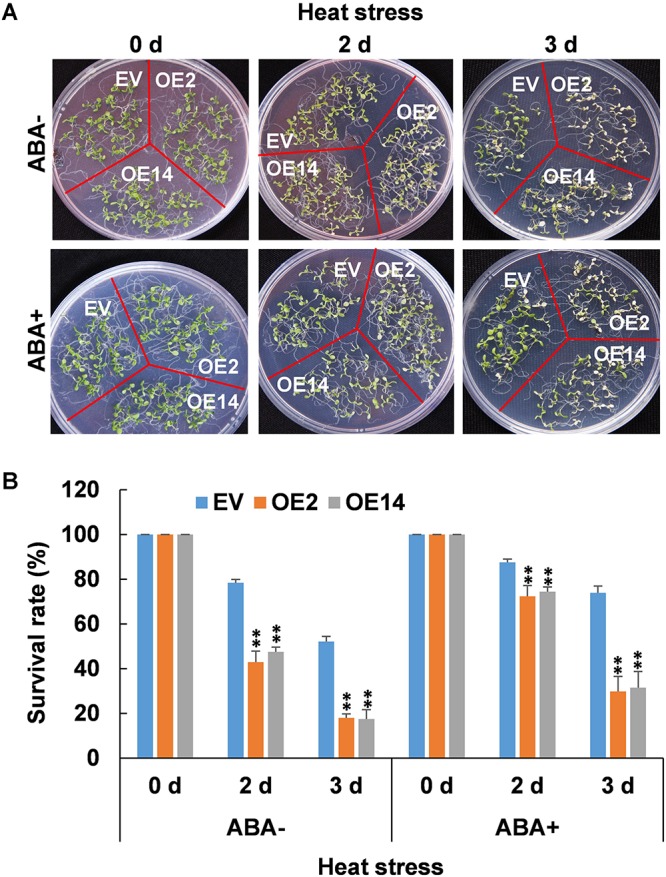
Thermotolerance of CaWAKL20-overexpressing Arabidopsis lines under ABA pretreatment. (A) Phenotypes and (B) survival rates of Arabidopsis seedlings grown on plates under heat stress (45°C in a water bath) for 50 min, and then recovery for 2 or 3 days. OE2 and OE14, transgenic Arabidopsis lines overexpressing CaWAKL20; EV, control transgenic Arabidopsis line with the empty vector. Error bars represent standard deviations for three replicates, and each replicate contained five plates; ** indicates a significant difference at the 0.01 level.
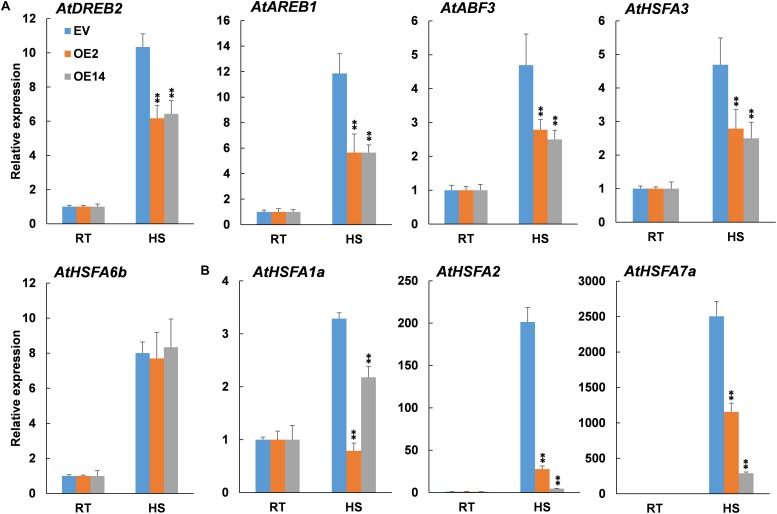
Heat Stress-Induced Expression of ABA-Responsive Genes Was Reduced in Arabidopsis CaWAKL20-OE Lines
To confirm the involvement of ABA in CaWAKL20-mediated thermotolerance, we further examined the expression of several heat-related and simultaneously ABA-responsive genes in Arabidopsis EV line and CaWAKL20-OE lines under heat stress; the genes included AREB (Fujita et al., 2005), ABF (Choi et al., 2013), HSFA6b (Huang et al., 2016), DREB, and HSFA3 (Schramm et al., 2008). All the genes were induced by heat stress in both the EV line and CaWAKL20-OE lines; however, except for AtHSFA6b, the expression levels were clearly lower in the CaWAKL20-OE lines than in the EV line (Figure 7A).
