Abstract
Diurnal changes in Chordc1 mRNA were recently described in mouse hypothalamus. This report shows that Chordc1 mRNA changes rhythmically throughout the entire adult brain with highest expression levels occurring around the dark–light transition. The rhythmic cycling pattern of Chordc1 was retained under various light–dark schedules and analysis of adult whole brain revealed diurnal patterns that were different than young animals (postnatal day (P) 6). Analysis of adult hippocampus, prefrontal cortex and cerebellum confirmed these observations and a comparison between adult and P6 animals using in situ hybridization indicated that Chordc1 underwent coordinated but altered diurnal changes in mRNA abundance during development. Further, a developmental profile of Chordc1 expression beginning at embryonic day 17 revealed a regional distribution of Chordc1 consistent with its adult pattern. These results suggest that Chordc1 mRNA is under complex and widespread transcriptional regulation during development and implicate Chordc1 in circadian and/or homeostatic mechanisms in mammalian brain.
Keywords: Circadian, Sleep, Chp-1, p23, RAR1, Hsp90
Introduction
The cysteine and histidine-rich domain (CHORD)-containing, zinc binding protein 1 (Chordc1, also known as Chp-1) is a member of a highly conserved protein family that include the plant protein, RAR1 (also known as PPHB SUSCEPTIBLE 2; [1] and the mammalian protein, Melusin [2]. CHORD proteins share two CHORD Zn2+-binding motifs in tandem organization (designated CHORD-I and CHORD-II) in the N-terminal portion of the protein [1]. Metazoan CHORD-proteins that include Chordc1 share an additional CS (CHORD-SGT1) domain toward the C-terminal end that has high sequence similarity to the Hsp90 co-chaperones p23 and SGT1 (suppressor of the G2 allele of Skp1).
The plant CHORD family member, RAR1 has been shown to interact with SGT1, and is a component of the Hsp90-mediated pathogen response system in plants [3–5]. SGT1 contains a CS domain that has also been shown to interact with Hsp90 [4, 6] and together with RAR1, is thought to mediate downstream signaling cascades for host defense mechanisms [7]. In mammals, Chordc1 has been shown to associate with Hsp90 [8], perhaps reflecting a conserved mechanism for CHORD protein family members. Melusin, an integrin-binding protein specific to muscle tissue, is important in the mechanical stress response and has been characterized in signaling pathways that leads to cardiomyocyte hypertrophy [9, 10]. While the functions of these CHORD-containing proteins are beginning to be identified in other species and systems, the function of Chordc1 in mammals, and in particular, the central nervous system, is poorly understood.
In a microarray screen for novel diurnally regulated genes, we identified rhythmic changes in Chordc1 mRNA expression in mammalian hypothalamus [11]. Given the function of CHORD-containing proteins in the modulation of various signaling events in other systems, we were interested in extending the analysis of Chordc1 mRNA regulation to adult and developing murine brain. Further, since Chordc1 may be involved in signaling pathways during development, we also examined the ontogenic expression profile of Chordc1 mRNA in mouse brain.
Materials and methods
Subjects and handling
Mice (C57BL/6) used in this study were either purchased directly from Harlan Sprague-Dawley (adult) or taken at specific ages from a breeding colony. Adult animals were entrained to a 12 h light, 12 h dark or 14 h light, 10 h dark schedule for at least 5 weeks. For tissue collection, animals were sacrificed by decapitation, their brains dissected, flash frozen at −30°C in 2-methylbutane, and stored at −80°C. Alternate hemispheres at each time point were used for either RNA isolation or cryostat sectioning. Animals at various postnatal ages were sacrificed in the same manner between zeitgeber time (ZT) 5 and 10, when Chordc1 expression was low in adult animals. For collection of tissue from embryos, timed-pregnant mice (designated embryonic day 0.5 the morning after breeding) were quickly sacrificed, embryos removed, brains dissected and frozen as described above. All animal care and use procedures were in strict accordance with University of Wisconsin IACUC and National Institute of Health guidelines for the humane treatment of laboratory animals.
In situ hybridization
Cryostat sections (20 μm) from brains collected as described above were placed onto Superfrost Plus (Fisher Scientific, Hampton, NH) slides, and stored at −80°C. Prior to hybridization, slides were placed into 4% paraformaldehyde for 1.5 h at 4°C. Slides were then washed three times in 2 × SSC (1 × SSC = 150 mM NaCl, 15 mM sodium citrate, pH 7.0) and then 10 min at 37°C in 100 mM Tris–HCl, 0.5 M EDTA, pH 8 (protease buffer) containing Proteinase K (Promega, Madison, WI). The concentration of protease used was 0.2 μg/ml for postnatal day (P) 10, P14, P25 and adult animals, 0.02 μg/ml for P1 and P5 tissue and was not added for embryonic tissue. Following protease digestion, slides were then washed in protease buffer without Proteinase K for 2 min and placed in 0.1 M TEA containing 0.25% acetic anhydride for 10 min. Following a 5 min rinse in 2 × SSC, slides were dehydrated in an ethanol series.
Antisense or sense riboprobe (35S-labeled) was made from Chordc1 PCR templates derived from T7-tagged reverse or forward primers, respectively. Forward primer: CATGATGCCCTAAAGGGTTG, reverse primer, AACCTCATGATGACCCCAAA. Chordc1 template used for in vitro transcription was generated using standard PCR conditions. T7 sequence tag (extending the nested reverse primer, ATCCCCTCGTGGAAAATAGG for antisense and the forward primer for sense template) was GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGC GG. In vitro transcription reactions were carried out as described by Promega Biotech (Madison, WI).
Slides were probed with 150 μl of 35S-labeled antisense riboprobe (10,000 cpms/μl) in hybridization buffer (3 × SSC, 1 × Denhardt’s, 50% formamide, 10% Dextran sulfate, 0.05% 1 M NaPO4 (pH 7.4), 200 μg/ml tRNA, 50 mM DTT), coverslipped, and incubated at 55°C for 16–20 h in a 50% formamide-saturated chamber. Following hybridization, slides were washed three times 10 min in 2 × SSC containing 2 mM DTT and 1 h in 20 μg/ml of pancreatic RNase A in 10 mM Tris–HCl, 0.5 M NaCl (pH 8.0) at 37°C. Slides were then washed in 1 × SSC (2 mM DTT) for 5 min, 0.5 × SSC (2 mM DTT) for 5 min, and 0.1 × SSC (2 mM DTT) at 70°C for 1 h. Sections were then dehydrated in a graded alcohol series, air dried, and exposed to a phosphoscreen for 6 days. Image analysis was performed using the Storm 860 and ImageQuant 5.2 software (Molecular Dynamics Sunnyvale, CA). For desitometric analysis of in situ hybridization data, specific regions from four sections were averaged per animal per timepoint. Images were color rendered in ImageQuant to enhance visualization of labeled regions.
For emulsion autoradiography, sections processed for in situ hybridization were dipped in NTB3 liquid emulsion (Eastman Kodak, Rochester, New York) under safelight conditions and stored at 4°C for 6 weeks. Developing times were as described by the manufacturer. Following development, sections were lightly stained in cresyl violet (0.1%), dehydrated in an ethanol series and mounted.
Northern Blot analysis
Northern blotting was performed essentially as described previously [12]. Briefly, brains were dissected and total RNA was isolated using TRIZOL (Invitrogen, Carlsbad, CA), according to the manufacturer’s specifications, and stored at −80°C. Prior to loading, each sample was incubated with sample buffer (7.5% Formaldehyde, 43% Formamide, 12% 10× MOPS, 0.12% Ethidium Bromide) for 5 min at 56°C, and then cooled on ice for 5 min. Northern Blots consisted of 1 μg of total RNA per lane and were electrophoresed on a 1.2% agarose/1× MOPS/7.5% formaldehyde gel for 2.5 h at 60 V in 1× MOPS buffer. Each gel was washed 4 × 15 min in distilled H2O with gentle agitation. RNA was transferred overnight onto a sheet of GeneScreen Plus nitrocellulose (NEN Life Science Products, Boston, MA) using the Stratagene PosiBlot 30-30 Pressure Blotter with 10× salt sodium citrate (SSC). The blots were then crosslinked using a 1800 UV Stratalinker (Stratagene, La Jolla, CA), covered in clear plastic wrap and stored at −20°C until hybridization. Prior to probing, blots were pre-hybridized in 10 ml of Hybrisol (Intergen, Burlington, MA) at 42°C for 1 h. DNA templates generated by PCR (see primers above) were labeled with 32P using the Megaprime DNA labeling system (Amersham Biosciences, England), mixed with hybrisol, and incubated overnight at 42°C. Blots were washed 3× with 100 ml of 2× SSC 1% SDS at RT, then 2 × 30 min with 100 ml of 2× SSC 1% SDS at 50°C, and finally 1 × 30 min with 100 ml of 0.5× SSC 1% SDS at 60°C. Following washes, blots were exposed to a phosphoscreen for 3 days and image analysis was performed using the Storm 860 and ImageQuant 5.2 software (Molecular Dynamics, Sunnyvale, CA). A 983-bp PCR product for glyceraldehyde-3-phosphate dehydrogenase (gapdh) was generated as described previously [13].
Results
Diurnal changes in the expression of Chordc1 mRNA in adult mouse brain
We previously identified Chordc1 in a screen for mRNAs in hypothalamus that underwent diurnal changes in expression [11]. To expand these observations, we examined the regulation of Chordc1 mRNA in additional brain regions using in situ hybridization and phosphorimage densitometry. In parallel with previous results in hypothalamus, Chordc1 was found to undergo diurnal regulation in all brain regions examined including hippocampus, prefrontal cortex and cerebellum (Fig. 1A, B). Data from each region were subjected to one-way ANOVA and a statistically significant diurnal variation was observed (hippocampus P < 0.001; prefrontal cortex P < 0.001; cerebellum P < 0.001; versus ZT12, post hoc Bonferroni). Peak levels of expression were evident at the transition between the dark and light period (ZT18 and ZT0).
Fig. 1.