FIGURE 7.
Expression of (A) ABA-responsive genes and (B) key regulator genes for thermotolerance in CaWAKL20-overexpressing Arabidopsis lines after heat stress treatment. RT, room temperature; HS, heat stress induction at 45°C for 3 h. AtActin2 was used to normalize gene expression, and the expression level under 0 h after treatment was taken as 1.0. Error bars represent standard deviations for three replicates, and each replicateconsisted of 12 seedlings. ** indicates significant difference at the 0.01 level.
In addition, the expression of several key regulator genes for plant thermotolerance, HSFA1a, HSFA2, and HSFA7a (Yoshida et al., 2011), was also assessed under heat stress. The results showed that in line with those of ABA-responsive genes, the transcription of all tested thermotolerance-regulator genes was enhanced by heat stress, whereas this enhancement was inhibited by the overexpression of CaWAKL20 in both OE2 and OE4 (Figure 7B).
Discussion
In a previous study, we isolated a heat-responsive gene Capana12g000852 from the pepper plant (data not shown). Further conserved domain analysis identified all the necessary WAK/WAKL domains in the deduced Capana12g000852 amino acid sequence (Supplementary Figures S1A,B). In addition, Capana12g000852 showed a similar domain distribution and closer phylogenetic relationship to Arabidopsis AtWAKL20 (Supplementary Figures S1A,C). From these data, we suggest that Capana12g000852 is a pepper homolog of AtWAKL20 and rename it CaWAKL20. The WAK/WAKL properties of CaWAKL20 are also supported by its subcellular localization in onion epidermal cells (Figure 1).
Although WAKs/WAKLs participate in plant responses to various biotic and abiotic stresses as a plasma membrane localized receptor-like kinase (Anderson et al., 2001), their roles in thermotolerance are unclear. Here, we showed that CaWAKL20 expression was downregulated in a manner that was dependent on the duration of the heat stress treatment (Figure 2B). When CaWAKL20 was silenced, pepper plant thermotolerance was enhanced as evidenced by the smaller increase in MDA content and the lower decline in the Fv/Fm value, compared to the plants transformed with the empty TRV2:00 vector (Figure 3). In contrast, the Arabidopsis CaWAKL20-OE lines displayed reduced thermotolerance in terms of seedling survival rate and ROS accumulation (Figure 4). These data suggest that CaWAKL20 negatively modulates plant thermotolerance.
The induced expression of WAK/WAKL genes is widely believed to be a requirement for plant survival during pathogen infection or heavy metal stress (He et al., 1998; Sivaguru et al., 2003); however, some contradictory results have also been reported. Harkenrider et al. (2016) found that overexpression of OsWAK25 increased rice susceptibility to necrotrophic fungal pathogens, although it enhanced rice resistance to hemibiotrophic pathogens. OsWAK14, OsWAK91, and OsWAK92 positively regulate rice resistance to blast fungus, while OsWAK112d functions as a negative regulator (Delteil et al., 2016). When the AtWAKL4 promoter was impaired, Arabidopsis tolerance to K+, Na+, Cu2+, and Zn2+ was reduced, but its tolerance to Ni2+ was enhanced (Hou et al., 2005). Therefore, WAKs/WAKLs may have differential roles in plant tolerance against biotic and/or abiotic stresses, and/or the roles may be stress type-dependent. As far as CaWAKL20, its functional model in the pepper plant response to heat stress remains to be further elucidated.
When contending with adverse environments, plants activate their protective mechanisms that, conversely, often suppress plant growth to focus energy on defending against the stress. Therefore, the ability to switch from growth to defense is crucial for plants’ survival under stressed conditions (Wang and Wang, 2014; Albrecht and Argueso, 2017). Several hubs for tuning plant stress signaling and development have been reported in plants, such as CDPKs (Calcium-Dependent Protein Kinases) (Schulz et al., 2013) and WAKs/WAKLs (Kohorn, 2016). Zhang et al. (2017) found that maize ZmWAK promotes cell growth in the absence of pathogens but switches to a protective role when the maize plant is attacked by Sporisorium reilianum. In our study, after heat stress treatment, CaWAKL20 expression in the pepper thermotolerant line declined continuously (Figure 2B); similarly, Giarola et al. (2016) also observed that CpWAK1 transcription in Craterostigma plantagineum, a plant species with tolerance to extreme desiccation, was downregulated during dehydration treatment. These results suggest that maintaining a lower level of CaWAKL20 expression to slow cell growth is beneficial for pepper tolerance to injury from heat stress. This is also supported by our transgenic data (Figures 3, 4) as well as the predicted interaction between CaWAKL20 and the growth regulator EGR2 (Supplementary Figure S1D), although the functional mechanisms involved require further detailed study.