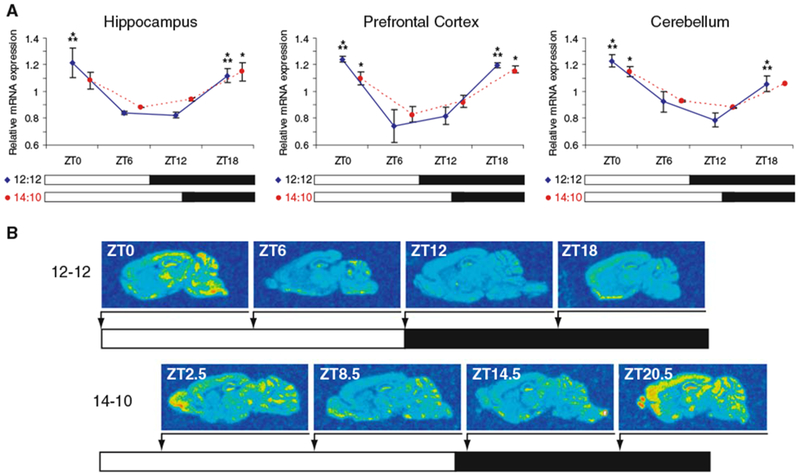
Diurnal expression of Chordc1 mRNA in adult mouse brain during two photic-entrainment schedules. (A) Densitometric analysis of Chordc1 mRNA at four timepoints in a 12:12 light-dark (LD) schedule (blue diamonds and blue solid lines; ZT0, ZT6, ZT12, ZT18) and four timepoints in a 14:10 LD schedule (red circles and red dashed lines; ZT2.5, ZT8.5, ZT14.5, ZT20.5) using in situ hybridization. Each value represents the average ± S.E.M. N = 2 per timepoint. Data from each region on 12:12 LD schedule were subjected to one-way ANOVA and a statistically significant diurnal variation was observed (Hippocampus P < 0.001; Prefrontal Cortex P < 0.001; Cerebellum P < 0.001). *** P < 0.001 versus ZT12 (post hoc Bonferroni). Data from each region on 14:10 LD schedule were also subjected to one-way ANOVA and a statistically significant diurnal variation was observed (Hippocampus P < 0.05; Prefrontal Cortex P < 0.05; Cerebellum P < 0.05). * P < 0.05 versus ZT8.5 (post hoc Bonferroni). (B) Representative sagittal sections from adult mouse brain hybridized to Chordc1 cRNA probe and analyzed for densitometry as described in A and Methods. N = 2 per timepoint in each 12:12 and 14:10 LD schedule
To determine whether a change in lighting conditions influenced the diurnal expression of Chordc1, we altered the entrainment schedule of mice from 12-12 light–dark (LD) to 14-10 LD and found that a change in lighting schedule did not alter the diurnal expression pattern of Chordc1 in the three regions analyzed (Fig. 1A, one-way ANOVA; hippocampus P < 0.05; prefrontal cortex P < 0.05; cerebellum P < 0.05 versus ZT8.5, post hoc Bonferroni). In addition, we examined the olfactory bulb, an important sensory nuclei in rodents, and found a statistically significant diurnal pattern of Chordc1 expression that was similar to the other brain regions (range of relative mRNA abundance was 1.1 (ZT2.5 and 20.5) to 0.8 (ZT14.5), Student t test, P < 0.01 versus ZT14.5). Further, Chordc1 mRNA levels in each of the brain regions examined were not significantly different between ZT 8.5 and 14.5 (30 min into the dark period) suggesting that Chordc1 expression is not immediately induced following dark onset.
Comparison of Chordc1 expression in postnatal day 6 and adult brain
To determine whether Chordc1 undergoes rhythmic changes in expression during early postnatal development, we used Northern blotting to compare Chordc1 mRNA levels in adult and early postnatal brain (P6) at four time points across the light–dark cycle (Fig. 2). Diurnal changes in Chordc1 mRNA levels in adult whole brain paralleled in situ hybridization results and higher levels of Chordc1 were evident at the transition between the dark and light period (ZT 20.5 and ZT2.5; Fig. 2A). A diurnal analysis of Chordc1 mRNA levels in P6 brain revealed a somewhat different pattern of expression compared to adult animals (Fig. 2B) with lowest Chordc1 expression levels observed at ZT 8.5 and highest levels at ZT 14.5 (Student t-test, P = 0.065; Fig. 2B).
Fig. 2.
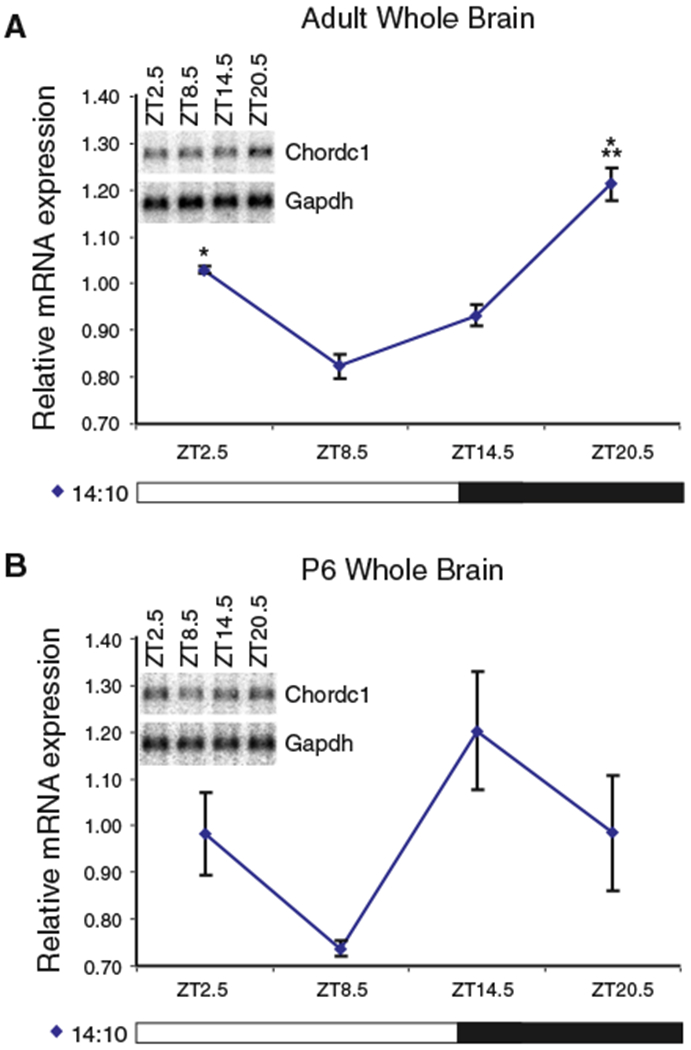
Diurnal rhythm of Chordc1 mRNA is differentially regulated in P6 compared to adult mouse whole brain. (A) Chordc1 mRNA levels from adult mouse whole brain expressed relative to the average value of all samples normalized to glyceraldehyde-3-phoshate dehydrogenase (Gapdh) from four timepoints on 14:10 LD schedule (ZT2.5, ZT8.5, ZT14.5, ZT20.5). N = 2 per timepoint. Data were subjected to one-way ANOVA and a statistically significant diurnal effect was revealed (P = 0.002). *P < 0.05, ***P = 0.001 (post hoc Bonferroni). The inset panel illustrates representative diurnal Northern blots of Chordc1 and Gapdh mRNA from adult mouse whole brain. Chordc1, 2200 bp; Gapdh, 1240 bp. (B) Chordc1 mRNA levels from P6 mouse whole brain expressed relative to the average value of all samples normalized to Gapdh from four timepoints on 14:10 LD schedule (ZT2.5, ZT8.5, ZT14.5, ZT20.5). N = 3 per ZT2.5 and ZT14.5 timepoints, N = 2 per ZT8.5 and ZT20.5 timepoints. Data were subjected to one-way ANOVA and a statistically significant diurnal effect was not observed (P = 0.114). t-test of highest and lowest expression points (ZT14.5 and ZT8.5, respectively) approached significance (P = 0.065). The inset panel illustrates representative diurnal Northern blots of Chordc1 and Gapdh mRNA from P6 mouse whole brain. Chordc1, 2200 bp; Gapdh, 1240 bp
To validate the Northern blotting results and further examine the expression of Chordc1 mRNA in specific regions of P6 brain, we used in situ hybridization and densitometric analysis on the three regions that were examined in adult brain (Fig. 3A, B). In parallel with the trend observed in Northern blots, we found highest Chordc1 mRNA levels at ZT14.5 in all three brain regions (Fig. 3A). Further, a significant diurnal effect was observed in each region (one-way ANOVA, hippocampus, P = 0.011; prefrontal cortex, P < 0.01; cerebellum, P < 0.05, versus ZT8.5, post hoc Bonferroni). However, we identified more variation in Chordc1 expression between different brain regions in young animals than we observed in adults. For example, Chordc1 mRNA levels in P6 olfactory bulb peaked at ZT14.5 but unlike other brain regions, high levels of expression continued through ZT20.5 (Student t-test, P < 0.05 versus ZT2.5, Fig. 3B). These results suggest that Chordc1 undergoes a more complex diurnal expression pattern in young animals.