Abscisic acid plays important roles in plant responses to a range of environmental stresses, including heat stress. ABA is thought to perform a number of cellular functions, such as controlling the production of protective enzymes and regulating the transfer of water, to keep plant cells alive from heat stress (reviewed by Vishwakarma et al., 2017). Heat acclimation triggers an increase in endogenous ABA content (Liu et al., 2006); conversely, exogenous ABA enhances thermotolerance by upregulating the expression of heat-shock proteins (HSPs), including transcription factors (Wang et al., 2017). In this study, CaWAKL20 was predicted to interact with three ABA signaling components – ABI2, HAI1, and AHG1 (Supplementary Figure S1D) – and CaWAKL20 expression was rapidly induced by ABA treatment (Figure 2C). Meanwhile, Arabidopsis CaWAKL20-OE lines showed a lower sensitivity to ABA than the EV line for seed germination, seedling survival, and root growth (Figure 5), and the enhanced thermotolerance of Arabidopsis seedlings by ABA pre-treatment was compromised in CaWAKL20-OE lines (Figure 6). These results suggest that CaWAKL20 functions in an ABA-related pathway. In addition, the heat-induced expression of several ABA-responsive genes, AREB, ABF, DREB, and HSFA3, and some key regulator genes for plant thermotolerance, HSFA1a, HSFA2, and HSFA7a, was reduced in Arabidopsis CaWAKL20-OE lines compared to the EV line (Figure 7). Interestingly, similar phenomena were observed in the plastid casein kinase 2 (CK2) knockout mutant; CK2 is a major serine/threonine-specific kinase in the chloroplast stroma, and the authors argued that CK2 positively regulated retrograde signaling from the plastid to the nucleus during plant responses to ABA and heat stress (Wang et al., 2014). These results indicate that CaWAKL20 negatively modulates pepper plant thermotolerance by repressing the expression of ABA-responsive genes.
As universal signals, ROS might integrate with hormone signaling, including ABA signaling, to tailor the cellular homeostasis under stress conditions (Suzuki and Katano, 2018). In our study, the expression was also induced by exogenous H2O2 with a pattern similar to that of ABA (Figure 2D). Suzuki et al. (2013) reported that ROS was responsible for the spread of heat signal from the initial site to the entire plant, and ABA specifically regulated plant acclimation to heat stress. Thus, it can be hypothesized that CaWAKL20 functions to link the ROS signal and ABA pathway. In this model, both ABA and H2O2 induce
the expression of CaWAKL20, and CaWAKL20 expression further negatively regulates ABA signaling by decreasing the ABA sensitivity of plant growth and inhibiting the induced-expression of ABA-responsive genes and key regulator genes for thermotolerance under heat stress. However, which node in the ABA signal pathway, such as perception, biosynthesis, degradation, or signaling, is regulated by CaWAKL20 needs more investigation to verify.
Conclusion
In this study, we report that pepper CaWAKL20 possesses the conserved domains of the WAK/WAKL family and is localized to the plasma membrane and linked to the cell wall. The expression of CaWAKL20 was downregulated by heat stress but upregulated by ABA treatment. Silencing CaWAKL20 expression enhanced pepper thermotolerance, but CaWAKL20 overexpression decreased Arabidopsis tolerance to heat stress. In addition, overexpressing CaWAKL20 reduced Arabidopsis sensitivity to ABA and decreased the heat-induced expression of ABA-responsive genes. Therefore, we suggest that CaWAKL20 negatively modulates plant thermotolerance by inhibiting ABA-responsive gene expression. Our results lay a foundation for further understanding of the functional mechanisms of WAKs/WAKLs during plant adaptation to environmental stress.
Author Contributions
HW and MLu designed the research. HW, HN, MLi, YZ, and WH performed the experiments. HW and HN analyzed the data and drafted the manuscript. QD, YD, and MLu revised the manuscript and contributed reagents, materials, and analysis tools.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by the National Natural Science Foundation of China (31572114, 31872091) and Agricultural Key Science and Technology Program of Shaanxi Province, China (2016NY-063, 2018NY-029).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00591/full#supplementary-material
References
- Albrecht T., Argueso C. T. (2017). Should I fight or should I grow now? The role of cytokinins in plant growth and immunity and in the growth-defence trade-off. Ann. Bot. 119 725–735. 10.1093/aob/mcw211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C. M., Wagner T. A., Perret M., He Z. H., He D., Kohorn B. D. (2001). WAKs: cell wall-associated kinases linking the cytoplasm to the extracellular matrix. Plant Mol. Biol. 47 197–206. 10.1023/A:1010691701578 [DOI] [PubMed] [Google Scholar]
- Choi Y. S., Kim Y. M., Hwang O. J., Han Y. J., Kim S. Y., Kim J. I. (2013). Overexpression of Arabidopsis ABF3 gene confers enhanced tolerance to drought and heat stress in creeping bentgrass. Plant Biotechnol. Rep. 7 165–173. 10.1007/s11816-012-0245-0 [DOI] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Dang F. F., Wang Y. N., Yu L., Eulgem T., Lai Y., Liu Z. Q., et al. (2013). CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 36 757–774. 10.1111/pce.12011 [DOI] [PubMed] [Google Scholar]
- Delteil A., Gobbato E., Cayrol B., Estevan J., Michel-Romiti C., Dievart A., et al. (2016). Several wall-associated kinases participate positively and negatively in basal defense against rice blast fungus. BMC Plant Biol. 16:17. 10.1186/s12870-016-0711-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhindsa R. S., Dhindsa P. P., Thorpe T. A. (1981). Leaf senescence: correlated with increased levels of membrane permeability and lipid peroxidation, and decreased levels of superoxide dismutase and catalase. J. Exp. Bot. 32 93–101. 10.1093/jxb/32.1.93 [DOI] [Google Scholar]
- Diener A. C., Ausubel F. M. (2005). Resistance to fusarium oxysporum 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics 171 305–321. 10.1534/genetics.105.042218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driedonks N., Xu J., Peters J. L., Park S., Rieu I. (2015). Multi-level interactions between heat shock factors, heat shock proteins, and the redox system regulate acclimation to heat. Front. Plant Sci. 6:999. 10.3389/fpls.2015.00999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finka A., Cuendet A. F., Maathuis F. J., Saidi Y., Goloubinoff P. (2012). Plasma membrane cyclic nucleotide gated calcium channels control land plant thermal sensing and acquired thermotolerance. Plant Cell 24 3333–3348. 10.1105/tpc.112.095844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Fujita M., Satoh R., Maruyama K., Parvez M. M., Seki M., et al. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17 3470–3488. 10.1105/tpc.105.035659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovesi V., Fornalé S., Fry S. C., Ruel K., Ferrer P., Encina A., et al. (2008). ZmXTH1, a new xyloglucan endotransglucosylase/hydrolase in maize, affects cell wall structure and composition in Arabidopsis thaliana. J. Exp. Bot. 59 875–889. 10.1093/jxb/ern013 [DOI] [PubMed] [Google Scholar]
- Giarola V., Krey S., von den Driesch B., Bartels D. (2016). The Craterostigma plantagineum glycine-rich protein CpGRP1 interacts with a cell wall-associated protein kinase 1 (CpWAK1) and accumulates in leaf cell walls during dehydration. New Phytol. 210 535–550. 10.1111/nph.13766 [DOI] [PubMed] [Google Scholar]
- Harkenrider M., Sharma R., De Vleesschauwer D., Tsao L., Zhang X., Chern M., et al. (2016). Overexpression of rice wall-associated kinase 25 (OsWAK25) alters resistance to bacterial and fungal pathogens. PLoS One 11:e0147310. 10.1371/journal.pone.0147310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z. H., He D., Kohorn B. D. (1998). Requirement for the induced expression of a cell wall associated receptor kinase for survival during the pathogen response. Plant J. 14 55–63. 10.1046/j.1365-313X.1998.00092.x [DOI] [PubMed] [Google Scholar]