Fig. 3.
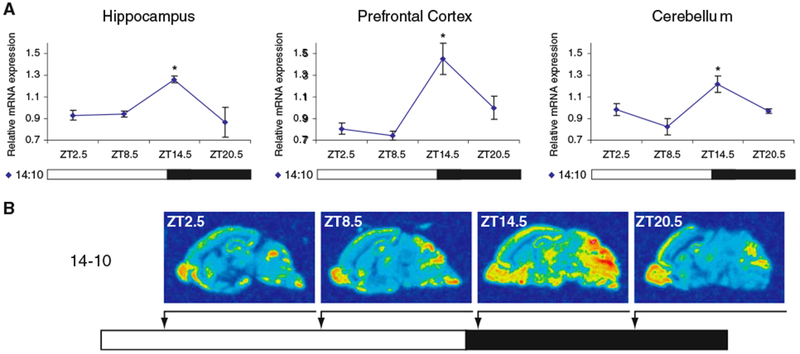
Diurnal expression of Chordc1 mRNA in P6 mouse brain. (A) Densitometric analysis of Chordc1 mRNA at four timepoints in a 14:10 LD schedule (ZT2.5, ZT8.5, ZT14.5, ZT20.5) using in situ hybridization. Each value represents the average ± S.E.M. N = 3 per ZT2.5 and ZT14.5 timepoints, N = 2 per ZT8.5 and ZT20.5 timepoints. Data from each region on 14:10 LD schedule were also subjected to one-way ANOVA and a statistically significant diurnal variation was observed (Hippocampus P = 0.011; Prefrontal Cortex P < 0.01; Cerebellum P < 0.05). *P < 0.05 versus ZT8.5 (post hoc Bonferroni). (B) Representative sagittal sections from mouse brain hybridized to Chordc1 cRNA probe and analyzed for densitometry as described in A and Methods. N = 3 per ZT2.5 and ZT14.5 timepoints, N = 2 per ZT8.5 and ZT20.5 timepoints
Chordc1 mRNA distribution during mouse brain development
We also examined the profile of Chordc1 mRNA expression during mouse brain development. Sagittal brain sections hybridized to cRNA probe to Chordc1 revealed a wide distribution pattern that was evident as early as postnatal day (P) 1 and that continued throughout postnatal development in agreement with in situ hybridization data on Chordc1 displayed in Gene Expression Nervous System Atlas (GENSAT; Fig. 4). Animals used for developmental analysis were collected between ZT5 and ZT10. Robust Chordc1 mRNA expression was evident in the cortical plate (CP) at embryonic day 17, the earliest time point examined (Fig. 5A). Lower levels of expression were also evident in intermediate (IZ) and subventricular (SVZ) zones at this age. A more diffuse expression pattern was evident in early postnatal cortex with only slight differences in intensity throughout cortical layers (Fig. 5B). By P14, a more laminated Chordc1 mRNA distribution pattern became evident with a punctate distribution apparent in deep cortical layers (Fig. 5C). The adult pattern of Chordc1 expression in cortex consisted of low expression in layer 1 and moderate expression in other cortical layers (Fig. 5D).
Fig. 4.
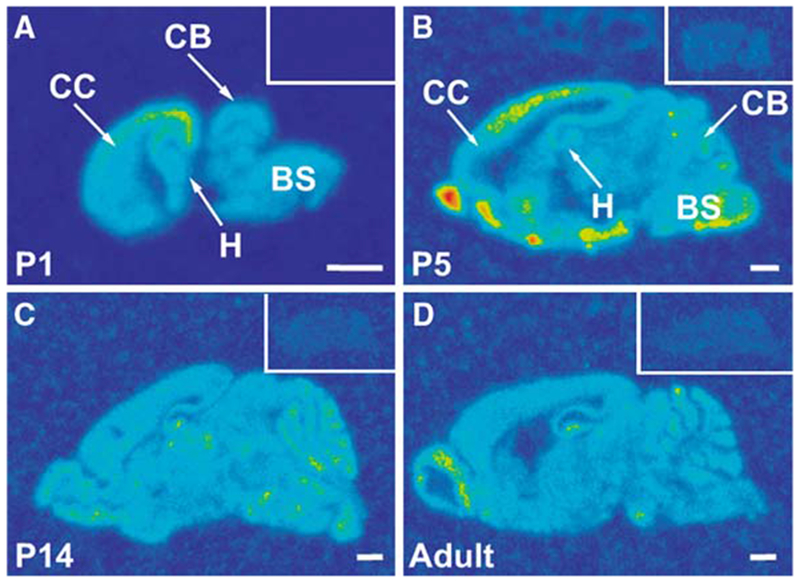
Distribution of Chordc1 mRNA in developing mouse brain In situ hybridization with 35S-labeled antisense riboprobe was used to detect the distribution of Chordc1 mRNA in sagittal sections from (A) postnatal day (P) 1, (B) P5, (C) P14 and (D) adult mouse brain. Insets show control brain sections at each age hybridized to 35S-labeled sense riboprobe. Images were color rendered to enable visualization of labeling (red, highest and blue, lowest levels). Bar in A–D = 1 mm. BS, brain stem; CB, cerebellum; CC, cerebral cortex; H, hippocampus; P, postnatal day
Fig. 5.
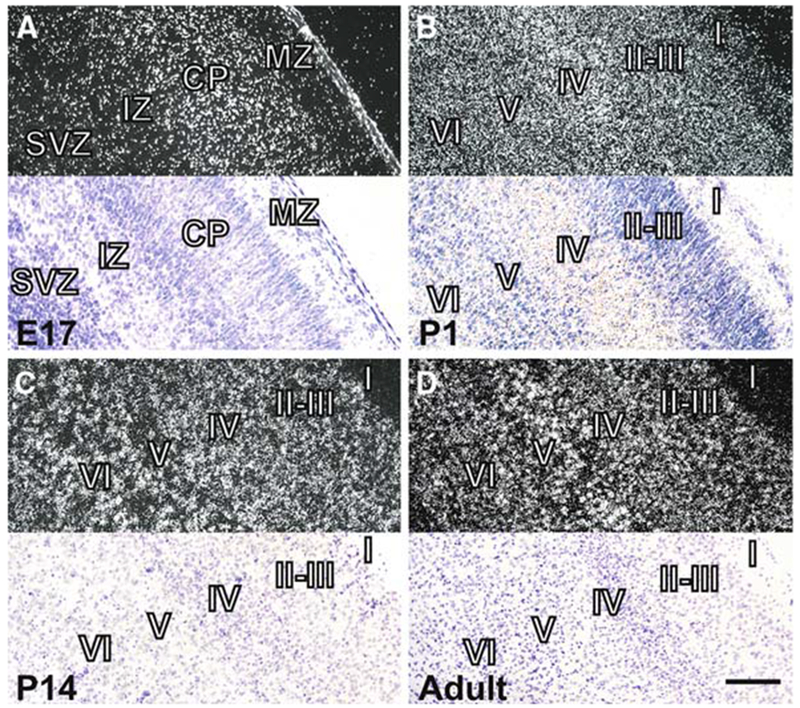
Distribution of Chordc1 mRNA in developing cerebral cortex. (A) Dark field photomicrographs (top panel) of coronal sections from embryonic day 17 mouse brain depicting Chordc1 mRNA labeling primarily within the cortical plate (CP) with lighter labeling in the subventricular zone (SVZ) and intermediate zone (IZ). Bottom panel is a light field micrograph of the same region counterstained with cresyl violet. (B) Dark field micrograph (top) of postnatal day (P1) cortex revealing a disperse pattern of Chordc1 labeling throughout the cortical layers. Bottom panel is section counterstained with cresyl violet. (C) By P14, label to Chordc1 mRNA was present throughout the cortex in a pattern suggesting the presence of Chordc1 mRNA in individual cells. Top, dark field, bottom, counterstained light field micrograph. (D) In adult brain, a more punctuate distribution of Chordc1 was evident and heavy relative labeling was present in large foci suggestive of pyramidal cells in layer V. Top, dark field, bottom, counterstained light field micrograph. Bar in A, B = 100 μm; in C, D = 200 μm. E, embryonic; P, postnatal
Chordc1 was also expressed diffusely throughout P1 cerebellum in a pattern consistent with Chordc1 mRNA in both migrating and post-migratory cell populations (Fig. 6A). By P5, less intense expression of Chordc1 was evident in more distal cell regions (external granule cell layer, EGL) while more intense silver grain accumulation was evident in more proximal layers such as Purkinje/internal granule cell layer (PCL/IGL, Fig. 6B). A similar labeling pattern was present at P10 with only sparse signal corresponding to Chordc1 mRNA in molecular layer (ML) and more concentrated labeling evident in Purkinje/granule cell layer (P, GL, Fig. 6C). By P14 (Fig. 6D) and into adulthood (not shown), distinct accumulations of labeling were evident over individual Purkinje cells (P) and within the granule cell layer (GL).
Fig. 6.