- Hofmann N. R. (2009). The plasma membrane as first responder to heat stress. Plant Cell 21:2544 10.1105/tpc.109.210912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Tong H., Selby J., Dewitt J., Peng X., He Z. H. (2005). Involvement of a cell wall-associated kinase, WAKL4, in Arabidopsis mineral responses. Plant Physiol. 139 1704–1716. 10.1104/pp.105.066910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. C., Niu C. Y., Yang C. R., Jinn T. L. (2016). The heat stress factor HSFA6b connects ABA signaling and ABA-mediated heat responses. Plant Physiol. 172 1182–1199. 10.1104/pp.16.00860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurni S., Scheuermann D., Krattinger S. G., Kessel B., Wicker T., Herren G., et al. (2015). The maize disease resistance gene Htn1 against northern corn leaf blight encodes a wall-associated receptor-like kinase. Proc. Natl. Acad. Sci. U.S.A. 112 8780–8785. 10.1073/pnas.1502522112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPCC (2014). Climate Change 2014: Synthesis Report. Geneva: IPCC, 55–74. [Google Scholar]
- Kohorn B. D. (2016). Cell wall-associated kinases and pectin perception. J. Exp. Bot. 67 489–494. 10.1093/jxb/erv467 [DOI] [PubMed] [Google Scholar]
- Li H., Zhou S. Y., Zhao W. S., Su S. C., Peng Y. (2009). A novel wall-associated receptor-like protein kinase gene, OsWAK1, plays important roles in rice blast disease resistance. Plant Mol. Biol. 69 337–346. 10.1007/s11103-008-9430-5 [DOI] [PubMed] [Google Scholar]
- Liu H. T., Liu Y. Y., Pan Q. H., Yang H. R., Zhan J. C., Huang W. D. (2006). Novel interrelation-ship between salicylic acid, abscisic acid, and PIP2-specific phospholipase C in heat acclimation-induced thermotolerance in pea leaves. J. Exp. Bot. 57 3337–3347. 10.1093/jxb/erl098 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Mittler R., Finka A., Goloubinoff P. (2012). How do plants feel the heat? Trends Biochem. Sci. 37 118–125. 10.1016/j.tibs.2011.11.007 [DOI] [PubMed] [Google Scholar]
- Ohama N., Sato H., Shinozaki K., Yamaguchi-Shinozaki K. (2017). Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 22 53–65. 10.1016/j.tplants.2016.08.015 [DOI] [PubMed] [Google Scholar]
- Rosli H. G., Zheng Y., Pombo M. A., Zhong S., Bombarely A., Fei Z., et al. (2013). Transcriptomics-based screen for genes induced by flagellin and repressed by pathogen effectors identifies a cell wall-associated kinase involved in plant immunity. Genome Biol. 14:R139. 10.1186/gb-2013-14-12-r139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rütgers M., Muranaka L. S., Schulz-Raffelt M., Thoms S., Schurig J., Willmund F., et al. (2017). Not changes in membrane fluidity but proteotoxic stress triggers heat shock protein expression in Chlamydomonas reinhardtii. Plant Cell Environ. 40 2987–3001. 10.1111/pce.13060 [DOI] [PubMed] [Google Scholar]
- Saidi Y., Finka A., Muriset M., Bromberg Z., Weiss Y. G., Maathuis F. J., et al. (2009). The heat shock response in moss plants is regulated by specific calcium-permeable channels in the plasma membrane. Plant Cell 21 2829–2843. 10.1105/tpc.108.065318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintenac C., Lee W. S., Cambon F., Rudd J. J., King R. C., Marande W., et al. (2018). Wheat receptor-kinase-like protein Stb6 controls gene-for-gene resistance to fungal pathogen Zymoseptoria tritici. Nat. Genet. 50 368–374. 10.1038/s41588-018-0051-x [DOI] [PubMed] [Google Scholar]
- Sangwan V., Orvar B. L., Beyerly J., Hirt H., Dhindsa R. S. (2002). Opposite changes in membrane fluidity mimic cold and heat stress activation of distinct plant MAP kinase pathways. Plant J. 31 629–638. 10.1046/j.1365-313X.2002.01384.x [DOI] [PubMed] [Google Scholar]
- Schramm F., Larkindale J., Kiehlmann E., Ganguli A., Englich G., Vierling E., et al. (2008). A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J. 53 264–274. 10.1111/j.1365-313X.2007.03334.x [DOI] [PubMed] [Google Scholar]
- Schulz P., Herde M., Romeis T. (2013). Calcium-dependent protein kinases: hubs in plant stress signaling and development. Plant Physiol. 163 523–530. 10.1104/pp.113.222539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S. H., Bleecker A. B. (2001). Plant receptor-like kinase gene family: diversity, function, and signaling. Sci. STKE 2001:re22. 10.1126/stke.2001.113.re22 [DOI] [PubMed] [Google Scholar]
- Sivaguru M., Ezaki B., He Z. H., Tong H., Osawa H., Baluska F., et al. (2003). Aluminum-induced gene expression and protein localization of a cell wall-associated receptor kinase in Arabidopsis. Plant Physiol. 132 2256–2266. 10.1104/pp.103.022129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Katano K. (2018). Coordination between ROS regulatory systems and other pathways under heat stress and pathogen attack. Front. Plant Sci. 9:490. 10.3389/fpls.2018.00490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Miller G., Salazar C., Mondal H. A., Shulaev E., Cortes D. F., et al. (2013). Temporal-spatial interaction between reactive oxygen species and abscisic acid regulates rapid systemic acclimation in plants. Plant Cell 25 3553–3569. 10.1105/tpc.113.114595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaballa A., Pasentsis K., Darzentas N., Tsaftaris A. S. (2011). Multiple evidence for the role of an Ovate-like gene in determining fruit shape in pepper. BMC Plant Biol. 11:46. 10.1186/1471-2229-11-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verica J. A., Chae L., Tong H., Ingmire P., He Z. H. (2003). Tissue-specific and developmentally regulated expression of a cluster of tandemly arrayed cell wall-associated kinase-like kinase genes in Arabidopsis. Plant Physiol. 133 1732–1746. 10.1104/pp.103.028530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verica J. A., He Z. H. (2002). The cell wall-associated kinase (WAK) and WAK-like kinase gene family. Plant Physiol. 129 455–459. 10.1104/pp.011028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwakarma K., Upadhyay N., Kumar N., Yadav G., Singh J., Mishra R. K., et al. (2017). Abscisic acid signaling and abiotic stress tolerance in plants: a review on current knowledge and future prospects. Front. Plant Sci. 8:161. 10.3389/fpls.2017.00161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Wang Z. Y. (2014). At the intersection of plant growth and immunity. Cell Host Microbe 15 400–402. 10.1016/j.chom.2014.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhuang L., Shi Y., Huang B. (2017). Up-regulation of HSFA2c and HSPs by ABA contributing to improved heat tolerance in tall fescue and Arabidpsios. Int. J. Mol. Sci. 18:1981. 10.3390/ijms18091981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Chang H., Hu S., Lu X., Yuan C., Zhang C., et al. (2014). Plastid casein kinase 2 knockout reduces abscisic acid (ABA) sensitivity, thermotolerance, and expression of ABA- and heat-stress-responsive nuclear genes. J.Exp. Bot. 65 4159–4175. 10.1093/jxb/eru190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., Yin S. J., Zhang K. L., Shi X. T., Lian C. L., Zhang H. S., et al. (2018). OsWAK11, a rice wall-associated kinase, regulates Cu detoxification by alteration the immobilization of Cu in cell walls. Environ. Exp. Bot. 150 99–105. 10.1016/j.envexpbot.2018.03.005 [DOI] [Google Scholar]
- Yang P., Praz C., Li B., Singla J., Robert C. A. M., Kessel B., et al. (2018). Fungal resistance mediated by maize wall-associated kinase ZmWAK-RLK1 correlates with reduced benzoxazinoid content. New Phytol. 221 976–987. 10.1111/nph.15419 [DOI] [PubMed] [Google Scholar]
- Ye Y., Ding Y., Jiang Q., Wang F., Sun J., Zhu C. (2017). The role of receptor-like protein kinases (RLKs) in abiotic stress response in plants. Plant Cell Rep. 36 235–242. 10.1007/s00299-016-2084-x [DOI] [PubMed] [Google Scholar]
- Yoshida T., Ohama N., Nakajima J., Kidokoro S., Mizoi J., Nakashima K., et al. (2011). Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol. Genet. Genomics 286 321–332. 10.1007/s00438-011-0647-7 [DOI] [PubMed] [Google Scholar]
- Zhang N., Zhang B., Zuo W., Xing Y., Konlasuk S., Tan G., et al. (2017). Cytological and molecular characterization of ZmWAK-mediated head-smut resistance in maize. Mol. Plant Microbe Inter. 30 455–465. 10.1094/MPMI-11-16-0238-R [DOI] [PubMed] [Google Scholar]
- Zhu J. K. (2016). Abiotic stress signaling and responses in plants. Cell 167 313–323. 10.1016/j.cell.2016.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.