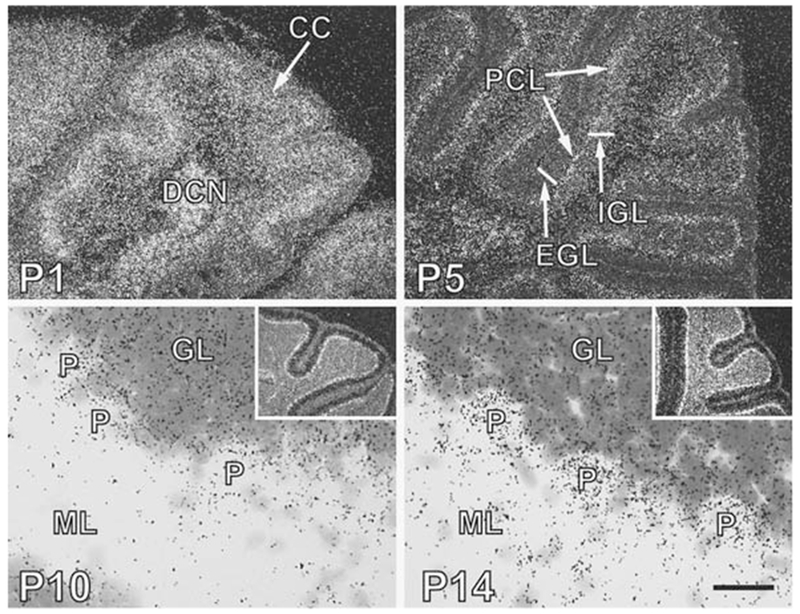
Chordc1 mRNA is expressed in both pre- and post-migratory cells in developing cerebellum. (A) In situ hybridization using Chordc1 riboprobe to postnatal day (P) 1 sagittal brain sections revealed robust labeling throughout cerebellum that did not distinguish layers in the cerebellar cortex (CC). DCN, deep cerebellar nuclei. Image is a dark field micrograph. (B) At P5, label corresponding to Chordc1 was present in the premigratory external granule cell layer (EGL) and the postmigratory Purkinji cell (PCL) and internal granule cell (IGL) layer. Note that in this dark field micrograph heavier Chordc1 labeling was present in PCL. (C) By P10, Chordc1 mRNA (dark sliver grains) was evident in the Purkinji (P) and granule cell layer (GL) but was absent from the cell-sparse molecular layer (ML). Section was counterstained with cresyl violet. Inset shows low magnification dark field micrograph with highest intensity of labeling in PCL. (D) In sagittal sections from P14 brain, silver grain accumulation could be identified over individual Purkinji cells (P). Chordc1 mRNA was also present in GL and in cells within the ML. Inset shows low magnification dark field micrograph. Bar in A = 200 μm; B = 250 μm; C = 30 μm; D = 25 μm
The expression of Chordc1 in developing hippocampus followed the maturation of neuronal layers within this region. At P1, diffuse labeling corresponding to Chordc1 mRNA was evident in pyramidal layers of the hippocampus (CA1, CA3) with only light labeling in the developing dentate gyrus (DG; Fig. 7A). By P5, Chordc1 mRNA expression clearly defined the major hippocampal and dentate gyral neuronal cell layers (Fig. 7B). Silver grain distribution was concentrated over somatic layers by P14 (Fig. 7C) and this pattern continued into adulthood (Fig. 7D).
Fig. 7.
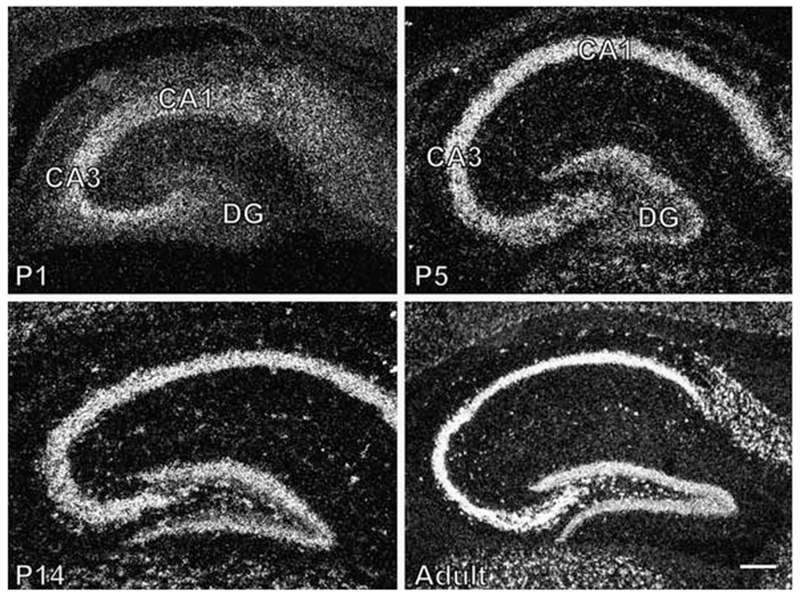
Expression of Chordc1 mRNA in developing hippocampus. (A) A diffuse labeling pattern corresponding to Chordc1 mRNA was evident in dark field micrographs from P1 hippocampus suggestive of Chordc1 in hippocampal neurons (CA1, CA3) in the final stages of migration and maturation. Only faint labeling was present in dentate gyrus (DG). (B) Chordc1 mRNA labeling was more concentrated in pyramidal cell layers of hippocampus (CA1, CA3) and granule cells of dentate gyrus (DG) at P5 (dark field micrograph). (C) By P14, labeling corresponding to Chordc1 mRNA followed the major cell layers of hippocampus and by (D) adulthood, Chordc1 labeling followed a neuronal cell-like pattern with intense labeling in CA1–3 pyramidal cells of the hippocampus. Bar in A, B = 100 μm; C = 230 μm; D = 200 μm
Discussion
The zinc-binding protein, Chordc1 has recently been identified in mammalian brain where it has been found to undergo diurnal [11] and circadian [14] changes in expression in hypothalamus. The current report extends these initial observations and indicates that Chordc1 is diurnally expressed throughout the brain in a coordinated pattern. Under two entrainment schedules (12:12 LD and 14:10 LD), Chordc1 reaches highest levels during the transition from dark to light (ZT18–ZT2.5). In young animals (P6), Chordc1 was also found to undergo diurnal changes in expression. However, the diurnal pattern of Chordc1 expression differed from adults and peak levels of Chordc1 were evident immediately following dark onset. Finally, analysis of Chordc1 mRNA in development revealed broad expression throughout the brain beginning during embryonic development, in agreement with data reported for Chordc1 on GENSAT. Chordc1 is therefore a broadly expressed gene that first appears early in brain development and undergoes complex changes in rhythmic mRNA expression as the brain matures.
Daily behavioral and physiological rhythms are synchronized by the circadian pacemaker in the suprachiasmatic nucleus (SCN), which likely influences the rhythmic expression of Chordc1. It is interesting, however, that the rhythmic pattern of expression of Chordc1 mRNA differed between young and adult animals. Whereas the highest levels of Chordc1 mRNA were evident at the end of the dark period and the beginning of the light period in adult animals, highest Chordc1 expression levels in P6 animals were evident immediately following dark onset. Since the SCN in rodent brain is not synaptically mature until P10 [15], the age-related difference in diurnal rhythmicity of Chordc1 may be due to an immature pacemaker in P6 animals. This would suggest that although rhythmic expression of Chordc1 is beginning to appear at this age, it is not yet synchronized to the SCN. The larger variability in Chordc1 mRNA rhythmicity that we observed between animals and brain regions at P6 supports this notion. However, SCN rhythmicity is already present in late rodent embryonic life [16, 17] and the expression of genes like Per1, Per2, Cry1 and Bmal, which comprise the molecular mechanisms governing circadian rhythmicity in SCN, are expressed in a circadian pattern by the end of the first postnatal week [18, 19]. An alternative explanation, therefore, is that Chordc1 rhythmicity is not tied to a central pacemaker but is driven by other homeostatic mechanisms such as sleep or feeding. In young animals, maternal cues appear to have a more potent influence on early postnatal circadian entrainment than other zeitgebers including light [15]. Most dramatically, maternal separation during the period when pups normally feed can reverse the circadian rhythms of Per1 and Per2 [20]. Furthermore, the behavioral rhythms of neonatal mice synchronize to maternal behavior so that the highest levels of activity in pups occur at dark onset when the mother ceases to nurse and leaves the litter to feed [21, 22]. It is likely, therefore, that the rhythmicity of Chordc1 expression at P6 is synchronized to maternal behavior since elevated Chordc1 mRNA levels are evident in young animals immediately following dark onset and within the period that a nursing dam would feed and pups would be most active. Whether the rhythmic expression of Chordc1 is influenced by activity in adult animals remains to be determined.
Although the precise function of Chordc1 in mammalian systems is unknown, Chordc1 has been found to associate with the chaperone protein, heat shock protein (Hsp) 90 [8]. Since Hsp90 and p23 act as co-chaperones, it is interesting that Chordc1 also contains a p23 domain. p23 (also known as prostaglandin E synthase) has been implicated in circadian and sleep mechanisms and has been shown to synthesize, in association with Hsp90, prostaglandin E2, a metabolite of arachidonic acid that is thought to promote wakefulness in many species [23–25]. Hsp90 and p23 are also involved in aryl hydrocarbon receptor (AhR) signaling, which, like the CHORD-containing RAR1/Hsp90 complex in plants, is important in host-defense mechanisms [26–30]. Further, AhR is a member of the basic helix-loop-helix PER-ARNT-SIM (bHLH/PAS) transcription factor family, which constitute central players in circadian rhythmicity. The aryl hydrocarbon receptor-like, Arnt3 (also known as MOP3 or BMAL), for example, heterodimerizes with CLOCK protein and activates the transcription of downstream circadian genes [31] altering baseline sleep architecture [32]. The rhythmic expression of Chordc1, its association with Hsp90 and the presence of a p23 domain in its carboxy terminus may implicate Chordc1 in one or all of these processes, perhaps as a competitor to p23. Further work will be required to definitively determine the function of Chordc1 in homeostatic mechanisms.
These results, therefore, delineate the complex regulation of a broadly expressed CHORD-containing, zinc-binding protein that interacts with the chaperone protein, Hsp90 and that may play a role in circadian or homeostatic mechanisms. The presence of Chordc1 in developing brain may further implicate the protein in mechanisms related to neural cell maturation since silencing of the nematode CHORD-containing gene, chp, results in semisterility and embryo lethality [1]. It will be important to study Chordc1 expression changes at the protein level to determine if protein expression and turnover mirror changes observed in mRNA expression. That the mRNA for Chordc1 undergoes complex changes in diurnal rhythmicity in developing brain may suggest that Chordc1 expression is influenced by factors such as activity, however further study will be required to determine the precise role of Chordc1 in developing and adult brain.
Acknowledgments
This work is in honor of Tony and Celia Campagnoni, my mentors and close friends. We would like to thank Andrew M. Brienen and Quentin Bremer for excellent technical assistance. This work was supported by National Institute of Heath grant 5 P50 CA084724-04 to J.R.G. and DA13780 to C.F.L.
Footnotes
Special issue dedicated to Anthony Campagnoni.
Contributor Information
Jason R. Gerstner, Neuroscience Training Program and Department of Psychiatry, University of Wisconsin-Madison, Madison, WI 53719, USA
Charles F. Landry, Neuroscience Training Program and Department of Psychiatry, University of Wisconsin-Madison, Madison, WI 53719, USA; Department of Psychiatry, University of Wisconsin-Madison, 6001 Research Park Blvd., Madison, WI 53711, USA
References
- 1.Shirasu K, Lahaye T, Tan MW et al. (1999) A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell 99:355–366 [DOI] [PubMed] [Google Scholar]
- 2.Brancaccio M, Menini N, Bongioanni D et al. (2003) Chp-1 and melusin, two CHORD containing proteins in vertebrates. FEBS Lett 551:47–52 [DOI] [PubMed] [Google Scholar]
- 3.Azevedo C, Sadanandom A, Kitagawa K et al. (2002) The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295:2073–2076 [DOI] [PubMed] [Google Scholar]
- 4.Takahashi A, Casais C, Ichimura K et al. (2003) HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in arabidopsis. Proc Natl Acad Sci USA 100:11777–11782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holt BF 3rd, Belkhadir Y, Dangl JL (2005) Antagonistic control of disease resistance protein stability in the plant immune system. Science 309:929–932 [DOI] [PubMed] [Google Scholar]
- 6.Lee YT, Jacob J, Michowski W et al. (2004) Human Sgt1 binds HSP90 through the CHORD-Sgt1 domain and not the tetratricopeptide repeat domain. J Biol Chem 279:16511–16517 [DOI] [PubMed] [Google Scholar]
- 7.Hahn JS (2005) Regulation of Nod1 by Hsp90 chaperone complex. FEBS Lett 579:4513–4519 [DOI] [PubMed] [Google Scholar]
- 8.Wu J, Luo S, Jiang H et al. (2005) Mammalian CHORD-containing protein 1 is a novel heat shock protein 90-interacting protein. FEBS Lett 579:421–426 [DOI] [PubMed] [Google Scholar]
- 9.Brancaccio M, Guazzone S, Menini N et al. (1999) Melusin is a new muscle-specific interactor for beta(1) integrin cytoplasmic domain. J Biol Chem 274:29282–29288 [DOI] [PubMed] [Google Scholar]
- 10.Brancaccio M, Fratta L, Notte A et al. (2003) Melusin, a muscle-specific integrin beta1-interacting protein, is required to prevent cardiac failure in response to chronic pressure overload. Nat Med 9:68–75 [DOI] [PubMed] [Google Scholar]
- 11.Gerstner JR, Vander Heyden WM, Lavaute TM et al. (2006) Profiles of novel diurnally regulated genes in mouse hypothalamus: expression analysis of the cysteine and histidine-rich domain-containing, zinc-binding protein 1, the fatty acid-binding protein 7 and the GTPase, ras-like family member 11b. Neuroscience 139:1435–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schiltz CA, Kelley AE, Landry CF (2005) Contextual cues associated with nicotine administration increase arc mRNA expression in corticolimbic areas of the rat brain. Eur J Neurosci 21:1703–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roseboom PH, Coon SL, Baler R et al. (1996) Melatonin synthesis: analysis of the more than 150-fold nocturnal increase in serotonin N-acetyltransferase messenger ribonucleic acid in the rat pineal gland. Endocrinology 137:3033–3045 [DOI] [PubMed] [Google Scholar]
- 14.Ueda HR, Chen W, Adachi A et al. (2002) A transcription factor response element for gene expression during circadian night. Nature 418:534–539 [DOI] [PubMed] [Google Scholar]
- 15.Weinert D (2005) Ontogenetic development of the mammalian circadian system. Chronobiol Int 22:179–205 [DOI] [PubMed] [Google Scholar]
- 16.Reppert SM, Schwartz WJ (1984) The suprachiasmatic nuclei of the fetal rat: characterization of a functional circadian clock using 14C-labeled deoxyglucose. J Neurosci 4:1677–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shibata S, Moore RY (1987) Development of neuronal activity in the rat suprachiasmatic nucleus. Brain Res 431:311–315 [DOI] [PubMed] [Google Scholar]
- 18.Shimomura H, Moriya T, Sudo M et al. (2001) Differential daily expression of Per1 and Per2 mRNA in the suprachiasmatic nucleus of fetal and early postnatal mice. Eur J Neurosci 13:687–693 [DOI] [PubMed] [Google Scholar]
- 19.Sladek M, Sumova A, Kovacikova Z et al. (2004) Insight into molecular core clock mechanism of embryonic and early postnatal rat suprachiasmatic nucleus. Proc Natl Acad Sci USA 101:6231–6236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohta H, Honma S, Abe H et al. (2003) Periodic absence of nursing mothers phase-shifts circadian rhythms of clock genes in the suprachiasmatic nucleus of rat pups. Eur J Neurosci 17:1628–1634 [DOI] [PubMed] [Google Scholar]
- 21.Viswanathan N, Chandrashekaran MK (1985) Cycles of presence and absence of mother mouse entrain the circadian clock of pups. Nature 317:530–531 [DOI] [PubMed] [Google Scholar]
- 22.Viswanathan N (1999) Maternal entrainment in the circadian activity rhythm of laboratory mouse (C57BL/6J). Physiol Behav 68:157–162 [DOI] [PubMed] [Google Scholar]
- 23.Hayaishi O (1991) Molecular mechanisms of sleep-wake regulation: roles of prostaglandins D2 and E2. FASEB J 5:2575–2581 [PubMed] [Google Scholar]
- 24.Tanioka T, Nakatani Y, Semmyo N et al. (2000) Molecular identification of cytosolic prostaglandin E2 synthase that is functionally coupled with cyclooxygenase-1 in immediate prostaglandin E2 biosynthesis. J Biol Chem 275:32775–32782 [DOI] [PubMed] [Google Scholar]
- 25.Tanioka T, Nakatani Y, Kobayashi T et al. (2003) Regulation of cytosolic prostaglandin E2 synthase by 90-kDa heat shock protein. Biochem Biophys Res Commun 303:1018–1023 [DOI] [PubMed] [Google Scholar]
- 26.Kazlauskas A, Poellinger L, Pongratz I (1999) Evidence that the co-chaperone p23 regulates ligand responsiveness of the dioxin (aryl hydrocarbon) receptor. J Biol Chem 274:13519–13524 [DOI] [PubMed] [Google Scholar]
- 27.Cox MB, Miller CA 3rd (2002) The p23 co-chaperone facilitates dioxin receptor signaling in a yeast model system. Toxicol Lett 129:13–21 [DOI] [PubMed] [Google Scholar]
- 28.Carlson DB, Perdew GH (2002) A dynamic role for the ah receptor in cell signaling? insights from a diverse group of ah receptor interacting proteins. J Biochem Mol Toxicol 16:317–325 [DOI] [PubMed] [Google Scholar]
- 29.Shetty PV, Bhagwat BY, Chan WK (2003) P23 enhances the formation of the aryl hydrocarbon receptor-DNA complex. Biochem Pharmacol 65:941–948 [DOI] [PubMed] [Google Scholar]
- 30.Mandal PK (2005) Dioxin: a review of its environmental effects and its aryl hydrocarbon receptor biology. J Comp Physiol [B] 175:221–230 [DOI] [PubMed] [Google Scholar]
- 31.Bunger MK, Wilsbacher LD, Moran SM et al. (2000) Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103:1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laposky A, Easton A, Dugovic C et al. (2005) Deletion of the mammalian circadian clock gene BMAL1/Mop3 alters baseline sleep architecture and the response to sleep deprivation. Sleep 28:395–409 [DOI] [PubMed] [Google Scholar]


